Abstract
The E3 ubiquitin ligase RNF8 plays critical roles in maintaining genomic stability by promoting the repair of DNA double-strand breaks (DSBs) through ubiquitin signaling. Abnormal activation of Notch signaling and defective repair of DSBs promote breast cancer risk. Here, we found that low expression of the full-length RNF8 correlated with poor prognosis for breast cancer patients. Our data revealed that in addition to its role in the repair of DSBs, RNF8 regulated Notch1 signaling and cell-fate determination of mammary luminal progenitors. Mechanistically, RNF8 acted as a negative regulator of Notch signaling by ubiquitylating the active NOTCH1 protein (N1ICD), leading to its degradation. Consistent with abnormal activation of Notch signaling and impaired repair of DSBs in Rnf8-mutant mammary epithelial cells, we observed increased risk of mammary tumorigenesis in mouse models for RNF8 deficiency. Notably, deficiency of RNF8 sensitized breast cancer cells to combination of pharmacological inhibitors of Notch signaling and poly(ADP-ribose) polymerase (PARP), suggesting implications for treatment of breast cancer associated with impaired RNF8 expression or function.
Keywords: Cell Biology, Oncology
Keywords: Signal transduction, Tumor suppressors
Introduction
Regulation of Notch signaling and repair of double-strand breaks (DSBs) is critical for maintaining tissue homeostasis and suppression of cancer. Notch signaling is important for the control of cell-fate determination of mammary epithelial cell (MEC) progenitors, and its abnormal activation is associated with human and mouse breast tumorigenesis (1–3). Defective DSB signaling promotes genomic instability and increases the risk for various human neoplasms, including breast cancer (4).
Notch signaling and DSB repair are highly regulated by posttranslational modifications including ubiquitylation (2, 5, 6). The E3 ligase RNF8 ubiquitylates histone H1 at DSB flanking sites, triggering an orchestrated recruitment of signaling and repair proteins (e.g., the breast cancer susceptibility protein BRCA1) to facilitate the repair of the break sites (7–13). Despite impaired DSB repair signaling in the absence of RNF8, and the reported RNF8 function in the activation of TWIST and cancer metastasis (14), whether deficiency of RNF8 promotes breast cancer initiation and development has yet to be examined. Here, we aimed to address the effect of Rnf8 mutation on mammary tumorigenesis. Notably, we also uncovered novel functions of RNF8 in the regulation of Notch signaling and cell-fate determination of MECs. Given the importance of DSB repair and Notch signaling in breast cancer, identification of mechanisms that coregulate these pathways will not only reveal novel key cancer drivers, but also can potentially provide novel, more effective, and precise therapeutic strategies.
Results
RNF8 deficiency promotes mouse mammary tumorigenesis.
To examine whether RNF8 deficiency fosters mammary tumorigenesis, we first examined mammary tissues of Rnf8–/– female mice. While Rnf8–/– females frequently developed hyperplastic lesions in mammary tissues at 13 months of age (60%; 12 of 20), no hyperplasia was observed in WT littermates (n = 19; Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI120401DS1). Next, monitoring the cohorts of Rnf8–/– females and WT controls for over 600 days, we observed that in addition to thymomas and/or lymphomas, 18% of Rnf8–/– females (6 of 33) also developed mammary adenocarcinomas (Figure 1, A and B).
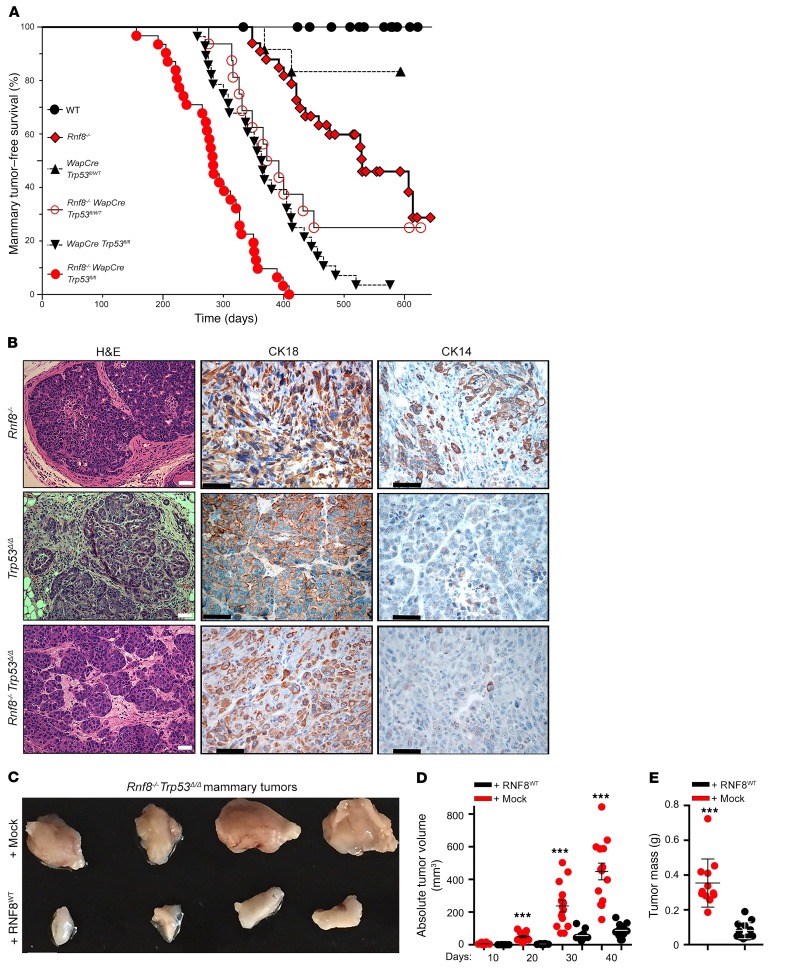
Figure 1. RNF8 deficiency promotes spontaneous mammary tumorigenesis in mouse models.
(A) Kaplan-Meier mammary tumor-free survival curves of cohorts of WT (n = 23), Rnf8–/– (n = 33), WapCre Trp53fl/fl (n = 28), WapCre Trp53fl/WT (n = 12), Rnf8–/– WapCre Trp53fl/WT (n = 16), and Rnf8–/– WapCre Trp53fl/fl (n = 31) females. Log-rank tests indicate statistically significant differences between Rnf8–/– and WT curves (P = 0.005), Rnf8–/– and Rnf8–/– WapCre Trp53fl/fl curves (P < 0.0001), Rnf8–/– WapCre Trp53fl/fl and WapCre Trp53fl/fl curves (P < 0.0001), and Rnf8–/– WapCre Trp53fl/WT and WapCre Trp53fl/WT curves (P < 0.003). (B) Representative H&E and cytokeratin (CK18 and CK14) immunohistochemical staining of Rnf8–/–, WapCre Trp53fl/fl (Trp53Δ/Δ), and Rnf8–/– WapCre Trp53fl/fl (Rnf8–/– Trp53Δ/Δ) mammary adenocarcinomas. At least 3 tumors per genotype were examined. Scale bars: 50 μm. (C) Representative photographs of Rnf8–/– Trp53Δ/Δ mammary tumors, either mock- or RNF8-reconstituted (as indicated), 40 days after orthotopic injection of these tumor cells into inguinal fat pads of NSG mice (n = 10–12 each). (D) Dot plots showing volume of tumors (n = 10–12 each; mean ± SEM) monitored for 40 days using external caliper. ***P < 0.001, 2-way ANOVA followed by Tukey’s test. (E) Dot plots depict average mass (mean ± SEM) of tumors resected from NSG mice (n = 10–12) following 40 days of outgrowth. ***P < 0.001, 2-sided Student’s t test.
Human TP53, and its mouse ortholog TRP53, are tumor suppressors important for the DNA damage response (DDR) (15, 16). Examination of Rnf8–/– mammary glands indicated upregulated expression of TRP53 and its transcriptional targets P21 and BAX, suggesting abnormally activated DDR in the absence of RNF8 (Supplemental Figure 1C). Based on the frequent mutations of TP53 in human breast cancer (17), we sought to examine the effect of inactivation of TRP53 in mammary tumorigenesis associated with Rnf8 mutation. Mice constitutively lacking RNF8 and TRP53 (Rnf8–/– Trp53–/–) died by approximately 12 weeks of age due to thymomas (18), precluding the examination of their risk for mammary tumorigenesis. To circumvent this limitation, Rnf8–/– and WapCre Trp53fl/fl mice were crossed to generate Rnf8–/– females in which Trp53 is conditionally deleted in the mammary epithelium following pregnancy. Monitoring cohorts of Rnf8–/– WapCre Trp53fl/fl, WapCre Trp53fl/fl, Rnf8–/–, and WT females, we observed that Trp53 deletion in the MECs of Rnf8–/– females significantly accelerated the incidence of mammary adenocarcinomas (Figure 1, A and B, and Supplemental Figure 1D). Interestingly, Rnf8–/– females lacking 1 copy of Trp53 in their MECs (Rnf8–/– WapCre Trp53fl/WT) also displayed elevated risk for developing mammary tumors, and these malignant lesions showed loss of heterozygosity for Trp53 (Figure 1A and Supplemental Figure 1E).
Mammary tumors developed by Rnf8–/–, Rnf8–/– WapCre Trp53fl/fl (referred to as Rnf8–/– Trp53Δ/Δ), and WapCre Trp53fl/fl (Trp53Δ/Δ) females exhibited predominant positivity for the luminal marker cytokeratin 18 (CK18) (Figure 1B). Rnf8–/– Trp53Δ/Δ mammary tumors were also positive for estrogen receptor (ER) (Supplemental Figure 1F), and exhibited metastasis to other organs (e.g., lung, lymph nodes, and brain) as confirmed by pan-CK staining (Supplemental Figure 1F). Moreover, Rnf8–/–, Rnf8–/– Trp53Δ/Δ, Rnf8–/– Trp53Δ/WT, and Trp53Δ/Δ mammary tumors were Lin– (lineage-negative: CD45–CD31–TER119–) and enriched for CD49floCD24+CD61+, markers of luminal progenitorsubpopulation (Supplemental Figure 1G).
Next, we sought to examine the effect of complementation of Rnf8-deficient mammary tumor cells with Rnf8WT on their in vivo growth capacity. We observed that relative to empty-vector controls, RNF8-reconstituted Rnf8–/– Trp53Δ/Δ tumor cells showed reduced growth when engrafted in inguinal fat pads of NOD scid gamma (NSG) mice as measured by tumor volume and mass (Figure 1, C–E, and Supplemental Figure 1H). Examination of the corresponding tumors revealed that RNF8WT reconstitution significantly restrained cancer cell proliferation as assessed with Ki67 staining (Supplemental Figure 1, I and J). Collectively, these data highlight the importance of RNF8 expression, and its collaboration with TRP53 function, in suppressing mammary tumorigenesis.
Low expression of the full-length RNF8 correlates with poor prognosis of breast cancer patients.
