Abstract
The M2 isoform of pyruvate kinase (PKM2) is highly expressed in most cancer cells, and has been studied extensively as a driver of oncogenic metabolism. In contrast, the role of PKM2 in nontransformed cells is little studied, and nearly nothing is known of its role, if any, in quiescent cells. We show here that endothelial cells express PKM2 almost exclusively over PKM1. In proliferating endothelial cells, PKM2 is required to suppress p53 and maintain cell cycle progression. In sharp contrast, PKM2 has a strikingly different role in quiescent endothelial cells, where inhibition of PKM2 leads to degeneration of tight junctions and barrier function. Mechanistically, PKM2 regulates barrier function independently of its canonical activity as a pyruvate kinase. Instead, PKM2 suppresses NF-kB and its downstream target, the vascular permeability factor angiopoietin 2. As a consequence, loss of endothelial cell PKM2 in vivo predisposes mice to VEGF-induced vascular leak, and to severe bacteremia and death in response to sepsis. Together, these data demonstrate new roles of PKM2 in quiescent cells, and highlight the need for caution in developing cancer therapies that target PKM2.
Keywords: Metabolism, Vascular Biology
Keywords: endothelial cells
Introduction
Endothelial cells (ECs) perform numerous critical functions in vertebrate biology, broadly separated into 2 activities. On the one hand, quiescent ECs line the vascular wall and provide a barrier to solutes, regulate nutrient transport, and maintain bloodstream homeostasis. On the other hand, in response to stimuli such as a wound, ECs escape dormancy, rapidly proliferate, and invade hypoxic and ischemic tissues in order to reestablish vascular support, a process known as angiogenesis. In this latter function, ECs resemble cancer cells. As with cancer cells, metabolism is emerging as a key regulator of ECs, in particular angiogenesis (1, 2). Glycolysis especially has been recognized as a driving force of EC proliferation and angiogenesis. ECs have higher glycolysis rates compared with other noncancer types of cells (3), akin to the Warburg effect observed in cancer cells, and the importance of rate-limiting glycolytic enzymes such as PFKFB3 (3) or HK2 (4) on angiogenesis has been reported.
Pyruvate kinase (PK) catalyzes the third and last rate-limiting step in glycolysis, transferring a phosphate group from phospho-enol pyruvate to ADP, thereby generating pyruvate and ATP. Two different genes express PK. The PKLR gene encodes PKL and PKR, from 2 different promoters, in liver and red blood cells, respectively. The PKM gene is expressed in most other tissues. The PKM transcript can be alternatively spliced to form either PKM1 or PKM2, selecting either exon 9 or 10, respectively (Figure 1A). Cancer cells almost invariably express the PKM2 isoform, and in this context, PKM2 has been studied extensively and shown to be critical for tumor growth in cell culture and in vivo (5, 6). Therapies that target PKM2 are therefore actively being sought. However, the potential untoward effects of systemic pharmaceutical PKM2 inhibition on normal cells have received little attention, presenting uncertainties about potential on-target side effects of PKM2 modulation. Indeed, the role of PKM isoforms in nontransformed or quiescent cells has been little studied (7).
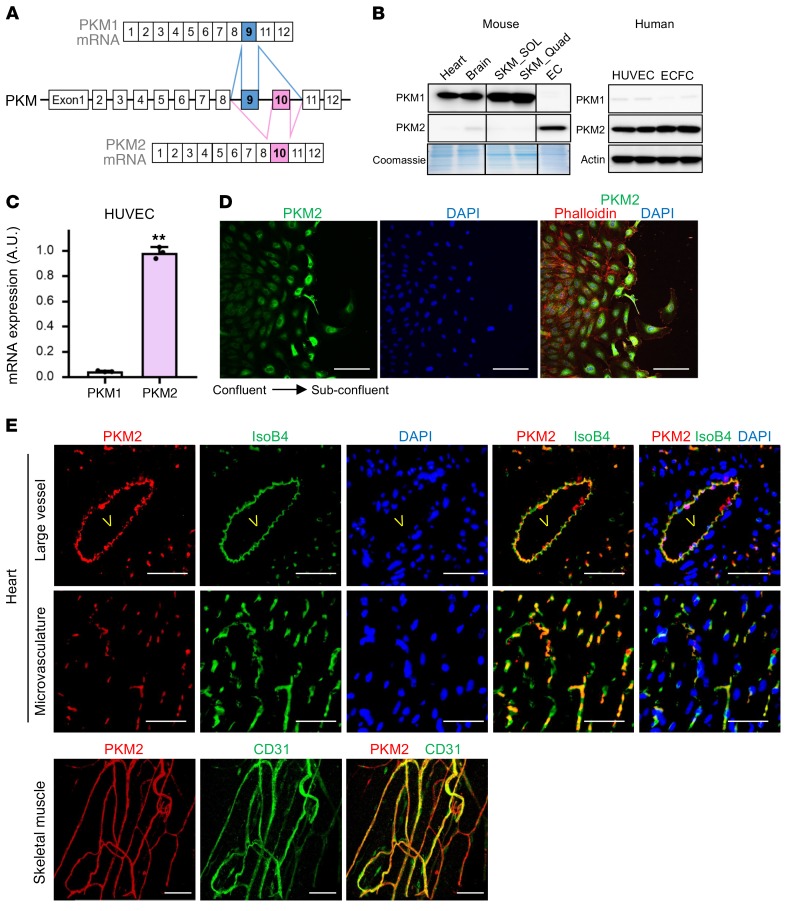
Figure 1. ECs predominantly express PKM2 over PKM1.
(A) Schematic illustration of alternative splicing of PKM1 and PKM2. The mutually exclusive exons for PKM1 (exon 9) and PKM2 (exon 10) are indicated in blue and pink, respectively. (B) Western blot analysis of PKM1 and PKM2 in various tissues from mouse and 2 different human-derived primary ECs. CD31+ ECs from mouse were directly lysed in RIPA buffer immediately after isolation, without cell culture. SKM, skeletal muscle; SOL, soleus; Quad, quadriceps; HUVEC, human umbilical vein endothelial cells; ECFC, endothelial colony forming cells. (C) qPCR analysis of PKM1 and PKM2 mRNA expression in subconfluent HUVECs (n = 3). (D) Staining of endothelial PKM2 in a scratch assay setting. HUVECs were cultured at full confluency for 3 days and scratch was made. PKM2 (green) was costained with phalloidin (red) and DAPI (blue) after 24 hours. Scale bar, 100 μm. (E) ECs lining both large vessels and the microvasculature in mouse heart and skeletal muscle express PKM2, in sharp contrast to adjacent cardiac and skeletal muscle cells that exclusively express PKM1 (shown in B). Cross-section of mouse heart was immuno-stained with PKM2 (red), IsoB4 (green), and DAPI (blue). v, lumen of a large vessel. Whole-mount staining of ECs lining microvasculature in the skeletal muscle of mouse is shown on bottom. PKM2 (red), CD31 (green). Scale bar, 50 μm. All data are mean ± SD. **P < 0.01, by 2-tailed Student’s t test.
PKM1 and PKM2 differ in important ways biochemically. PKM1 functions as a homo-tetramer with constitutively high PK activity. In contrast, PKM2 is regulated by multiple allosteric modulators and posttranslational modifications, each promoting either the less active dimer or the more active PKM1-like tetrameric form. PK catalyzes the last step of glycolysis, 1 of 3 rate-limiting steps, and is thus an ideal enzyme to regulate glycolytic flux. If the PK activity is low, upper glycolytic intermediates can accumulate and be channeled to various synthetic pathways, including the pentose phosphate pathway necessary for nucleotide synthesis and glycogen synthesis. The less active dimeric PKM2 is thus widely thought to promote proliferation by promoting shunting of glucose carbons to creation of biomass. Consistent with this notion, PKM2 is highly expressed in proliferating cells such as embryonic or cancer cells, whereas PKM1 is usually favored in nonproliferating and terminally differentiated types of cells. TEPP-46, a small molecule activator of PKM2 pyruvate activity, binds to a pocket at the PKM2 subunit interface and promotes PKM2 subunits to form stable tetramers. Thus, treatment of TEPP-46 mimics the properties of PKM1 in PKM2-expressing cells. In line with the hypothesis that low PK activity promotes anabolic metabolism to cancer cell proliferation, TEPP-46 has been shown to suppress cancer cell proliferation in vitro and xenograft tumor growth in vivo (8–10).
Although the importance of glycolysis in ECs has been well-established, the role of PKM isoforms in endothelial biology has not been thoroughly examined. One study suggested that JMJD8 regulates endothelial sprouting by interacting with PKM2 (11), and another study briefly showed that PKM2 knockdown does not decrease viability in endothelial cells, whereas it does decrease viability in multiple cancer cell lines (12). No study has investigated the role of PKM isoforms in confluent and quiescent monolayers of quiescent ECs, as are found throughout the healthy adult body. In this study, we report that PKM2 is exclusively expressed over PKM1 in both growing and confluent ECs. We thoroughly examined the role of endothelial PKM2 during proliferation and migration in vitro and angiogenesis in vivo and elucidate a novel PK-independent PKM2/p53 axis as a proproliferative mechanism. Most strikingly, we also report a novel role of PKM2 on barrier function in quiescent ECs, mediated via regulation of NF-kB and angiopoietin 2. The implications for the ongoing development of PKM2 cancer therapies are discussed.
