Abstract
Commensal and pathogenic bacteria have evolved efficient enzymatic pathways to feed on host carbohydrates, including protein-linked glycans. Most proteins of the human innate and adaptive immune system are glycoproteins where the glycan is critical for structural and functional integrity. Besides enabling nutrition, the degradation of host N-glycans serves as a means for bacteria to modulate the host’s immune system by for instance removing N-glycans on immunoglobulin G. The commensal bacterium Cutibacterium acnes is a gram-positive natural bacterial species of the human skin microbiota. Under certain circumstances, C. acnes can cause pathogenic conditions, acne vulgaris, which typically affects 80% of adolescents, and can become critical for immunosuppressed transplant patients. Others have shown that C. acnes can degrade certain host O-glycans, however, no degradation pathway for host N-glycans has been proposed. To investigate this, we scanned the C. acnes genome and were able to identify a set of gene candidates consistent with a cytoplasmic N-glycan-degradation pathway of the canonical eukaryotic N-glycan core. We also found additional gene sequences containing secretion signals that are possible candidates for initial trimming on the extracellular side. Furthermore, one of the identified gene products of the cytoplasmic pathway, AEE72695, was produced and characterized, and found to be a functional, dimeric exo-β-1,4-mannosidase with activity on the β-1,4 glycosidic bond between the second N-acetylglucosamine and the first mannose residue in the canonical eukaryotic N-glycan core. These findings corroborate our model of the cytoplasmic part of a C. acnes N-glycan degradation pathway.
Introduction
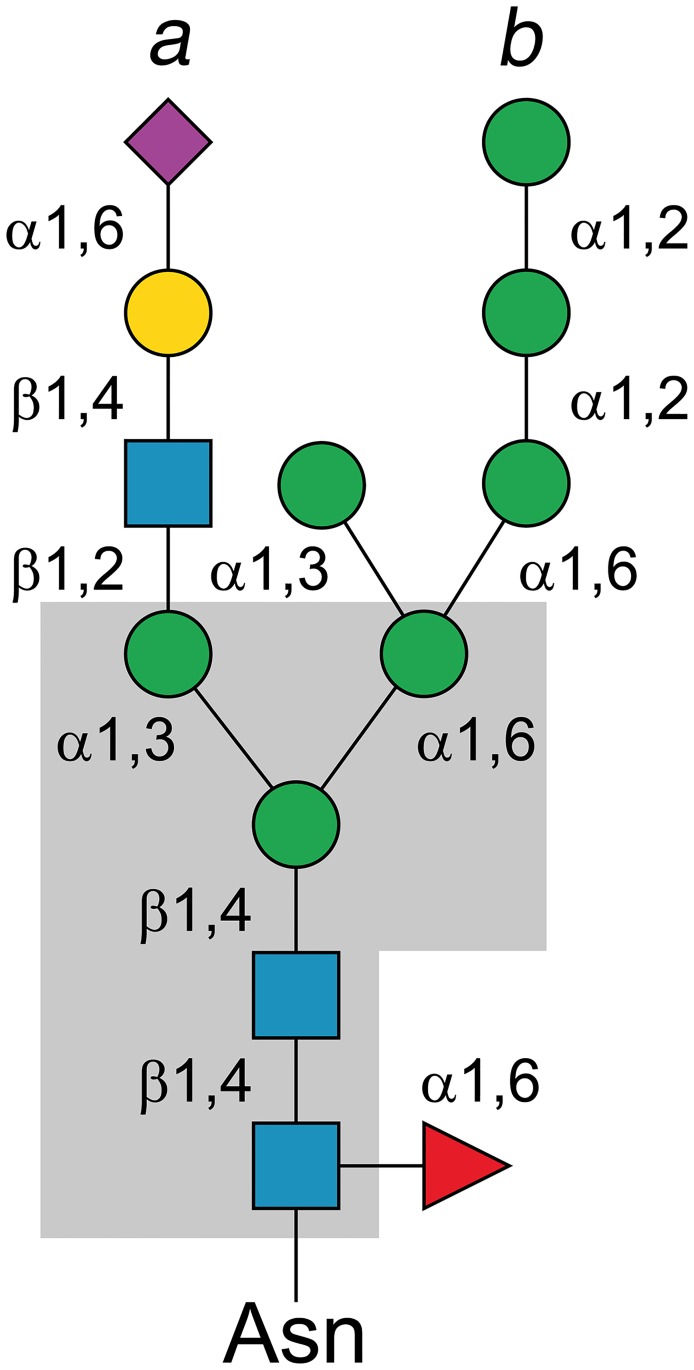
More than two-thirds of all human proteins are predicted to be glycosylated [1], which makes glycosylation one of the most common post-translational protein modifications [2,3]. Protein glycosylation can alter the biophysical properties and function of native proteins that are coded by the genomic sequence [3,4], as for example in the case of proteins of the immune system, e.g., immunoglobulins, where N-glycosylation offers structural and functional diversity beyond that provided by genetic V(D)J recombination [5,6]. In eukaryotes, the N-glycan precursors will be step-wise assembled at the endoplasmic reticulum (ER) through the successive transfer of sugar units (glucose, Glc; mannose, Man; N-acetylglucosamine, GlcNAc) catalyzed by glycosyltransferases [4,7]. The assembled precursor will be transferred “en bloc” to a protein asparagine (present in the sequon Asn-X-Thr/Ser) by the enzyme oligosaccharyltransferase (OST) in the ER lumen [8–10]. All three known N-glycans: high-mannose, complex and hybrid, share an identical five-sugar core structure with the sequence Man3-GlcNAc2 (Fig 1) [11].
