Abstract
A complex DNA repair machinery has evolved to protect genomic integrity in the face of a myriad of DNA damage sources. When DNA repair fails, this damage can lead to carcinogenesis and tumor genomic instability. Indeed, many heritable cancer predisposition syndromes are attributable to germline defects in DNA repair pathways. On the other hand, these defects may also portend particular vulnerabilities of the cancer and may be exploited therapeutically. Most recently this has been demonstrated in the case of mismatch repair-deficient cancers, in which the immune checkpoint inhibitors have been demonstrated to be highly active. This observation has paved the way for further research investigating other sources of genomic instability that may serve as biomarkers to select patients for immunotherapy.
Preservation of the integrity of the genetic code requires a complex machinery to defend against both endogenous and exogenous sources of damage. Multiple repair pathways have been described and are generally divided into those involved in single-strand repair of damaged or mispaired bases (e.g., base excision repair, nucleotide excision repair, direct repair, and mismatch repair pathways), and those involved in repair of DNA double-strand breaks (e.g., homology-dependent recombination, nonhomologous end joining, and the Fanconi anemia pathway). When DNA repair processes fail, damage resulting in somatic mutations in tumor suppressor genes or oncogenes can lead to uncontrolled proliferation and carcinogenesis. Several cancer susceptibility syndromes have been identified that are linked to heritable defects in specific DNA repair pathways and proteins; however, cancers associated with defective DNA repair may occur sporadically as well and defective DNA repair has a critical role in tumor progression (1). Defective DNA repair is highly prevalent in cancers, with a recent analysis of DNA damage repair deficiency across 33 cancer types reporting somatic alterations in approximately one-third of cancers analyzed (2).
The presence of such defects in DNA repair pathways in cancers may have therapeutic implications, indicating a potential vulnerability to certain DNA-damaging therapies, such as platinum chemotherapies and inhibitors of poly (ADP-ribose) polymerase (PARP) in the case of BRCA1/2-mutated breast and ovarian cancers and the alkylating agent temozolomide in MGMT-methylated glioblastoma multiforme (3–7). More recently, a role for DNA damage repair pathways in selection of patients for immunotherapy has emerged, and this review will summarize what is known regarding the association between DNA repair defects and the host immune response and tumor susceptibility to immunotherapy.
Tumor immunogenicity and response to immunotherapy
The immune checkpoint inhibitors (CPIs) are a class of drugs targeting negative inhibitory receptors on host T lymphocytes, such as cytotoxic T lymphocyte antigen 4 (CTLA-4) and programmed death 1 (PD-1), receptors that are frequently hijacked by tumors to prevent an effective antitumor immune response. Monoclonal antibodies blocking these checkpoints have been developed and have shown remarkable ability to induce deep and durable responses in certain advanced, treatment-refractory cancers, leading to regulatory approvals in a wide range of cancer subtypes, including melanoma, non–small cell lung cancer (NSCLC), head and neck cancer (HNSCC), renal cell carcinoma (RCC), urothelial carcinoma (UC), and Hodgkin lymphoma. Despite the dramatic responses seen in these cancers, clinically significant activity appears limited to a minority of patients, and the ability to predict response and better select patients for these therapies through the development of reliable biomarkers remains a critical need.
As the target of PD-(L)1 inhibitors, expression of PD-1’s ligand, PD-L1, is a natural potential biomarker of response and this has been explored extensively in numerous studies with conflicting results. A sensitivity analysis of PD-L1 expression and response to PD-(L)1 inhibitors in melanoma, NSCLC, and genitourinary cancers reported an overall response rate of 34.1% in PD-L1–positive and 19.9% in PD-L1–negative cancers, with a differential effect based on tumor type and specific CPI (8). A lack of standardization for testing as well as heterogeneity of expression in the tumor itself have likely contributed to these inconsistent results, and the true predictive value of this biomarker across tumor types remains to be determined. Other features of the tumor microenvironment, such as effector immune cell infiltration and inflammatory gene expression signatures, can also suggest a more immunologically active, inflamed phenotype and are being explored as potential biomarkers for selection of patients for CPI therapy (9–13).
Emerging evidence suggests tumor-intrinsic genomic instability as the basis for the enhanced immunogenicity seen in some cancers, and tumor mutation burden (TMB) in particular has emerged as a promising potential biomarker for immunotherapy. This began with the observation that several cancers in which CPIs are highly active are associated with mutagenic exposures (e.g., smoking in lung cancer, UV light in melanoma). Additionally, measures of carcinogen exposure, including pack-year smoking history and frequency of C>A transversions (a molecular signature of smoking) in the case of lung cancer, have been correlated with clinical benefit from CPI (14, 15). Resultant high TMB was thought to link mutagenic exposure with enhanced immunogenicity in these cancers, and numerous studies of patients with melanoma and lung cancer receiving CTLA-4 and/or PD-(L)1 inhibitors have since confirmed the association between TMB and likelihood of response (16–19).
TMB is an attractive biomarker given the increasing ease and availability of sequencing techniques; however, similarly to PD-L1 expression, it appears limited in its predictive value, with responses in intermediate-TMB/low-TMB tumors as well as nonresponders among the high-TMB tumors. A recent analysis of immunotherapy-treated patients who had undergone TMB assessment reported a response rate of 58% in high-TMB cases (≥20 mutations per Mb) versus 20% in those with intermediate and low TMB; however, this relationship was less prominent in patients treated with dual CPI (PD-(L)1 plus CTLA-4 inhibitor) (20). Two recent studies in NSCLC have examined its relationship with respect to PD-L1 expression as a biomarker of response. One study demonstrated superiority of combination CPI over chemotherapy in the first line in patients with high-TMB NSCLC tumors irrespective of PD-L1 expression status (21). A separate retrospective analysis in NSCLC suggested the two biomarkers may be complementary in patient selection for CPI, with potentially greatest benefit to combined CTLA-4/PD-(L)1 inhibition in high-TMB, high-PD-L1–expressing tumors (22).