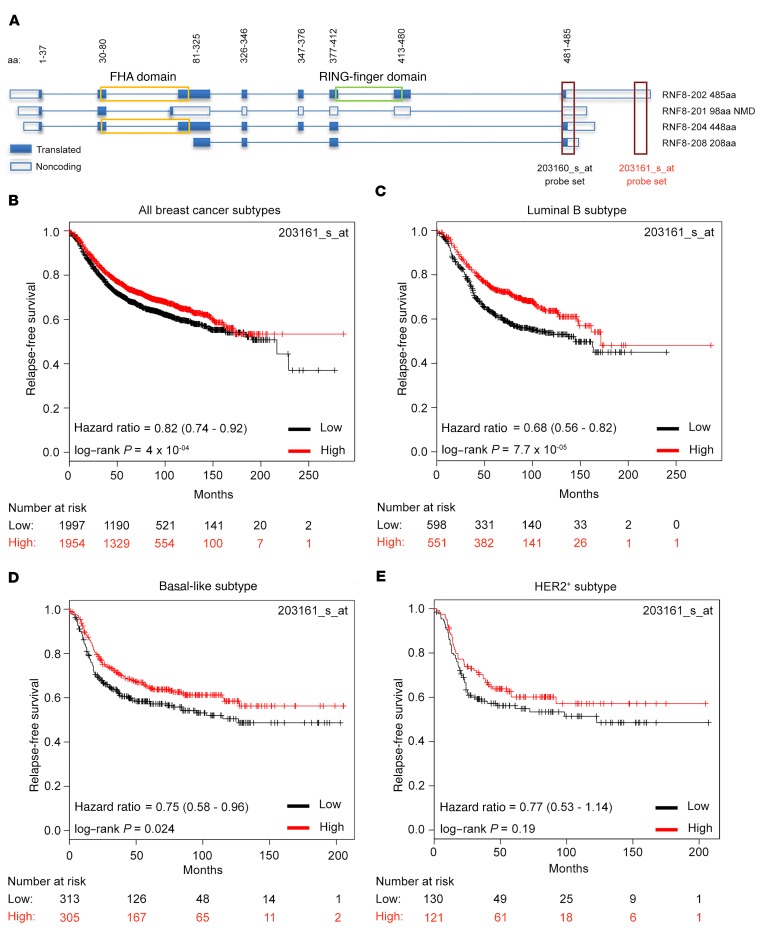
To examine the clinical relevance of RNF8 expression levels in human breast cancer subtypes, we used the Kaplan-Meier Plotter (KM Plotter) tool to analyze an integrated data set including 5,143 breast cancer cases of which 3,955 had relapse-free survival information (19). Two Affymetrix microarray probes for RNF8 are included in the application (Figure 2A). Disparity of results between microarray probes supposedly mapping on a single gene target has been previously recognized (e.g., BRCA1 and MYC) (20–22). Therefore, we first examined curated Ensembl annotations and observed that the human RNF8 locus includes at least 4 isoforms (Figure 2A). Examination of the 2 Affymetrix microarray probes for RNF8 indicated that they display different specificity toward these isoforms. While 203160_s_at recognizes all 4 isoforms, 203161_s_at only recognizes the transcript (RNF8-202) that encodes for the complete functional protein (according to UniProt, O76064_1), including its forkhead-associated (FHA) and RING-finger domains (Figure 2A), both required for RNF8 function in DSB repair (7–10). In contrast, the other RNF8 isoforms (201, 204, and 208) lack the FHA domain, the RING-finger domain, or both and therefore are predicted to be nonfunctional (Figure 2A). KM Plotter analysis of breast cancer patients was performed using the median expression level as the cutoff. Since the 203160_s_at probe set recognizes RNF8 transcripts lacking functional domains (Figure 2A), these analyses indicated a modest association between high RNF8 expression and reduced relapse-free survival of breast cancer patients (Supplemental Figure 2A; hazard ratio = 1.15 [1.03–1.28], log-rank P = 0.013). However, the same analysis using the functionally relevant RNF8 probe set (203161_s_at), specific to the full transcript encoding the functional RNF8, revealed that low RNF8 expression significantly associates with poorer outcome (Figure 2B; hazard ratio = 0.82 [0.74–0.92], log-rank P = 4 × 10–04). The strongest associations using RNF8 203161_s_at were observed for luminal B and basal-like breast cancer subtypes (Figure 2, C and D). The HER2+ subtype showed an effect in the same direction, although with lower significance due to the substantially smaller sample size in this setting (Figure 2E).
Figure 2. Low expression of the full-length RNF8 correlates with poor prognosis for breast cancer patients.
(A) Schematic of the different human RNF8 isoforms and the position of the 2 Affymetrix microarray probes 203160_s_at and 203161_s_at included in the KM Plotter application. The 203160_s_at probe recognizes all 4 RNF8 isoforms; the 203161_s_at probe only recognizes the full-length transcript (RNF8-202) that encodes for the full-length functional RNF8 protein containing both its FHA and its RING-finger domains. (B–E) Analysis of relapse-free survival of breast cancer patients using KM Plotter and the median expression level of the RNF8 203161_s_at probe to classify breast cancer cases as expressing either high or low levels of the full-length isoform. The analysis included all breast cancer patients (n = 3,955) and patients with luminal B (n = 1,149), basal-like (n = 618), or HER2+ (n = 251) subtypes. Hazard ratio and 95% confidence intervals of the Cox regression analysis and log-rank P values are shown. NMD, nonsense-mediated decay.
RNF8 deficiency promotes genomic instability in MECs and tumors.
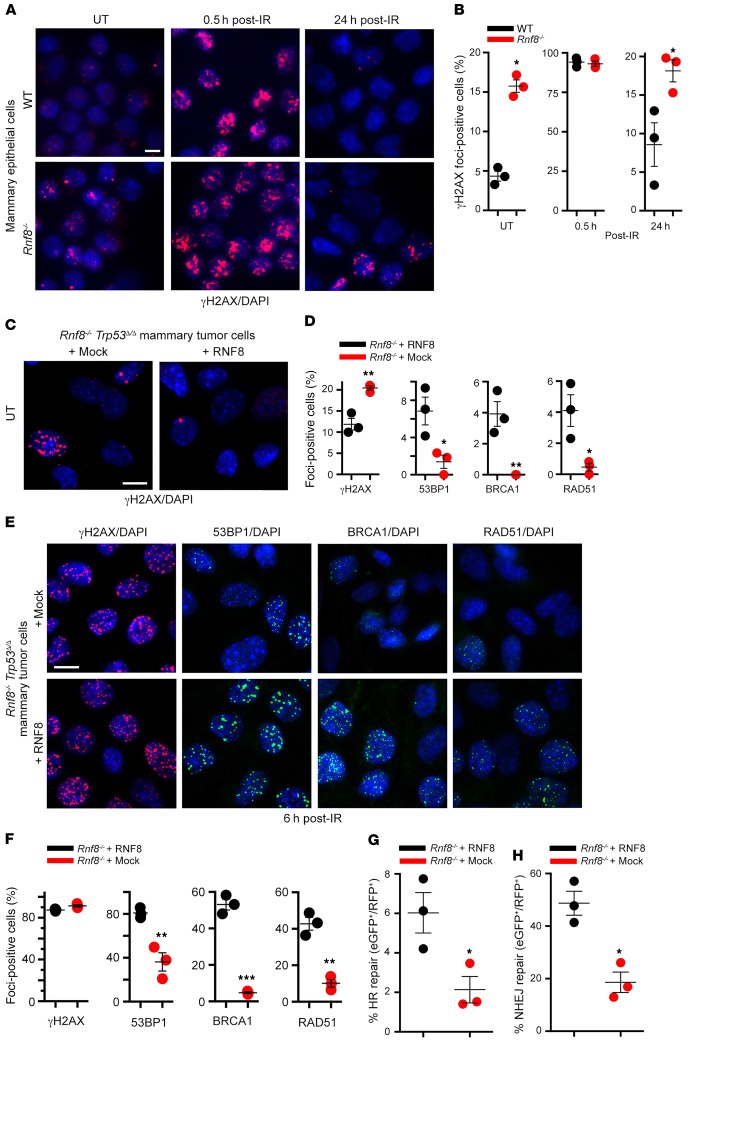
Given the established role of genomic instability in driving cancer (23), we examined the effect of RNF8 deficiency on DSB signaling, repair, and genomic integrity in MECs. Examination of γH2AX foci, a marker for DSBs (24), indicated elevated levels of spontaneous and γ-irradiation–induced (IR-induced) DSBs in Rnf8–/– MECs compared with WT controls (Figure 3, A and B). We also examined DSB levels in Rnf8–/– Trp53Δ/Δ mammary tumor cell lines reconstituted with mock or RNF8WT. We observed that RNF8WT‑reconstituted Rnf8–/– Trp53Δ/Δ mammary tumor cells displayed significantly lower levels of spontaneous DSBs, and their resolution of IR-induced DSBs was more efficient, compared with their mock-reconstituted controls (Figure 3, C–E). Reconstitution of Rnf8–/– Trp53Δ/Δ mammary tumor cells with RNF8WT also restored the ability of these tumor cells to recruit homologous recombination (HR) and nonhomologous end joining (NHEJ) repair factors BRCA1, RAD51, and 53BP1 to DSB sites (Figure 3, D–F). Consistent with these data, complementation of RNF8-deficient tumor cells with RNF8 significantly increased their efficiency of HR- and NHEJ-mediated DSB repair (Figure 3, G and H). Collectively these data underscore the importance of RNF8 for maintaining genomic stability in mammary cancer cells and tumors.
Figure 3. Deficiency of mouse RNF8 promotes genomic instability in MECs and mammary tumors.
(A) Purified MECs from WT and Rnf8–/– littermate females at estrus phase were either left untreated (UT) or irradiated (5 Gy) and fixed at the indicated time points after IR. Cells were stained with anti-γH2ax antibody and counterstained with DAPI. (B) Quantitative analysis of γH2AX subnuclear foci in WT and Rnf8–/– MECs from A. Cells with more than 3 foci were counted as foci-positive. Dot plots show mean ± SEM of 3 independent experiments, and more than 100 cells for each condition and genotype were counted. (C) Representative images of spontaneous γH2AX subnuclear foci in Rnf8–/– Trp53Δ/Δ mammary tumor cells reconstituted with RNF8 or mock (empty vector). (D) Dot plots depicting quantification of the spontaneous subnuclear foci formation for γH2AX, 53BP1, BRCA1, and RAD51 observed in Rnf8–/– Trp53Δ/Δ mammary tumor cells reconstituted with RNF8 or mock. Three independent experiments were performed, and more than 100 cells were counted for each condition and genotype (mean ± SEM). (E) Representative images of γH2AX, 53BP1, BRCA1, and RAD51 subnuclear foci formation 6 hours after IR (5 Gy) of the indicated mammary tumor cells. (F) Dot plots depicting quantifications of subnuclear foci formation observed in E. Cells harboring at least 10 foci were counted as foci-positive. Three independent experiments were performed, and more than 100 cells were counted for each condition and genotype (mean ± SEM). (G and H) Dot plots depicting efficiencies of HR-mediated (G) and NHEJ-mediated (H) repair of I-SceI–induced DSBs in Rnf8 Trp53Δ/Δ mammary tumor cells reconstituted as indicated (mean ± SEM). Three independent experiments were performed. Two-sided Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001. Scale bars: 20 μm.
Aberrant expansion of the mammary luminal progenitors in the absence of RNF8.
Deregulated lineage commitment of MECs has been shown to be associated with increased breast cancer risk (1, 25). Therefore, we also investigated the role of RNF8 in mammary gland development and its link to tumorigenesis. First, analysis of Rnf8 expression in mammary cell subpopulations indicated a significantly lower level in luminal progenitors compared with luminal differentiated and basal cells (Supplemental Figure 3A). Next, mammary epithelial subpopulations of 10- to 12-week-old, tumor-free, Rnf8–/– females and WT littermates were examined for potential differences. Notably, studied females were in estrus to alleviate hormonal effects on homeostasis of MECs (26, 27). Flow cytometry analyses indicated aberrant expansion of the luminal lineage in Rnf8–/– females compared with WT littermates (Figure 4, A–C; Lin– luminal cells: P < 0.05, 36.2% ± 1.8% vs. 20.5% ± 0.4%; Lin– basal cells: P > 0.05, 9.3% ± 1.2% vs. 13% ± 0.3%). In addition, there was an expansion of the luminal progenitor subpopulation (CD29loCD24+CD61+) in Rnf8–/– females compared with WT littermates (Figure 4, D–F; P < 0.05, 56% ± 3.7% vs. 42% ± 1.6%), whereas the luminal differentiated (CD29loCD24+CD61–) and basal lineages (CD29hiCD24+) were found to be unaffected (Figure 4, D–F). Similar results were obtained using the marker CD49f instead of CD29 (Supplemental Figure 3, B and C).
Figure 4. Aberrant expansion of the luminal lineage in the mammary glands of Rnf8-mutant females.