Fig 1. Schematic structure of an N-glycan.
The N-glycans contain different carbohydrates depending on the type, but share a common core structure (gray box): branch a, complex N-glycan consisting of two to four branches with mixed sugar residues; branch b, high-mannose N-glycan contains only mannose residues beyond the core structure; mixed a+b, a combination of complex and high-mannose type N-glycan. Color scheme: blue square, N-acetylglucosamine; green circle, mannose; yellow circle, galactose; purple diamond, sialic acid; red triangle, fucose.
Considering the magnitude of the human glycoproteome, and the importance of simple sugars as nutrients for microorganisms, it is not surprising that commensal and pathogenic bacteria have developed efficient systems to degrade and utilize the abundant sugar pool offered by protein-linked glycans [12,13]. By degrading immunoglobulin-linked N-glycans, a bacterial pathogen can alter the host’s immune response to its best advantage [14–20]. For example, in the case of the pathogen Streptococcus pneumoniae, efficient virulence requires an endo-β-N-acetylglucosaminidase that hydrolyzes the β-1,4-glycosidic bond between the two GlcNAc residues in the N,N-diacetylchitobiose (GlcNAc2) core, as well as an α-1,2-mannosidase to trim the terminal α-1,2-linked mannose residues of high-mannose N-glycans down to the Man3-GlcNAc core [21].
The gram-positive bacterium Cutibacterium acnes (formerly Propionibacterium acnes) belongs to the phylum Actinobacteria, and is one of the major inhabitants of the sebaceous follicles of the human skin. Some individuals develop acne vulgaris, i.e., a typically chronic, inflammation of the pilosebaceous unit resulting from C. acnes colonization of the sebaceous glands connected to the hair follicles in the pilosebaceous unit. The disease is multifactorial, however, the androgen-mediated increase in sebum production in adolescents and young adults in combination with C. acnes colonization of the sebaceous glands is a typical disease trigger. Furthermore, C. acnes can become particularly problematic in immunosuppressed transplant patients where 20–25% are affected by acne infections. Recent results from genome-wide association studies have also shown that complex patterns of genetic predisposing factors can trigger pathological acne infection [22–24].
C. acnes has been shown to degrade O-linked glycans, and there are indications that the bacterium can cleave off sugar components from glycolipids [25–27]. Although the first sequenced C. acnes genome was released in 2004, and the current number of available C. acnes genomes is close to a hundred [28,29], there are thus far no reports of attempts to identify N-glycan-degrading enzymes. With the aim to arrive at a more detailed picture of the host-microbe interactions associated with C. acnes in general, and clinically relevant acne infection in particular, we have, to this end, analyzed the C. acnes genome for genes coding for glycoside hydrolases (GHs) that are candidate enzymes in an N-glycan-degradation (NGD) pathway. In this work, we propose a degradation pathway for the cytoplasmic path of host N-glycans by C. acnes, as well as a detailed structural and functional characterization of the exo-β-1,4-mannosidase, a member of glycoside hydrolase family 5 subfamily 18 (GH5_18), that catalyzes hydrolysis of the β-1,4-glycosidic bond between the second N-acetylglucosamine and the first mannose residue in the canonical eukaryotic N-glycan core.
Material and methods
Identification of candidate genes for N-glycan degradation
The genome analyses were based on data available in the carbohydrate-active enzymes (CAZy) database (www.cazy.org) [30], the C. acnes strain 266 genome (CP002409) [31], as well as other C. acnes genomes publicly available in the NCBI database. Operons were investigated using MicrobesOnline Operon Predictions [32]. One unique gene, AEE72695, coding for an enzyme classified in CAZy as belonging to family 5, subfamily 18 (GH5_18) was selected for a more detailed phylogenetic analysis, and we hereafter refer to this gene product as CaMan5_18. Sequences belonging to GH5_18 were retrieved from CAZy, or identified using the Basic Local Alignment Search Tool (BLAST). A total of 55 sequences that represented the diversity of the subfamily were trimmed and used as input for multiple amino-acid sequence alignment using MUSCLE [33]. PhyML was used to create a maximum likelihood tree [34] available at (http://www.phylogeny.fr) [35]. The statistical significance of the branches was assessed with the Approximate Likelihood-Ratio Test (aLRT). The phylogenetic tree was displayed using the Interactive Tree Of Life (iTOL) v3. The presence of possible signal peptides was analyzed using SignaIP 4.1 [36]. ESPript 3 was used to display the sequence alignment (http://espript.ibcp.fr/ESPript/cgi-bin/ESPript.cgi) [37] using pre-aligned sequences from Clustal X.