Results
As reported previously, most terminally differentiated mouse cell types express PKM1 (Figure 1B). Surprisingly, on the other hand, ECs isolated from tissues express predominantly PKM2 (Figure 1B), despite their quiescent state, with doubling times of weeks to months (13, 14). These ECs are rapidly isolated from mice by bead-based capture and undergo no culture in vitro; thus, they have not had time to change to a proliferative phenotype. Two different human-derived ECs, HUVECs and ECFCs (Figure 1, B and C), also express predominantly PKM2. PKM2 expression in cultured ECs is strong both in actively replicating and confluent contact-inhibited cells (Figure 1D), although interestingly the expression of PKM2 increases during the process of activation in response to a scratch wound in an EC monolayer (Figure 1D). In vivo, ECs of both large vessels and of the microvasculature express PKM2, as seen in heart and skeletal muscle (Figure 1E). These data demonstrate that in cell culture and in vivo, mice and humans, replicating and quiescent cells, and large and micro vasculature, PKM2 is the predominant PK isoform in ECs.
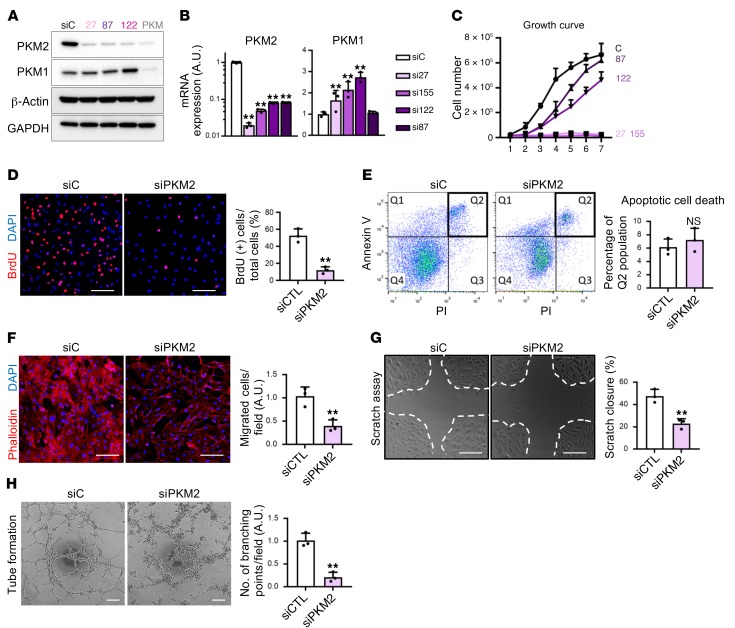
To investigate the role of PKM2 in ECs, we generated several siRNAs that target PKM2 specifically, while not targeting PKM1 (12) (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI120912DS1). The different siRNAs achieved variable suppression of PKM2 mRNA and protein expression (Figure 2, A and B). PKM1 expression was mildly or not at all increased (Figure 2, A and B). Strikingly, suppression of PKM2 led to dose-dependent inhibition of cellular proliferation, in proportion to the extent of PKM2 suppression (Figure 2C). siRNA27, the most efficient at suppressing PKM2 (labeled siPKM2 from here on), led to complete growth arrest (Figure 2C) and a near-complete block of BrdU incorporation (Figure 2D), demonstrating a block of entry into S phase. Other duplex siRNAs, si87 and si155, had similar effects on BrdU incorporation (Supplemental Figure 1B). siPKM2 did not, however, promote cell death (Figure 2E), indicating a block specifically in cell proliferation. ECs have various unique properties germane to their ability to form new blood vessels, including a strong capacity for migration and spontaneous tube formation. Both these properties were profoundly suppressed by siPKM2 (and si87 and si155), as shown by transwell migration assay (Figure 2F and Supplemental Figure 1C), scratch closure assay (Figure 2G and Supplemental Figure 1D), and tube formation assay (Figure 2H and Supplemental Figure 1E). VEGF is known to affect all of these phenotypes, but VEGF did not affect PKM2 expression (Supplemental Figure 1F).
Figure 2. PKM2 is crucial for EC proliferation and migration in vitro.
(A) Western blot analysis of PKM2 and PKM1 protein expression in HUVECs demonstrates that the siRNAs used in this study give PKM2-specific and efficient knockdown. qPCR analysis of PKM2 and PKM1 mRNA expression. Note that the y axis for PKM2 is a log scale (n = 3). (C) Growth curve of HUVECs with various siRNAs targeting PKM2. The efficiency of knockdown shown in B correlates with the effect on proliferation (n = 5). (D) Percentage of BrdU+ cells is reduced in siPKM2 ECs (n = 3). (E) Apoptotic cell death, assessed by Annexin V and PI staining, is not induced in siPKM2 ECs (n = 3). (F) Reduction in transwell migration of ECs by siPKM2. Cells that migrated across the transwell membrane were visualized by staining with phalloidin (red) and DAPI (blue) (n = 3). (G) Scratch closure was retarded in ECs with siPKM2 (n = 3). (H) Tube formation on Matrigel was impaired in ECs with siPKM2 (n = 3). Scale bars, 100 μm. All data are mean ± SD. **P < 0.01, by 2-tailed Student’s t test.
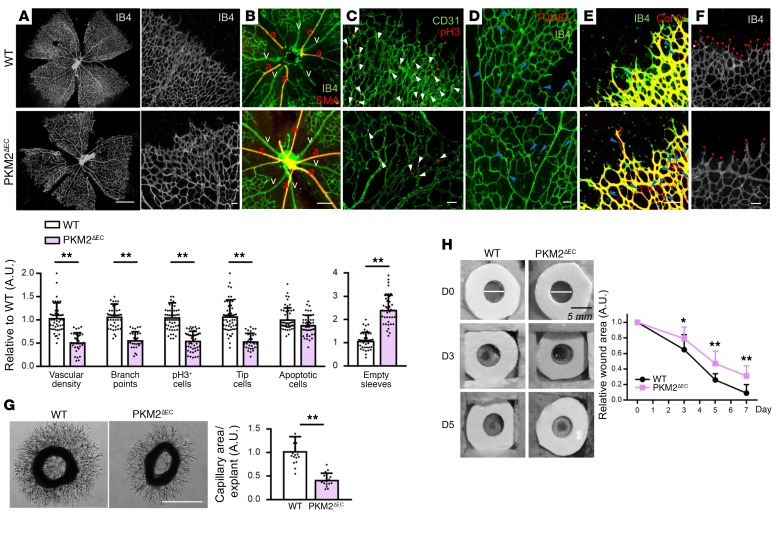
To test the role of PKM2 in EC biology in vivo, we generated mice that lack exon 10 of the PKM gene specifically in ECs (PKM2ΔEC mice), using mice bearing floxed alleles of PKM (15) and a transgene encoding for Cre recombinase driven by the inducible EC-specific VE-cadherin promoter (16), leading to efficient EC-specific deletion of PKM2 in vivo (Supplemental Figure 2A). Deletion of exon 10 led to alternative splicing and reciprocal expression of EC PKM containing exon 9 (i.e., PKM1, Supplemental Figure 2, B and C), as has been observed before (15). Analysis of retinal vasculature at P7 revealed marked reduction in vessel density and branch points in both angiogenic front and rear plexus in PKM2ΔEC mice compared with control mice, but no change in large vessels (Figure 3, A and B). EC proliferation was significantly blunted in PKM2ΔEC mice, as evidenced by pH3 staining (Figure 3C). Apoptotic cell death was not altered in PKM2ΔEC mice as analyzed by TUNEL staining (Figure 3D). Consistent with the impaired proliferation, vessel regression was induced in PKM2ΔEC mice, as evidenced by empty sleeves with IsoB4-negative and collagen IV–positive staining (Figure 3E). The number of tip cells in PKM2ΔEC mice was profoundly reduced (Figure 3F), consistent with the observed blunted migration in cell culture (Figure 2, F–H and Supplemental Figure 1, C–E). In addition, capillary sprouting from aortic rings derived from PKM2ΔEC mice was significantly impaired, compared with controls (Figure 3G). Despite these defects in developmental angiogenesis, PKM2ΔEC mice survive to adulthood and appeared grossly normal, though trending to be slightly smaller (Supplemental Figure 2D). In the adult, however, angiogenesis is also required for pathophysiological processes, such as recovery after wound infliction. Wound healing was dramatically slowed in PKM2ΔEC mice (Figure 3H). Together, these data demonstrate a profound defect in both developmental and pathophysiological angiogenesis in PKM2ΔEC mice.
Figure 3. PKM2 is crucial for angiogenesis in vivo.
(A–F) Staining of WT versus PKM2ΔEC mouse retina at P7. (A) The entire retinal vasculature is visualized by IB4 staining. Scale bar, 1 mm. (B) Arterio-venous patterning is analyzed by IB4 and SMA staining. a, artery; v, vein. (C) The number of pH3+ ECs (white arrows) is analyzed by costaining of pH3 and CD31. (D) Apoptosis was analyzed by TUNEL staining. (E) Vascular regression is assessed by empty sleeves (blue arrows) with IB4-negative and collagen IV–positive staining. (F) Number of tip cells (red arrows) is visualized via IB4 staining. Quantifications are at bottom of figure. Scale bars, 50 μm (B–F). Data are mean ± SD of 4–8 different images per mouse (n = 8). **P < 0.01, by 2-tailed Student’s t test. (G) Capillary sprouting of aortic explants from WT versus PKM2ΔEC mouse. At day 3 of aortic ring explant incubation, each explant was photographed and the area of capillary outgrowth was quantified using Image J. Scale bar, 1 mm. Data are mean ± SD; n = 6 segments per aorta from 3 mice each group. **P < 0.01, by 2-tailed Student’s t test. (H) Wound healing assay in WT versus PKM2ΔEC mouse. Two wounds (5 mm) were made on the back of each mouse by using biopsy punch and wound size was monitored every 2–3 days for a week. Data are mean ± SD (n = 5). *P < 0.05, **P < 0.01, by 2-tailed Student’s t test.