(A) Representative FACS plots showing Lin– (CD45–CD31–TER119–) luminal (L: CD29loCD24+) and basal (B: CD29hiCD24+) cells of inguinal mammary glands from 10- to 12-week-old WT and Rnf8–/– females. Data are representative of 3 independent experiments. (B) Dot plots depicting percentages of mammary luminal and basal populations from inguinal mammary glands from females in A (n = 3; mean ± SEM). (C) Dot plots depicting the absolute numbers of MECs and luminal and basal cells per inguinal mammary gland from WT and Rnf8–/– females (n = 3; mean ± SEM). (D) CD24+ MECs from A were analyzed for luminal progenitor (LP; CD29loCD24+CD61+) and luminal differentiated (LD; CD29loCD24+CD61–) cells. Representative FACS images of 3 independent experiments are shown. (E and F) Dot plots depicting percentages (E) and absolute numbers (F) of LP and LD subpopulations from inguinal mammary glands of 10- to 12-week-old WT and Rnf8–/– females (n = 3; mean ± SEM). (G) Dot plots depicting the clonogenic potentials (mean ± SEM) in Matrigel of MECs from WT and Rnf8–/– littermate females (n = 4 each). (H) Representative images of 3 independent experiments showing Ki67/DAPI staining of mammary glands from 6.5-month-old WT and Rnf8–/– female littermates (scale bar: 100 μm), and dot plots depicting quantification of Ki67+ cells from mammary glands of WT and Rnf8–/– females (n = 3; mean ± SEM). A minimum of 1,000 cells per gland were counted. (I) Representative FACS plots showing luminal and basal cells of mammary glands from 6.5-month-old females as indicated (n = 4 each) that had undergone 4 pregnancies and involution to induce Cre-mediated deletion of Trp53. All females were at estrus phase. Two-sided Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001.
Next, we examined the clonal growth potential of Rnf8–/– MECs using a 3D colony-forming cell (CFC) assay (28). The number of CFCs was found to be double and the colonies were larger in size in Rnf8–/– relative to WT cell controls (Figure 4G and Supplemental Figure 3D). Moreover, Rnf8–/– mammary glands exhibited a 4-fold increase in the number of proliferating cells as indicated by Ki67 positivity (Figure 4H). Taken together, these data indicate that RNF8 deficiency promotes aberrant expansion of luminal progenitor cells in the mouse mammary epithelium.
Following on the previous observations and considering that TRP53 deficiency increased the risk of breast cancer in Rnf8–/– females, the expansion of luminal progenitors in Rnf8 and Trp53 double-mutant background was also examined. Flow cytometry analyses of MECs were performed from inguinal mammary glands of 6.5-month-old WT, Rnf8–/–, WapCre Trp53fl/fl, and Rnf8–/– WapCre Trp53fl/fl females that had undergone 4 pregnancies and involutions, permitting complete deletion of Trp53-floxed alleles in the luminal lineage (Supplemental Figure 3E). The expansion of the luminal lineage associated with RNF8 deficiency persisted in 6.5-month-old females and was exacerbated by the additional deletion of Trp53 (Figure 4I; Lin– luminal cells: WT, 34.8% ± 1.7%; Rnf8–/–, 45.5% ± 1.5%; WapCre Trp53fl/fl, 31.5% ± 2.3%; and Rnf8–/– WapCre Trp53fl/fl, 54.8% ± 2%; P < 0.05 Rnf8–/– vs. WT and Rnf8–/– vs. Rnf8–/– WapCre Trp53fl/fl; Supplemental Figure 3, F and G). Whole-mount staining of the axillary mammary glands from these 6.5-month-old females showed no sign of hyperplasia (Supplemental Figure 3H), precluding contribution of hyperplasia to the observed aberrant expansion of luminal progenitors in the absence of RNF8. These data unveil a novel function for RNF8 in the regulation of luminal lineage cell fate.
RNF8 negatively regulates Notch signaling in murine mammary luminal progenitors and tumors.
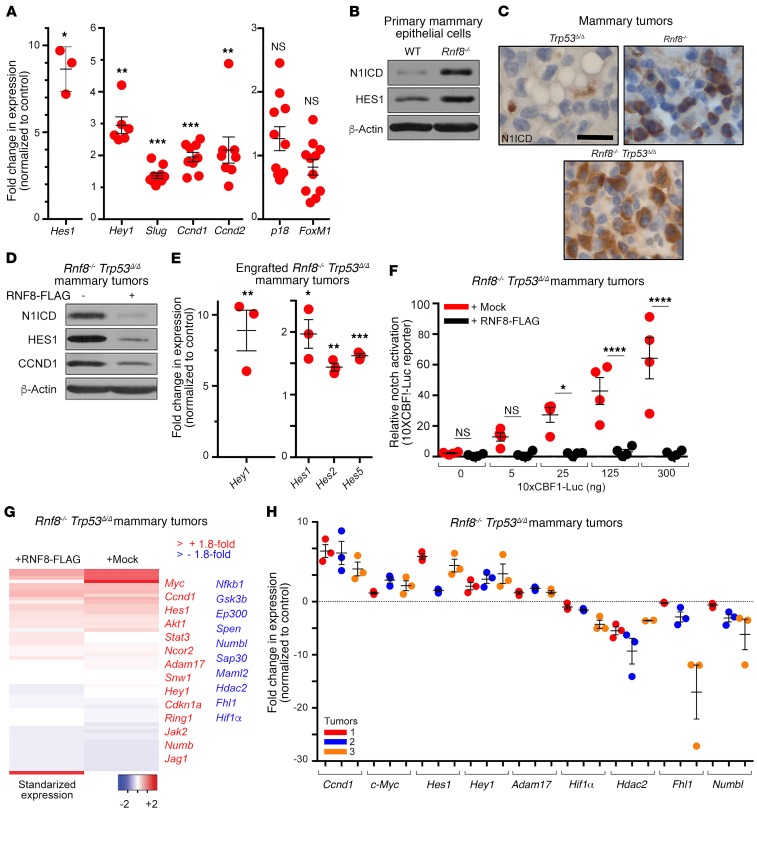
On the basis of the aberrant expansion of luminal progenitors in Rnf8-mutant females, we hypothesized that RNF8 regulates signaling pathways required for epithelial cell-fate determination. Therefore, we examined mammary cell subpopulations sorted from Rnf8–/– and WT females for expression of genes whose alterations are known to promote luminal expansion. Notably, the Notch signaling that plays important roles in the luminal cell-fate commitment (1, 29, 30) was found to be activated in Rnf8–/– luminal progenitors as indicated by the significantly increased expression of the Notch-target genes Hes1, Hey1, Slug, and Ccnd1 (Figure 5A). The expression level of the proliferation marker Ccnd2 was also found to be increased in Rnf8–/– luminal progenitors, while expression of p18INK4c and FoxM1, factors involved in regulating mammary luminal cell fate (31, 32), remained unaffected by RNF8 deficiency (Figure 5A). Next, given that interaction of NOTCH1 receptor with its ligands triggers its proteolytic cleavage and the release of its activated intracellular domain (N1ICD) (5), the effect of RNF8 deficiency on N1ICD expression level was analyzed in MECs. Rnf8–/– and Rnf8–/– Trp53Δ/Δ primary MECs were found to display elevated protein levels of N1ICD and its canonical target HES1 (Figure 5B and Supplemental Figure 4A). Overall, these data support increased NOTCH transcriptional activity in Rnf8–/– mammary luminal progenitors, thus providing a possible mechanism for their aberrant expansion.
Figure 5. RNF8 negatively regulates Notch signaling in murine mammary luminal progenitors and mammary tumors.
(A) Quantitative reverse transcriptase PCR (RT-qPCR) showing the expression changes of Notch targets and other genes in luminal progenitors from mammary glands of Rnf8–/– females at estrus phase compared with WT littermates. (B) Immunoblot analysis of N1ICD and HES1 in the indicated cells. (C) Representative immunohistochemical analysis of N1ICD levels in the indicated mammary adenocarcinomas (n = 3 each; scale bar: 25 μm). (D) Immunoblot showing expression of the indicated proteins in Rnf8–/– Trp53Δ/Δ mammary tumor cells reconstituted with mock or RNF8-FLAG. (E) RT-qPCR showing increased expression of indicated Notch1 targets in mock-reconstituted, compared with RNF8WT-reconstituted, Rnf8–/– Trp53Δ/Δ mammary tumor cells engrafted for 40 days in NSG mice (see Figure 1C). (F) Notch reporter 10xCBF1-Luc showing relative activation of Notch signaling in mock-reconstituted Rnf8–/– Trp53Δ/Δ mammary tumor cells compared with RNF8WT-reconstituted controls. *P < 0.05, ****P < 0.0001, 2-way ANOVA followed by Tukey’s test. (G) Heatmap showing the standardized expression of Notch targets and/or Notch pathway elements in Rnf8–/– Trp53Δ/Δ mammary tumor cells reconstituted with mock or RNF8-FLAG. (H) Notch targets differentially expressed in RNA-Seq analysis in G were validated by RT-qPCR in 3 different Rnf8–/– Trp53Δ/Δ mammary tumor cell lines and their RNF8-reconstituted controls. Data in B and D are representative of at least 3 experiments. A, E, F, and H: At least 3 experiments were performed, and the means ± SEM are shown. A, E, and H: 2-sided Student’s t test, Rnf8–/– Trp53Δ/Δ mammary tumor cells mock-reconstituted compared with RNF8WT-reconstituted controls. *P < 0.05, **P < 0.01, ***P < 0.001. H: All P values were less than 0.05.
Given that gain-of-function mutations of NOTCH1 are associated with human breast cancer (2, 3), and that Notch signaling is activated in Rnf8–/– luminal progenitors, we examined the level of this signaling in Rnf8–/– Trp53Δ/Δ mammary tumors. Immunohistochemical staining indicated increased expression of N1ICD in Rnf8–/– and Rnf8–/– Trp53Δ/Δ primary mammary tumors relative to Trp53Δ/Δ tumors (Figure 5C). Consistent with these data, Rnf8–/– Trp53Δ/Δ mammary tumor cells displayed elevated expression of N1ICD and its targets (HES1 and CCND1), and the activation of Notch1 signaling was restrained following RNF8WT reconstitution (Figure 5D and Supplemental Figure 1H). In addition, expression of NOTCH targets was higher in mock-reconstituted Rnf8–/– Trp53Δ/Δ mammary tumor cells engrafted into NSG mice, compared with the corresponding RNF8WT-reconstituted controls (Figure 5E). Moreover, a reporter for Notch signaling activity (10xCBF1-Luc) (33) confirmed RNF8-mediated negative regulation of NOTCH transcriptional activity in RNF8WT-reconstituted Rnf8–/– Trp53Δ/Δ cells (Figure 5F).
To further investigate the functional relevance of RNF8 to the regulation of Notch signaling, we compared RNA sequencing data obtained from Rnf8–/– Trp53Δ/Δ mammary tumor cells and RNF8WT-reconstituted controls. Transcriptional upregulation of several NOTCH-target genes and components of the pathway were observed in RNF8-deficient cells (Figure 5G). Downregulation of a few Notch pathway elements was also observed (e.g., Numbl and Maml2; Figure 5G), perhaps reflecting the complex regulation of this signaling pathway and that, in addition to positive feedback, activation of Notch signaling also promotes negative feedback regulation (5). Quantitative expression analyses performed on 3 different Rnf8–/– Trp53Δ/Δ mammary tumors reconstituted with either mock or RNF8WT confirmed the changes in expression of NOTCH-target genes associated with RNF8 deficiency (Figure 5H). These data reveal novel functions of RNF8 as a negative regulator of Notch signaling in mouse mammary luminal progenitors and tumors.
RNF8 mediates negative regulation of Notch signaling in human breast cancer.