Cloning and site-directed mutagenesis of CaMan5_18
The gene (AEE72695) coding for CaMan5_18 from Cutibacterium acnes strain 266 was synthesized and codon-optimized for expression in Escherichia coli by Gen9 Inc. The gene was subcloned into the ligation-independent cloning (LIC) vector pNIC-CH2 adding a C-terminal, non-cleavable hexa-histidine tag (https://ki.se/en/mbb/protein-production-platform) using the following forward and reverse primers:
LIC_fwd: 5’-AGAGGAGATAATTAATGAAGATTGGCGCCAATTACAC-3’
LIC_rev: 5’-AATGGTGGTGATGATGGTGCGCGTGGGCTCGAGCACTGG-3’
Site-directed mutagenesis to generate a catalytically deficient CaMan5_18 mutant (E140Q/E259Q) was performed using the following forward and reverse primers:
E140Q_fwd (5’-ATGACGGTGGGCAACCAGTTCCCGCAGTACGCA-3’)
E140Q_rev (5’-TGCGTACTGCGGGAACTGGTTGCCCACCGTCAT-3’)
E259Q_fwd (5’-CCACTCTGGCTGCAGCAGGTAGGAGCACCTCGA-3’)
E259Q_rev (5’-TCGAGGTGCTCCTACCTGCTGCAGCCAGAGTGG-3’)
The PCR samples contained 50 ng plasmid DNA, 2 U Phusion High-Fidelity DNA polymerase (Thermo Fisher), 230 nM of each primer, 500 μM of each dNTP, and 1 x HF Phusion buffer (Thermo Fisher). For mutagenic PCR the following conditions were used: 95°C for 30 s, 30 cycles of 95°C for 30 s; 68°C for 9 min, with a final incubation at 68°C for 10 min. The resulting PCR products were treated with 10 U of DpnI (Thermo Fisher) to degrade the methylated template-DNA. The remaining PCR products were purified using the GeneJET PCR Purification Kit (Thermo Fisher), and transformed into chemically competent E. coli DH5α cells (Invitrogen) followed by plating on Luria-Bertani (LB) agar supplemented with 50 μg mL-1 kanamycin at 37°C for 17 h.
Production of recombinant CaMan5_18
The recombinant plasmid was transformed into E. coli BL21(DE3)-T1 competent cells. The bacteria were cultured in Terrific Broth (TB) medium supplemented with 50 μg mL-1 kanamycin. After reaching an OD600 value in the range 0.8 to 1.0, recombinant gene expression was induced by adding β-D-1-thiogalactopyranoside (IPTG) to a final concentration of 0.2 mM, and the culture further incubated at 17°C for 16 h. The cells were harvested by centrifugation at 5,000 r.p.m. (Beckman Coulter JA-10 fixed-angle rotor, 4,424×g). The bacterial pellet was resuspended in 25 mM K2HPO4 (pH 7.2), 150 mM NaCl, 5% (v/v) glycerol, with one tablet of complete protease inhibitor cocktail (Roche). The resuspended pellet was homogenized using an AVESTIN Emulsiflex-C3 system, and centrifuged at 10,000 r.p.m. (Beckman Coulter JA-25.50 fixed-angle rotor, 12,096×g) using an Avanti J20XP centrifuge (Beckman Coulter) for 20 min at 4°C. The resulting supernatant was mixed with Ni-NTA agarose resin (Invitrogen), packed in Bio-Rad Econo-Pac columns, and set aside to incubate at 4°C for 1 h. The columns were washed with 25 mM K2HPO4 (pH 7.2), 150–300 mM NaCl, 30–50 mM imidazole, 0–10% (v/v) glycerol. Bound protein was eluted with buffer containing 500 mM imidazole. The eluted protein was concentrated using Vivaspin20 centrifugal concentrators (polyethersulfone filter; molecular weight cut-off, MWCO, 30 kDa), and loaded onto a HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare Life Sciences) equilibrated with 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.5, 150 mM NaCl, and 0–10% (v/v) glycerol. The recovered fractions containing CaMan5_18 were pooled and further concentrated to 15–20 mg L-1 using a Vivaspin20 centrifugal spin concentrator (MWCO, 30 kDa). The same procedure as described above for the wild type was used for the E140Q/E259Q mutant.
SEC analysis of oligomeric state
A size-exclusion chromatography (SEC) HiLoad 16/60 Superdex 200 prep grade column (GE Healthcare Life Sciences) was equilibrated in 25 mM K2HPO4 (pH 7.2), 150 mM NaCl, and calibrated with five protein standards: ferritin (440,000 Da), aldolase (158,000 Da), conalbumin (75,000 Da), ovalbumin (43,000 Da) and ribonuclease (13,700 Da); after which CaMan5_18 was subjected to SEC to determine the retention volume. The logarithm of the molecular weights (log10MW) of the protein standards were plotted versus the retention volume using GraphPad Prism 7 (GraphPad Software, San Diego California USA) to obtain a standard curve (R2 = 0.9974) for extrapolation of the log10MW for CaMan5_18.
Structure determination and model refinement
Crystals of CaMan5_18 were grown by vapor diffusion in sitting drops at 4°C in the presence of metal ion and substrate. Successful crystallization was achieved by mixing 1 μL 25 mg mL-1 protein in 20 mM HEPES pH 7.5, 300 mM NaCl, 10% (v/v) glycerol, 0.5 mM tris(2-carboxyethyl)phosphine (TCEP) with 0.5 μL reservoir solution containing 0.2 M 3-(N-morpholino) propane sulfonic acid (MOPS) buffer (pH 6.5), 0.2 M magnesium acetate, 20% (w/v) polyethylene glycol (PEG) 8000 and 5 mM D-mannose. For experimental phasing, a solution of gold cyanide, Au(CN)2, was added to a final concentration of 10 mM in the drop.