The mechanism for the enhanced immunogenicity of high-TMB cancers is believed to be through the production of so-called mutation-associated neoantigens (MANAs), mutant proteins resulting from nonsynonymous mutations, which are more strongly immunogenic than their nonmutant counterparts (23). This has been supported by the demonstrated association of MANA-specific T cell responses with outcomes including improved objective responses, durable clinical benefit, progression-free survival, and overall survival (23–26), as well as by neoantigen loss as a mechanism of acquired resistance to CPIs (27).
It remains controversial whether it is quantity of neoantigens or the presence of certain high-quality neoantigens that is necessary to generate an antitumor immune response. In a study examining predictors of response to CTLA-4 inhibition in metastatic melanoma, high TMB was associated with clinical benefit but was not sufficient for response, with some high-TMB tumors among the nonresponders (28). The study identified a specific neoepitope signature that appeared more strongly associated with clinical benefit. Interestingly, these neoepitopes demonstrated sequence homology to more viral and bacterial antigens than neoepitopes that were not linked with clinical benefits. Others have similarly provided evidence that neoepitope quality, and particularly homology to microbial antigens, may be a more important factor than quantity in the antitumor immune response (29). On the other hand, another study of CTLA-4 inhibitors in melanoma confirmed the association of neoantigen load with clinical benefit, but was unable to identify a recurrent peptide sequence predictive of response (30). A trial is ongoing to prospectively evaluate treatment with pembrolizumab in patients with high-TMB tumors and may provide additional insight into the utility of TMB as a biomarker of response (NCT01876511).
Independent of neoantigen production, damaged DNA may also directly activate the immune system via the stimulator of interferon genes (STING) pathway (31). In this pathway, cytosolic double-stranded DNA is sensed by cyclic GMP-AMP synthase (cGAS), leading to activation of the transmembrane protein STING and subsequent recruitment and activation of transcription factors and upregulation of IFN-β and related genes (32). While first recognized for its role in the immune response to DNA viruses, there is accumulating evidence for a role for this pathway in response to host cytosolic DNA and in particular in the immune response to DNA damage repair-deficient (DDRD) tumors. For example, studies of mice deficient in ataxia-telangiectasia mutated (ATM), a crucial DNA damage signaling protein, and studies of patients with ataxia-telangiectasia (congenital ATM deficiency) demonstrated that ATM loss was associated with enhanced type I interferon production resulting from the accumulation of cytosolic DNA and activation of the STING pathway (33). In preclinical models, a functional cGAS pathway has been demonstrated to be necessary for response to PD-L1 inhibitor, and therapeutic activation of the cGAS pathway was shown to potentiate the activity of the CPI (34). Therefore, the upregulation of the cGAS/STING pathway in DDRD tumors may suggest a tumor primed for response to CPIs.
The presence of defects in DNA replication and repair in the tumor are a major source of genomic instability, with mutations in mismatch repair genes and DNA polymerases accounting for 13.5% of high-TMB tumors in a recent large-scale analysis (35). These defects may present another opportunity to optimally select patients for CPI therapy, as has been demonstrated in the case of cancers associated with mismatch repair deficiency.
DNA repair pathway defects and immunogenicity
Mismatch repair deficiency.
To date, deficiency in DNA mismatch repair (dMMR) remains the only validated tissue-agnostic biomarker of response to CPI therapy. dMMR cancers harbor impaired expression of one of the genes involved in the MMR pathway (MSH2, MSH6, MLH1, and PMS2), which corrects base mispairs and small loops in repetitive sequence DNA. Tumors with dMMR accumulate exceptionally high numbers of mutations, particularly at more vulnerable short tandem repeat sequences, termed microsatellites, leading to a microsatellite instability-high (MSI-H) phenotype. This can occur in the setting of the heritable Lynch syndrome due to a germline mutation in a gene encoding one of the DNA mismatch repair proteins, or sporadically through epigenetic alterations leading to loss of expression of these proteins. While certain cancers such as endometrial and gastrointestinal are enriched for these defects, MSI-H has been observed at lower levels across at least 24 different tumor types, and it is estimated that the annual incidence of new MSI-H cancers is in the thousands (36).