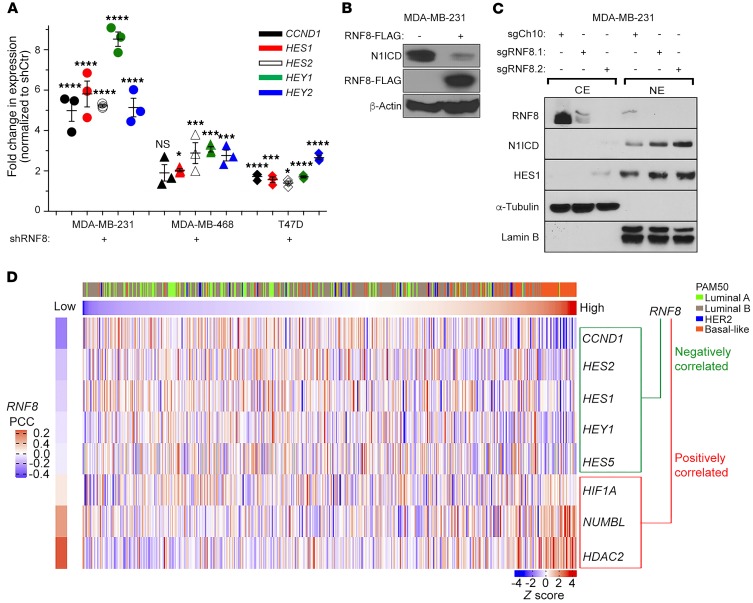
To extend our studies of the effect of RNF8 on Notch signaling to relevant human breast cancer cells, we first performed quantitative expression analysis in RNF8-depleted breast cancer cell lines. Our data indicate a significant increase in the expression of NOTCH targets (HES1, HES2, HEY1, HEY2, and CCND1) in RNF8-depleted breast cancer cell lines compared with controls (Figure 6A and Supplemental Figure 4B). Consistent with these findings, overexpression of ectopic RNF8WT in the breast cancer cell line MDA-MB-231 significantly reduced the protein level of N1ICD (Figure 6B), while CRISPR/Cas9–mediated knockout of RNF8 in MDA-MB-231 and MCF7 cells led to increased protein expression of N1ICD and its downstream target HES1 (Figure 6C and Supplemental Figure 4C). Further supporting the link between RNF8 and Notch signaling in human breast cancer cells, analysis of data from The Cancer Genome Atlas (TCGA) revealed significant negative correlations between the expression of RNF8 and several NOTCH-target genes (HES1, HES2, HES5, HEY1, and CCND1), whereas positive correlations were found with HDAC2, HIF1A, and NUMBL (Figure 6D).
Figure 6. RNF8 mediates negative regulation of Notch signaling in human breast cancer.
(A) RT-qPCR analysis of the fold change in the expression of Notch targets in the indicated RNF8-knockdown (shRNF8) human breast cancer cell lines compared with their controls (shCtr). Dot plots show mean ± SEM of 3 independent experiments. *P < 0.05, ***P < 0.001, ****P < 0.0001, 1-way ANOVA followed by Tukey’s test. (B) Immunoblot analysis of the indicated proteins in MDA-MB-231 cells transfected with FLAG empty vector (–) or RNF8-FLAG. (C) Immunoblot analysis of cytoplasmic (CE) and nuclear (NE) extracts from indicated cells. (D) Supervised clustering (from low to high RNF8 expression) of the profiles of genes with a significant negative or positive correlation (Pearson’s correlation coefficients [PCCs]) with RNF8 in the TCGA breast cancer (BRCA) data set. The breast cancer subtypes based on PAM50 are indicated. Data in B and C are representative of at least 3 experiments.
To further assess the RNF8-NOTCH link and examine whether it is influenced by TP53 mutations, the TCGA data were analyzed for pathway associations using RNF8 coexpression ranks from all tumors or from those with somatic TP53 mutations. RNF8 expression was found to be negatively correlated with the Pathway Interaction Database (PID) NOTCH set in both settings (Supplemental Figure 4D). These data are consistent with the observed activation of Notch signaling in both RNF8-deficient MCF7 (TP53-WT) and MDA-MB-231 (TP53-mutant) breast cancer cell lines (Figure 6C and Supplemental Figure 4C). Taken together, these results indicate that, as in murine mammary luminal progenitors and mammary tumors, RNF8 negatively regulates Notch signaling in human breast cancer.
RNF8 ubiquitylates N1ICD to control its turnover.
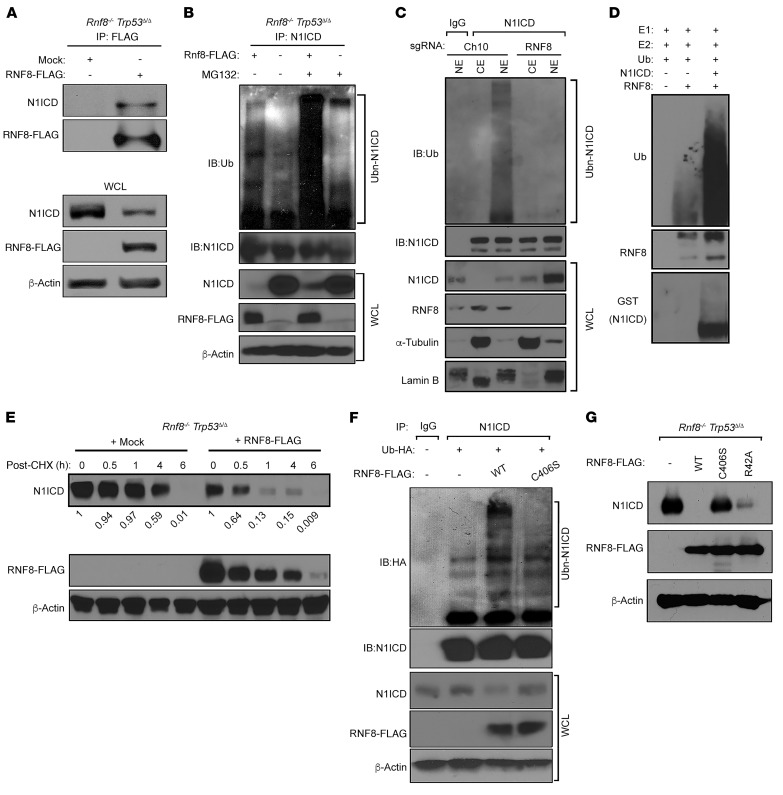
To understand the molecular mechanisms underlying the observed negative regulation of Notch signaling, we first examined whether RNF8 interacts with components of the corresponding pathway. N1ICD was detected in FLAG-tagged RNF8 immunoprecipitates from Rnf8–/– Trp53Δ/Δ mammary tumors reconstituted with RNF8-FLAG (Figure 7A). Notably, no interaction was detected between RNF8 and RBPJ, a Notch transcriptional effector (2), and deficiency of RNF8 had no effect on RBPJ expression (Supplemental Figure 5A).
Figure 7. RNF8 polyubiquitylates N1ICD to promote its turnover.
(A) N1ICD interacts with RNF8. Immunoblot (IB) analysis of N1ICD and RNF8-FLAG following anti-FLAG immunoprecipitation (IP) from RNF8-deficient mammary tumor cells and their RNF8-restored isogenic controls. WCL, whole cell lysate. (B) Ubiquitylation of N1ICD depends on RNF8 in vivo. RNF8-deficient mammary tumor cells complemented with empty FLAG vector (–) or RNF8-FLAG (+) were either left untreated or treated with the proteasome inhibitor MG132 and subjected to IP with anti-N1ICD. Immunoblot analysis was performed with the indicated antibodies. (C) Ubiquitylation of endogenous N1ICD by RNF8. Nuclear (NE) and cytoplasmic (CE) extracts prepared from MDA-MB-231 cells deleted of RNF8 (sgRNF8) and their controls (sgCh10) were subjected to IP with anti-N1ICD or IgG, followed by immunoblot as indicated. (D) In vitro ubiquitylation of recombinant N1ICD in the presence of recombinant RNF8, UBE1 (E1), Ubc13 (E2), and Ub proteins. (E) Cycloheximide (CHX) chase analysis of N1ICD protein half-lives in RNF8-deficient mammary tumor cells and their RNF8-restored isogenic controls. Cells were incubated in the presence of CHX for the indicated times, and immunoblots were performed using the indicated antibodies. N1ICD levels were quantified by densitometry using ImageJ and normalized with β-actin level. N1ICD fold decrease is indicated. (F) Requirement of RNF8 catalytic activity for N1ICD ubiquitylation. HEK293T cells, transfected with empty FLAG vector (–), RNF8WT-FLAG, RNF8C406S-FLAG (catalytically inactive), and Ub-HA as indicated, were subjected to IP with anti-N1ICD followed by immunoblot for HA and N1ICD. WCL was analyzed by immunoblot as indicated. (G) FHA domain of RNF8, responsible for its recruitment to DSB sites, is dispensable for N1ICD turnover. Immunoblots showing expression of N1ICD and RNF8-FLAG in RNF8-deficient mammary tumor cells reconstituted with FLAG-RNF8WT, FLAG-RNF8C406S, FLAG-RNF8R42A (FHA domain mutant), or empty FLAG vector (–) as indicated. Data in A–G are representative of at least 3 experiments.
Ubiquitylation of N1ICD is critical for the regulation of its turnover (2). Examination of the expression of FBW7, an E3 ligase important for N1ICD ubiquitylation and degradation (2, 5), indicated that it was unaffected by RNF8 deficiency (Supplemental Figure 5B). Next, we sought to determine whether RNF8 ubiquitylates N1ICD. First, we performed an in vivo ubiquitylation assay and examined the effect of RNF8 deficiency on the ubiquitylation of endogenous N1ICD. Immunoprecipitated N1ICD from RNF8-deficient mammary tumor cells reconstituted with RNF8-FLAG, and left untreated or treated with the proteasome inhibitor MG132, migrated on SDS-PAGE gels as higher–molecular weight smears that were reactive to anti-ubiquitin (Ub) (Figure 7B). In contrast, these N1ICD Ub smears were greatly reduced in mock-reconstituted RNF8-deficient mammary tumor cells (Figure 7B). Next, we examined the effect of RNF8 on the ubiquitylation of the endogenous human N1ICD and observed that deficiency of RNF8 in MDA-MB-231 cells remarkably decreased the ubiquitylation level of nuclear N1ICD (Figure 7C).
Ubiquitylation assays performed in HEK293T cells further supported RNF8-mediated polyubiquitylation of N1ICD, as coexpression of exogenous RNF8 and Ub in these cells produced robust Ub smears of N1ICD compared with controls (Supplemental Figure 5C). Furthermore, examination of RNF8-catalyzed Ub linkages for N1ICD polyubiquitylation indicated that RNF8 mediates both K63 and K48 ubiquitylation of N1ICD (Supplemental Figure 5C). Finally, in vitro ubiquitylation assays using recombinant proteins (RNF8, N1ICD, E1, E2, and Ub) confirmed that N1ICD is a novel and direct ubiquitylation substrate for RNF8 (Figure 7D).
The importance of ubiquitylation in protein turnover (34, 35) and the elevated N1ICD levels in RNF8-deficient mammary cells and tumors prompted us to perform cycloheximide chase experiments and examine the regulatory role of RNF8 on N1ICD. A strong reduction of the half-life of N1ICD was observed in RNF8-deficient mammary tumors upon their complementation with RNF8WT (Figure 7E).
RNF8 RING-finger and FHA domains are required for its function in DSB signaling (7–10). Therefore, we examined the contribution of these domains to RNF8-mediated negative regulation of Notch signaling. Similarly to its inability to stimulate DSB signaling and repair (8), a catalytic dead form of RNF8 that carries C406S substitution in the RING-finger domain (RNF8C406S) was defective in mediating N1ICD ubiquitylation and degradation (Figure 7, F and G). However, in contrast to the inability of the FHA mutant RNF8R42A to activate the repair cascade of DSBs (8, 9), this RNF8 mutant form effectively mediated N1ICD degradation (Figure 7G). These findings highlight the importance of the E3 ligase activity of RNF8 for N1ICD ubiquitylation and turnover, and indicate that RNF8 negatively regulates Notch signaling in a manner independent of its recruitment to DSB sites.