X-ray intensity data for non-derivatized and derivatized crystals were recorded using synchrotron radiation, followed by data merging and scaling using the XDS package [38]. The crystals belong to space group P21 with four molecules in the asymmetric unit, and an approximate solvent content of 50%. Heavy-atom substructure determination and initial phasing were performed on data from the gold-derivatized crystal by single-wavelength anomalous dispersion (Au-SAD) as implemented in the program autoSHARP [39] included in the SHARP package [40]. The initial SAD phases were improved and extended to 1.8 Å resolution by solvent flattening using SOLOMON as implemented in SHARP.
An initial model for CaMan5_18 was built by alternating manual model building using COOT [41], and refinement with PHENIX [42] guided by σA-weighted 2Fo-Fc electron density maps. Refinement with PHENIX included refinement of x,y,z coordinates, real-space refinement, and refinement of individual atomic displacement parameters. The gold contribution to the Fourier amplitudes induced undesirable effects in the electron density near the sites of substitution, which obstructed map interpretation and completion of the model in areas close to the heavy-atom sites. Calculated phases from the partial model were therefore used to phase the Fourier amplitudes from a native, non-derivatized crystal. The resulting 2Fo-Fc electron density map was of high quality and a complete CaMan5_18 model was generated. Refinement statistics are given in Table 1.
Table 1. Data collection, phasing and refinement statistics.
| Data collection | CaMan5_18 | CaMan5_18 derivative |
|---|---|---|
| Protein variant | Native | Au(CN)2 |
| Cell constants a, b, c (Å); β(°) | 84.18, 104.37, 112.75; 90.27 | 83.65, 101.44, 111.86; 90.13 |
| Space group / molecules per a.s.u. | P21 / 4 | P21 / 4 |
| Beamline, λ (Å) | SOLEIL PROXIMA1, 1.07175 | SOLEIL PROXIMA1, 1.03927 |
| Resolution range, nominal (Å) | 49.60–1.70 (1.80–1.70) |
48.98–2.00 (2.10–2.00) |
| Unique reflections | 207,504 (31,843) | 217,603 (16,499) |
| Multiplicity | 6.8 (6.9) | 3.3 (2.9) |
| Completeness (%) | 97.1 (94.9) | 87.5 (48.7) |
| <I / σI> | 8.8 (0.9) | 10.9 (1.69) |
| Rsym | 0.111 (2.74) | 0.076 (0.692) |
| Rmeas | 0.121 (2.96) | 0.090 (0.846) |
| CC(1/2) | 99.8 (44.0) | 99.7 (62.9) |
| CC(1/2)ano | N/A | 0.39 (0.12) |
| Wilson B factor (Å2) | 29.3 | 25.9 |
| SAD phasing | ||
| Resolution (Å) | 2.00 | |
| Number of sites | 17 | |
| PPano (acen) | 0.773 | |
| Rcullisano (acen) | 0.889 | |
| FOM (acen/cen) | 0.248/0.082 | |
| Solomon E2/contrast (solvfrac) | 2.1895 (0.506) | |
| Crystallographic refinement | ||
| Resolution range (Å) | 49.601–1.800 (1.845–1.800) |
|
| Completeness, all % (outer bin) | 97.5 (96.0) | |
| Rfactor/work reflns, all | 0.195 / 175,603 | |
| Rfree/free reflns, all | 0.226 / 2001 | |
| Number of amino-acid residues | 1,575 | |
| Non-hydrogen atoms | 13,584 | |
| Mean B (Å2) protein all | 33.5 / 12,748 | |
| Mean B (Å2) solvent / N°. mol. | 37.2 / 836 | |
| Rmsd bond lengths (Å), angles (°) | 0.007, 0.86 | |
| Ramachandran: favored (%) / allowed (%) / Outliers | 97.8 / 100 / 0 | |
| PDB accession code | 6GVB |
ThermoFluor stability assay
Thermal stability to unfolding of CaMan5_18 was analyzed using ThermoFluor screening [43]. The effect of pH on the melting temperature, Tm, was investigated. Reaction mixtures of 100 μL contained 2 μL of 10 mg mL-1 CaMan5_18, 2 μL SYPRO Orange dye (25x stock solution diluted 1:200 with water), and 97 μL 50 mM buffer (sodium acetate pH 5.0–5.5, sodium phosphate pH 6.0–7.5, and Tris-HCl pH 8.5–9.0), and were added to the wells of a 96-well thin-wall PCR plate (Bio-Rad). The pH value was varied from 5 to 9 in intervals of 0.5 pH units. The plates were sealed with Optical-Quality Sealing Tape (Bio-Rad), and heated in an iCycler iQ Real Time PCR Detection System (Bio-Rad) from 20–90°C in steps of 0.2°C. The changes in fluorescence were monitored with a charge-coupled device (CCD) camera. The wavelengths for excitation and emission were 490 nm and 575 nm, respectively.
Substrate screening
The substrate specificity of CaMan5_18 was investigated by screening a range of p-nitrophenyl β-D-glycosides and polysaccharides: p-nitrophenyl-β-D-mannopyranoside (pNP-βMan), p-nitrophenyl-β-D-xylopyranoside (pNP-βXyl), p-nitrophenyl-β-D-fucopyranoside (pNP-βFuc), p-nitrophenyl-α-L-fucopyranoside (pNP-αFuc), p-nitrophenyl-β-D-galactopyranoside (pNP-βGal), p-nitrophenyl-β-D-glucopyranoside (pNP-βGlc), and p-nitrophenyl-β-D-cellobioside (pNP-βCel) were purchased from Sigma Chemical Co. (St. Louis, MO); ivory nut mannan, konjac glucomannan, birchwood xylan, barley β-glucan, carboxymethylcellulose (CMC, low viscosity), and cellobiose were purchased from Megazyme (Bray, Ireland).