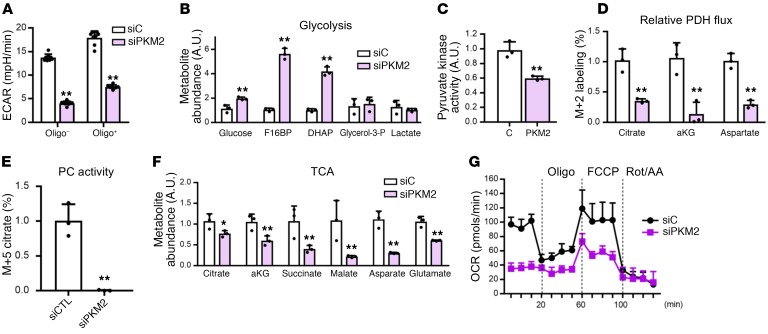
To investigate mechanisms by which PKM2 controls EC growth and function, we first turned to the role of PKM2 in glycolysis. PKM2 catalyzes 1 of 3 rate-limiting reactions of glycolysis. Consistent with this, siPKM2 significantly suppressed glycolytic flux in ECs, as revealed by reduced extracellular acidification (Figure 4A) and dramatic accumulation of intermediates upstream of PK (Figure 4B and Supplemental Figure 3, A and B). PKM2 is thus crucial for glycolysis in ECs. PKM1, though mildly induced (Figure 2, A and B), is insufficient to compensate the cellular PK activity (Figure 4C). In addition to being secreted as lactate, the pyruvate generated by PK can also enter the tricarboxylic acid (TCA) cycle in mitochondria via pyruvate dehydrogenase (PDH). 13C labeling of TCA intermediates in the presence of [U-13C] glucose was profoundly suppressed by siPKM2 (Figure 4D), indicating reduced relative flux through PDH. In addition, glucose-derived anaplerosis of TCA via pyruvate carboxylase (PC), as reflected in M+5 13C labeling of citrate, was entirely blocked by siPKM2 (Figure 4E). Consistent with these findings, the levels of nearly all TCA intermediates were reduced in siPKM2 cells (Figure 4F), and the oxygen respiration capacity of the cells was concomitantly greatly suppressed (Figure 4G and Supplemental Figure 3E). Together, these data indicate that PKM2 in ECs controls flux of carbons through glycolysis and into the TCA cycle. Despite these changes, however, siPKM2 cells were not energetically depleted, as shown by ATP/ADP ratio, energy charge, NADH/NAD+ ratio, and mitochondrial membrane potential (Supplemental Figure 3, C and D).
Figure 4. Glycolysis is impaired in PKM2 knockdown ECs.
(A) Extracellular acidification rate (ECAR) of HUVECs with si-control/scrambled RNA (siC) versus siPKM2 in the absence and presence of oligomycin (Oligo) demonstrating basal and maximal glycolytic rate, respectively (n = 8). (B) Relative abundance of glycolysis intermediates in HUVECs with siC versus siPKM2 were quantified by mass spectrometry after 24 hours of incubation (n = 3). (C) Pyruvate kinase activity of HUVECs with siC versus siPKM2 (n = 6). (D) PDH activity estimated by incorporation of [U-13C] glucose–derived carbons into M+2 citrate, αKG, and aspartate in HUVECs with siC versus siPKM2 (n = 3). (E) Pyruvate carboxylase (PC) activity estimated by incorporation of [U-13C] glucose–derived carbons into M+5 citrate (n = 3). (F) Relative abundance of TCA cycle intermediates in HUVECs with siC versus siPKM2 were quantified by mass spectrometry after 24 hours of incubation (n = 3). (G) Oxygen consumption rate (OCR) of HUVECs with siC versus siPKM2 was measured by Seahorse (n = 8). All data are mean ± SD. *P < 0.05, **P < 0.01, by 2-tailed Student’s t test.
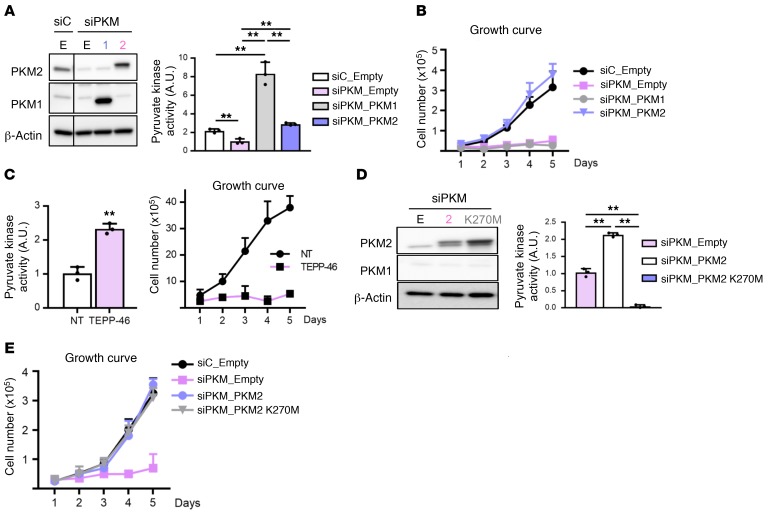
In tumor cells and in mouse embryo fibroblasts (MEFs), the growth arrest caused by PKM2 suppression is in part mediated by the reciprocal induction of PKM1, and is rescued by cosuppression of PKM1 (10). In contrast, in ECs, cosuppression of PKM1 with siRNA targeting both isoforms (siPKM, Supplemental Figure 3F) failed to rescue proliferation (Supplemental Figure 3G). siPKM led to metabolic effects identical to siPKM2, including reduced cellular PK activity (Supplemental Figure 3H), accumulation of upstream glycolytic metabolites (Supplemental Figure 3, I and J), reduced relative PDH and PC flux (Supplemental Figure 3, K and L), and lower abundance of TCA intermediates (Supplemental Figure 3M), consistent with the dominant role of PKM2 in these cells. These data suggested that loss of glycolytic flux in siPKM2 and siPKM cells may be driving the observed growth arrest. To test this notion, we overexpressed PKM1 in siPKM, siPKM2, or WT nontransfected cells, yielding a marked increase in cellular PK activity even above that achieved with PKM2 overexpression (Figure 5A and Supplemental Figure 4, A and B), consistent with the known higher PK activity of PKM1 compared with PKM2 (17, 18). Elevated PK activity, however, failed to rescue proliferation of siPKM2 or siPKM ECs (Figure 5B and Supplemental Figure 4, A and B). Interestingly, activation of PK activity with TEPP-46, a PKM2 activator, increased cellular PK activity (8) but completely suppressed proliferation, even in the absence of siPKM (Figure 5C). Together, these data demonstrate that rescuing cellular PK activity is insufficient for rescue of proliferation. To test if PK activity is necessary for rescue of proliferation, we next overexpressed a PKM2 mutant (K270M) that lacks PK activity (19), leading in a dominant-negative fashion to complete inhibition of measurable PK activity in the cells (Figure 5D). Consistent with the lack of PK activity, PKM2 K270M did not rescue the elevated ECAR, reduced OCR, and blunted glycolytic flux seen in siPKM2 cells (Supplemental Figure 4, C and D). Expression of PKM2 K270M, however, completely rescued cellular proliferation (Figure 5E, and Supplemental Figure 4E). Together, these data surprisingly clearly dissociate PKM2 PK enzymatic activity from its ability to promote EC proliferation.
Figure 5. Pyruvate kinase activity of PKM2 is dispensable for EC proliferation.
(A) Western blot and pyruvate kinase activity analysis of empty vector (E), PKM1 (1), or PKM2 (2) overexpression in the presence of siPKM in HUVECs (n = 3). (B) Growth curve demonstrating complete or no rescue of proliferation with PKM2 or PKM1 overexpression, respectively, in the presence of siPKM (n = 4). (C) Pyruvate kinase activity assay and growth curve in the presence or absence of 10 μM PKM2 activator (TEPP-46) (n = 4). (D) Western blot and pyruvate kinase activity analysis of empty vector (E), PKM2 WT (2), or PKM2 PEP binding mutant (K270M) overexpression in the presence of siPKM in HUVECs (n = 3). (E) Growth curve demonstrating complete rescue of proliferation with PKM2 PEP binding mutant (K270M) overexpression (n = 4). All data are mean ± SD. **P < 0.01, by 2-tailed Student’s t test.
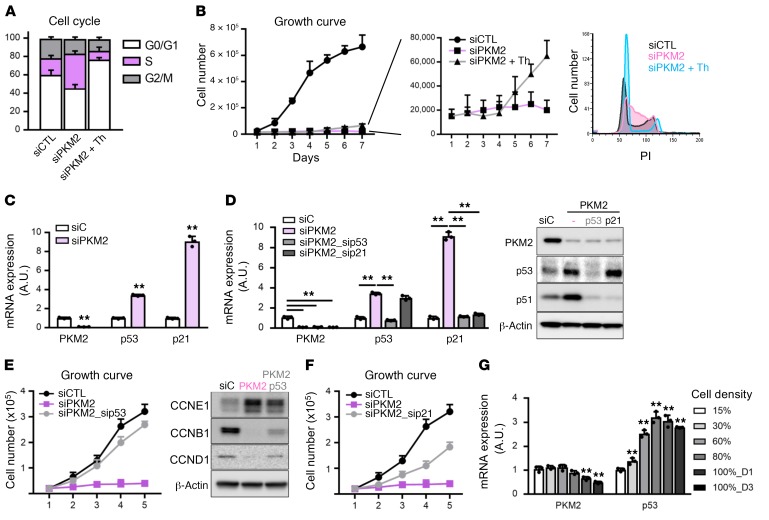
Others have shown that thymine supplementation can rescue proliferation in MEFs lacking PKM2 (10). Consistent with this idea, we find that siPKM2 and siPKM cells accumulate in S-phase, compared with controls (Figure 6A). And indeed, supplementation with thymine partially rescued EC proliferation (Figure 6B). However, the rescue of proliferation was only partial, began only at day 4 of culture, and was quite slow compared with rescue with PKM2 (Figure 5B, Figure 6B and Supplemental Figure 4A). Moreover, after thymine supplementation, siPKM cells accumulated in both G1 and G2 (Figure 6B). These data indicated to us that multiple blocks in the cell cycle must be elicited in ECs by the absence of PKM2.
