Supplemental Digital Content is available in the text
Keywords: ion channel, single nucleotide polymorphism, sudden cardiac arrest, sudden cardiac death, sudden unexplained death
Abstract
Background:
We sought to identify common ion channel single nucleotide polymorphisms (SNPs) associated with the occurrence of sudden cardiac death (SCD) to predict the incidence of SCD in clinical settings.
Methods:
This study involved a systematic review and meta-analysis of ion channel SNPs and risk of SCD in adults. We searched public databases for studies published up to September 19, 2017. We examined relationships between SNPs in common ion channel genes and the incidence of SCD.
Results:
We collected data for 22 trials that included a total of 4149 patients who experienced SCD or had a high risk of SCD and assessed these data in our meta-analysis. An allelic model showed that rs11720524 in SCN5A clearly protected against SCD (odds ratio [OR]: 0.76; 95% confidence interval [95% CI]: 0.67–0.85; P < .001). Subgroup analysis showed that rs11720524 in SCN5A protected against SCD in Europeans and Caucasians but not in Koreans. The allelic model indicated that rs12296050 in KCNQ1 also had significant protective effects against SCD (OR: 0.85; 95% CI: 0.76–0.96; P = .007). Moreover, this model demonstrated that rs2283222 in KCNQ1 had a significant negative relationship with SCD (OR: 0.73; 95% CI: 0.62–0.85; P < .001). Rs12296050 in KCNQ1 protected against SCD in Koreans and Americans. Our results also showed that rs790896 in RYR2 was negatively associated with SCD in a dominant model (OR: 0.66; 95% CI: 0.45–0.97; P = .033).
Conclusions:
Rs11720524 in SCN5A is negatively related to SCD in Europeans and Caucasians, and rs12296050 and rs2283222 in KCNQ1 and rs790896 in RYR2 clearly have protective effects against SCD.
1. Introduction
Sudden cardiac death (SCD) is defined as an unexpected death that usually occurs within 1 hour of symptom onset and is not accompanied by premonitory symptoms.[1] SCD accounts for approximately 20% of deaths in the general population.[2] The incidence of SCD differs among different races and ethnic groups.[3] In Europe and North America, 50 to 100 sudden unexpected cardiac deaths per 100,000 persons occur every year. In adults older than 35, the incidence of SCD is as high as 1 per 1000 persons.[4] The high incidence of SCD, as well as the sudden, unexpected manner in which it occurs and the low rate with which affected individuals are successfully resuscitated, makes SCD a major unsolved problem in clinical cardiology, emergency medicine, and public health.[5]
Among older adults, sudden death is often caused by atherosclerotic coronary artery disease (CAD), terminal ventricular fibrillation (VF), cardiomyopathy, cardiac rhythm disturbances, or hypertensive heart disease. Approximately half of sudden deaths are attributable to ventricular tachycardia or VF.[4] Ion channel genes may be closely related to cardiac arrhythmias and SCD.[6] The movement of ions across specific channels embedded in the membranes of individual cardiomyocytes is crucial for the generation and propagation of cardiac electric impulses.[7] Therefore, we speculated that some single nucleotide polymorphisms (SNPs) in ion channel genes may be associated with the incidence of SCD.
A previous study showed that Asian populations carry more SNPs causing SCD-related genetic ion channelopathies than other populations; however, the relationships between the carriage of specific SNPs and the incidence of SCD remain unclear.[8] Furthermore, although Asians carry more high-risk SNPs than other ethnic groups, there is no evidence indicating that the incidence of SCD in Asian populations is significantly higher than that in other populations. A meta-analysis of other high-risk SNPs showed that variations in 9q21 genes increase the risk of CAD and SCD in European populations.[9] Lahtinen et al[10] found an association between the SCN5A rs41312391 mutation and SCD incidence in Finnish subjects using an adjusted logistic regression model (RR: 1.27; 95% confidence interval [95% CI]: 1.11–1.45; P = 3.4 × 10−4).
Determining the risk of sudden death and devising an effective means of preventing sudden death are the main issues facing clinicians caring for patients with genetic and cardiac abnormalities. Genetic analysis may improve the risk stratification of the general population and enable the prediction and prevention of SCD. It is important to note, however, that there is no means of predicting the risk of SCD in specific individuals in the general population. We aimed to identify the common ion channel SNPs associated with the occurrence of SCD by meta-analysis to predict the incidence of SCD in the clinic.
2. Methods
We used the Meta-analysis Of Observational Studies in Epidemiology (MOOSE) guidelines in this study.[11] Because prior studies provided the basis for performing our study, ethical approval was not required.
2.1. Data sources and search strategy
Two authors independently performed a literature search using PubMed, Embase, the Cochrane Library, and the following Chinese public databases: the China National Knowledge Infrastructure, the China Science Periodical Database (Wanfang Database), the VIP Journal Integration Platform, and the China Biology Medicine Database. The authors searched for studies published up to July 10, 2018. The following terms were used in the searches: “sudden cardiac death,” “sudden cardiac arrest,” “heart arrest,” “cardiopulmonary arrest,” “circulatory arrest,” “sudden arrhythmic death,” “sudden unexplained death,” “sudden unexpected death,” “single nucleotide polymorphism,” “SNP,” “genetic variation,” “genetic polymorphism,” “DNA polymorphism,” “sodium channel,” “ion channel,” and “channel.” The search strategy is presented in Supplemental Table 1. We also conducted manual searches of the reference lists of relevant review articles to identify additional eligible studies.