Combined pharmacological inhibition of Notch signaling and PARP function promotes killing of RNF8-deficient breast tumors.
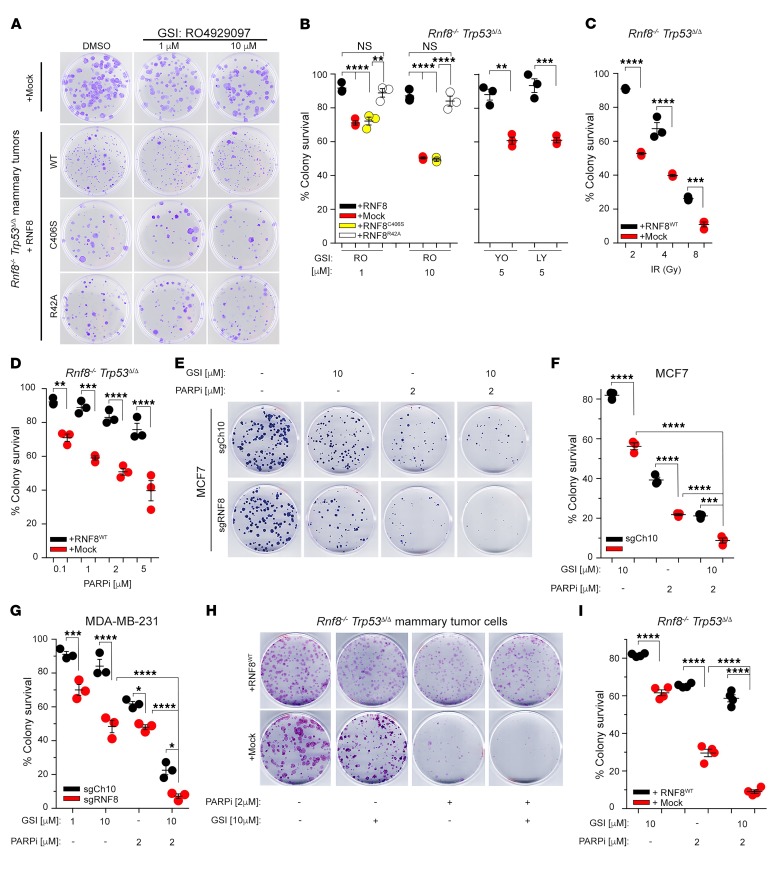
Based on the importance of RNF8 in Notch signaling and DSB repair, we examined whether impaired RNF8 function in breast cancer could be exploited therapeutically. First, exposure to different γ-secretase inhibitors (GSIs; RO4929097, YO-01027, and LY411575), which block Notch signaling, led to a significant growth impairment of Rnf8–/– Trp53Δ/Δ mammary tumor cells compared with their RNF8WT-reconstituted controls (Figure 8, A and B). Consistent with the requirement of the FHA domain of RNF8 for DSB repair (refs. 7–10 and Supplemental Figure 6A), but not for negative regulation of N1ICD (Figure 7G), Rnf8–/– Trp53Δ/Δ tumor cells reconstituted with the FHA mutant RNF8R42A displayed reduced sensitivity to GSI (Figure 8, A and B). In contrast, the E3 ligase dead RNF8C406S, which exhibits defective ability to mediate DSB signaling/repair (ref. 8 and Supplemental Figure 6, A and B) together with impaired ubiquitylation and regulation of N1ICD (Figure 7, F and G), failed to rescue hypersensitivity of Rnf8–/– Trp53Δ/Δ tumor cells to GSI (Figure 8, A and B).
Figure 8. RNF8-deficient breast tumors are sensitive to combination of pharmacological inhibitors of Notch signaling and PARP.
(A) Representative dish images of 3 independent experiments in triplicate showing clonogenic survival under GSI (RO4929097) treatments of Rnf8–/– Trp53Δ/Δ mammary tumor cells reconstituted with mock or either WT or mutant forms of RNF8 as indicated. Cells were fixed and stained with crystal violet at day 12 of culture. (B) Dot plots depicting percentages of clonogenic survival of Rnf8–/– Trp53Δ/Δ mammary tumor cells and their reconstituted controls (as indicated) after 12 days of treatment with different GSIs (RO, RO4929097; YO, YO-01027; LY, LY411575). (C and D) Dot plots depicting clonogenic survival of Rnf8–/– Trp53Δ/Δ mammary tumor cells, reconstituted with mock or RNF8, 12 days after IR (C) or treatment with the PARPi KU0058948 (D). (E) Representative dish images showing clonogenic survival of MCF7 cells deficient for RNF8 (sgRNF8) and their controls (sgCh10) 25 days after treatment with GSI (RO4929097), PARPi (olaparib), or their combination as indicated. Images are representative of 3 independent experiments in triplicate. (F and G) Dot plots depicting percentages of clonogenic survival of MCF7 cells (F) and MDA-MB-231 cells (G) deficient for RNF8 (sgRNF8) and their controls (sgCh10) after treatment with GSI (RO4929097), PARPi (olaparib), or their combination as indicated. (H) Representative images of clonogenic survival of the indicated mammary tumor cells 12 days after treatment with GSI (RO4929097), PARPi (olaparib), or their combination as indicated. (I) Dot plots depicting clonogenic survival of Rnf8–/– Trp53Δ/Δ mammary tumor cells reconstituted with mock or RNF8 after treatment (12 days) with GSI (RO4929097), PARPi (olaparib), or their combination as indicated. B–D, F, G, and I: n = 3; mean ± SEM, normalized to DMSO-treated controls except for C, in which normalization was to nonirradiated controls. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, 2-way ANOVA followed by Tukey’s test.
Defective DSB repair increases cancer radiosensitivity, while poly(ADP-ribose) polymerase inhibitors (PARPi) promote synthetic lethality of cancer cells defective for HR-mediated repair (36). Considering the inherent defects of DSB signaling and HR-mediated repair associated with RNF8 deficiency (Figure 3), we examined the response of Rnf8–/– Trp53Δ/Δ mammary tumor cells, complemented with empty vector or RNF8WT, to IR and PARPi (KU0058948 and olaparib). In accordance with their impaired DSB repair and defective homologous recombination pathway, RNF8-deficient mammary tumor cells were highly sensitive to both IR and PARPi compared with their control counterparts (Figure 8, C and D).
Next, the above therapeutic opportunity was assessed in human breast cancer cells. Examination of RNF8-deficient MDA-MB-231 and MCF7 breast cancer cells showed impaired DSB repair as indicated by accumulation of unresolved γH2AX foci 24 hours after IR (Supplemental Figure 6, C–E). Consistent with their impaired repair of DSBs and increased activation of Notch1 signaling, and similarly to mouse cell models, RNF8-deficient MCF7 and MDA-MB-231 cells displayed increased sensitivity to GSI and PARPi (Figure 8, E–G). Importantly, both human cell models deficient for RNF8 (Figure 8, E–G), as well as Rnf8–/– Trp53Δ/Δ mouse mammary tumor cells (Figure 8, H and I), displayed higher sensitivity to the combination of GSI and PARPi compared with the respective controls. In summary, evidence from human and mouse breast cancer models highlights that combination of inhibitors of Notch signaling and PARP provides a precise therapeutic strategy for breast tumors with defective RNF8 function.
Discussion
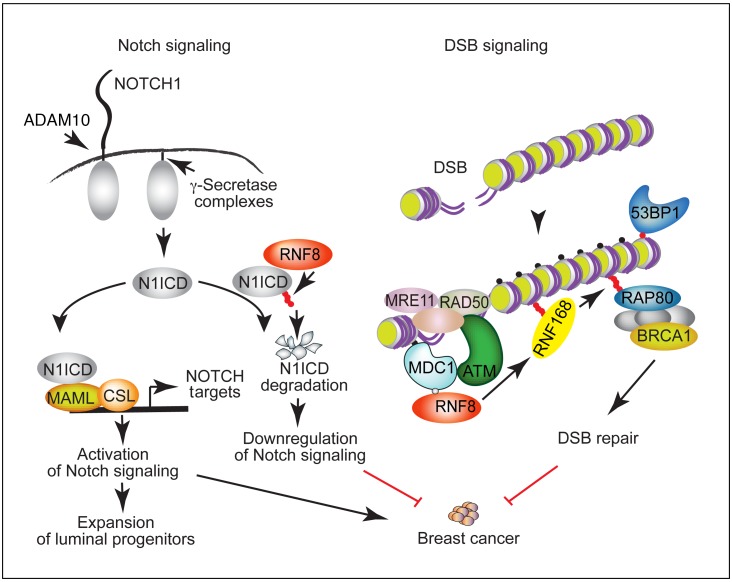
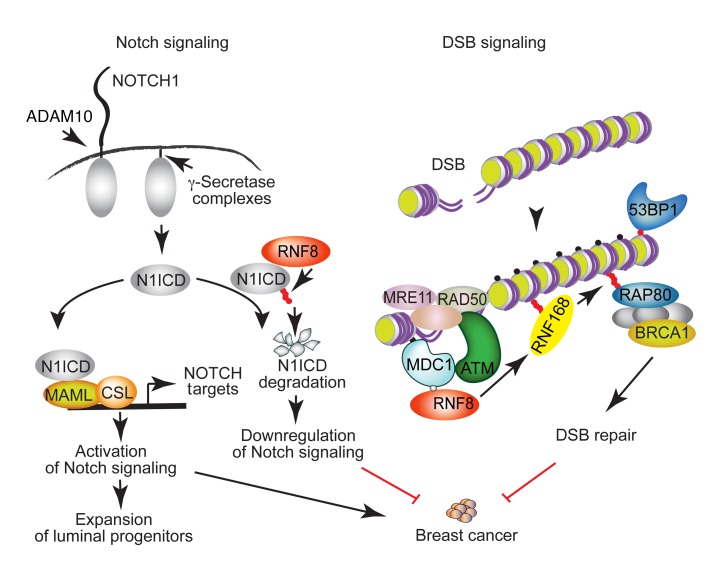
Notch signaling and DSB signaling are fundamental cellular processes, and their regulation is critical for minimizing breast cancer risks (2, 3, 23). Although these pathways are highly regulated, the mechanisms that may mediate their coregulation remain poorly understood. This work reveals that RNF8 mediates this coregulation of Notch and DSB signaling in the mammary epithelium and, if perturbed, promotes breast cancer (Figure 9).
Figure 9. A schematic diagram depicting a model for RNF8-mediated coregulation of Notch signaling and DSB repair and the breast cancer–suppressive function of RNF8.
RNF8 is recruited to DSBs, where it ubiquitylates histones at the flanking DNA damage sites. This ubiquitylation triggers recruitment of downstream DSB signaling and repair proteins (e.g., RNF168 and BRCA1) to DNA damage sites, allowing their repair. Loss of RNF8 expression or function impairs DSB repair, leading to genomic instability and increased breast cancer risk. Our data reveal an important role for RNF8 in mediating negative regulation of Notch signaling. RNF8 ubiquitylates the active form of NOTCH1 (N1ICD) to promote its turnover. Thus, impaired RNF8 expression or function results in constitutive activation of Notch signaling in mammary luminal progenitors, a process that promotes expansion of these progenitors and increases their risk for malignant transformation. We propose that RNF8 coregulation of Notch signaling and DSB repair is critical for its suppressor function in breast cancer. Small filled circles indicate γH2ax (black); phosphorylation (gray); and Ub (red).