Figure 6. PKM2 suppresses p53-mediated growth arrest.
(A) Flow cytometry analysis of cell cycle with propidium iodide (PI) staining in siCTL, siPKM2, and siPKM2 with 500 μM thymine (Th) supplementation (n = 3). (B) Growth curve demonstrating mild rescue of siPKM2 HUVECs by thymine supplementation. Cell cycle analysis with PI staining shows block at both G0/G1 and G2/M phases by thymine supplementation in HUVECs with siPKM2 (n = 3). (C) qPCR analysis of mRNA expression in HUVECs with siC versus siPKM2 (n = 3). (D) qPCR analysis of mRNA expression and Western blot analysis of protein expression with double knockdown of p53 or p21 in the presence of siPKM2 (n = 3). (E) Growth curve demonstrating proliferation rescue of siPKM2 HUVECs with double knockdown of p53. Western blot analysis of cyclin proteins (CCNE1, late G1 phase; CCNB1, M phase; and CCND1, early G1 phase) (n = 4). (F) Growth curve demonstrating proliferation rescue of siPKM2 HUVECs with double knockdown of p21, downstream of p53 (n = 4). (G) qPCR analysis of mRNA expression at indicated density (15% to 100%) of HUVECs. D1 and D3 indicate day 1 and day 3 after reaching 100% confluency, respectively (n = 3). All data are mean ± SD. **P < 0.01, by 2-tailed Student’s t test.
To identify which pathways PKM2 targets to promote proliferation in ECs, we performed a genome-wide CRISPR/Cas9-based loss-of-function proliferation rescue screen in siPKM2 cells (Supplemental Figure 5A). The tumor suppressor p53 gene scored among the top 1% of hits among the over 20,000 genes tested (Supplemental Figure 5, A and B). p53 is known to block cell cycle progression at multiple points, including S-phase, G1/S, and G2/M (20), mimicking the multiple blocks in cell cycle that we observed in the absence of PKM2 (Figure 6B). We therefore tested the role of p53 in siPKM2-induced growth arrest. Treating ECs with siPKM2 robustly induced the expression of p53 and its downstream target p21 (CDKN1A), known to mediate p53-induced growth arrest (Figure 6C), whereas loss of only PKM1 did not alter p53 expression (Supplemental Figure 5C) (21). Suppression of p53 with siRNA completely rescued the siPKM2-induced growth arrest (Figure 6, D and E), consistent with the result from CRISPR screen (Supplemental Figure 5B). Suppression of p21 also rescued proliferation (Figure 6, D and F). Both approaches rescued proliferation much more robustly than did thymine supplementation (Figure 6B). Consistent with the rescue of proliferation, sip53 rescued the loss of cyclin B and D in siPKM2 cells, indicators of passage through S-phase and M-phase respectively, and rescued the accumulation of cyclin E, an indicator of G1/S arrest (22) (Figure 6E). Similar findings were noted in ECs isolated from the lungs of WT versus PKM2ΔEC mice, including elevated expression of p53 and p21 and reduced expression of cyclin B (Supplemental Figure 5D). Suppression of p53 also rescued the impaired migration of siPKM2 ECs in the scratch closure assay (Supplemental Figure 5E). In addition, while PKM2 expression fell as cells became confluent and contact-inhibited, consistent with the data shown in Figure 1D, a concomitant reciprocal induction of p53 and expression was seen (Figure 6G). Together, these data demonstrate that PKM2 in proliferating ECs suppresses p53 expression and function in order to prevent p53-induced growth arrests at multiple points in the cell cycle.
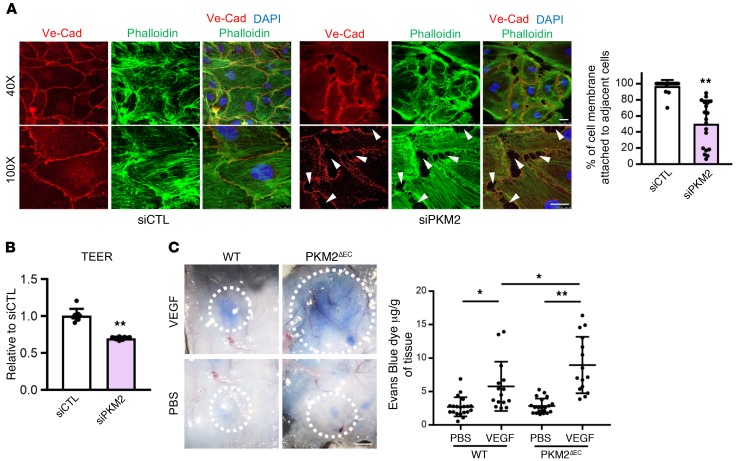
These observations led us to investigate the role of PKM2 in confluent contact-inhibited ECs. Unlike most cancer cells, ECs arrest proliferation when reaching confluence in a monolayer. This process is accompanied by solidification of cell-cell contacts into adherent and tight junctions, a process required for ECs to transition to their key in vivo role of maintaining a barrier between the plasma and the surrounding parenchyma. Despite the reduced levels of PKM2 in contact-inhibited ECs, further suppression of PKM2 with siRNA greatly interfered with the process of cell-cell contact solidification, as reflected in the inability to achieve continuous cell-cell contact and the persistence of cytoplasmic membranes not physically connected to adjoining cells (Figure 7A). Consistent with these effects, the trans-endothelial electrical resistance (TEER) across a monolayer of ECs, a measure of its barrier function (23), was greatly reduced by siPKM2 (Figure 7B). In vivo, barrier function can be measured by the extent of extravasation of cytoplasmic dye after a stimulus, such as subcutaneous injection of VEGF (24). In PKM2ΔEC mice, this response was markedly exaggerated (Figure 7C), demonstrating that PKM2 is required for maintaining barrier function in vivo. In addition, PKM2ΔEC mice demonstrated increased basal pulmonary microvasculature permeability (Supplemental Figure 6).
Figure 7. PKM2 is required for vascular barrier function in confluent contact-inhibited ECs.
(A) Disrupted formation of VE-cadherin (VE-Cad) adherent junction with siPKM2. Two days after siRNA transfection, HUVECs were trypsinized and same number of cells was reseeded and allowed to form junctions. At 24 hours after reseeding, cells were immunostained with VE-Cad (red), phalloidin (green), and DAPI (blue). Confocal analysis demonstrates continuous VE-Cad distribution around the entire periphery of the cells with siCTL whereas siPKM2 cells show discontinuous and unstable junctions with intercellular gaps (white arrows). Quantification of cell membrane (%) attached to adjacent cells in siCTL versus siPKM2 HUVECs. siPKM2 cells have less attachment to adjacent cells compared with siCTL (n = 6). Scale bar, 10 μm. (B) Changes in trans-endothelial electrical resistance (TEER) was measured in siCTL versus siPKM2 HUVECs on an electric cell-substrate impedance sensor (ECIS) in 8W1E+ plate at 4,000 Hz (n = 8). (C) Acute vascular hyperpermeability was assessed in WT versus PKM2ΔEC mouse via miles assay. Mice were injected intravenously with Evans Blue dye and were subsequently injected intradermally with PBS and VEGF (100 ng). Representative skin images in response to PBS or VEGF intradermal injections. Scale bar, 50 mm. Quantification of extracted Evans Blue dye normalized to tissue weight (μg of Evans Blue dye / g of tissue; n = 15 each group). All data are mean ± SD. *P < 0.05, **P < 0.01, by 2-tailed Student’s t test.
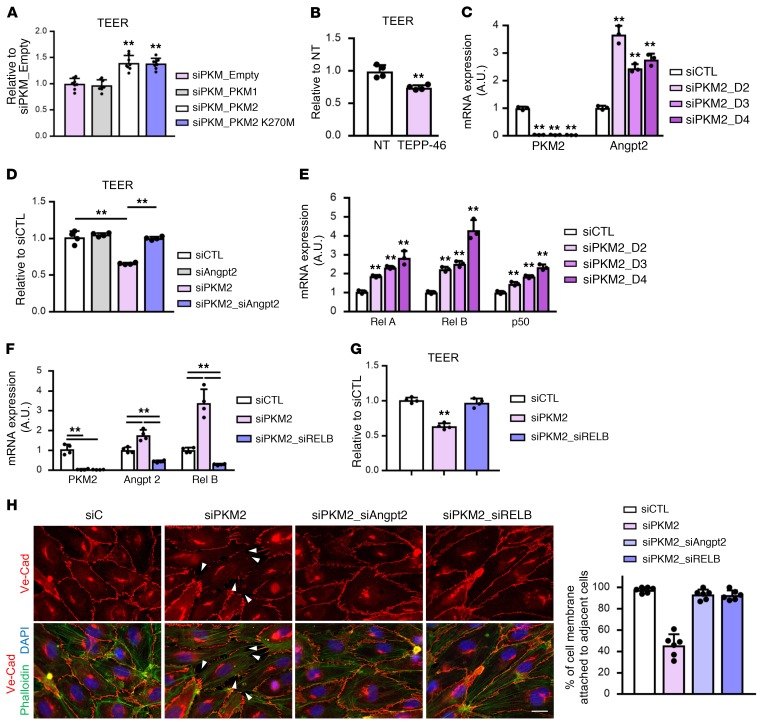
To test whether siPKM2-induced vascular leak is due to the loss of PK activity, we overexpressed either empty, PKM1, PKM2, or mutant PKM2 (K270M) in siPKM ECs. We found that PKM1 failed to rescue the loss of TEER seen in siPKM2, while PKM2 did rescue. Strikingly, the PK-dead PKM2 K270M mutant also completely rescued TEER (Figure 8A). Thus, as with the effect of PKM2 on proliferation, the maintenance of barrier function by PKM2 is independent of its activity as a pyruvate kinase. In fact, activation of PK activity with TEPP-46, which promotes the tetrameric form of PKM2, impaired TEER even in the absence of siPKM (Figure 8B), suggesting that the dimeric form of PKM2 is the active form in this context. Together these data demonstrate that rescuing cellular PK activity is neither sufficient nor necessary for the rescue of vascular leak in siPKM2 cells.