2.2. Selection criteria
The following studies were included in our meta-analysis: case–control or cohort studies comparing populations comprising individuals who suffered sudden cardiac/unexplained death (including patients who were found to have suffered SCD on autopsy or patients who suffered cardiac arrest and survived after cardiopulmonary resuscitation) with other populations; studies assessing the associations between common ion channel genetic polymorphisms and SCD; and studies comparing the frequencies of specific alleles or the effect sizes of individual genotypes between cases and controls. The following studies were excluded from the analysis: noncase–control studies, such as case reports; case–control studies investigating the risk factors for SCD that lacked data pertaining to victims of SCD or survivors of SCD whose episodes were attributable to ventricular tachyarrhythmia, Brugada syndrome (BrS), or long QT syndrome; studies of sudden infant deaths; studies of familial SCD; studies pertaining to topics other than ion channel gene-related SNPs; and studies that did not report data pertaining to allelic frequencies or effect sizes. Conference reports, editor comments, and academic dissertations were also excluded from the analysis.
2.3. Data extraction and quality assessment
Two authors independently extracted data regarding the following information from each eligible study: the first author's name, the publication year, the population source, the sample size, the definition of death, the control population, and the genes of interest. Disagreements were resolved by discussion. The Newcastle–Ottawa Scale (NOS), a validated tool for evaluating the quality of observational studies included in meta-analyses, was used to evaluate the methodological quality of the included studies. The NOS rates studies according to their scores on the following 3 subscales: selection (4 items, 4 stars), comparability (1 item, 2 stars), and exposure (3 items, 3 stars).[12]
2.4. Statistical analysis
The Hardy–Weinberg equilibrium of the included the populations was calculated to assess the consistencies of allele frequencies between generations.[13] Association analysis was performed using 5 genetic models, namely, allelic (W vs. M), dominant (WW + WM vs. MM), recessive (WW vs. WM + MM), heterozygous (WM vs. MM), and homozygous (WW vs. MM) models. In these models, W represents the major wild-type allele and M represents the minor mutant-type allele.[14] The effect size was estimated by calculating the summary odds ratio (OR) and its 95% CI. The I2 statistic was used to estimate the degree of heterogeneity among the studies. If I2 was ≥50%, the random-effects model was used; otherwise, the fixed-effects model was used. Subgroup analysis was also performed to determine the risk of SCD according to the ethnicities and locations of the population. The risk of false-positive errors (positive errors) may arise in a meta-analysis, especially a meta-analysis featuring a small sample. To correct for the incremental risk of type I errors, we used trial sequential analysis (TSA) to determine whether the findings of the cumulative meta-analysis were dependable and conclusive. The TSA was conducted to maintain a type I error rate of 5% with a power of 80%. The relative risk reduction (RRR) was set as 0.15 to calculate the accrued information size.[15] All tests were 2-tailed, and a P value < .05 was deemed statistically significant. We analyzed study data using R (version 3.3.1; R Foundation for Statistical Computing, Vienna, Austria) and STATA (version 14.0; Stata Corporation, College Station, TX).
3. Results
After the removal of duplicate articles, 1193 and 2916 articles were identified via searches of the aforementioned English-language and Chinese public databases, respectively. A total of 4001 articles were excluded after their titles and abstracts were screened. The full texts of 93 English-language articles and 15 Chinese articles were assessed. The following articles were excluded from the analysis: studies involving non-SCD or high-risk populations (24), noncase–control (cohort) studies (18), nonion channel gene-related studies (14), conference papers (14), reviews (8), studies lacking data regarding the numbers of SNP carriers in each research group (5), duplicate studies (2), and studies of patients with concomitant cardiac diseases (tetralogy of Fallot) (1). Ultimately, 22 articles that involved a total of 4149 patients who experienced SCD or had a high risk of SCD were included in our systematic review (Table 1).[16–37]
Table 1.
Characteristics of the included studies.