Notch signaling is essential for luminal cell-fate determination in the mammary epithelium (1, 29). Constitutive activation of Notch signaling in mammary luminal progenitors promotes the expansion of these progenitors and increases the risk for their malignant transformation (1, 29, 30). In this study, we demonstrate that RNF8 is a potent negative regulator of Notch signaling and its deficiency in MECs causes sustained activation of this signaling, likely contributing to the aberrant expansion of mammary luminal progenitors. Our study reveals that RNF8 negatively regulates the Notch1 signaling pathway through its ubiquitylation of N1ICD, and its subsequent targeting for degradation. By restraining intracellular levels of N1ICD, RNF8 restrains transcriptional activity of NOTCH-target genes to promote tissue homeostasis. Consistent with the elevated levels of N1ICD and activation of Notch signaling in mammary epithelium of Rnf8-mutant female mice, human breast cancer cell lines deficient for RNF8 also displayed elevated levels of N1ICD and Notch signaling. Gene expression profile analyses using TCGA breast cancer data support the link between low RNF8 expression and abnormal Notch signaling.
The existence of several RNF8 isoforms prompted us to examine whether a correlation exists between breast cancer relapse–free survival and the expression level of the full-length isoform that encodes for functional RNF8. These analyses revealed that low expression of the full-length RNF8 isoform that contains both its FHA and RING-finger domains correlates with poor prognosis for breast cancer patients. These data suggest a prognostic role for the integral function of RNF8 in breast cancer. This link may be independent of an impact on breast cancer initiation; however, our results examining female mice carrying mutation of Rnf8 indicate that loss of RNF8 increases the risk of mammary tumorigenesis. Nonetheless, studies in different cancer settings may be warranted to further decipher the proposed dual impact of RNF8 on breast cancer.
Given the novel role of RNF8 as a negative regulator of Notch signaling, we hypothesized that when RNF8 is absent or nonfunctional, sustained Notch1 signaling drives aberrant proliferation of luminal progenitor cells. This aberrant proliferation, in conjunction with increased genomic instability, in the absence of RNF8 results in spontaneous mammary tumorigenesis. Consistent with elevated levels of DSBs and TRP53 activation in RNF8-deficient MECs, the risk of spontaneous mammary tumorigenesis in Rnf8–/– females was further increased in the presence of homozygous or heterozygous Trp53 mutations in mammary epithelium. Our observation that tumors isolated from Rnf8–/– Trp53Δ/WT mice often displayed a loss of heterozygosity for Trp53 suggests that activated TRP53 plays a critical role in the suppression of RNF8-associated mammary tumorigenesis.
The RING-finger domain of RNF8 is critical for its E3 ligase function and for the positive regulation of DSB signaling cascade (7–10). Our study demonstrates that the RNF8 RING-finger domain is also essential to mediate negative regulation of Notch1 signaling. In contrast, the phosphothreonine-binding FHA domain of RNF8 responsible for recruitment of RNF8 to DSB sites (7–10) is dispensable for RNF8-mediated downregulation of Notch signaling. Therefore, despite coregulation of Notch signaling and DSB repair, recruitment of RNF8 to chromatin at DSBs, or its possible binding to phosphorylated proteins through its FHA domain, is not required for ubiquitylation of N1ICD. Therefore, the initial mechanisms by which RNF8 engages its regulation of Notch1 signaling and DSB repair cascade appear to be different.
Combination therapy offers the advantage of targeting different molecular processes to enhance tumor cell death and reduce the risk of cancer recurrence. The simultaneous alteration of Notch signaling and DSB repair associated with defective RNF8 expression in breast cancer provides a unique therapeutic opportunity using specific inhibitors of Notch and PARP. Indeed, we observed increased sensitivity of mouse and human breast cancer cells deficient for RNF8 to individual treatment with GSI or PARPi. Notably, this sensitivity of RNF8-deficient breast cancer cells was further increased in response to combination of pharmacological inhibitors of Notch signaling and PARP.
In summary, our data uncover an unanticipated role for RNF8 in the regulation of cell-fate determination of mammary luminal progenitors and demonstrate a novel function for RNF8 in the regulation of Notch1 signaling through the control of N1ICD turnover. We also provide evidence that RNF8 deficiency increases the risk of mammary tumorigenesis. Thus, while RNF8 is an integral component of DSB signaling and, as such, its deficiency or loss of function can potentially contribute to increased breast cancer risk, we propose that the novel role of RNF8 in fine-tuning Notch1 signaling is not only important for cell-fate determination of luminal progenitors, but also contributes to protection from breast cancer development (Figure 9). The high efficacy of combination treatment of RNF8-deficient breast tumors with GSI and PARPi suggests that such combination may have therapeutic benefit for patients with defective expression or function of RNF8. Elucidation of the mechanisms that modulate RNF8’s negative regulation of Notch1 signaling and its impact on the fate and transformation of MECs sheds new light on the mechanisms of breast cancer development and uncovers a novel avenue for targeted therapy.
Methods
Mice.
Rnf8–/– mice (AS0574 strain) were previously reported (11). Trp53fl/fl conditional mutant mice (37) (NCI Mouse Repository) were intercrossed with WapCre transgenic mice (The Jackson Laboratory) to obtain WapCre Trp53fl/fl mice. Subsequent crosses of Rnf8–/– mice with WapCre Trp53fl/fl mice generated Rnf8–/– WapCre Trp53fl/fl females. Mice in this study were on a mixed 129/J × C57BL/6 genetic background and were genotyped by PCR for Rnf8WT (forward 5′-TGATGACACCTGGGCATGT-3′; reverse 5′-TCTTTGAGACAGCGCCTGG-3′), Rnf8 mutant (forward 5′-TCAAAGGTTTGCCCTCTGAT-3′; reverse 5′-CGGAGCGGATCTCAAACTCT-3′), Trp53fl (forward 5′-CAGCCTCTGTTCCACATACACT-3′; common 5′-TGGATGGTGGTATACTCAGAGC-3′), Trp53WT (forward 5′-AGGCTTAGAGGTGCAAGCTG-3′; common 5′-TGGATGGTGGTATACTCAGAGC-3′), and Cre (forward 5′-CCATCTGCCACCAGCCAG-3′; reverse 5′-TCGCCATCTTCCAGCAGG-3′). All mice were housed in a pathogen-free mouse facility at the Princess Margaret Cancer Centre. Ten- to 12-week-old female mice were used for experiments, unless indicated otherwise in the figure legends. To rule out the effect of hormonal changes on MECs (26, 27), all experimental females were followed to determine their phases of estrus cycle for at least 4 continuous days. Females at the estrus cycle were used for all experiments. Survival cohorts of female mice were monitored for mammary tumor onset by palpation as well as signs of other tumors twice a week for over 600 days. Power analysis was performed to determine the minimum number of animals needed to have a statistical power of 80% for comparing Rnf8–/– WapCre Trp53fl/fl and WapCre Trp53fl/fl survival analysis. Log-rank test was used for statistical analysis of survival curves, and was performed using OriginLab Pro 8 software. For all animal experiments, only females were included, and block randomization was used to warrant a balance in sample size across the different genotypes. We did not exclude any animals from the experiments. Investigators were not blinded to the genotype identity of mice.
Southern blot analysis for loss of heterozygosity.
Genomic DNA from Rnf8–/– WapCre Trp53fl/WT mammary tumors and respective tails of the tumor-bearing females was digested with BglII followed by blotting and hybridization using standard procedures.
PCR analysis of WapCre-mediated Trp53 deletion.
Genomic DNA was isolated from cell-sorted Lin– mammary epithelial subpopulations (CD49floCD24+CD61– luminal differentiated cells; CD49floCD24+CD61+ luminal progenitor cells; CD49fhiCD24+ basal cells) of WapCre Trp53fl/fl females at estrus phase that had undergone zero pregnancies (16 weeks old), 2 pregnancies (4 months old), and 4 pregnancies (6.5 months old) and involution. Touchdown PCR was performed on equal amounts of genomic DNA to detect the presence of Trp53WT, Trp53fl, and Trp53Δ2–10 alleles as previously described (37).
Notch reporter assay.
RNF8-deficient mammary tumor cells (passage 5) and their respective isogenic cells reconstituted with RNF8 were cotransfected with the reporter construct 10xCBF1-Luc (33) and Renilla luciferase using PolyJet DNA in vitro transfection reagent (SignaGen Laboratories). Forty-eight hours later, transfected cells were counted and plated at 2.5 × 105 cells per 75 μl DMEM per well of flat-bottomed opaque 96-well tissue culture plates (Greiner CellStar). Luciferase activities of the transfected cells were measured using the Dual Glo Luciferase assay kit (Promega) with firefly luciferase–based reporter gene activity normalized to the Renilla luciferase internal control reporter activity. Background autoluminescence from nontransfected cells was subtracted from the autoluminescence of the transfected cells.
Transfection and virus transduction.
HEK293T cells (ATCC) and the human breast cancer cell line MDA-MB-231 (ATCC) were transfected using PolyJet DNA in vitro transfection reagent (SignaGen Laboratories). Mouse mammary tumors were processed into single-cell suspensions following the same procedure for mammary gland single-cell suspension described above. Then, mammary tumor cells were analyzed by flow cytometry for Lin– expression of CD49f, CD24, and CD61, and were cultured in vitro into stable cell lines and validated by Western blot, PCR, and Southern blot for their respective mutations. All tumor cells were cultured in DMEM supplemented with 10% FBS, 5 × 10–5 M 2-ME, 100 U/ml penicillin, and 100 μg/ml streptomycin (complete DMEM), and were maintained in a humidified atmosphere at 37°C and 5% CO2. To complement Rnf8–/– Trp53Δ/Δ mammary tumor cell lines with WT and mutant forms of RNF8, cDNA for mouse Rnf8 (WT, C406S, and R42A) was subcloned into MSCV-FLAG. Rnf8 retroviral constructs and empty MSCV-FLAG were transfected to Phoenix cells, and virus supernatants were collected 48 and 72 hours after transfection. Three independent mammary tumor cell lines were transduced with the harvested retroviral supernatants in the presence of Polybrene, and selected for puromycin resistance. Human breast cancer cell lines MDA-MB-231, MDA-MB-468, and T47D (ATCC) were transduced with PLKO-puro lentiviruses encoding shRNAs targeting human RNF8 (shRNA#1: TRCN0000003438, 5′-TGGAGCAACTAGAGAAGACTT-3′; and shRNA#2: TRCN0000003441, 5′-CCAAAGAATGACCAAATGATA-3′). As control, cells were transduced with lentiviral particles carrying the empty pLKO.1-puro vector (Addgene, 8453). The expression of RNF8 was examined by Western blotting. Human breast cancer cell lines MDA-MB-231 and MCF7 (ATCC) were also transduced with lentiCRISPR v2 (Addgene, 52961) lentiviruses encoding short-guide RNAs (sgRNAs) targeting human RNF8 (sgRNF8.1: 5′-CGGGGTCGAGTAGGCGATGG-3′; sgRNF8.2: 5′-CCATCGCCTACTCGACCCCG-3′) or a control lentiCRISPR v2 vector loaded with a sgRNA targeting a noncoding region of chromosome 10 (sgCh10: 5′-CAATACACCCATAGTTGAGC-3′). All cell lines were tested to be mycoplasma-free.
DSB repair assays.
To determine HR and NHEJ efficiency, HR and NHEJ reporter constructs (38) were linearized with the I-SceI restriction enzyme and transfected to mammary tumor cell lines using GenJet in vitro DNA transfection kit (SignaGen Laboratories). RFP construct was cotransfected as internal control. Transfected cells were examined 48 hours later by flow cytometry for their expression of eGFP and RFP. Repair efficiencies of HR and NHEJ were expressed as eGFP+/RFP+ ratios.
Flow cytometry analysis and sorting of MECs.