Figure 8. PKM2 promotes vascular barrier function by suppressing NF-kB and ANGPT2 expression independently of PK activity.
(A) TEER demonstrating no rescue of vascular leak with PKM1 overexpression and complete rescue of vascular leak with PKM2 K270M in the presence of siPKM. TEER assay was performed 2 days after siPKM transfection (n = 8). (B) TEER demonstrating impaired barrier function in the presence of 10 μM TEPP-46. TEER assay was performed immediately after treating TEPP-46 to WT HUVECs in suspension (n = 4). (C) qPCR analysis of Angpt2 mRNA expression with knockdown of PKM2 for indicated amount of time (n = 3). (D) TEER demonstrating fully rescued vascular leak by double knockdown of Angpt2 and PKM2 (n = 4). (E) qPCR analysis of mRNA expression on NF-kB transcription factors with PKM2 knockdown for indicated amount of time (n = 3). (F) qPCR analysis demonstrating normalized Angpt2 expression by double knockdown of RELB and PKM2 (n = 4). (G) TEER demonstrating fully rescued vascular leak by double knockdown of RELB (n = 4). (H) Disrupted formation of VE-cadherin (VE-Cad) adherent junction with siPKM2 is fully rescued by double knockdown of either ANGPT2 or RELB. VE-Cad (red), phalloidin (green), and DAPI (blue). White arrows indicate intercellular gaps. Percentage (%) of cell membrane attached to adjacent cells are quantified and compared with siCTL (n = 6). Scale bar, 10 μm. All data are mean ± SD. **P < 0.01, by 2-tailed Student’s t test.
Angiopoietin 2 (ANGPT2) is a well-established promoter of vascular leak, and is induced in high vascular permeability states such as sepsis (25). We found that knockdown of PKM2 in confluent ECs significantly induced ANGPT2 expression (Figure 8C). Strikingly, cosuppression of ANGPT2 by siRNA in siPKM2 cells completely rescued the increase in vascular leak, measured by TEER (Figure 8D). We conclude that PKM2 suppresses vascular leak via suppression of ANGPT2. The expression of ANGPT2 is regulated in part by the inflammatory response, including the transcription factor complex NF-kB. We found that PKM2 knockdown elevates gene expression and nuclear localization of NF-kB subunits RELA, RELB, and p50 in ECs (Figure 8E and Supplemental Figure 7A) independently of ROS production (Supplemental Figure 7, B and C). As a consequence, canonical NF-kB target genes were upregulated in siPKM2 ECs (Supplemental Figure 7D). Knockdown of RELB completely blocked the induction of ANGPT2 (Figure 8F) and fully rescued the impaired TEER in siPKM2 cells (Figure 8G). Furthermore, knockdown of either ANGPT2 or RELB fully rescued the impaired formation of adherent junction in siPKM2 cells (Figure 8H). Together, these data demonstrate that in quiescent contact-inhibited cells PKM2 is critical to achieve complete barrier function in cell culture and in vivo, and does so via suppressing the NF-kB–mediated induction of ANGPT2, a potent inducer of vascular leak.
Interestingly, the regulation of proliferation and barrier function by PKM2 in ECs appear mechanistically distinct. Knockdown of RELB or ANGPT2 failed to rescue the impaired proliferation of siPKM2 ECs (Supplemental Figure 7E), despite rescuing the impaired barrier function in siPKM2 confluent ECs (Figure 8, D, G, and H). Conversely, knockdown of p53 failed to rescue the impaired TEER in confluent siPKM2 ECs (Supplemental Figure 7F), despite rescuing the impaired proliferation of siPKM2 ECs (Figure 6E).
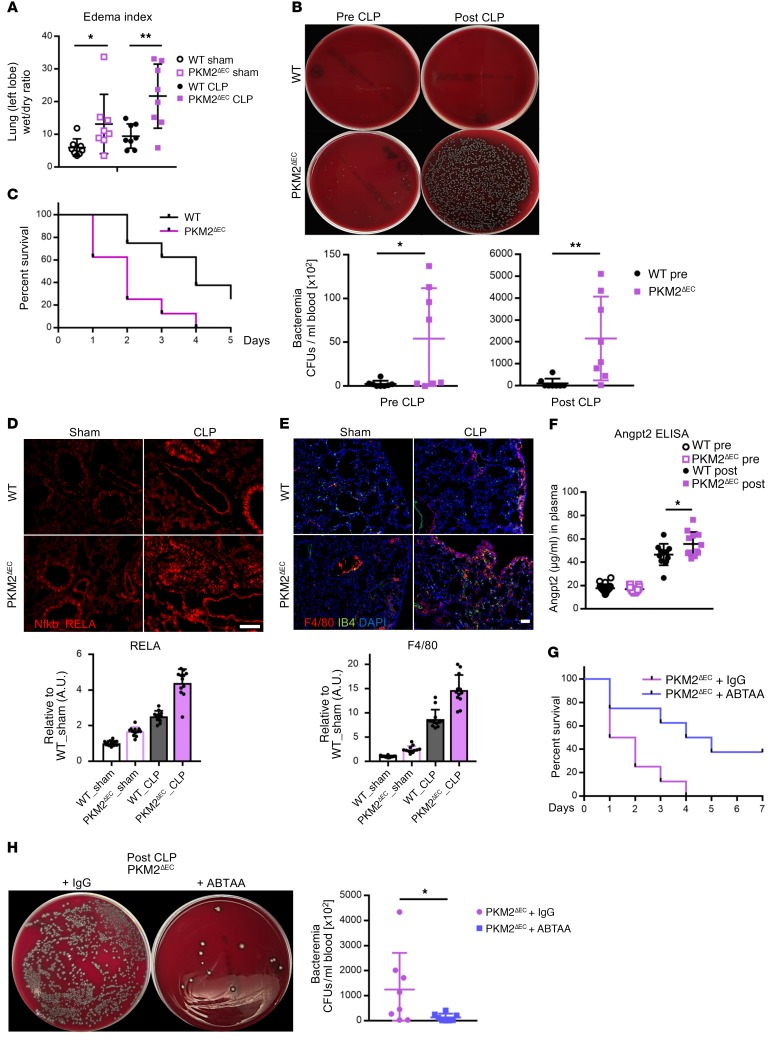
Failure to maintain a tight barrier in the vasculature can lead to exaggerated pathology in settings where barrier function is challenged, such as sepsis. To test the contribution of PKM2 to barrier function in pathological states, we subjected PKM2ΔEC mice to a cecal ligation and puncture (CLP), a widely used model of experimental sepsis. Even in the absence of the challenge, PKM2ΔEC mice showed defective vascular permeability, as seen by increased pulmonary edema, and this defect was markedly pronounced by CLP (Figure 9A). Similarly, even in the absence of CLP, inappropriate intravasation of gut organisms could be seen in PKM2ΔEC mice, and this bacteremia was markedly worsened after CLP (Figure 9B). As a consequence, PKM2ΔEC mice had markedly accelerated mortality compared with littermate controls (Figure 9C). Consistent with the mechanism elucidated in vitro, we found upregulated inflammatory responses in the PKM2ΔEC mice, including marked induction of NF-kB expression (Figure 9D), excess macrophage infiltration into lungs, visualized by F4/80 staining (Figure 9E), and increase of ANGPT2 in the plasma of PKM2ΔEC mice (Figure 9F). Treating animals with ANGPT2-neutralizing antibodies (ABTAA) (26) rescued the mortality (Figure 9G) and bacteremia (Figure 9H) seen in PKM2ΔEC mice after CLP. We conclude from these data that the ANGPT2-mediated instability of barrier function in PKM2ΔEC mice leads to a striking propensity to succumb from a septic insult.
Figure 9. Deletion of endothelial PKM2 exacerbates sepsis-induced responses and mortality in CLP model.
(A) Lung edema was measured as the ratio of the wet weight to the dry weight of the left lobe 20 hours after sham or CLP surgery. Each group used 12- to 14-week-old mice (n = 8 each group). (B) PKM2ΔEC mice have higher bacteremia before and after CLP. Each group used 20- to 22-week-old mice. Representative images of bacterial growth on blood agar and quantification of number of bacterial CFUs in blood (n = 8 each group) are shown. (C) Survival rates of WT and PKM2ΔEC mice after CLP surgery. Each group used 20- to 22-week-old mice (n = 8 each group). (D) Representative immunohistochemistry images of NF-kB RELA subunit (red) 20 hours after sham or CLP surgery. Mice 12- to 14-weeks-old were used. Scale bar, 100 μm. (E) Representative immunohistochemistry images of macrophages (F4/80 in red), endothelial cells (IB4 in green), and nucleus (DAPI in blue). Each group used 12- to 14-week-old mice (n = 12 each group). Scale bar, 50 μm. (F) Plasma levels of ANGPT2 before and after CLP as determined by ELISA analysis. Each group used 12- to 14-week-old mice (n = 12 each group). (G) Survival rates of PKM2ΔEC mice with IgG or ABTAA injection after CLP. Each group used 8- to 10-week-old mice (n = 8 each group). (H) Rescued bacteremia by ABTAA administration in PKM2ΔEC mice after CLP. Representative images of bacterial growth on blood agar. Quantification of number of bacterial CFUs in blood. Each group used 8- to 10-week-old mice (n = 8 each group). All data are mean ± SD. *P < 0.05, **P < 0.01, by 2-tailed Student’s t test.