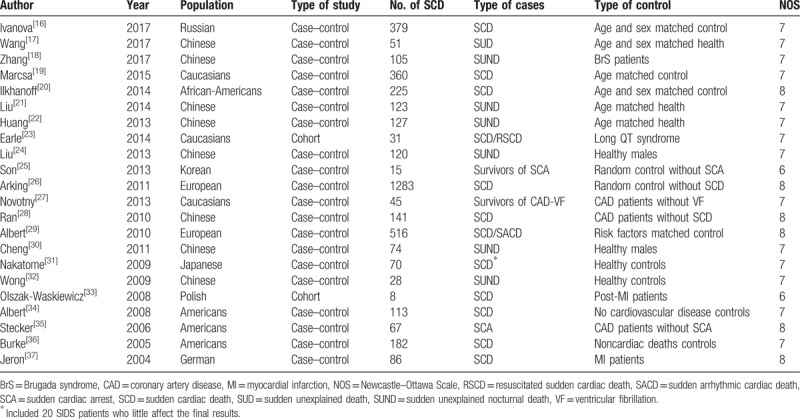
The included studies were published from 2004 to 2017, and the populations of interest were mainly from Europe, America, and East Asia. Three articles pertained to Caucasian populations.[19,23,27] Four articles pertained to survivors of SCD who were successfully resuscitated.[23,25,27,35] The common ion channel-related genes that were assessed in the studies included SCN5A, SCN10A, KCNQ1, KCNH2, KCNE1, and RYR2. The NOS scores of the included studies ranged from 6 to 8. The overall quality of the included studies was generally ideal; however, several factors may have biased the results of the studies. The included studies included only victims and survivors of SCD; thus, the patient inclusion criteria were relatively consistent across the studies. However, there are many causes of SCD; thus, it was difficult to identify the cause of SCD in each person included in the analysis, a phenomenon that may have affected the results. In addition, the characteristics of the control groups were not consistent across the studies, nor were they clearly described in the studies. Moreover, each study used different selection criteria to select its control population. Therefore, in this study, we avoided performing comparisons of the SCD (which included SCD survivors) and control populations.
Rs1805124 in SCN5A was not significantly related to SCD in the allelic model (OR: 1.05; 95% CI: 0.92–1.19; P = .51) or the other models. Subgroup analysis showed that there was no significant relationship between rs1805124 in SCN5A and SCD in European and Caucasian (OR: 1.09; 95% CI: 0.95–1.25; P = .227) or Chinese populations (OR: 0.64; 0.28–1.47; P = .293). The allelic model showed that rs11720524 in SCN5A clearly protected against SCD (OR: 0.76; 95% CI: 0.67–0.85; P < .001). Subgroup analysis showed that rs1805124 in SCN5A protected against SCD in Europeans and Caucasians in the allelic model (OR: 0.76; 95% CI: 0.67–0.86; P < .001), the heterozygous model (OR: 0.67; 95% CI: 0.49–0.91; P = .011), the homozygous model (OR: 0.57; 95% CI: 0.34–0.94; P = .029), and the dominant model (OR: 0.65; 95% CI: 0.48–0.87; P = .005). However, there was no significant relationship between rs1805124 in SCN5A and SCD in Koreans in the allelic model (OR: 0.47; 95% CI: 0.13–1.69; P = .25). The allelic model showed that rs7430407 in SCN5A was not significantly related to SCD (OR: 1.40; 95% CI: 0.63–3.12; P = .415). Similar results were noted in African Americans in the allelic model (OR: 1.40; 95% CI: 0.63–3.12; P = .415). The results for the Chinese population could not be analyzed because the number of patients in whom zero events occurred was high (Fig. 1).
Figure 1.
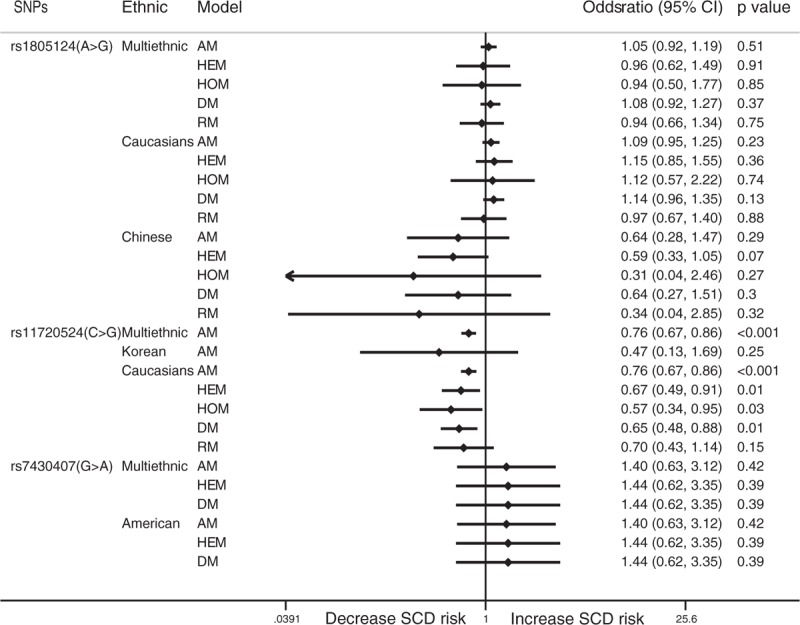
Forest plot for the meta-analysis of associations between SNPs in SCN5A and SCD. SCD = sudden cardiac death, SNPs = single nucleotide polymorphisms.
The allelic model showed that rs6795970 in SCN10A is not related to SCD (OR: 1.10; 95% CI: 0.75–1.63; P = .616), while the recessive model showed that rs6795970 in SCN10A may be related to SCD. However, the relationship between the 2 variables was not significant (OR: 2.19; 95% CI: 0.93–5.18; P = .074). Subgroup analysis showed that there was no significant relationship between rs6795970 in SCN10A and SCD in Caucasian (OR: 0.98; 95% CI: 0.50–1.91; P = .954) or Chinese populations (OR: 1.17; 95% CI: 0.73–1.89; P = .511) (Fig. 2).