The enzyme reactions were performed in final volumes of 210 μL. The reaction mixtures contained 200 μL 1 mM pNP sugar in 50 mM sodium phosphate buffer (pH 6.0) and were pre-heated at 50°C for 5 min before adding 10 μL 10 mg/mL CaMan5_18. Following incubation at 50°C for 10 min, 20 μL 2 M Na2CO3 was added to inactivate the enzyme and stabilize the chromophore in its anionic 4-nitrophenolate form. The released p-nitrophenol was measured spectrophotometrically at 410 nm. The amount of pNP released was measured at A410 (ε410 = 18.3 mM-1 cm-1). For cellobiose, CMC, glucomannan, mannan, barley β-glucan and xylan, 1% substrate was used, and the reaction was stopped by heating the sample at 100°C for 5 min and liberated glucose was measured by the dinitrosalicylic (DNS) colorimetric method. The data were analyzed with SigmaPlot (Systat Software, San Jose, CA).
Kinetic analysis with pNP-βMan as substrate
The enzyme kinetics was determined for hydrolysis of pNP-βMan at various substrate concentrations (0.5 to 20 mM). The enzyme reaction was performed as described above for pNP sugars. Enzyme and substrate-free controls were prepared and assayed under the same conditions. One unit of enzyme activity was defined as the amount of enzyme required to release 1 μmol of pNP min-1 mL-1 under the standard assay conditions. The data were analyzed with GraphPad Prism 7 (GraphPad Software, San Diego California USA). All enzyme assays were performed in triplicates.
Effect of pH and temperature on enzyme activity on pNP-βMan
The optimal pH (pHopt) and temperature (Topt) for enzyme activity were investigated by incubating 10 μg of CaMan5_18 with pNP-βMan at 50°C for 10 min. For the determination of optimal pH for enzyme activity, the pH range 4.0 to 8.5 (in intervals of 0.5) was analyzed using the following buffer systems: 50 mM sodium acetate, pH 4.0 to 5.5; 50 mM sodium phosphate, pH 6.0 to 7.5; 50 mM Tris-HCl, pH 8.0 to 8.5. For determination of optimal reaction temperature, the enzyme activity was measured at pH 6.0 for the temperature range 20–80°C (in intervals of 10°C). The activity obtained at the Topt or pHopt was used to calculate the relative percentage of enzyme activity at different temperature and pH values.
The kinetic stability was determined by incubating 10 μg of CaMan5_18 at 40°C, 50°C, 60°C and 70°C for 0 to 40 h (samples were taken at the time points: 0, 5, 10, 20, 30, 40, 60, 80 min; and 2, 4, 6, 12, 24, 36, 40 h). Following incubation, residual activity was immediately measured for the samples, and the half-life time of irreversible thermal inactivation (at) was determined by non-linear regression using the equation of exponential decay:
Data were collected in triplicate, and analyzed using SigmaPlot (Systat Software, San Jose, CA).
TLC analysis of reaction products
The products resulting from CaMan5_18 hydrolysis of mannooligosaccharides, and the candidate for natural substrate, Man-GlcNAc, were analyzed by thin-layer chromatography (TLC). Briefly, 1 μL of 10 mg/mL enzyme was incubated with buffer solutions containing 20 μL of 10 mM mannooligosaccharide or 10 μL of 10 mM Man-GlcNAc. The mannooligosaccharides tested were mannotriose (M3), mannotetraose (M4), mannopentaose (M5), and mannohexaose (M6). The reactions were carried out at 40°C for 24 h with samples withdrawn at different time points (0, 1, 3, 6, 9, 12, 24 h), and terminated by heating the samples at 100°C in a water bath for 10 min. After centrifugation at 10,000×g for 10 min, the samples were spotted on silica 60 F254 plates and air-dried. TLC was performed using a mobile solvent containing butanol:propanol:ethanol:water (1:2:2:1). The silica plates were developed by spraying with thymol solution and heating at 120°C for 10 min. The control samples were treated identically to the experimental samples, with the exception of adding CaMan5_18 that had been heat-inactivated at 100 °C for 10 min.
MALDI-TOF MS analysis of reaction products
Matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-ToF MS, Applied Biosystems, CA, USA) was used to identify the products resulting from hydrolysis of Man-GlcNAc catalyzed by CaMan5_18. For sample preparation, control samples or reaction product (2.5 μL) were mixed with 10 mM NaCl (6 μL) and 2,5-dihydroxybenzoic acid (DHB) (10 mg/mL, 10 μL) in 50% (v/v) acetonitrile. Then 2 μL of the mixture was spotted onto a stainless steel plate and rapidly dried under vacuum for homogeneous crystallization. MALDI-TOF mass-spectrometry analysis was performed using an accelerating voltage of 20,000 V with a delay time of 200 ns, and operating the instrument under the reflectron mode.