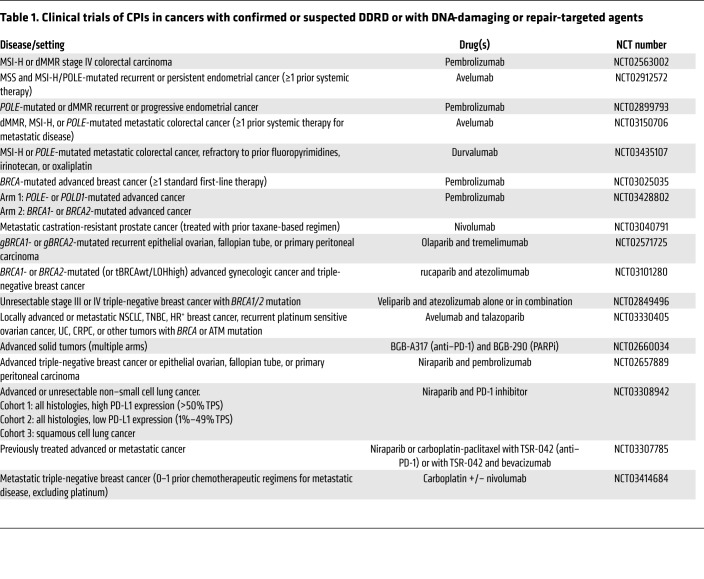
In addition to high TMB, MSI-H cancers bear other hallmarks of an immunogenic cancer, including prominent immune cell infiltration and increased expression of PD-1/PD-L1 (37, 38). There was therefore a strong rationale for testing CPIs in MSI-H cancers, and a phase II trial of the PD-1 inhibitor pembrolizumab in patients with metastatic treatment-refractory dMMR or MSI-H cancers demonstrated significant activity. In a cohort of 86 patients across 12 different cancer types, an objective response rate (ORR) of 53% was observed, including 21% complete responses; an additional 23% achieved stable disease for a disease control rate (DCR) of 77% (36, 39). Median disease-free and overall survivals had not been reached at a median follow-up time of 12.5 months. Similarly, in a phase II study in previously treated advanced dMMR/MSI-H colorectal cancers, treatment with the PD-1 inhibitor nivolumab yielded an ORR of 31.1% and a DCR at 12 weeks of 69%, with the median duration of response not yet reached (40). Nivolumab plus ipilimumab demonstrated substantial activity in the same population, with ORR of 55%, DCR at 12 weeks of 80%, and progression-free survival and overall survival rates at 12 months of 71% and 85%, respectively (41). These findings have led to the accelerated approval of pembrolizumab (KEYTRUDA) in advanced dMMR/MSI-H cancers and of nivolumab (OPDIVO) alone or in combination with ipilimumab (YERVOY) in advanced dMMR colorectal cancers (CRCs). Several trials are ongoing to evaluate the role of combination immunotherapy in dMMR cancers, as well as investigating CPIs in the first-line metastatic and nonmetastatic settings (Table 1).
Table 1. Clinical trials of CPIs in cancers with confirmed or suspected DDRD or with DNA-damaging or repair-targeted agents.
The identification of dMMR as the first tissue-agnostic biomarker of response to CPIs has led to an expansion of interest in other DNA repair defects and their potential implications for immunotherapy.
Homology-dependent recombination deficiency.
Among the most well-studied of DNA repair defects with regard to tumor immunogenicity is the defect in homology-dependent recombination (HR). Mutations in HR pathway proteins have been long recognized for their role in cancer susceptibility, in particular the hereditary breast and ovarian cancer syndromes associated with germline mutations in BRCA1 and BRCA2. HR is an important pathway of repair of DNA double-strand breaks (DSBs). In addition to BRCA1/2, the HR pathway includes other genes that may be altered in cancer, including partner and localizer of BRCA2 (PALB2). Defective HR leads to enhanced reliance on the alternative pathway of nonhomologous end-joining, which is associated with a unique genetic signature of frequent deletions flanked by short tandem repeat sequences (42). Both germline and somatic mutations in genes in the HR pathway have been since observed in a wide range of cancer subtypes (43), and these alterations may lead to a “BRCA-ness” phenotype that resembles germline BRCA1/2-mutant cancers, specifically in its enhanced susceptibility to platinum chemotherapy and PARP inhibitors (42).
Accumulating evidence suggests enhanced immunogenicity of HR-deficient (HRD) cancers and a potential role for CPIs. In high-grade serous ovarian cancers (HGSOCs), the presence of BRCA1/2 mutation was associated with higher predicted neoantigen load, increased tumor-infiltrating lymphocytes (TILs), enhanced PD-1/PD-L1 expression, and a gene expression signature associated with tumor cytotoxicity, when compared to HR-proficient HGSOCs (44). Enhanced TILs have also been observed in breast cancer associated with DNA repair defects (45, 46). In pancreatic ductal adenocarcinoma (PDA), molecular profiling revealed that a DSB repair-defective subtype was associated with an antitumor immune gene signature (47). A recent large-scale analysis of 777 tumors across multiple types with next-generation sequencing demonstrated defects in at least one HR gene in 25%, and significantly higher TMB in the HRD group than in the non-HRD group (48). Breast cancer cells associated with BRCA/Fanconi anemia repair pathway loss also harbored increased cytosolic DNA associated with activation of the cGAS/STING/TBK1/IRF3 pathway (49).
Preclinical and clinical data regarding the use of CPIs in HRD cancers has generated mixed results. In a preclinical model of BRCA1-associated triple-negative breast cancer, mice treated with cisplatin plus combined anti–PD-1 and anti-CTLA4 blockade demonstrated an enhanced intratumoral and systemic immune response and improved outcomes when compared to cisplatin alone, cisplatin with either CPI, or to CPI alone (45). Among patients with melanoma treated with PD-1 inhibitor, it was reported that responding tumors were enriched for BRCA2 mutations, while TMB was not a statistically significant predictor of response (50). In a phase II multicenter study of the Bruton tyrosine kinase inhibitor acalabrutinib with or without pembrolizumab in metastatic PDA, 3 of 32 patients receiving the combination achieved a partial response; of these three, one carried a germline BRCA2 variant of uncertain significance and the other two patients had a strong family history of pancreatic or breast cancer (51). On the other hand, a phase Ib trial of the PD-L1 inhibitor avelumab for recurrent or refractory ovarian cancer reported no responses in nine subjects with tumors confirmed or suspected to be BRCA-mutated (52).
Base excision repair pathway defects.
Biallelic mutations in MUTYH, encoding a DNA glycosylase involved in the base excision repair pathway, may also yield a “Lynch-like syndrome,” termed MUTYH-associated polyposis (MAP), with predisposition to CRC with predominant proximal location. These tumors have been reported to harbor increased TILs (53) and frequent loss of HLA class I expression, suggestive of immune-driven selective pressure (54). Sequencing of these tumors reveals a modestly increased mutational burden over pMMR CRC, with an average of 5.3 mutations per Mb, and a mutational signature characterized by prominent G:C>T:A transversions (55). The incidence of MAP in the general population is rare, accounting for approximately 0.4%–1% of CRC cases (56, 57), and the clinical activity of CPI in these patients has not been described.