Figure 2.
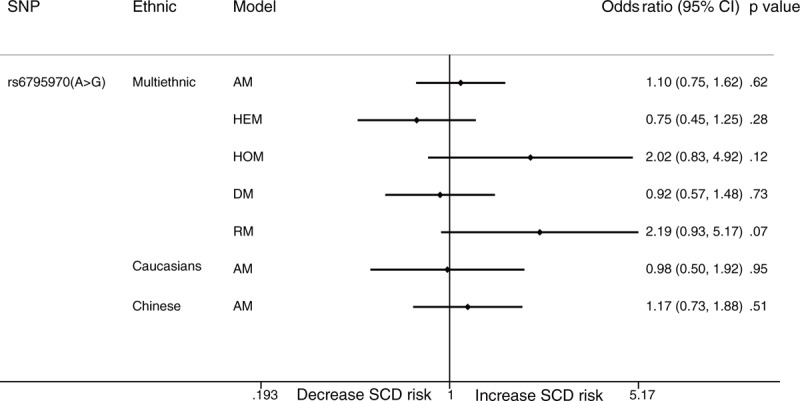
Forest plot for the meta-analysis of associations between SNPs in SCN10A and SCD. SCD = sudden cardiac death, SNPs = single nucleotide polymorphisms.
The dominant model showed that rs1805123 in KCNH2 was not significantly related to the incidence of SCD (OR: 0.91; 95% CI: 0.75–1.12; P = .487). Rs12296050 in KCNQ1 had a significant protective effect against SCD in the allelic model (OR: 0.85; 95% CI: 0.76–0.96; P = .007). Similar results were noted in Europeans (OR: 0.85; 95% CI: 0.76–0.96; P = .006). The allelic model showed that rs2283222 in KCNQ1 was significantly negatively related to SCD (OR: 0.73; 95% CI: 0.62–0.85; P < .001). The clear protective effects of rs2283222 in KCNQ1 were also noted in Koreans (OR: 0.25; 95% CI: 0.07–0.87; P = .03) and Americans (OR: 0.74; 95% CI: 0.63–0.86; P < .001) (Fig. 3). We also analyzed the relationships between rs790896, rs41315858, and rs3766871 in RYR2 and SCD. The results showed that only rs790896 was negatively associated with SCD in the dominant model (OR: 0.66; 95% CI: 0.45–0.97; P = .033). No other SNPs were related to SCD (Fig. 4). Sample size subgroup analysis according to the number of SCD events in each trial was performed (Supplemental Table 2). However, ethnicity and sample size were not considered to be sources of heterogeneity because heterogeneity was not explicitly reduced in subgroup analyses conducted to examine these factors.
Figure 3.
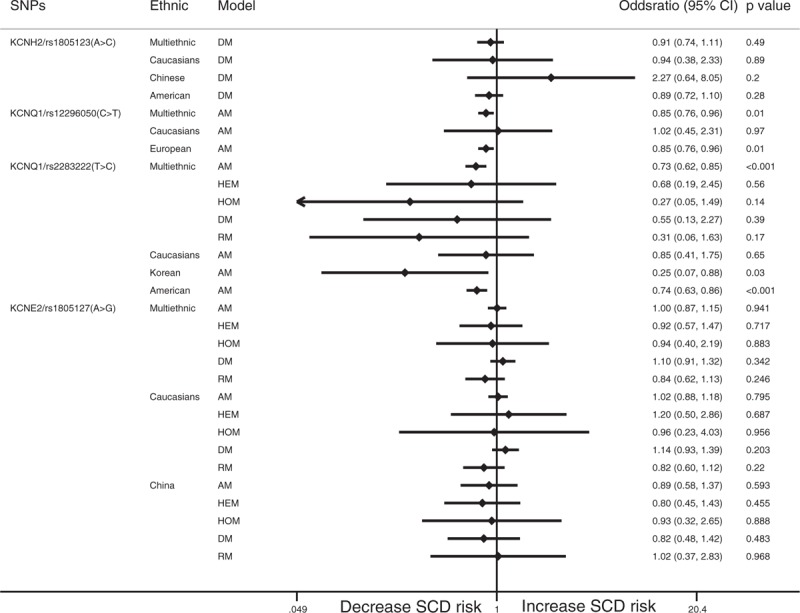
Forest plot for the meta-analysis of associations between SNPs in KCNs and SCD. SCD = sudden cardiac death, SNPs = single nucleotide polymorphisms.
Figure 4.