Results
Screening of candidate genes for N-glycan degradation
To identify a possible C. acnes degradation pathway for N-glycans, the genome of C. acnes strain 266 (phylogenetic group IA1), was screened for genes coding for carbohydrate-active enzymes (CAZymes) classified in the CAZy database. By applying the operon-prediction method by Price and co-workers [32], we identified an operon corresponding to a possible pathway for cytoplasmic trimming of the Man3-GlcNAc2 core. The operon contains a GH38 gene (AEE72694) with putative exo-α-1,3- or exo-α-1,6-mannosidase activity (EC 3.2.1.24), and a GH5 (subfamily 18) gene (AEE72695) coding for a predicted β-mannosidase (EC 3.2.1.25). These two enzyme activities are essential for deconstructing the fundamental Man3-GlcNAc2 core structure, and the absence of a predicted signal peptide suggests that these gene products are located in the cytoplasm. The GH38 and GH5_18 genes cluster together in all C. acnes strains, beyond which variation occurs in accessory functions. In addition, we note that this gene pair is present in genomes of other bacteria associated with the human microflora, such as Bifidobacterium breve, Bifidobacterium longum, Dermabacter vaginalis and Arcanobacterium haemolyticum.
In the C. acnes strain 266, the GH38/GH5_18 locus contains two additional GH genes (Fig 2): (i) a GH20 (AEE72696) predicted to be an exo-β-1,4-N-acetylhexoaminidase, which would be able to cleave between the GlcNAc residues in the chitobiose core, and (ii) a GH18 (AEE72704) that may act as an endo-β-N-acetylglucosaminidase (EC 3.2.1.96). However, as shown by comparative genomics [44], the GH20 and GH18 genes are part of a genomic deletion in C. acnes strains belonging to the phylogenetic groups IB, II and III (S1 Fig), showing that the gene organization observed in strain 266 is not conserved. In addition to the GH genes, the locus contains relevant genes coding for putative sugar ABC transporter permeases and ABC transporter substrate-binding proteins.
Fig 2. Proposed C. acnes 266 N-glycan-processing locus 1.
Gene organization of the proposed N-glycan processing locus in the genome of C. acnes 266. GH genes predicted as mannosidases are colored green, and GH genes with predicted N-acetylhexosaminidase activity are blue. Other associated genes are colored light gray and include: predicted sugar ABC-transporter substrate-binding protein (SBP), sugar ABC-transporter permease (PERM), transcriptional-regulator gene (REG), and ABC-transporter ATP-binding protein (ATPB). Accession numbers (GenBank, or RefSeq when GenBank was not available) are shown below each gene.
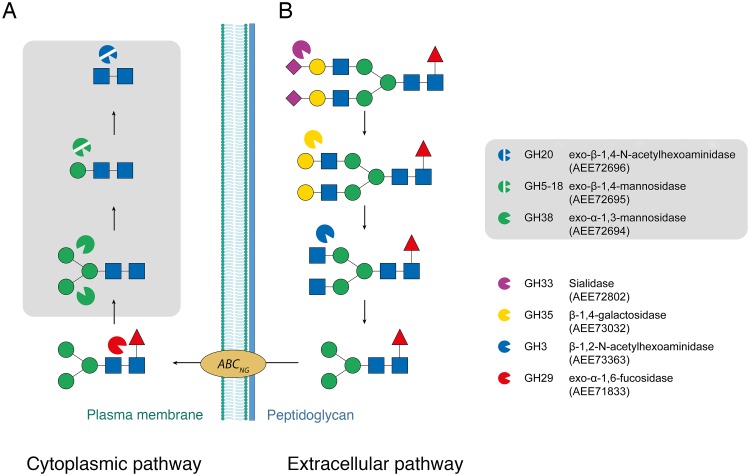
Provided that the enzyme activities predicted by CAZy are correct, the locus containing the triad GH38/GH5_18/GH20 would indeed be sufficient to depolymerize the Man3-GlcNAc2 core into monosaccharide components (Fig 3A). Taken together, these observations strongly suggest that this gene cluster coincides with a carbohydrate-processing locus (Fig 2). Separate from this locus, we also identified a GH29 gene (AEE71833) that lacks secretion signal, and is predicted to code for an exo-α-1,6-fucosidase, an activity relevant for removal of fucose from N-glycans that are fucosylated at the chitobiose core.
Fig 3. Proposed degradation pathway for host N-glycans by C. acnes.
(A) Proposed cytoplasmic pathway involving the enzymes GH38, GH5_18 and GH20, and (B) a hypothetical extracellular pathway. Symbols: blue square: N-acetylglucosamine; green circle, mannose; yellow circle, galactose; purple diamond, sialic acid; red triangle, fucose.
Identification of candidate genes coding for secreted enzymes that could participate in the initial trimming of the N-glycan on the extracellular side is more speculative, but three GH genes that contain signal-peptide sequences deserve particular attention, namely genes coding for a GH33 (AEE72802), GH3 (AEE73363), and a GH35 (AEE73032) (Fig 3B). The GH33 gene (AEE72802) displays 59% sequence identity to the nedA gene from Micromonospora viridifaciens (UniProt Q02834), which codes for the enzyme NanH that catalyzes removal of terminal sialic acid in glycoconjugates (sialidase; EC 3.2.1.18) [45]. The C. acnes GH33-gene sequence corresponds mainly to the catalytic sialidase domain that folds as a six-bladed β-propeller [46], and lacks the accessory domains present in M. viridifaciens NanH (i.e., a linking NPCBM domain and a sugar-binding F5/8-type C domain). Importantly, all catalytically important amino acids, and the five bacterial neuraminidase repeats (BNR repeats; Asp-box motifs [47]) are present. The GH33 sequence in C. acnes 266 displays a frame shift at the C-terminus, however, the corresponding genes in high-quality genomes of the closely related C. acnes strains PA_12_1_L1 and PA_12_1_R1 are present as full-length versions suggesting that the 266 sequence is incorrect due to gene sequencing or assembly problems. Still, even after correcting for this error, the C. acnes GH33 sequence beyond the catalytic domain is different from that of NanH.