Nucleotide excision repair pathway defects.
The nucleotide excision repair (NER) pathway is critical to the repair of UV-associated damage, as well as removal of bulky adducts, crosslinks, and oxidized bases (58). Xeroderma pigmentosum (XP) is a rare autosomal-recessive cancer predisposition syndrome associated with defective NER, affecting approximately only one per million people in the United States (59). These patients harbor germline defects in one of eight genes in the NER pathway and are highly susceptible to the development of skin and mucous membrane cancers (60). Due to the extreme rarity of this syndrome, documentation of clinical activity of CPIs is limited to case reports; however, dramatic and durable responses have been reported in melanomatous and nonmelanomatous XP-associated skin cancers (61–65).
Somatic alterations in the NER pathway and particularly in the ERCC genes are commonly observed and may have implications for immunotherapy. Specifically, in NSCLC it has been reported that the presence of ERCC1 SNPs was associated with significantly higher ORR (62.5% vs. 6.9%) to nivolumab (66).
Error-prone DNA replication as a source of genomic instability
DNA polymerase mutations.
Most promising in the search for additional markers of genomic instability and potential responsiveness to immunotherapy are mutations in genes encoding the DNA proofreading proteins polymerase δ (POLD1) and polymerase ε (POLE). These mutations may yield a phenotype similar to dMMR, including similar cancer susceptibilities when present in the germline (67). These tumors are often ultramutated and similarly demonstrate enhanced T lymphocyte infiltration and activation of immune checkpoints (68–72). Preliminary clinical data suggest that these cancers may also be able to achieve deep and durable responses to CPI therapy. A trial of a PD-1 inhibitor in NSCLC noted durable clinical response in three patients whose tumors harbored POLD1 or POLE mutations (18). Additionally, anecdotal responses to PD-1 inhibition have been reported in POLE-mutant endometrial cancer (71) and POLE-mutant microsatellite stable colorectal cancer (68).
Enhanced endogenous mutator activity.
The APOBEC cytidine deaminases are a family of endogenous mutators important for innate antiviral immunity; enhanced APOBEC activity has been observed in several cancers (73). Using data from 8,475 cancers in the Cancer Genome Atlas (TCGA), Boichard et al. demonstrated that alterations in APOBEC3 and the associated “kataegis” mutational signature, characterized by localized hypermutation, were associated with PD-L1/PD-L2 expression, independent of mutation burden (74). These findings have been confirmed in separate studies in UC (75, 76) and HNSCC (77). In NSCLC, the APOBEC signature was noted to be associated with loss of heterozygosity in human leukocyte antigen (HLA), suggestive of enhanced immune pressure (78). Together these findings support the hypothesis that the kataegis mutational signature can promote antitumor immunity and provides further evidence for enhanced immunogenicity of some cancers that is linked to specific mutational signatures rather than TMB alone.
Therapeutic strategies to induce genomic instability
Combination of CPIs with other DNA damage–enhancing agents may be a viable strategy to enhance genomic instability and therefore activity of immunotherapy. Standard of care cytotoxic chemotherapy and radiation therapy may enhance the immunogenicity of cancers through a number of potential mechanisms, including DNA damage and increased cytosolic dsDNA as well as inducing immunogenic cell death and enhanced antigen presentation. In support of this, a secondary analysis of patients participating in a phase I trial of pembrolizumab noted significantly longer survival in patients previously treated with radiotherapy (79). Several clinical trials are investigating DNA-damaging treatments in combination with CPI, and a recent study in NSCLC demonstrated a significant survival benefit with the addition of CPI to chemotherapy in the first line, in further support of this strategy (80).
Novel agents targeting DNA repair proteins, including the PARP inhibitors and inhibitors of ATM, ATM and RAD3-related (ATR), and CHK1, may also have a role in CPI combinations. In particular, PARP inhibitors have been observed to have immunomodulatory effects in the tumor, including upregulation of PD-L1 expression in preclinical models and increased infiltration by CD8 and NK cells, suggesting a potential role in combination with immune checkpoint blockade (81, 82). In a BRCA1-null syngeneic model of ovarian carcinoma, the PARP inhibitor rucaparib was noted to increase CD8+ T cell infiltration and CD8/CD4 ratio and had synergistic activity when combined with PD-1 or PD-L1 inhibition (83). Clinical trials evaluating PD-(L)1 inhibitors combined with PARP inhibitors are ongoing to further evaluate the activity of the combination. Preliminary results of 49 patients treated on a phase I trial of the PD-1 inhibitor tislelizumab plus the PARP inhibitor pamiparib in advanced cancers likely to harbor DNA damage repair deficiencies demonstrated an ORR of 20% and clinical benefit rate of 39% (84). The ongoing phase II MEDIOLA trial is evaluating the PD-L1 inhibitor durvalumab with the PARP inhibitor olaparib in BRCA1/2-mutated cancers, and it recently reported an 80% DCR at 12 weeks in BRCA1/2-mutated HER2-negative metastatic breast cancer (85). Another phase I trial of the PD-L1 inhibitor durvalumab plus olaparib in women’s cancers reported an ORR of 17% (2 responses of 12) and a DCR of 83%; of note, 11 of the 12 tumors were confirmed negative for BRCA mutations and the remaining one tumor was unconfirmed (86).