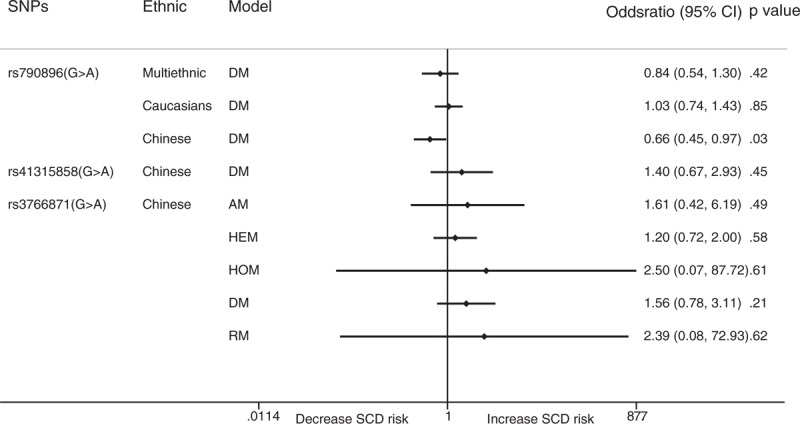
Forest plot for the meta-analysis of associations between SNPs in RYR2s and SCD. SCD = sudden cardiac death, SNPs = single nucleotide polymorphisms.
We performed TSA to determine whether the results pertaining to rs1805124 in SCN5A, which has been studied frequently, and rs11720524 in SCN5A and rs2283222 in KCNQ1, which have also been studied extensively, were robust. The TSA results for rs1805124 showed that the cumulative z-curve did not cross the trial sequential monitoring boundary or even the conventional test boundary (z = 1.96) when the RRR was 15% (Fig. 5A). The TSA results for rs11720524 showed that the cumulative z-curve crossed the trial sequential monitoring boundary when the RRR was 15% (Fig. 5B). However, the small number of studies included in the analysis may have limited the reliability of this result. The fixed-effects model showed that the results for rs2283222 were positive and that the heterogeneity was moderate (I2 = 33.4%) (Fig. 5C). However, in cases in which the random-effects model was used, the cumulative z-curve did not cross the conventional test boundary (P = .067) (Fig. 5D). Therefore, the results of the analysis are unclear, and more studies regarding the relationships between specific SNPs and SCD are needed.
Figure 5.
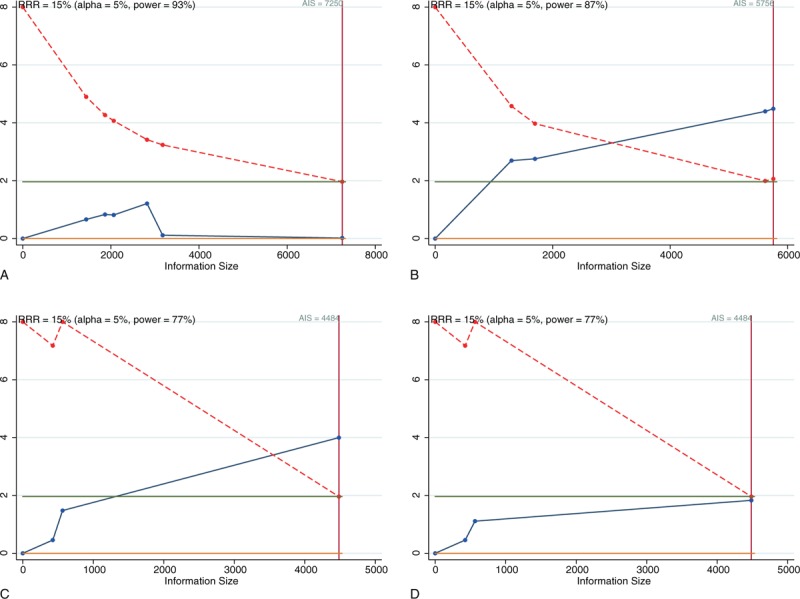
Trial sequential analyses of the cumulative meta-analyses with the allelic models of SCN5A/rs1805124 (A), SCN5A/rs11720524 (B), and KCNQ1/rs2283222 in fixed- (C) and random-effects models (D).
4. Discussion
In our meta-analysis, we comprehensively analyzed the associations between the incidence of SCD and common ion channel gene SNPs. Because SCD is caused by various phenomena, this study did not analyze the risk factors associated with the disease; rather, this study evaluated the above associations only in SCD victims and SCD survivors who were resuscitated. We excluded infants who experienced SCD from the analysis because in young people, genetic heart diseases are more common than coronary heart disease. Because of negative selection and premature deaths, malignant genetic polymorphisms were not widely distributed in the study population, and the mutant gene allele frequency maintenance rates were low. Additionally, there were a large number of SNPs in the ion channel genes of interest. Therefore, we analyzed only common SNPs that have been investigated in more than 2 studies.
The results showed that rs1805124 and rs7430407 have no relationship with SCD. In addition, rs11720524 has a negative relationship with SCD in Europeans and Caucasians. Rs6795970 in SCN10A was not significantly associated with SCD. Additional results showed that rs12296050 and rs2283222 in KCNQ1 and rs790896 in RYR2 have clear protective effects against SCD.
The relationship between rs1805124 and SCD has been studied frequently. Our results indicated that this SNP has no relationship with SCD. The SNP causes a histidine-to-arginine mutation; however, the mutation does not affect ion channel properties. Another study found that rs1805124 reduces SCN5A promoter methylation and increases SCN5A expression in cardiac tissue. In addition, the mutation also prevents VF in patients with BrS.[38] However, the mutation has not been shown to be associated with cardiac repolarization abnormalities, such as T-wave alternans.[39] These results indicate that rs1805124 is substitutable or does not directly affect the incidence of SCD.