Mammary glands from female mice at estrus phase were processed into mouse EpiCult-B medium supplemented with 3% FBS (Wisent), 750 U/ml collagenase, and 250 U/ml hyaluronidase, and incubated at 37°C for 2.5 hours. Subsequently, mammary organoids were subjected to red blood cell lysis in NH4Cl, followed by further dissociation in 0.25% trypsin for 2 minutes, and then in a mixture of 5 mg/ml dispase and 0.1 mg/ml DNase I for 2 minutes. After filtering through a 40-μm mesh, mammary single-cell suspensions were obtained. All the reagents described above were from STEMCELL Technologies, and all antibodies described below were from Thermo Fisher Scientific unless otherwise indicated. Mammary cell suspensions were blocked with Fc receptor antibody, followed by incubation with biotinylated anti-CD31 (14-0311), anti-CD45 (13-0451; clone 30-F11), and anti-TER119 (13-5921). CD45+ immune cells, TER119+ red blood cells, and CD31+ endothelial cells were further conjugated with streptavidin-PE-Cy7 (25-4317) and were excluded using flow cytometry. Propidium iodide (Sigma-Aldrich) was used to stain dead cells, which were excluded from analysis. MEC subpopulations were identified using anti-CD49f–FITC (11-0495; clone GoH3), anti-CD24–PE (12-0242; clone M1/69), anti-CD29–FITC (11-0291; clone ebioHMb1-1), and anti-CD61–APC (MCD6105; Invitrogen) using a FACSCalibur (BD Biosciences) or Canto (BD Biosciences) and analyzed using FlowJo software (Tree Star Inc.). Cell sorting of mammary epithelial subpopulations was performed on a FACSAria (BD Biosciences), and the purity of sorted populations was routinely greater than 97%.
Whole-mount staining.
Mammary glands from females were mounted on glass slides and fixed with Carnoy’s formula 2 fixative (10% glacial acetic acid, 30% chloroform, and 60% absolute ethanol) for 12 hours. Fixed mammary glands were processed through a hydration series using descending concentrations of ethanol (70%, 50%, and 20%; 15 minutes each) and distilled water for 10 minutes, followed by staining in a carmine-alum mix (0.2% carmine [Sigma-Aldrich] and 0.5% aluminum potassium sulfate [Sigma-Aldrich]) for 16 hours. Subsequently, the stained mammary glands were dehydrated through ascending concentrations of ethanol (25%, 50%, 70%, and 100%; 15 minutes each) and cleared in xylene overnight, and then the stained mammary glands were mounted with Permount medium (Fisher Scientific).
3D Matrigel mammary colony-forming assay.
Mammary single-cell suspensions were prepared as described above from inguinal mammary glands of 10- to 12-week-old females at estrus phase. Five thousand total mammary cells were seeded into 50 μl of growth factor–reduced Matrigel (BD Biosciences) and cultured as a droplet on the center of 6-cm dishes containing mouse EpiCult-B medium supplemented with 5% FBS, cytokines EGF and FGF, and EpiCult-B proliferation supplements (STEMCELL Technologies) as previously described (26). Colonies were blindly scored after 12 days of culture under a light microscope, and tiled image composites spanning Matrigel droplets were then taken.
In vivo growth of Rnf8-deficient mammary tumor cells in NSG mice.
Logarithmically growing Rnf8–/– WapCre Trp53fl/fl tumor cells and their RNF8-complemented counterparts were harvested, resuspended in PBS, and combined in a 1:1 ratio with Matrigel (Corning) at a concentration of 1.0 × 107 cells/ml. One hundred microliters of tumor cell suspensions were orthotopically injected into the inguinal fat pads of female NSG mice (6–7 weeks old; The Jackson Laboratory). Tumor volume was measured externally using a digital caliper. Tumor volume = (π × length × width2)/6, where length represents the largest tumor diameter and width represents the diameter perpendicular to the length.
Histological analysis and immunohistochemistry.
Paraffin sections of mammary glands and mammary tumors were stained with H&E for histological analysis as previously described (11). For immunohistochemistry, tissues and tumor sections were prepared according to standard protocols (11). Antibodies used were anti-CK18 (10R-10302; Fitzgerald), anti-CK14 (PRB-155P; Covance), anti-Ki67 (NB500-170; Novus Biologicals), and anti-N1ICD (4147, Val1744 [D3B8]; Cell Signaling Technology). The investigators were blinded during the scoring of Ki67+ cells.
Immunofluorescence.
Staining for subnuclear foci formation was performed as previously described (11). MECs from females at estrus phase were purified by negative depletion using magnetic beads (Dynabeads; Invitrogen). FACS analysis indicated that the purity of the isolated MECs was greater than 97%. Immunofluorescence staining for subnuclear foci was also performed on RNF8-deficient mammary tumors and human breast cancer cell lines MDA-MB-231 and MCF7. Primary antibodies used for immunofluorescence staining were anti-γH2AX (07-164, Millipore; 05-636 JBW301, Millipore), anti-53BP1 (A300-272; Bethyl), anti-RAD51 (sc-8349, clone H-92; Santa Cruz Biotechnology), and anti-BRCA1 (39) (homemade, raised against aa 831–845 of murine BRCA1). Anti-rabbit or anti-mouse IgG conjugated with Alexa Fluor 488 or 594 (Invitrogen, catalog A11008, A11037, A11059, A11032) was used for secondary staining. Subsequently, slides were counterstained with DAPI (Invitrogen) and mounted with Mowiol (Sigma-Aldrich). Images were taken using a fluorescence microscope (Leica) under ×100 magnifications. ImageJ software (NIH) was used to process the raw data. The investigators were blinded during the scoring of foci-positive cells.
Clonogenic survival assay.
Exponentially growing mouse mammary and human breast tumor cells were seeded at 200 cells per 6-cm dish in complete DMEM. Tumor cells were either left untreated, cultured in the presence of 0.1–5 μM of PARP inhibitor (KU0058948, Axon Medchem; or olaparib, Selleckchem), or subjected to γ-irradiation (2–8 Gy). For Notch inhibitor treatment, mammary tumor cells were either left untreated or treated with γ-secretase inhibitors (RO4929097, 1–10 μM; LY411575, 5 μM; YO-01027, 5 μM; Selleckchem). Olaparib (2 μM) and RO4929097 (10 μM) were used for combination treatments. Medium was changed every 4 days to replenish drug levels. Cells were allowed to grow for 12–25 days, depending on the tumor cells, before their fixation with methanol and staining with crystal violet. Numbers of colonies in each dish were counted, and pictures of the dishes were taken. All experiments were performed in triplicate. Data from treated samples were normalized to their respective untreated controls. The investigators were blinded during the scoring of colonies.
Protein analysis and cytoplasmic and nuclear extract preparation.
Immunoblot analyses were performed using standard protocols. For fractionation, 3 × 107 cells were washed with PBS and resuspended in 200 μl of buffer A (10 mM HEPES [pH 7.9], 50 mM NaCl, 0.5 M sucrose, 0.1 mM EDTA, 1 mM DTT, 0.5% Triton-X and protease inhibitor mixture [Roche Molecular Biochemicals]). The cells were incubated for 5 minutes on ice, and subsequently nuclei were collected in the pellet (P1) by low-speed centrifugation (1,000 g, 10 minutes, 4°C) and the supernatant collected as cytoplasmic extract. The pellet was washed once with buffer A; then nuclear extracts were prepared by addition of 200 μl of buffer B (10 mM HEPES [pH 7.9], 250 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.1% NP40 and protease inhibitor mixture), followed by 10 minutes of incubation on ice and subsequent solubilization using sonication. Antibodies used for immunoblotting were against N1ICD (Val1744; Cell Signaling Technology, 4147), HES1 (sc-25392, clone H-140; Santa Cruz Biotechnology), CCND1 (sc-717, clone C-20; Santa Cruz Biotechnology), TRP53 (sc-6243, clone FL393; Santa Cruz Biotechnology), P21 (sc-471, clone M19; Santa Cruz Biotechnology), BAX (sc-493, clone N-20; Santa Cruz Biotechnology), FLAG (F1804, clone M2; Sigma-Aldrich), HA (11583816001, clone 12CA5; Sigma-Aldrich), ubiquitin (NB300-130, clone Ubi-1; Novus Biologicals), RNF8 (sc-271462, clone B-2, Santa Cruz Biotechnology; and NB100-1176, Novus Biologicals), lamin B (sc-3739; Santa Cruz Biotechnology), α-tubulin (sc-53646; Santa Cruz Biotechnology), RBPJ (5442; Cell Signaling Technology), FBW7 (40-1500; Invitrogen), and β-actin (sc-47778, clone C4; Santa Cruz Biotechnology).
Quantitative reverse transcription PCR.
Total RNA from FACS-sorted primary mammary cell subpopulations, mammary tumors, and human breast cancer cell lines was extracted using Trizol (Invitrogen). RNA samples were treated with an RNase-free DNA digestion kit (Invitrogen), and were reverse transcribed into cDNA using the oligo-dT priming method of the First-Strand cDNA synthesis system kit (Invitrogen). Quantitative reverse transcriptase PCR (RT-qPCR) was performed using SYBR Green PCR Master Mix (Applied Biosystems) and the gene-specific primers as indicated in Supplemental Table 1. Assays were performed in triplicate using ABI PRISM 7900HT Sequence Detection System (Applied Biosystems), followed by data analysis using Sequence Detection System software version 2.1 (Applied Biosystems). The expression level of each target gene was normalized to either human or mouse endogenous β-actin and GAPDH transcripts.
RNA sequencing analysis.
Total RNA from mammary tumor cell lines was extracted using the RNeasy kit (Qiagen), and RNA quality was assessed by Agilent 2100 Bioanalyzer (Agilent Technologies). Illumina TruSeq Stranded mRNA Libraries for RNA-Seq were prepared according to Illumina TruSeq protocols. Samples were then sequenced on an Illumina HiSeq (100-bp paired-end). The RNA-Seq data have been deposited to the Gene Expression Omnibus (GEO; reference GSE76075; https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE76075). The RNA-Seq reads were trimmed for adaptors, masked for low-complexity or low-quality sequence, and subsequently mapped to the mouse genome (build mm10, GRCm38) using STAR (version 2.4.0f1) (40). The SAM/BAM files were further processed using SAMtools (version 0.1.18) (41), and read count quantitation was obtained using BEDTools multicov (version 2.25.0) (42). Normalization of read counts and differential expression analysis between genotypes were carried out using the DESeq2 R package in Bioconductor (release 2.13). The Notch signaling pathway annotation was obtained from WikiPathways (43).
In vivo ubiquitylation assay.
Rnf8–/– mammary tumor cells complemented with empty FLAG vector or RNF8-FLAG were either left untreated or treated with the proteasome inhibitor MG132 (20 μM; Selleckchem) for 6 hours, and ubiquitylation levels of N1ICD were examined as previously described (44). In vivo ubiquitylation of N1ICD in MDA-MB-231 cells deleted of endogenous RNF8, and their controls, was similarly examined.
Ubiquitylation assay.
HEK293T cells were transfected with Rnf8WT-FLAG or Rnf8C406S-FLAG, and constructs encoding Ub-WT (Addgene, pRK5-HA-Ub-WT [ID 17608]) or mutated Ub-HA (pRK5-HA-Ub-K48 [ID 17605], pRK5-HA-Ub-K63 [ID 17606]) as indicated. Forty-eight hours later, cell lysates were prepared, precleared with Protein A-Sepharose beads (Life Technologies), and subjected to immunoprecipitation using anti-N1ICD (Cell Signaling Technology). Subsequently, the beads were washed, and proteins released from the beads by boiling in 2× SDS-PAGE sample buffer. Immunoblot analysis was performed using the indicated antibodies.
In vitro ubiquitylation assay.
Recombinant RNF8 (45) was a gift from T.K. Sixma (Division of Biochemistry and Cancer Genomics Centre, Netherlands Cancer Institute, Amsterdam, the Netherlands). UBE1, Ubc13, and Ub recombinant proteins were gifts from C. Arrowsmith (Princess Margaret Cancer Centre, University Health Network and Department of Medical Biophysics, University of Toronto, Toronto, Ontario, Canada). Recombinant N1ICD (2 μg; Addgene, 47612), recombinant RNF8 (1 μg), UBE1 (0.1 μg; E1), Ubc13 (0.2 μg; E2), Ub (5 μg), and ATP (5 mM) were mixed, and the reactions were incubated at 30°C for 2 hours in buffer containing 50 mM Tris/HCl (pH 7.5), 5 mM MgCl2, and 1 mM DTT and examined by immunoblot as previously reported (39).