Summary and future directions
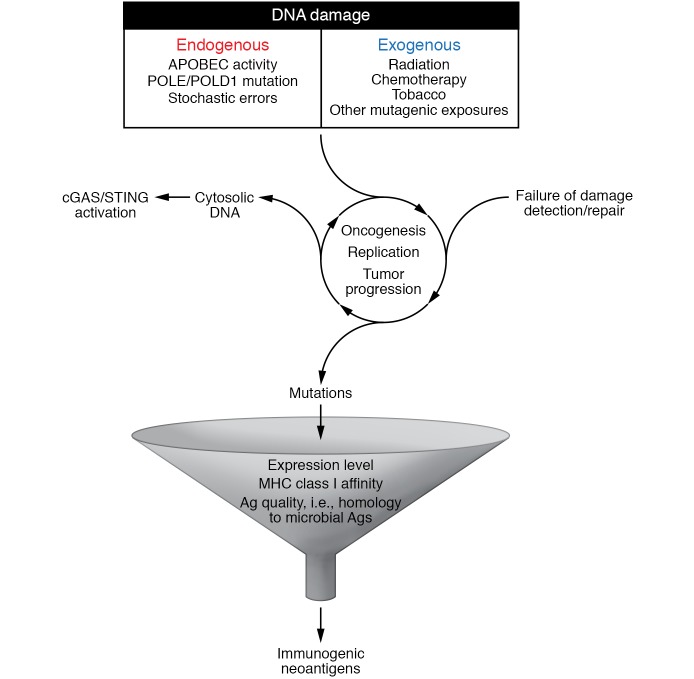
To date, dMMR remains the only confirmed genomic biomarker of response to CPIs, with deep and durable responses in a high proportion of these cancers. The exceptionally high TMB characteristic of these tumors likely underlies this immunogenicity and has opened the door to the exploration of other genomic biomarkers of response, and specifically other cancer-associated DNA repair pathway defects; however, how DNA damage and repair influences the immunogenicity of these cancers appears to be multifaceted and complex (Figure 1). In particular, how different signatures of DNA damage may impact tumor immunogenicity remains poorly understood and may influence the likelihood of response to CPI. In a study of 60 patients with advanced UC treated with PD-1 or PD-L1 inhibitor who underwent targeted exon sequencing on pretreatment tumor samples, the presence of a DDR alteration was associated with response rate of 67.9%, compared with 18.8% in those without; however, a differential effect on response was noted depending on affected pathway, with the largest effect seen in tumors with POLE mutations or defects in NER (87). Prospective studies are needed to confirm the relative contribution of specific pathway defects on likelihood of response to CPI.
Figure 1. DNA damage combined with failed DNA repair is an initiating step in oncogenesis, but also contributes to ongoing tumor progression with uncontrolled replication and accumulation of genomic errors and genomic instability.
Genomic instability may lead to enhanced tumor immunogenicity through (a) the accumulation of high numbers of mutations, possibly resulting in the expression of mutation-associated neoantigens, which can activate the adaptive immune response, and (b) the accumulation of cytosolic DNA, which can activate the innate immune response via the cGAS/STING pathway. Persistence/progression of these tumors thus relies on mechanisms of immune escape such as PD-(L)1. The addition of DNA-damaging therapies (e.g., chemotherapy, radiation, PARP inhibitors) may further enhance genomic instability and therefore synergize with CPIs in some tumors.
Additionally, for many of these defects, the optimal means of determination for its presence or absence also remains obscure. While clinically validated assays for dMMR via immunohistochemistry, polymerase chain reaction (PCR), or next-generation sequencing (NGS) exist, accurate assessment of HRD has proved more complex. Targeted sequencing requires thorough understanding of the relative contribution of involved proteins in the pathway and may miss altered expression through epigenetic alterations or promoter mutations. On the other hand, phenotypic assessment may be costly and may not reflect the current function of the pathway if intervening treatment occurred.
Finally, other DNA-damaging agents, including cytotoxic chemotherapy, radiation, and inhibitors of PARP, ATM, or ATR, may have the potential to exploit cancer-associated DNA repair defects and enhance genomic instability, and should be explored prospectively for their potential to increase susceptibility to immune attack and to synergize with CPI. Table 1 summarizes several ongoing trials evaluating immunotherapy in DDRD cancers and in combination with DNA-damaging therapies.
Acknowledgments
KMB and DTL are members of the Bloomberg Kimmel Institute for Cancer Immunotherapy. We thank Marian Novak for his critical review of the manuscript.
Version 1. 10/01/2018
Print issue publication
Footnotes
Conflict of interest: DTL receives research funding from Bristol-Meyers Squib, Merck, and Aduro Biotech, speaking honorarium from Merck, and serves on advisory boards at Bristol-Meyers Squib and Merck.
Reference information: J Clin Invest. 2018;128(10):4236–4242.
https://doi.org/10.1172/JCI122010.