Rs11720524 is located in intron 1 of SCN5A and protects against SCD. Another study found that the major allele, C, is associated with VF caused by ST-segment elevation myocardial infarction.[40] However, the mechanism underlying this association and the effect of rs11720524 in SCN5A in Asian populations are unclear. Rs6795970 in SCN10A causes a valine-to-alanine mutation that is significantly associated with changes in the PR interval, longer P-wave durations, and changes in the QRS duration.[41] However, this mutation may not be associated with the incidence of SCD. In addition, rs6795970 AA genotype carriers with multiple sclerosis have significantly worse cerebellar dysfunction than g carriers.[42]
Regarding 3 other mutations that may protect against SCD, rs12296050 in KCNQ1 is located in an intron that is associated with QT interval changes,[43] and rs2283222 in KCNQ1 is located in an intron region that does not affect amino acid sequences. However, this study suggests that these mutations are negatively correlated with SCD. Rs790896 in RYR2 does not change an amino acid sequence that has been shown to protect against ventricular arrhythmias.[28]
Patients with a substantial risk of sudden death usually need an implantable cardioverter defibrillator (ICD) to reduce the risk of sudden cardiac arrest and mortality.[44] However, not all patients at risk for SCD are eligible for or have access to ICD implantation. Amiodarone has been shown to reduce the incidence of SCD by 29%.[45] In addition, a meta-analysis showed that statin treatment reduced the risk of SCD by approximately 10%.[46] Angiotensin-converting enzyme inhibitors can reduce the risk of SCD by approximately 20% in patients with acute MI.[47]
It is necessary to consider psychosociological factors in SCD risk assessments. SCD is often caused by an unrecognized underlying heart condition, and screening has been proposed as a method for preventing sudden death. However, thus far, screening for SNPs or other risk factors has not been shown to significantly reduce the frequency of SCD events. Fifty to three hundred thousand of every 1 million people have been identified to be at risk for SCD. These people require more cardiovascular tests, which cause unnecessary anxiety and stress, and face restrictions with respect to participating in sports. Their conditions may also have an impact on their insurance and employment.[48] Therefore, future studies must strike a balance between protecting against SCD and causing psychosociological stress.
Further study is needed to improve the specificity with which SNPs and other risk factors predict SCD. In addition, the relationships between many other genes, such as the genes encoding NOS1AP, ACE, IL18, and β-adrenoceptors, and SCD need to be investigated. We can then perform comparisons of, for example, populations with and without malignant arrhythmia after MI or myocardial ischemia to identify the specific diseases that lead to SCD.
Our analysis had several limitations. First, this study was performed at the study level but not at the individual level. Second, some sudden deaths are caused by epilepsy, and autopsy could not definitively exclude noncardiac causes in some patients who experienced sudden death. Thus, there is a risk of bias associated with the selection of populations comprising individuals who have experienced sudden death. Third, because large numbers of SNPs in ion channel genes appear to cause SCD, we may have overlooked some key SNPs by excluding SNPs that not have been studied extensively.
Author contributions
Conceptualization: Xiaoli Liu.
Data curation: Jianli Shi.
Formal analysis: Xiaoli Liu.
Funding acquisition: Peilin Xiao.
Investigation: Xiaoli Liu.
Methodology: Xiaoli Liu.
Resources: Jianli Shi.
Software: Jianli Shi.
Validation: Jianli Shi.
Visualization: Jianli Shi.
Writing – original draft: Xiaoli Liu, Peilin Xiao.
Writing – review & editing: Peilin Xiao.
Supplementary Material
Footnotes
Abbreviations: BrS = Brugada syndrome, CAD = coronary artery disease, CI = confidence interval, ICD = implantable cardioverter defibrillator, NOS = Newcastle–Ottawa Scale, OR = odds ratio, RRR = relative risk reduction, SCD = sudden cardiac death, SNPs = single nucleotide polymorphisms, TSA = trial sequential analysis, VF = ventricular fibrillation.
This study was supported by the National Natural Science Foundation of China (Youth Foundation, No. 81600215; http://www.nsfc.gov.cn/).
The authors have no conflicts of interest to disclose.
Supplemental Digital Content is available for this article.