Cycloheximide chase analysis.
Rnf8–/– Trp53Δ/Δ mammary tumor cells reconstituted with empty vector (mock) or RNF8 were seeded onto 6-cm dishes overnight. Cells were then either left untreated or treated with 150 μM of cycloheximide for 0.5–6 hours, and immunoblot analysis was performed to determine levels of N1ICD, RNF8-FLAG, and β-actin.
Bioinformatics.
To characterize clinical relevance of RNF8 in human breast cancer, we used KM Plotter (http://kmplot.com) (19), 2017 version, with data from 5,142 breast cancer samples, of which 3,955 had patient survival information, downloaded from GEO (Affymetrix microarrays only), the European Genome-Phenome Archive (EGA), and TCGA. We plotted relapse-free survival using the median expression level of RNF8 (probes 203160_s_at and 203161_s_at) to classify breast cancer cases as having either high or low RNF8 expression. The specificity of the 2 RNF8 probes available in the KM Plotter was analyzed using Ensembl (www.ensembl.org). Isoforms of RNF8 were inspected using Ensembl and UniProt (http://www.uniprot.org).
Statistics.
Two-tailed unpaired Student’s t test was used to compare 2 means. Multiple comparisons were performed using 1-way or 2-way ANOVA followed by Tukey’s multiple-comparisons test or Dunnett’s test as indicated in the figure legends. Statistical significance is denoted as follows: *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. The investigators were blinded to the identity of the specific sample during the experiment and when assessing the outcome, whenever the nature of the experiments permitted it. Groups that were statistically compared exhibited a similar variance, as shown in the figures. The log-rank test was used to compare survival curves from mouse models. GraphPad Prism 6 was used for statistical analysis.
Study approval.
All experimental procedures using mice were reviewed and approved by the Princess Margaret Cancer Centre animal care committee. All experiments involving viruses were approved by the University Health Network biosafety committee.
Author contributions
LL, KKNG, BG, PSP, MM, AS, FM, HWJ, HB, LP, AG, DN, MK, CP, IJ, EBA, RK, BR, MAJ, OS, MAP, AH, and RH designed and performed the experiments and analyzed data. LL, KKNG, BG, PSP, MAP, AH, and RH wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by Worldwide Cancer Research, the Canadian Institute of Health Research, and the Canadian Breast Cancer Foundation (to RH). This study was also supported by the “Asociación Española Contra el Cáncer” (AECC), the Generalitat de Catalunya (SGR 2014-364), the “Instituto de Salud Carlos III” (ISCIII), Spanish Ministry of Economy and Competitiveness, “Fondo Europeo de Desarrollo Regional (FEDER)” (grants PIE13/00022-ONCOPROFILE, FIS PI12/01528, and PI15/00854 and RTICC RD12/0036/0008) (to MAP). The authors thank D. Durocher, S. Benchimol, K. Mekhail, V. Stambolic, and L. Salmena for helpful discussions and for reviewing the manuscript, and T.K. Sixma and C. Arrowsmith for providing reagents. We also thank Y. Sheng, P.A. Bissey, and J. Jeon for help with experiments.
Version 1. 09/17/2018
Electronic publication
Version 2. 10/01/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information: J Clin Invest. 2018;128(10):4525–4542.https://doi.org/10.1172/JCI120401.
Contributor Information
Li Li, Email: johnnyinphoton@gmail.com.
Kiran Kumar Naidu Guturi, Email: kirankumarndkm@gmail.com.
Brandon Gautreau, Email: brandon.gautreau@mail.utoronto.ca.
Parasvi S. Patel, Email: parasvi.patel@gmail.com.
Amine Saad, Email: salakisca@msn.com.
Mayako Morii, Email: mayako@gipc.akita-u.ac.jp.
Francesca Mateo, Email: fmateo1980@gmail.com.
Luis Palomero, Email: lpalomerol@gmail.com.
Haithem Barbour, Email: haithem.barbour@gmail.com.
Antonio Gomez, Email: antgomo@gmail.com.
Deborah Ng, Email: deb.ng@mail.utoronto.ca.
Max Kotlyar, Email: maxk.email@gmail.com.
Chiara Pastrello, Email: chiara.pastre@gmail.com.
Hartland W. Jackson, Email: hartlandjackson@gmail.com.
Rama Khokha, Email: Rama.Khokha@uhnresearch.ca.
Igor Jurisica, Email: juris@ai.utoronto.ca.
El Bachir Affar, Email: el.bachir.affar@umontreal.ca.
Brian Raught, Email: braught@uhnres.utoronto.ca.
Otto Sanchez, Email: otto.sanchez@uoit.ca.
Moulay Alaoui-Jamali, Email: moulay.alaoui-jamali@mcgill.ca.
Miguel A. Pujana, Email: miguelangel.pujana@gmail.com.
Anne Hakem, Email: ahakem@uhnres.utoronto.ca.
Razq Hakem, Email: rhakem@uhnres.utoronto.ca.
References
- 1.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: current status and perspectives. Genes Dev. 2014;28(11):1143–1158. doi: 10.1101/gad.242511.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling — are we there yet? Nat Rev Drug Discov. 2014;13(5):357–378. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]
- 3.Ntziachristos P, Lim JS, Sage J, Aifantis I. From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell. 2014;25(3):318–334. doi: 10.1016/j.ccr.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bray SJ. Notch signalling in context. Nat Rev Mol Cell Biol. 2016;17(11):722–735. doi: 10.1038/nrm.2016.94. [DOI] [PubMed] [Google Scholar]
- 6.Panier S, Durocher D. Push back to respond better: regulatory inhibition of the DNA double-strand break response. Nat Rev Mol Cell Biol. 2013;14(10):661–672. doi: 10.1038/nrm3659. [DOI] [PubMed] [Google Scholar]
- 7.Huen MS, et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell. 2007;131(5):901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolas NK, et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science. 2007;318(5856):1637–1640. doi: 10.1126/science.1150034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mailand N, et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell. 2007;131(5):887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 10.Wang B, Elledge SJ. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci U S A. 2007;104(52):20759–20763. doi: 10.1073/pnas.0710061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li L, et al. Rnf8 deficiency impairs class switch recombination, spermatogenesis, and genomic integrity and predisposes for cancer. J Exp Med. 2010;207(5):983–997. doi: 10.1084/jem.20092437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santos MA, et al. Class switching and meiotic defects in mice lacking the E3 ubiquitin ligase RNF8. J Exp Med. 2010;207(5):973–981. doi: 10.1084/jem.20092308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thorslund T, et al. Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature. 2015;527(7578):389–393. doi: 10.1038/nature15401. [DOI] [PubMed] [Google Scholar]
- 14.Lee HJ, et al. The DNA damage transducer RNF8 facilitates cancer chemoresistance and progression through twist activation. Mol Cell. 2016;63(6):1021–1033. doi: 10.1016/j.molcel.2016.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bieging KT, Mello SS, Attardi LD. Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer. 2014;14(5):359–370. doi: 10.1038/nrc3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donehower LA, Lozano G. 20 years studying p53 functions in genetically engineered mice. Nat Rev Cancer. 2009;9(11):831–841. doi: 10.1038/nrc2731. [DOI] [PubMed] [Google Scholar]
- 17.Hainaut P, Pfeifer GP. Somatic TP53 mutations in the era of genome sequencing. Cold Spring Harb Perspect Med. 2016;6(11):a026179. doi: 10.1101/cshperspect.a026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Halaby MJ, et al. Synergistic interaction of Rnf8 and p53 in the protection against genomic instability and tumorigenesis. PLoS Genet. 2013;9(1):e1003259. doi: 10.1371/journal.pgen.1003259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Györffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 20.ElShamy WM, Livingston DM. Identification of BRCA1-IRIS, a BRCA1 locus product. Nat Cell Biol. 2004;6(10):954–967. doi: 10.1038/ncb1171. [DOI] [PubMed] [Google Scholar]
- 21.Solé X, et al. Genetic and genomic analysis modeling of germline c-MYC overexpression and cancer susceptibility. BMC Genomics. 2008;9:12. doi: 10.1186/1471-2164-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alvarez MJ, Sumazin P, Rajbhandari P, Califano A. Correlating measurements across samples improves accuracy of large-scale expression profile experiments. Genome Biol. 2009;10(12):R143. doi: 10.1186/gb-2009-10-12-r143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.O’Connor MJ. Targeting the DNA damage response in cancer. Mol Cell. 2015;60(4):547–560. doi: 10.1016/j.molcel.2015.10.040. [DOI] [PubMed] [Google Scholar]
- 24.Schwertman P, Bekker-Jensen S, Mailand N. Regulation of DNA double-strand break repair by ubiquitin and ubiquitin-like modifiers. Nat Rev Mol Cell Biol. 2016;17(6):379–394. doi: 10.1038/nrm.2016.58. [DOI] [PubMed] [Google Scholar]
- 25.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23(22):2563–2577. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joshi PA, et al. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465(7299):803–807. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- 27.Asselin-Labat ML, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465(7299):798–802. doi: 10.1038/nature09027. [DOI] [PubMed] [Google Scholar]
- 28.Stingl J, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439(7079):993–997. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 29.Bouras T, et al. Notch signaling regulates mammary stem cell function and luminal cell-fate commitment. Cell Stem Cell. 2008;3(4):429–441. doi: 10.1016/j.stem.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Rodilla V, et al. Luminal progenitors restrict their lineage potential during mammary gland development. PLoS Biol. 2015;13(2):e1002069. doi: 10.1371/journal.pbio.1002069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pei XH, et al. CDK inhibitor p18(INK4c) is a downstream target of GATA3 and restrains mammary luminal progenitor cell proliferation and tumorigenesis. Cancer Cell. 2009;15(5):389–401. doi: 10.1016/j.ccr.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carr JR, et al. FoxM1 regulates mammary luminal cell fate. Cell Rep. 2012;1(6):715–729. doi: 10.1016/j.celrep.2012.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKenzie GJ, et al. Nuclear Ca2+ and CaM kinase IV specify hormonal- and Notch-responsiveness. J Neurochem. 2005;93(1):171–185. doi: 10.1111/j.1471-4159.2005.03010.x. [DOI] [PubMed] [Google Scholar]
- 34.Hoeller D, Hecker CM, Dikic I. Ubiquitin and ubiquitin-like proteins in cancer pathogenesis. Nat Rev Cancer. 2006;6(10):776–788. doi: 10.1038/nrc1994. [DOI] [PubMed] [Google Scholar]
- 35.Kravtsova-Ivantsiv Y, Ciechanover A. Non-canonical ubiquitin-based signals for proteasomal degradation. J Cell Sci. 2012;125(pt 3):539–548. doi: 10.1242/jcs.093567. [DOI] [PubMed] [Google Scholar]
- 36.Lord CJ, Tutt AN, Ashworth A. Synthetic lethality and cancer therapy: lessons learned from the development of PARP inhibitors. Annu Rev Med. 2015;66:455–470. doi: 10.1146/annurev-med-050913-022545. [DOI] [PubMed] [Google Scholar]
- 37.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 38.Mao Z, Jiang Y, Liu X, Seluanov A, Gorbunova V. DNA repair by homologous recombination, but not by nonhomologous end joining, is elevated in breast cancer cells. Neoplasia. 2009;11(7):683–691. doi: 10.1593/neo.09312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guturi KK, et al. RNF168 and USP10 regulate topoisomerase IIα function via opposing effects on its ubiquitylation. Nat Commun. 2016;7:12638. doi: 10.1038/ncomms12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kutmon M, et al. WikiPathways: capturing the full diversity of pathway knowledge. Nucleic Acids Res. 2016;44(D1):D488–D494. doi: 10.1093/nar/gkv1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bohgaki M, et al. RNF168 ubiquitylates 53BP1 and controls its response to DNA double-strand breaks. Proc Natl Acad Sci U S A. 2013;110(52):20982–20987. doi: 10.1073/pnas.1320302111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mattiroli F, et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150(6):1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.