References
- 1.Jeggo PA, Pearl LH, Carr AM. DNA repair, genome stability and cancer: a historical perspective. Nat Rev Cancer. 2016;16(1):35–42. doi: 10.1038/nrc.2015.4. [DOI] [PubMed] [Google Scholar]
- 2.Knijnenburg TA, et al. Genomic and molecular landscape of DNA damage repair deficiency across The Cancer Genome Atlas. Cell Rep. 2018;23(1):239–254.e6. doi: 10.1016/j.celrep.2018.03.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mylavarapu S, Das A, Roy M. Role of BRCA mutations in the modulation of response to platinum therapy. Front Oncol. 2018;8:16. doi: 10.3389/fonc.2018.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hegi ME, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 5.Hegi ME, et al. Correlation of O6-methylguanine methyltransferase (MGMT) promoter methylation with clinical outcomes in glioblastoma and clinical strategies to modulate MGMT activity. J Clin Oncol. 2008;26(25):4189–4199. doi: 10.1200/JCO.2007.11.5964. [DOI] [PubMed] [Google Scholar]
- 6.Robson M, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377(6):523–533. doi: 10.1056/NEJMoa1706450. [DOI] [PubMed] [Google Scholar]
- 7.Kaufman B, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol. 2015;33(3):244–250. doi: 10.1200/JCO.2014.56.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carbognin L, et al. Differential activity of nivolumab, pembrolizumab and MPDL3280A according to the tumor expression of programmed death-ligand-1 (PD-L1): sensitivity analysis of trials in melanoma, lung and genitourinary cancers. PLoS One. 2015;10(6):e0130142. doi: 10.1371/journal.pone.0130142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khunger M, et al. Incidence of pneumonitis with use of programmed death 1 and programmed death-ligand 1 inhibitors in non-small cell lung cancer: a systematic review and meta-analysis of trials. Chest. 2017;152(2):271–281. doi: 10.1016/j.chest.2017.04.177. [DOI] [PubMed] [Google Scholar]
- 10.Tumeh PC, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515(7528):568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ji RR, et al. An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother. 2012;61(7):1019–1031. doi: 10.1007/s00262-011-1172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ayers M, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Prat A, et al. Immune-related gene expression profiling after PD-1 blockade in non-small cell lung carcinoma, head and neck squamous cell carcinoma, and melanoma. Cancer Res. 2017;77(13):3540–3550. doi: 10.1158/0008-5472.CAN-16-3556. [DOI] [PubMed] [Google Scholar]
- 14.Gettinger SN, et al. Overall survival and long-term safety of nivolumab (anti-programmed death 1 antibody, BMS-936558, ONO-4538) in patients with previously treated advanced non-small-cell lung cancer. J Clin Oncol. 2015;33(18):2004–2012. doi: 10.1200/JCO.2014.58.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hellmann M, Rizvi N, Wolchok JD, Chan TA. Genomic profile, smoking, and response to anti-PD-1 therapy in non-small cell lung carcinoma. Mol Cell Oncol. 2016;3(1):e1048929. doi: 10.1080/23723556.2015.1048929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Johnson DB, et al. Targeted next generation sequencing identifies markers of response to PD-1 blockade. Cancer Immunol Res. 2016;4(11):959–967. doi: 10.1158/2326-6066.CIR-16-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carbone DP, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N Engl J Med. 2017;376(25):2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adusumilli PS, et al. New cancer immunotherapy agents in development: a report from an associated program of the 31stAnnual Meeting of the Society for Immunotherapy of Cancer, 2016. J Immunother Cancer. 2017;5:50. doi: 10.1186/s40425-017-0253-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodman AM, et al. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hellmann MD, et al. Nivolumab plus ipilimumab in lung cancer with a high tumor mutational burden. N Engl J Med. 2018;378(22):2093–2104. doi: 10.1056/NEJMoa1801946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellmann MD, et al. Tumor mutational burden and efficacy of nivolumab monotherapy and in combination with ipilimumab in small-cell lung cancer. Cancer Cell. 2018;33(5):853–861.e4. doi: 10.1016/j.ccell.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riaz N, Morris L, Havel JJ, Makarov V, Desrichard A, Chan TA. The role of neoantigens in response to immune checkpoint blockade. Int Immunol. 2016;28(8):411–419. doi: 10.1093/intimm/dxw019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gubin MM, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–581. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown SD, et al. Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res. 2014;24(5):743–750. doi: 10.1101/gr.165985.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGranahan N, et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351(6280):1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anagnostou V, et al. Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov. 2017;7(3):264–276. doi: 10.1158/2159-8290.CD-16-0828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Snyder A, et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–2199. doi: 10.1056/NEJMoa1406498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balachandran VP, et al. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 2017;551(7681):512–516. doi: 10.1038/nature24462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Van Allen EM, et al. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–211. doi: 10.1126/science.aad0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Corrales L, McWhirter SM, Dubensky TW, Gajewski TF. The host STING pathway at the interface of cancer and immunity. J Clin Invest. 2016;126(7):2404–2411. doi: 10.1172/JCI86892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corrales L, Gajewski TF. Molecular pathways: targeting the stimulator of interferon genes (STING) in the immunotherapy of cancer. Clin Cancer Res. 2015;21(21):4774–4779. doi: 10.1158/1078-0432.CCR-15-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Härtlova A, et al. DNA damage primes the type I interferon system via the cytosolic DNA sensor STING to promote anti-microbial innate immunity. Immunity. 