The C. acnes 266 GH3 gene (AEE73363) is predicted to code for a β-1,2-N-acetyl-hexosaminidase (EC 3.2.1.52), which would be a suitable candidate for cleavage of GlcNAc from an N-glycan. The GH3 gene shows 36% sequence identity to NagZ (YbbD) from Bacillus subtilis strain 168 (UniProt P40406), and also contains the required catalytic Asp-His dyad [48]. In line with our hypothesis, this GH3 enzyme has been shown to be part of the C. acnes secretome [26].
The third required activity in the extracellular path of complex and hybrid N-glycan degradation by C. acnes is the exo-β-1,4-galactosidase (EC 3.2.1.23) to remove β-1,4-linked galactose. The only relevant gene candidate with a signal-peptide sequence (AEE73032) belongs to family GH35. The gene is annotated as a fragment in the CAZy database, but closer inspection of the sequence reveals that it is 32% identical to the eukaryotic N-terminal β-galactosidase (β-Gal) TIM-barrel domain of Penicillium sp. (PDB code 1XC6 [49], residues 41–265), with the active site and most galactose-binding residues conserved. The GH35 gene is absent in a few C. acnes strains, but when present, including in high-quality genomes [29], it always occurs in this short version. Hence, the observed shorter GH35 sequence is not likely due to a sequencing error. We find several examples in various C. acnes genomes of a gene combination with a short GH35 module and a GH20 module. There is also considerable variation in the genomic location of the GH35 gene across C. acnes strains, and occasionally, a second short GH35 gene is encountered, sometimes with one or two GH85 genes that encode fragments or full-length proteins. We thus hypothesize that C. acnes strains employ a minimal structural β-Gal framework for the purpose of trimming off galactose units from N-glycans. It should be noted that while all three proposed extracellular enzymes in C. acnes 266 contain a secretion signal, neither has a typical C-terminal cell-wall anchoring LPxTG motif. However, this motif does not seem to be consistently present in secreted glycan-processing enzymes. For instance, the GH35 β-galactosidase BgaC from Streptococcus pneumoniae has been shown to localize to the cell surface although the protein lacks a typical signal peptide and LPxTG sequence motif [50], which highlights differences in sorting mechanisms among Gram-positive bacteria.
Phylogenetic analysis of the CaMan5_18 sequence
The AEE72695 gene coding for the GH5_18 enzyme (408 amino acids) was predicted to be a β-mannosidase based on the GH5-subfamily classification in CAZy [51]. A BLAST search with the AEE72695 sequence against the Protein Data Bank did not return any sequence hits for proteins with known three-dimensional structure. We speculated that this gene product might be responsible for cleaving off Man from the Man-GlcNAc2 core, since this bond is the only mannose-linking glycosidic bond in human N-glycans in β configuration. In fact, the β-1,4 bond in Man-GlcNAc is the only conceivable substrate for a β-mannosidase involved in host N-glycan metabolism.
Although subfamily GH5_18 includes 178 actinobacterial sequences (as of May 2018), no biochemically confirmed substrate specificity has been reported for any of the enzymes [51]. There is one exo-mannosidase characterized that belongs to the most closely related subfamily GH5_19 (EC 3.2.1.25) [52], as well as other mannan-active enzymes that are members of subfamilies GH5_7, GH5_10, GH5_17 and GH5_31. Together with GH5_18 and GH5_19, these make up one of the three major clades of the GH5 family, and it has been proposed that members of these subfamilies are likely to act on mannose-containing carbohydrates [51].
Subfamily GH5_18 is dominated by sequences from three genera: Cutibacterium (21 sequences), Bifidobacterium (51 sequences) and Streptomyces (51 sequences). All available genomes of C. acnes strains present in the CAZy database contain a GH5_18 gene. A phylogenetic tree of GH5_18 sequences shows that CaMan5_18 falls in a clade with other enzymes from various C. acnes strains, as well as sequences from C. avidum and Cutibacterium sp. oral taxon 193 (S2 Fig). In addition, this clade contains proteins from Acidipropionibacterium acidipropionici ATCC 4875 and Tessaracoccus bendigoensis DSM 12906. The latter species belong to the same family, Propionibacteriaceae, as the Cutibacterium genus. There are minor sequence variations between different C. acnes phylotypes (S3 Fig), where the largest discrepancy is observed at the C-terminus in different strains. The C-terminus in our CaMan5_18 structure (residues 396–408) is flexible and lacks interpretable electron density. This region is located far away from the active site and dimer interface, which means that it does not play a direct role in substrate hydrolysis or dimer formation in either our CaMan5_18 (IA strain), or in the homologous enzymes of type IB, II and III strains. The most consistent difference among sequences of the Cutibacterium clade and other GH5_18 proteins is a short insert of four residues (149PGHH152, CaMan5_18 numbering) downstream of the catalytic residue Glu140 (S4 Fig).