Discussion
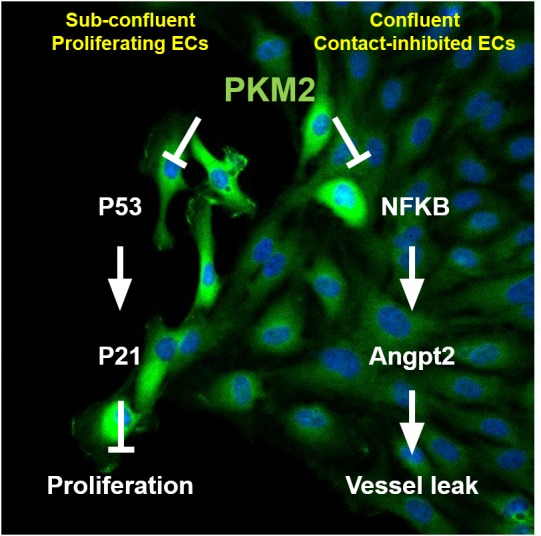
Our data uncover novel dual roles for PKM2 in proliferating and quiescent ECs. Proliferating ECs, though primary cells, display a number of features of cancer cells, including capacity for rapid proliferation, migration, and invasion of hypoxic and low-nutrient environments. As with cancer cells, we show that ECs rely on PKM2 for these attributes. Mechanistically, however, our observations in ECs differ from those reported in cancer cells. The suppression of tumor cell growth in the absence of PKM2 appears to be mediated in many cases via reciprocal induction of PKM1 (6, 15), but we found PKM1 in ECs to be dispensable. We found that the suppression of EC proliferation in the absence of PKM2 occurs via the induction of p53, a process that is surprisingly independent of the canonical role of PKM2 as a pyruvate kinase. The reliance on p53 explains the observation that siPKM2 cells arrest in multiple points of the cell cycle, as observed by others (10), because p53 is known to block cell cycle progression in G1/S, S, and G2/M (20). There have been reports in cancer cells of PK-independent PKM2 functions, including a report of interaction with p53 upon DNA damage in human tumor cells that include MCF7 breast cancer, HCT116 colon cancer, and U2OS osteosarcoma cells (27). However, the report indicated that PKM2 binds to and inhibits p53-dependent transactivation of p21, whereas we found that PKM2 regulates p53 gene expression. Moreover, the report showed that PKM2 suppresses p53 by translocating to the nucleus, whereas we have not observed PKM2 in the nucleus of ECs under any condition that we have tested thus far. It will be of interest in future work to understand precisely how cytoplasmic PKM2 modulates nuclear p53 function. One conclusion we can make solidly from our observations is that the active form of PKM2 in this case is almost certainly the dimeric form, because tetrameric PKM1 fails to rescue the absence of PKM2, and because promoting PKM2 tetramer formation with TEPP-46 arrests cells, despite activating PKM2 PK activity. It will also be of interest to determine if the PKM2-p53-p21 pathway discovered here, via which PKM2 regulates proliferative transition through multiple steps in the cell cycle, also plays a role in at least certain types of proliferating cancer cells.
In vivo, in the absence of insult, most ECs are quiescent rather than proliferative. In contrast to proliferating ECs, quiescent ECs differ remarkably from cancer cells: they have exited the cell cycle, become quiescent, and are contact-inhibited. Despite this striking difference from cancer cells, we find that confluent, contact-inhibited, and nonproliferating ECs also express PKM2 dominantly over PKM1. ECs arose 700 million years ago when high-pressure circulatory systems developed, thus necessitating an osmotic barrier capable of counterbalancing the high pressure system and keeping solutes within circulation. Formation by ECs of a tight barrier is thus crucial to systemic health. Here, we show that PKM2 is indispensable for this critical function. And again, strikingly, this role of PKM2 is independent of its canonical PK activity. In this case, mechanistically, PKM2 in quiescent ECs modulates an inflammatory pathway, dependent on NF-kB and ANGPT2, exposing new links between basic metabolism and blood vessel maturation. PKM2 thus vitally modulates both fundamental physiological roles of ECs: barrier function in their quiescent phase and angiogenesis in their proliferative phase.
Our data suggest that the role of PKM2 in ECs differs not only from that reported in cancer cells but also from that in other normal cells, in the few instances it which it has been studied. Loss of PKM2 in primary MEFs leads to growth arrest via reciprocal upregulation of PKM1 (10), as in many cancer cells, but distinct from what we found here in ECs. Moreover, the growth arrest in MEFs was mediated in part via metabolic suppression of nucleotide synthesis, while we found that this only marginally contributes to the growth arrest in ECs. A number of reports have noted a role for PKM2 in circulating immune cells, which like ECs are typically highly glycolytic, and in which PKM2 promotes cytokine production and PD-L1 expression, often independently of its glycolytic role (28, 29). In these contexts, however, suppression of PKM2 does not appear to slow growth, in contrast to our findings in ECs. Moreover, the biological effects of PKM2 in immune cells appears to be largely mediated by nuclear effects (e.g., via HIF-1α or β-catenin), while we found no evidence of a nuclear role of PKM2 in ECs. PKM2 has also been proposed to play a role in hepatocyte steatosis, again via modulation of HIF-1α (30), and in kidney podocytes and diabetic glomerulonephropathy via nuclear mechanism (31). Both of these cell types are highly oxidative, in contrast to ECs. In cardiomyocytes, which overwhelmingly express PKM1 and are also highly oxidative, there appears to be a partial induction of PKM2 expression in response to ischemic or other insults, often correlating with a relative increase in glycolysis (32, 33), although the biological significance of this observation is not clear. To summarize, the response of ECs to loss of PKM2 differs dramatically from other normal cells where it has been investigated.
The loss of vascular barrier function in mice lacking endothelial PKM2 leads to a profound susceptibility to septic bacteremia and death. These findings have important clinical implications, because various pharmaceutical agencies are developing anticancer agents targeting PKM2. Cancer patients are typically already heavily immunocompromised by their cancer and even more so by their chemotherapeutic regimens. Superimposing an agent that predisposes to bacteremia may therefore have significant untoward effects. Our work predicts that sepsis may be a serious complication of PKM2-targeting adjuvant therapy, which needs to be followed closely.
In summary, we unveil here in cell culture and in vivo a critical dual role for PKM2 in proliferating and quiescence ECs: control of cell cycle progression in the former and of vascular barrier function in the latter. Both of these critical functions are mediated independently of canonical PK activity of PKM2, instead mediated via suppression of p53 and NF-kB, respectively. Targeting PKM2 as an adjuvant antineoplastic therapy might therefore be predicted to predispose already immunocompromised patients to sepsis.
Methods
Culture of human endothelial cells.
Pooled HUVECs were purchased from Lonza and used between passage 3 and 5. Cells were grown in EBM containing EGM supplements (Lonza, CC-3121 and CC-4133) with 10% FBS.
siRNA transfection.
Lipofectamine RNAimax reagent (Invitrogen) was used for siRNA transfection. For PKM2-specific knockdown, 4 different siRNAs designed in exon 10 of PKM were used: si27 (AGGCAGAGGCUGCCAUCUA), si87 (UUACCAGCGACCCCACAGA), si122 (GCCGUGGAGGCCUCCUUCA), and si155 (GCCAUAAUCGUCCUCACCA) (12). See Supplemental Figure 1E for detail. For knockdown of both PKM2 and PKM1, siPKM (SASI_Hs01_00024880) that targets exon 2 of PKM was used. Other siRNAs used were siAngpt2 (SASI_Hs02_00327020) and siRELB (SASI_Hs01_00103187). All siRNAs are from Sigma-Aldrich.
qPCR.
mRNA was isolated and cDNAs were synthesized by using TurboCapture mRNA kit (Qiagen) according to manufacturer’s instructions. qPCR was performed on the CFX384 Bio-Rad Real-Time PCR detection system by using SYBR green (iQ SYBR Green super mix). The sequences of the primers used in this study are as follows: PKM (forward: ATAACGCCTACATGGAAAAGTGT, reverse: TAAGCCCATCATCCACGTAGA), PKM1 (forward: ATAGCTCGTGAGGCTGAGGCAGCCATGTT, reverse: ACTCCGTCAGAACTATCAAAGCTGCTGCTA), PKM2 (forward: TGAGGCAGAGGCTGCCATCTACCACTT, reverse: TGCCAGACTTGGTGAGGACGATTATGGC), p53 (forward: TTTGCGTGTGGAGTATTTGGAT, reverse: CAACCTCAGGCGGCTCATA), p21 (forward: TGTCCGTCAGAACCCATGC, reverse: AAAGTCGAAGTTCCATCGCTC), ANGPT2 (forward: AATGCAGTACAGAACCAGACG, reverse: TTAACTTCCGCGTTTGCTCAG), RELA (forward: ATGTGGAGATCATTGAGCAGC, reverse: CCTGGTCCTGTGTAGCCATT), RELB (forward: CAGCCTCGTGGGGAAAGAC, reverse: GCCCAGGTTGTTAAAACTGTGC), p50 (forward: AACAGAGAGGATTTCGTTTCCG, reverse: TTTGACCTGAGGGTAAGACTTCT).
Western blot.