2015;42(2):332–343. doi: 10.1016/j.immuni.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Wang H, et al. cGAS is essential for the antitumor effect of immune checkpoint blockade. Proc Natl Acad Sci U S A. 2017;114(7):1637–1642. doi: 10.1073/pnas.1621363114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chalmers ZR, et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017;9(1):34. doi: 10.1186/s13073-017-0424-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Le DT, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357(6349):409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smyrk TC, Watson P, Kaul K, Lynch HT. Tumor-infiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer. 2001;91(12):2417–2422. doi: 10.1002/1097-0142(20010615)91:12<2417::AID-CNCR1276>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 38.Llosa NJ, et al. The vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5(1):43–51. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Le DT, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overman MJ, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (checkMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overman MJ, et al. Durable clinical benefit with nivolumab plus ipilimumab in dna mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 42.Lord CJ, Ashworth A. BRCAness revisited. Nat Rev Cancer. 2016;16(2):110–120. doi: 10.1038/nrc.2015.21. [DOI] [PubMed] [Google Scholar]
- 43.Riaz N, et al. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat Commun. 2017;8(1):857. doi: 10.1038/s41467-017-00921-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strickland KC, et al. Association and prognostic significance of BRCA1/2-mutation status with neoantigen load, number of tumor-infiltrating lymphocytes and expression of PD-1/PD-L1 in high grade serous ovarian cancer. Oncotarget. 2016;7(12):13587–13598. doi: 10.18632/oncotarget.7277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nolan E, et al. Combined immune checkpoint blockade as a therapeutic strategy for BRCA1-mutated breast cancer. Sci Transl Med. 2017;9(393):eaal4922. doi: 10.1126/scitranslmed.aal4922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Green AR, et al. Clinical impact of tumor DNA repair expression and T-cell infiltration in breast cancers. Cancer Immunol Res. 2017;5(4):292–299. doi: 10.1158/2326-6066.CIR-16-0195. [DOI] [PubMed] [Google Scholar]
- 47.Connor AA, et al. Association of distinct mutational signatures with correlates of increased immune activity in pancreatic ductal adenocarcinoma. JAMA Oncol. 2017;3(6):774–783. doi: 10.1001/jamaoncol.2016.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li H, et al. Analysis of association between homologous recombination deficiency and tumor mutational burden in solid tumors. Cancer Res. 2018;78(13 supplement):1369. doi: 10.1158/1538-7445.AM2018-1369. [DOI] [Google Scholar]
- 49.Parkes EE, et al. Activation of STING-dependent innate immune signaling by S-phase-specific DNA damage in breast cancer. J Natl Cancer Inst. 2017;109(1):djw19. doi: 10.1093/jnci/djw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hugo W, et al. Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell. 2016;165(1):35–44. doi: 10.1016/j.cell.2016.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Overman MJ, et al. A randomized phase 2 study of the Bruton tyrosine kinase (Btk) inhibitor acalabrutinib alone or with pembrolizumab for metastatic pancreatic cancer (mPC) J Clin Oncol. 2016;34(15_suppl):4130. doi: 10.1200/JCO.2016.34.15_suppl.4130. [DOI] [Google Scholar]
- 52.Disis ML, et al. Avelumab (MSB0010718C; anti-PD-L1) in patients with recurrent/refractory ovarian cancer from the JAVELIN Solid tumor phase Ib trial: safety and clinical activity. J Clin Oncol. 2016;34(15_suppl):5533. doi: 10.1200/JCO.2016.34.15_suppl.5533. [DOI] [Google Scholar]
- 53.Nielsen M, et al. Colorectal carcinomas in MUTYH-associated polyposis display histopathological similarities to microsatellite unstable carcinomas. BMC Cancer. 2009;9:184. doi: 10.1186/1471-2407-9-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.de Miranda NF, et al. MUTYH-associated polyposis carcinomas frequently lose HLA class I expression — a common event amongst DNA-repair-deficient colorectal cancers. J Pathol. 2009;219(1):69–76. doi: 10.1002/path.2569. [DOI] [PubMed] [Google Scholar]
- 55.Viel A, et al. A specific mutational signature associated with DNA 8-oxoguanine persistence in MUTYH-defective colorectal cancer. EBioMedicine. 2017;20:39–49. doi: 10.1016/j.ebiom.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Croitoru ME, et al. Association between biallelic and monoallelic germline MYH gene mutations and colorectal cancer risk. J Natl Cancer Inst. 2004;96(21):1631–1634. doi: 10.1093/jnci/djh288. [DOI] [PubMed] [Google Scholar]
- 57.Cleary SP, et al. Germline MutY human homologue mutations and colorectal cancer: a multisite case-control study. Gastroenterology. 2009;136(4):1251–1260. doi: 10.1053/j.gastro.2008.12.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garfinkel DJ, Bailis AM. Nucleotide excision repair, genome stability, and human disease: new insight from model systems. J Biomed Biotechnol. 2002;2(2):55–60. doi: 10.1155/S1110724302201023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Walsh MF, et al. Recommendations for childhood cancer screening and surveillance in DNA repair disorders. Clin Cancer Res. 2017;23(11):e23–e31. doi: 10.1158/1078-0432.CCR-17-0465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cleaver JE, Lam ET, Revet I. Disorders of nucleotide excision repair: the genetic and molecular basis of heterogeneity. Nat Rev Genet. 2009;10(11):756–768. doi: 10.1038/nrg2663. [DOI] [PubMed] [Google Scholar]
- 61.Salomon G, et al. Efficacy of anti-programmed cell death-1 immunotherapy for skin carcinomas and melanoma metastases in a patient with xeroderma pigmentosum. Br J Dermatol. 2018;178(5):1199–1203. doi: 10.1111/bjd.16270. [DOI] [PubMed] [Google Scholar]
- 62.Kraemer KH, Tamura D, Khan SG. Pembrolizumab treatment of a patient with xeroderma pigmentosum with disseminated melanoma and multiple nonmelanoma skin cancers. Br J Dermatol. 2018;178(5):1009. doi: 10.1111/bjd.16525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hauschild A, et al. Regression of melanoma metastases and multiple non-melanoma skin cancers in xeroderma pigmentosum by the PD1-antibody pembrolizumab. Eur J Cancer. 2017;77:84–87. doi: 10.1016/j.ejca.2017.02.026. [DOI] [PubMed] [Google Scholar]