References
- [1].Semsarian C, Sweeting J, Ackerman MJ. Sudden cardiac death in athletes. BMJ 2015;350:h1218. [DOI] [PubMed] [Google Scholar]
- [2].Bezzina CR, Lahrouchi N, Priori SG. Genetics of sudden cardiac death. Circ Res 2015;116:1919–36. [DOI] [PubMed] [Google Scholar]
- [3].Narla V, Tseng ZH. Sudden cardiac death in multi-ethnic populations. Curr Cardiovasc Risk Rep 2016;10:27. [Google Scholar]
- [4].John RM, Tedrow UB, Koplan BA, et al. Ventricular arrhythmias and sudden cardiac death. Lancet 2012;380:1520–9. [DOI] [PubMed] [Google Scholar]
- [5].Zheng ZJ, Croft JB, Giles WH, et al. Sudden cardiac death in the United States, 1989 to 1998. Circulation 2001;104:2158–63. [DOI] [PubMed] [Google Scholar]
- [6].Obeyesekere MN, Leong-Sit P, Massel D, et al. Risk of arrhythmia and sudden death in patients with asymptomatic preexcitation: a meta-analysis. Circulation 2012;125:2308–15. [DOI] [PubMed] [Google Scholar]
- [7].Abriel H, Rougier JS, Jalife J. Ion channel macromolecular complexes in cardiomyocytes: roles in sudden cardiac death. Circ Res 2015;116:1971–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Kong T, Feulefack J, Ruether K, et al. Ethnic differences in genetic ion channelopathies associated with sudden cardiac death: a systematic review and meta-analysis. Ann Clin Lab Sci 2017;47:481–90. [PubMed] [Google Scholar]
- [9].Newton-Cheh C, Cook NR, VanDenburgh M, et al. A common variant at 9p21 is associated with sudden and arrhythmic cardiac death. Circulation 2009;120:2062–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lahtinen AM, Noseworthy PA, Havulinna AS, et al. Common genetic variants associated with sudden cardiac death: the FinSCDgen study. PLoS ONE 2012;7:e41675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stroup DF, Berlin JA, Morton SC, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. JAMA 2000;283:2008–12. [DOI] [PubMed] [Google Scholar]
- [12].Stang A. Critical evaluation of the Newcastle–Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 2010;25:603–5. [DOI] [PubMed] [Google Scholar]
- [13].Chen J, Chatterjee N. Exploiting Hardy–Weinberg equilibrium for efficient screening of single SNP associations from case-control studies. Hum Hered 2007;63:196–204. [DOI] [PubMed] [Google Scholar]
- [14].Areeshi MY, Mandal RK, Panda AK, et al. CD14-159 C>T gene polymorphism with increased risk of tuberculosis: evidence from a meta-analysis. PLoS ONE 2013;8:e64747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Liao LN, Chen CC, Wu FY, et al. Identified single-nucleotide polymorphisms and haplotypes at 16q22.1 increase diabetic nephropathy risk in Han Chinese population. BMC Genet 2014;15:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ivanova AA, Maksimov VN, Ivanoshchuk DE, et al. Association of polymorphism in SCN5A, GJA5, and KCNN3 gene with sudden cardiac death. Bull Exp Biol Med 2017;163:73–7. [DOI] [PubMed] [Google Scholar]
- [17].Wang S, Zhang Z, Yang Y, et al. An insertion/deletion polymorphism within 3′UTR of RYR2 modulates sudden unexplained death risk in Chinese populations. Forensic Sci Int 2017;270:165–72. [DOI] [PubMed] [Google Scholar]
- [18].Zhang L, Zhou F, Huang L, et al. Association of common and rare variants of SCN10A gene with sudden unexplained nocturnal death syndrome in Chinese Han population. Int J Legal Med 2017;131:53–60. [DOI] [PubMed] [Google Scholar]
- [19].Marcsa B, Denes R, Voros K, et al. A common polymorphism of the human cardiac sodium channel alpha subunit (SCN5A) gene is associated with sudden cardiac death in chronic ischemic heart disease. PLoS ONE 2015;10:e0132137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Ilkhanoff L, Arking DE, Lemaitre RN, et al. A common SCN5A variant is associated with PR interval and atrial fibrillation among African Americans. J Cardiovasc Electrophysiol 2014;25:1150–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu C, Tester DJ, Hou Y, et al. Is sudden unexplained nocturnal death syndrome in Southern China a cardiac sodium channel dysfunction disorder? Forensic Sci Int 2014;236:38–45. [DOI] [PubMed] [Google Scholar]
- [22].Huang L, Liu C, Tang S, et al. Postmortem genetic screening of SNPs in RyR2 gene in sudden unexplained nocturnal death syndrome in the southern Chinese Han population. Forensic Sci Int 2014;235:14–8. [DOI] [PubMed] [Google Scholar]
- [23].Earle N, Yeo Han D, Pilbrow A, et al. Single nucleotide polymorphisms in arrhythmia genes modify the risk of cardiac events and sudden death in long QT syndrome. Heart Rhythm 2014;11:76–82. [DOI] [PubMed] [Google Scholar]
- [24].Liu C, Zhao Q, Su T, et al. Postmortem molecular analysis of KCNQ1, KCNH2, KCNE1 and KCNE2 genes in sudden unexplained nocturnal death syndrome in the Chinese Han population. Forensic Sci Int 2013;231:82–7. [DOI] [PubMed] [Google Scholar]