Cells were lysed in RIPA buffer that contained phosphatase inhibitor (PhosSTOP, Roche) and proteinase inhibitor (Complete mini proteinase inhibitor cocktail, Roche). The insoluble cell debris or lipid fraction was removed by centrifugation. Protein concentration was measured by BCA protein assay kit (Thermo Fisher Scientific). The samples were then boiled in SDS sample buffer and loaded into 4%–20% gradient gel (Bio-Rad), transferred to PVDF membrane (Millipore), and analyzed by immunoblotting. The following antibodies were used: PKM1 (CST 7067), PKM2 (CST 4053), actin (CST 4097), GAPDH (CST 5714), p53 (CST 2524), p21 (CST 2947), CCNE1 (CST 4129), CCNB1 (CST 4138), CCND1 (CST 2978), Angpt2 (R&D Systems, MAB0983-100). Secondary antibodies were purchased from Cell Signaling Technology. Signal was detected using the ECL system (ImageQuant LAS 4000, Amersham Biosciences, GE Healthcare) according to the manufacturer’s instructions.
Cell viability assay.
Cell viability was analyzed by using the Dead Cell Apoptosis Kit with Annexin V Alexa Fluor 488 and propidium iodide (PI) (Thermo Fisher Scientific) according to manufacturer’s instructions and analyzed via flow cytometry.
Transwell migration assay.
Two days after siRNA transfection, cells were trypsinized and the same number of cells was applied to each of 8-μm pore transwells that were precoated with gelatin. The cells were allowed to migrate across the membrane for 12 hours. The cells that remained in the upper chamber were removed by using a cotton swab, and the migrated cells on the lower side of the membrane were stained with phalloidin and DAPI for visualization.
Scratch closure assay.
Scratch assay was conducted on a confluent HUVEC monolayer 2 days after siRNA transfection. Cells were pretreated with mitomycin C at 10 μg/ml for 2 hours prior to wound infliction to inhibit proliferation. The scratch closure was monitored for 12 hours in EGM media supplemented with 0.2% FBS. Scratch closure was quantified using Image J.
Tube formation assay.
Tube formation assay was performed on Matrigel to analyze more of an organized migration. Two days after siRNA transfection, the same number of HUVECs (10,000 cells) was seeded on top of Matrigel in each well of a 96-well plate and photographed 6 hours after incubation.
Mice.
We generated PKM2ΔEC mice that lack exon 10 of PKM gene specifically in ECs. Previously described PKM2fl/fl mice (15) were bred with transgenic mice expressing Cre recombinase under the tamoxifen-inducible VE-cadherin promoter (16) (provided by Ralf Adams, Max Planck Institute for Molecular Biomedicine, Muenster, Germany).
Retina staining.
For the analysis of retinal vasculature, tamoxifen was injected into P1–P3 pups for 3 consecutive days. Eyes were harvested at P7 and fixed in 4% paraformaldehyde (PFA) on ice for 30 minutes. Eyes were washed in PBS and retinas were dissected. Isolated retinas were incubated with blocking/permeabilization solution in 5% BSA and 0.3% Triton X-100 (in PBS) for 1 hour at room temperature. The retinas were incubated with primary antibody at 4°C overnight. Primary antibodies against the following proteins were used: isolectin IB4 Alexa Fluor 488 conjugate (Thermo Fisher Scientific), CD31 (Millipore), phosphorylated histone H3 (pH3) (Millipore), aSMA Cy3 conjugate (MilliporeSigma), collagen IV (Serotec). After washing in 0.3% Triton X-100, retinas were incubated with secondary antibodies at 4°C overnight. For IB4 staining, the retina was mounted after washing without secondary antibody incubation.
Aortic capillary sprouting assay.
Thoracic aorta segments (~500-um long, >6 segments per aorta) were dissected from 3 mice per group and implanted in Matrigel as previously described (34). Each explant was photographed 3 days later. The ex vivo angiogenesis was analyzed as the area of capillary outgrowth using Image J software.
Wound healing assay.
Wound healing assay was performed as a tissue injury model as previously described (35). Briefly, after administration of anesthesia (ketamine/xylazine), mice were shaved in the back, wiped with ethanol, and 5-mm wounds were made by using a biopsy punch. To monitor wound healing, each mouse was anesthetized every 2–3 days and digital pictures of wounds were acquired.
Pyruvate kinase activity assay.
Pyruvate kinase activity assay kit (MilliporeSigma, MAK072) was used to measure PK activity according to the manufacturer’s instruction.
Seahorse assay.
Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were measured using the Seahorse XF96 analyzer (Seahorse Bioscience) according to the manufacturer’s instruction. Briefly, 2 days after siRNA transfection, HUVECs were seeded on XF96 microplates precoated with gelatin. After overnight incubation, the media was changed to a nonbuffered assay medium and preincubated in a 37°C incubator without CO2 for an hour. For glycolysis stress test, glucose (10 mM), oligomycin (1 μM), and 2-DG (50 mM) were sequentially added to the assay medium that is supplemented with glutamine (4 mM). For mitochondria stress test, oligomycin (1 μM), FCCP (0.5 μM), and Rotenone/antimycin A (0.5 μM) were sequentially added to the assay medium supplemented with glucose (5.5 mM) and glutamine (4 mM).
Isotopic labeling and mass spectrometry analysis.
The tracer [U-13C] glucose was purchased from Cambridge Isotope Laboratories. Isotope-labeled glucose medium was prepared in DMEM-based media supplemented with EGM bullet kit. Cells were cultured for 24 hours in the presence of tracer, and metabolites were extracted by aspirating medium and immediately adding 80% methanol (MeOH) prechilled at –80°C. After 20 minutes of incubation on dry ice, the extract was scraped, collected into a centrifuge tube, and centrifuged at 10,000g for 5 minutes. Insoluble pellets were reextracted with 1 ml MeOH. The supernatants from 2 rounds of extraction were combined, dried under nitrogen gas, and analyzed by reverse-phase ion pairing chromatography coupled with negative mode ESI high-resolution MS on a stand-alone orbitrap (36).
Cell cycle analysis.
Cells were detached from vessel by trypsinization and fixed in ice-cold 70% ethanol. Fixed cells were then stained with propidium iodide (PI) and analyzed via flow cytometry.
Retroviral overexpression.
Mouse PKM1 (pLHCX-Flag-mPKM2, Addgene plasmid 42511), PKM2 (pLHCX-Flag-mPKM2, Addgene plasmid 42512), or various PKM2 mutants (generated by site-directed mutagenesis) were overexpressed in HUVECs via retroviral infection.
CRISPR screen.
To identify genes whose knockout rescues the impaired proliferation of PKM2 knockdown ECs, we performed a genome-wide CRISPR/Cas9 screen (schematic in Supplemental Figure 5A) by using the GeCKOv2 human library that contains 6 different gRNAs targeting each of over 20,000 genes in the lentiGuide-Puro vector (37). Briefly, HUVECs were infected with lentivirus that contains this library. Infected cells were selected with puromycin for 5 days and then transfected with scrambled RNA (siCTL) or siPKM2. These cells were cultured and harvested after short (2 days) or long (10 days) time points. Rescue in proliferation was observed from the siPKM2 cells cultured for 10 days, in contrast to those receiving siCTL, suggesting that knockout of some genes by CRISPR rescued the blocked proliferation by siPKM2. gRNA sequences were then amplified by PCR from these cells and sequenced on a HiSeq 2500 (Illumina). Two different comparisons were performed to identify enriched gRNAs: (a) the 10-day-cultured population compared with the 2-day-cultured population after transfecting with siPKM2, and (b) the 10-day-cultured population after siPKM2 compared with siCTL. Z scores were used to determine the enrichment of gRNAs between the 2 groups (38). gRNAs targeting AGO2 (a key regulator of RISC complex), an internal positive control of this screen because it is necessary for knocking PKM2 down with siRNA transfection, ranked number 1 in Z score.
Trans-endothelial electrical resistance (TEER).
Endothelial permeability was measured by TEER analysis in an endothelial monolayer using an 8-well gold microelectrode (8W10E+, 40 microelectrodes per well, Applied Biosciences) pretreated with 10 mM l-cysteine and 1% gelatin. Cells were counted and full confluency of cells was reseeded onto an 8W1E+ electrode 2 days after transfection. The 8W1E+ electrode was connected to an electric cell-substrate impedance sensor (ECIS) system (Applied BioPhysics) in a chamber at 37°C with 5% CO2, and resistance changes were measured at 4,000 Hz.
Miles assay.
Hair was shaved from the entire back of the mice 1 day prior to experiment. Mice were anesthetized with isoflurane and intravenously injected with Evans Blue dye at 50 mg/kg. After 10 minutes, recombinant human VEGF-A121 or PBS as a vehicle control was injected intradermally. After 20 minutes, organs and skin were harvested and photographed. Tissues were incubated in 1 ml formamide at 56°C for 48 hours to extract extravasated dye. Absorbance of extravasated dye was measured at 620 nm with a standard curve of known dye concentrations and normalized to tissue weight.
Cecal ligation and puncture surgery.
The abdominal area was disinfected after shaving. A midline incision was made and cecum was exposed out of the abdominal cavity. A 1-cm ligation was made and cecum was perforated with through-and-through puncture by using a 21-gauge needle. The cecum was gently squeezed to ensure the patency of holes. Cecum was then placed back into cavity and the incision was closed with sutures. Prewarmed saline (1 ml/20 g body weight) was subcutaneously administered after procedure. Sham group underwent the same procedures except for the ligation and puncture of the cecum. For ABTAA injection experiment, either ABTAA or IgG control (10 mg/kg) was intravenously administered to mice via tail vein 1 hour before CLP.
Lung edema measurement.
The left lobe was excised, all extrapulmonary tissues were cleared and gently dried with blotting paper, and wet weight was measured. The tissue was then dried at 60°C for 72 hours and dry weight was measured. The change in the ratio of wet to dry weight was used as an indicator of lung edema.
Analysis of bacteremia.
Tail blood was collected before and 20 hours after CLP from each mouse into heparin-coated tubes (Microvete CB 300, Sarstedt). Whole blood (10 μl) was plated on 5% sheep blood agar following dilution in saline. After 24 hours of bacterial culture at 37°C, CFUs were quantified.
Angpt2 ELISA assay.