- 64.Deinlein T, Lax SF, Schwarz T, Giuffrida R, Schmid-Zalaudek K, Zalaudek I. Rapid response of metastatic cutaneous squamous cell carcinoma to pembrolizumab in a patient with xeroderma pigmentosum: case report and review of the literature. Eur J Cancer. 2017;83:99–102. doi: 10.1016/j.ejca.2017.06.022. [DOI] [PubMed] [Google Scholar]
- 65.Chambon F, Osdoit S, Bagny K, Moro A, Nguyen J, Réguerre Y. Dramatic response to nivolumab in xeroderma pigmentosum skin tumor. Pediatr Blood Cancer. 2018;65(2):e26837. doi: 10.1002/pbc.26837. [DOI] [PubMed] [Google Scholar]
- 66.Aiello MM, Vigneri PG, Bruzzi P, Verderame F, Paratore S, Restuccia N. Excision repair cross complementation group 1 (ERCC-1) gene polymorphisms and response to nivolumab in advanced non-small cell lung cancer (NSCLC) J Clin Oncol. 2017;35(15_suppl):3032. doi: 10.1200/JCO.2017.35.15_suppl.3032. [DOI] [Google Scholar]
- 67.Buchanan DD, et al. Risk of colorectal cancer for carriers of a germ-line mutation in POLE or POLD1. Genet Med. doi: 10.1038/gim.2017.185. doi: 10.1038/gim.2017.185. [published online ahead of print November 9, 2017]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gong J, Wang C, Lee PP, Chu P, Fakih M. Response to PD-1 blockade in microsatellite stable metastatic colorectal cancer harboring a POLE mutation. J Natl Compr Canc Netw. 2017;15(2):142–147. doi: 10.6004/jnccn.2017.0016. [DOI] [PubMed] [Google Scholar]
- 69.van Gool IC, et al. POLE proofreading mutations elicit an antitumor immune response in endometrial cancer. Clin Cancer Res. 2015;21(14):3347–3355. doi: 10.1158/1078-0432.CCR-15-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Howitt BE, et al. Association of polymerase e-mutated and microsatellite-instable endometrial cancers with neoantigen load, number of tumor-infiltrating lymphocytes, and expression of PD-1 and PD-L1. JAMA Oncol. 2015;1(9):1319–1323. doi: 10.1001/jamaoncol.2015.2151. [DOI] [PubMed] [Google Scholar]
- 71.Mehnert JM, et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J Clin Invest. 2016;126(6):2334–2340. doi: 10.1172/JCI84940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Domingo E, et al. Somatic POLE proofreading domain mutation, immune response, and prognosis in colorectal cancer: a retrospective, pooled biomarker study. Lancet Gastroenterol Hepatol. 2016;1(3):207–216. doi: 10.1016/S2468-1253(16)30014-0. [DOI] [PubMed] [Google Scholar]
- 73.Rebhandl S, Huemer M, Greil R, Geisberger R. AID/APOBEC deaminases and cancer. Oncoscience. 2015;2(4):320–333. doi: 10.18632/oncoscience.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Boichard A, Tsigelny IF, Kurzrock R. High expression of PD-1 ligands is associated with kataegis mutational signature and APOBEC3 alterations. Oncoimmunology. 2017;6(3):e1284719. doi: 10.1080/2162402X.2017.1284719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mullane SA, et al. Correlation of Apobec Mrna expression with overall survival and PD-L1 expression in urothelial carcinoma. Sci Rep. 2016;6:27702. doi: 10.1038/srep27702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Glaser AP, et al. APOBEC-mediated mutagenesis in urothelial carcinoma is associated with improved survival, mutations in DNA damage response genes, and immune response. Oncotarget. 2018;9(4):4537–4548. doi: 10.18632/oncotarget.23344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Reike DT, et al. Association of an APOBEC mutational signature, mutational load, and BRCAness with inflammation and PD-L1 expression in HNSCC. J Clin Oncol. 2018;35(15_suppl):e14613 [Google Scholar]
- 78.McGranahan N, et al. Allele-specific HLA loss and immune escape in lung cancer evolution. Cell. 2017;171(6):1259–1271.e11. doi: 10.1016/j.cell.2017.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shaverdian N, et al. Previous radiotherapy and the clinical activity and toxicity of pembrolizumab in the treatment of non-small-cell lung cancer: a secondary analysis of the KEYNOTE-001 phase 1 trial. Lancet Oncol. 2017;18(7):895–903. doi: 10.1016/S1470-2045(17)30380-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gandhi L, et al. Pembrolizumab plus chemotherapy in metastatic non-small-cell lung cancer. N Engl J Med. 2018;378(22):2078–2092. doi: 10.1056/NEJMoa1801005. [DOI] [PubMed] [Google Scholar]
- 81.Jiao S, et al. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res. 2017;23(14):3711–3720. doi: 10.1158/1078-0432.CCR-16-3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sato H, et al. DNA double-strand break repair pathway regulates PD-L1 expression in cancer cells. Nat Commun. 2017;8(1):1751. doi: 10.1038/s41467-017-01883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Robillard L, et al. Preclinical evaluation of the PARP inhibitor rucaparib in combination with PD-1 PD-L1 inhibition in a syngeneic BRCA1 mutant ovarian cancer model. AACR Annual Meeting. 2017;77(13 Supplement):3640 [Google Scholar]
- 84.Friedlander M, et al. A phase 1b study of the anti-PD-1 monoclonal antibody BGB-A317 (A317) in combination with the PARP inhibitor BGB-290 (290) in advanced solid tumors. J Clin Oncol. 2017;35(15_suppl):3013. doi: 10.1200/JCO.2017.35.15_suppl.3013. [DOI] [Google Scholar]
- 85.Domchek SM, et al. An open-label, multitumor, phase II basket study of alaparib and durvalumab (MEDIOLA): results in germline BRCA-mutated (gBRCAm) HER2-negative metastatic breast cancer (MBC) Cancer Res. 2018;78(4 Supplement):PD6-11. doi: 10.1158/1538-7445.SABCS17-PD6-11. [DOI] [Google Scholar]
- 86.Lee JM, et al. Safety and clinical activity of the programmed death-ligand 1 inhibitor durvalumab in combination with poly (ADP-Ribose) polymerase inhibitor olaparib or vascular endothelial growth factor receptor 1-3 inhibitor cediranib in women’s cancers: a dose-escalation, phase I study. J Clin Oncol. 2017;35(19):2193–2202. doi: 10.1200/JCO.2016.72.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Teo MY, et al. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 Blockade in advanced urothelial cancers. J Clin Oncol. 2018;36(17):1685–1694. doi: 10.1200/JCO.2017.75.7740. [DOI] [PMC free article] [PubMed] [Google Scholar]