- [25].Son MK, Ki CS, Park SJ, et al. Genetic mutation in Korean patients of sudden cardiac arrest as a surrogating marker of idiopathic ventricular arrhythmia. J Korean Med Sci 2013;28:1021–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Arking DE, Junttila MJ, Goyette P, et al. Identification of a sudden cardiac death susceptibility locus at 2q24.2 through genome-wide association in European ancestry individuals. PLoS Genet 2011;7:e1002158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Novotny T, Kadlecova J, Raudenska M, et al. Mutation analysis ion channel genes ventricular fibrillation survivors with coronary artery disease. Pacing Clin Electrophysiol 2011;34:742–9. [DOI] [PubMed] [Google Scholar]
- [28].Ran Y, Chen J, Li N, et al. Common RyR2 variants associate with ventricular arrhythmias and sudden cardiac death in chronic heart failure. Clin Sci (Lond) 2010;119:215–23. [DOI] [PubMed] [Google Scholar]
- [29].Albert CM, MacRae CA, Chasman DI, et al. Common variants in cardiac ion channel genes are associated with sudden cardiac death. Circ Arrhythm Electrophysiol 2010;3:222–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Cheng J, Makielski JC, Yuan P, et al. Sudden unexplained nocturnal death syndrome in Southern China: an epidemiological survey and SCN5A gene screening. Am J Forensic Med Pathol 2011;32:359–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Nakatome M, Yamamoto T, Isobe I, et al. Diplotype analysis of the human cardiac sodium channel regulatory region in Japanese cases of sudden death by unknown causes. Leg Med (Tokyo) 2009;11:298–301. [DOI] [PubMed] [Google Scholar]
- [32].Wong CH, Koo SH, She GQ, et al. Genetic variability of RyR2 and CASQ2 genes in an Asian population. Forensic Sci Int 2009;192:53–5. [DOI] [PubMed] [Google Scholar]
- [33].Olszak-Waskiewicz M, Kubik L, Dziuk M, et al. The association between SCN5A, KCNQ1 and KCNE1 gene polymorphisms and complex ventricular arrhythmias in survivors of myocardial infarction. Kardiol Pol 2008;66:845–53. [PubMed] [Google Scholar]
- [34].Albert CM, Nam EG, Rimm EB, et al. Cardiac sodium channel gene variants and sudden cardiac death in women. Circulation 2008;117:16–23. [DOI] [PubMed] [Google Scholar]
- [35].Stecker EC, Sono M, Wallace E, et al. Allelic variants of SCN5A and risk of sudden cardiac arrest in patients with coronary artery disease. Heart Rhythm 2006;3:697–700. [DOI] [PubMed] [Google Scholar]
- [36].Burke A, Creighton W, Mont E, et al. Role of SCN5A Y1102 polymorphism in sudden cardiac death in blacks. Circulation 2005;112:798–802. [DOI] [PubMed] [Google Scholar]
- [37].Jeron A, Hengstenberg C, Holmer S, et al. KCNJ11 polymorphisms and sudden cardiac death in patients with acute myocardial infarction. J Mol Cell Cardiol 2004;36:287–93. [DOI] [PubMed] [Google Scholar]
- [38].Matsumura H, Nakano Y, Ochi H, et al. H558R, a common SCN5A polymorphism, modifies the clinical phenotype of Brugada syndrome by modulating DNA methylation of SCN5A promoters. J Biomed Sci 2017;24:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Koskela J, Kahonen M, Fan M, et al. Effect of common KCNE1 and SCN5A ion channel gene variants on T-wave alternans, a marker of cardiac repolarization, during clinical exercise stress test: the Finnish Cardiovascular Study. Transl Res 2008;152:49–58. [DOI] [PubMed] [Google Scholar]
- [40].Jabbari R, Glinge C, Jabbari J, et al. A common variant in SCN5A and the risk of ventricular fibrillation caused by first ST-segment elevation myocardial infarction. PLoS ONE 2017;12:e0170193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chambers JC, Zhao J, Terracciano CM, et al. Genetic variation in SCN10A influences cardiac conduction. Nat Genet 2010;42:149–52. [DOI] [PubMed] [Google Scholar]
- [42].Roostaei T, Sadaghiani S, Park MT, et al. Channelopathy-related SCN10A gene variants predict cerebellar dysfunction in multiple sclerosis. Neurology 2016;86:410–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yu W, Zhang F, Hu W, et al. Association between KCNQ1 genetic variants and QT interval in a Chinese population. Diabet Med 2013;30:1225–9. [DOI] [PubMed] [Google Scholar]
- [44].Lee DS, Green LD, Liu PP, et al. Effectiveness of implantable defibrillators for preventing arrhythmic events and death: a meta-analysis. J Am Coll Cardiol 2003;41:1573–82. [DOI] [PubMed] [Google Scholar]
- [45].Piccini JP, Berger JS, O’Connor CM. Amiodarone for the prevention of sudden cardiac death: a meta-analysis of randomized controlled trials. Eur Heart J 2009;30:1245–53. [DOI] [PubMed] [Google Scholar]
- [46].Rahimi K, Majoni W, Merhi A, et al. Effect of statins on ventricular tachyarrhythmia, cardiac arrest, and sudden cardiac death: a meta-analysis of published and unpublished evidence from randomized trials. Eur Heart J 2012;33:1571–81. [DOI] [PubMed] [Google Scholar]
- [47].Domanski MJ, Exner DV, Borkowf CB, et al. Effect of angiotensin converting enzyme inhibition on sudden cardiac death in patients following acute myocardial infarction. A meta-analysis of randomized clinical trials. J Am Coll Cardiol 1999;33:598–604. [DOI] [PubMed] [Google Scholar]
- [48].Van Brabandt H, Desomer A, Gerkens S, et al. Harms and benefits of screening young people to prevent sudden cardiac death. BMJ 2016;353:i1156. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