Tail blood was collected before and 20 hours after CLP from each mouse into MiniCollect TUBE with lithium heparin separator. Plasma samples were collected after centrifugation at 2,000g for 10 minutes at 4°C and immediately stored at –80°C until analyzed. Angpt2 ELISA assay was performed according to manufacturer instruction (Mouse/Rat Angiopoietin-2 kit, R&D Systems).
Statistics.
P values were calculated using the 2-tailed Student’s t test. For statistical comparisons between study groups, 1-way ANOVA was used followed by Bonferroni’s post hoc testing. P < 0.05 was considered statistically significant. All data are mean ± SD. Data from cell culture experiments are representative from a minimum of 3 independent experiments.
Study approval.
All mouse experiments were performed according to procedures approved by the University of Pennsylvania Institute for Animal Care and Use Committees (Philadelphia, PA).
Author contributions
BK, CJ, HD, and ZA designed the research studies. BK, CJ, HD, JL, MB, SY, and KL conducted experiments and acquired data. BK, CJ, HD, and ZA analyzed data. BK and ZA wrote the manuscript.
Supplementary Material
Acknowledgments
We thank Matt Vander Heiden for contributing the PKM2 floxed mice and Ralf Adams for the VeCad-ERTCre mice. We also thank Gou Young Koh for providing ABTAA antibody. This work was supported by fellowships from the American Heart Association and American Diabetes Association (to BK), a fellowship from the American Diabetes Association (to CJ), grants from the NIH (HL094499 and DK107667 to ZA), pilot funding and support from the DRC Regional Metabolomics Core (P30 DK19525), and an Established Investigator Award the American Heart Association (to ZA).
Version 1. 09/17/2018
Electronic publication
Version 2. 10/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(10):4543–4556.https://doi.org/10.1172/JCI120912.
Contributor Information
Boa Kim, Email: boakim@pennmedicine.upenn.edu.
Cholsoon Jang, Email: cholsoon@princeton.edu.
Harita Dharaneeswaran, Email: hdharane@gmail.com.
Jian Li, Email: lij2@mail.med.upenn.edu.
Mohit Bhide, Email: mobhide@sas.upenn.edu.
Steven Yang, Email: ysteven@sas.upenn.edu.
Kristina Li, Email: kli1@bu.edu.
References
- 1.Rohlenova K, Veys K, Miranda-Santos I, De Bock K, Carmeliet P. Endothelial cell metabolism in health and disease. Trends Cell Biol. 2018;28(3):224–236. doi: 10.1016/j.tcb.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Pircher A, Treps L, Bodrug N, Carmeliet P. Endothelial cell metabolism: a novel player in atherosclerosis? Basic principles and therapeutic opportunities. Atherosclerosis. 2016;253:247–257. doi: 10.1016/j.atherosclerosis.2016.08.011. [DOI] [PubMed] [Google Scholar]
- 3.De Bock K, et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell. 2013;154(3):651–663. doi: 10.1016/j.cell.2013.06.037. [DOI] [PubMed] [Google Scholar]
- 4.Yu P, et al. FGF-dependent metabolic control of vascular development. Nature. 2017;545(7653):224–228. doi: 10.1038/nature22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. 2011;43(7):969–980. doi: 10.1016/j.biocel.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 6.Christofk HR, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452(7184):230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 7.Dayton TL, Jacks T, Vander Heiden MG. PKM2, cancer metabolism, and the road ahead. EMBO Rep. 2016;17(12):1721–1730. doi: 10.15252/embr.201643300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anastasiou D, et al. Pyruvate kinase M2 activators promote tetramer formation and suppress tumorigenesis. Nat Chem Biol. 2012;8(10):839–847. doi: 10.1038/nchembio.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walsh MJ, et al. ML265: A potent PKM2 activator induces tetramerization and reduces tumor formation and size in a mouse xenograft model. In: Probe Reports from the NIH Molecular Libraries Program. Bethesda, MD: National Center for Biotechnology Information (US); 2010. [PubMed] [Google Scholar]
- 10.Lunt SY, et al. Pyruvate kinase isoform expression alters nucleotide synthesis to impact cell proliferation. Mol Cell. 2015;57(1):95–107. doi: 10.1016/j.molcel.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boeckel JN, et al. JMJD8 regulates angiogenic sprouting and cellular metabolism by interacting with pyruvate kinase M2 in endothelial cells. Arterioscler Thromb Vasc Biol. 2016;36(7):1425–1433. doi: 10.1161/ATVBAHA.116.307695. [DOI] [PubMed] [Google Scholar]
- 12.Goldberg MS, Sharp PA. Pyruvate kinase M2-specific siRNA induces apoptosis and tumor regression. J Exp Med. 2012;209(2):217–224. doi: 10.1084/jem.20111487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erben RG, Odörfer KI, Siebenhütter M, Weber K, Rohleder S. Histological assessment of cellular half-life in tissues in vivo. Histochem Cell Biol. 2008;130(5):1041–1046. doi: 10.1007/s00418-008-0470-3. [DOI] [PubMed] [Google Scholar]
- 14.Sholley MM, Cotran RS. Endothelial DNA synthesis in themicrovasculature of rat skin during the hair growth cycle. Am J Anat. 1976;147(2):243–254. doi: 10.1002/aja.1001470208. [DOI] [PubMed] [Google Scholar]
- 15.Israelsen WJ, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155(2):397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sörensen I, Adams RH, Gossler A. DLL1-mediated Notch activation regulates endothelial identity in mouse fetal arteries. Blood. 2009;113(22):5680–5688. doi: 10.1182/blood-2008-08-174508. [DOI] [PubMed] [Google Scholar]
- 17.Vander Heiden MG, et al. Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science. 2010;329(5998):1492–1499. doi: 10.1126/science.1188015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ikeda Y, Tanaka T, Noguchi T. Conversion of non-allosteric pyruvate kinase isozyme into an allosteric enzyme by a single amino acid substitution. J Biol Chem. 1997;272(33):20495–20501. doi: 10.1074/jbc.272.33.20495. [DOI] [PubMed] [Google Scholar]
- 19.Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. 2005;44(27):9417–9429. doi: 10.1021/bi0474923. [DOI] [PubMed] [Google Scholar]
- 20.Zilfou JT, Lowe SW. Tumor suppressive functions of p53. Cold Spring Harb Perspect Biol. 2009;1(5):a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9(10):749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaufmann WK, Paules RS. DNA damage and cell cycle checkpoints. FASEB J. 1996;10(2):238–247. doi: 10.1096/fasebj.10.2.8641557. [DOI] [PubMed] [Google Scholar]
- 23.Szulcek R, Bogaard HJ, van Nieuw Amerongen GP. Electric cell-substrate impedance sensing for the quantification of endothelial proliferation, barrier function, and motility. J Vis Exp. 2014;(85):51300. doi: 10.3791/51300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Radu M, Chernoff J. An in vivo assay to test blood vessel permeability. J Vis Exp. 2013;(73):50062. doi: 10.3791/50062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parikh SM. The angiopoietin-tie2 signaling axis in systemic inflammation. J Am Soc Nephrol. 2017;28(7):1973–1982. doi: 10.1681/ASN.2017010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han S, et al. Amelioration of sepsis by TIE2 activation-induced vascular protection. Sci Transl Med. 2016;8(335):335ra55. doi: 10.1126/scitranslmed.aad9260. [DOI] [PubMed] [Google Scholar]
- 27.Xia L, et al. A novel role for pyruvate kinase M2 as a corepressor for P53 during the DNA damage response in human tumor cells. J Biol Chem. 2016;291(50):26138–26150. doi: 10.1074/jbc.M116.737056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palsson-McDermott EM, et al. Pyruvate kinase M2 regulates Hif-1α activity and IL-1β induction and is a critical determinant of the warburg effect in LPS-activated macrophages. Cell Metab. 2015;21(1):65–80. doi: 10.1016/j.cmet.2014.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang YH, et al. Cell-state-specific metabolic dependency in hematopoiesis and leukemogenesis. Cell. 2014;158(6):1309–1323. doi: 10.1016/j.cell.2014.07.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ouyang X, et al. Digoxin suppresses pyruvate kinase M2-promoted HIF-1α transactivation in steatohepatitis. Cell Metab. 2018;27(5):339–350. doi: 10.1016/j.cmet.2018.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qi W, et al. Pyruvate kinase M2 activation may protect against the progression of diabetic glomerular pathology and mitochondrial dysfunction. Nat Med. 2017;23(6):753–762. doi: 10.1038/nm.4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams AL, et al. HIF1 mediates a switch in pyruvate kinase isoforms after myocardial infarction. Physiol Genomics. 2018;50(7):479–494. doi: 10.1152/physiolgenomics.00130.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rees ML, Subramaniam J, Li Y, Hamilton DJ, Frazier OH, Taegtmeyer H. A PKM2 signature in the failing heart. Biochem Biophys Res Commun. 2015;459(3):430–436. doi: 10.1016/j.bbrc.2015.02.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baker M, et al. Use of the mouse aortic ring assay to study angiogenesis. Nat Protoc. 2011;7(1):89–104. doi: 10.1038/nprot.2011.435. [DOI] [PubMed] [Google Scholar]
- 35.Dioufa N, et al. Acceleration of wound healing by growth hormone-releasing hormone and its agonists. Proc Natl Acad Sci U S A. 2010;107(43):18611–18615. doi: 10.1073/pnas.1013942107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu YF, Lu W, Rabinowitz JD. Avoiding misannotation of in-source fragmentation products as cellular metabolites in liquid chromatography-mass spectrometry-based metabolomics. Anal Chem. 2015;87(4):2273–2281. doi: 10.1021/ac504118y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parnas O, et al. A Genome-wide CRISPR screen in primary immune cells to dissect regulatory networks. Cell. 2015;162(3):675–686. doi: 10.1016/j.cell.2015.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.