Overall structure and active site of CaMan5_18
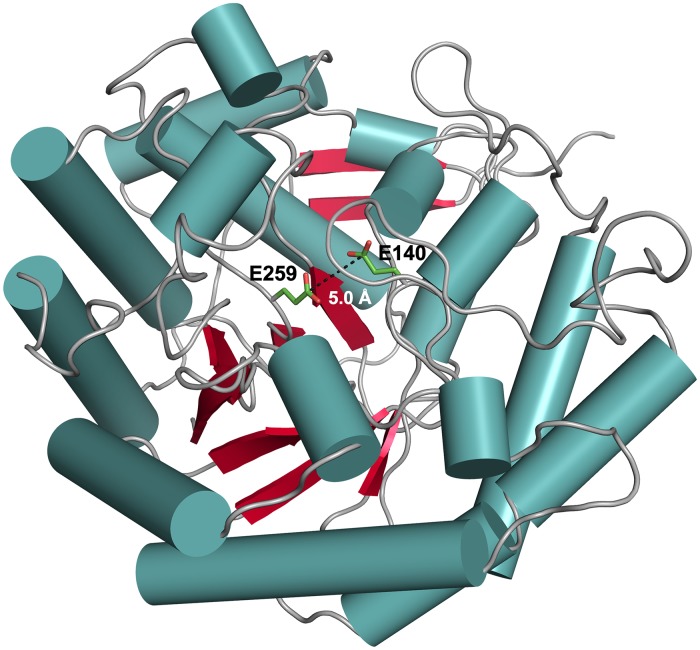
The crystal structure of CaMan5_18 was determined and refined in the unliganded state at a final resolution of 1.8 Å (Table 1). The enzyme was co-crystallized with mannose, however, no clear density for the sugar could be identified. The overall structure displays the, for GH5 enzymes typical, (β/α)8 TIM-barrel fold. Members of the GH5 family share two conserved glutamate residues at the active site, which are approximately 5 Å apart and buried some 12 Å below the protein surface (Fig 4). Using the DALI server (http://ekhidna2.biocenter.helsinki.fi/dali/) to identify structurally similar proteins, the top 20 three-dimensional structures show low sequence identities to CaMan5_18 in the range 10–19%, and high root-mean-square deviation (r.m.s.d) values for atomic positions in the range 2.9–3.3 Å. This confirms the initial, unsuccessful attempts to identify structural homologs using BLAST.
Fig 4. Ribbon representation of the CaMan5_18 subunit structure.
The subunit structure of CaMan5_18 features a (β/α)8 TIM-barrel fold typical for members of the GH5 family. The catalytic acid/base Glu140 and the nucleophile Glu259 are represented as stick models. Secondary-structure elements are represented as: α-helices, blue cylinders; β-sheets, red arrows; and loops, gray coils.
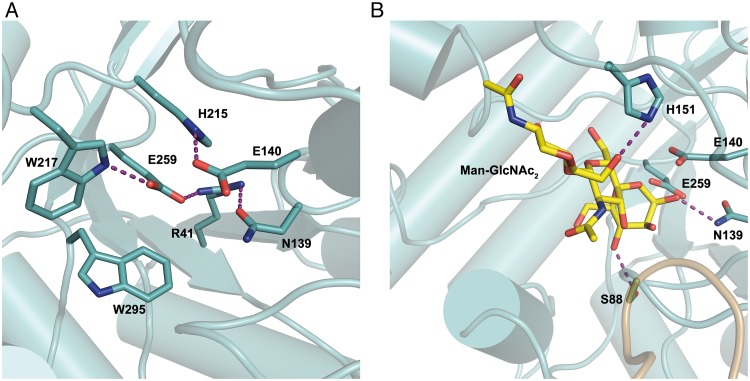
With the aim to capture a crystal complex of wild-type CaMan5_18 with the Man-GlcNAc substrate, we produced the E140Q/E259Q mutant where the catalytic glutamates had been replaced by their isosteric amide counterparts. Unfortunately however, the mutant did not crystallize. Furthermore, we were unsuccessful in capturing a complex of CaMan5_18 with mannose, probably because a disordered MOPS molecule is present in the active site of the crystal structure. Although the experimental data do not show bound ligand in the active site, we can derive from close homologs that Glu140 is acting as the acid/base catalyst and Glu259 as a nucleophile (S4 Fig). The conformation of Glu140 is supported by interactions with His215, and the neighboring Asn139 is kept in position through a possible hydrogen bond with Arg41. The orientation of the catalytic nucleophile Glu259 can be stabilized through interactions with Arg41 and Trp217 (Fig 5A).
Fig 5. Close up of the active site in CaMan5_18.
(A) Interactions made by the catalytic residues Glu140 and Glu259 with Arg41, Asn139, His215, Trp217 and Trp295. (B) Hypothetical binding of Man-GlcNAc2 and possible interactions with Ser88, Asn139 and His151. Purple dashed lines highlight interactions.
To analyze possible substrate-protein interaction the proposed substrate, Man-GlcNAc2 (see product analysis below), was docked manually in the active site of CaMan5_18 (Fig 5B). Briefly, the Man-GlcNAc2 compound was generated manually by using the coordinates for the corresponding sugar units in the crystal structure of a glycosylated IgG Fc (PDB code 5DI8, chain A [53]). Furthermore, the mannose in subsite –1 was modified to adopt the conformation observed for the corresponding mannose in the crystal structures of Rhizomucor miehei Man5 (PDB codes 4NRR and 4NRS [54]). Torsion angles of the GlcNAc units of the chitobiose core were adjusted manually in COOT [41] to avoid unfavorable contacts with proteins atoms in subsites +1 and +2. Based on this hypothetical binding mode, the side chains of Ser88 and Asn139 are predicted to interact with the 4-hydroxyl and 2-hydroxyl oxygen of a mannose residue in subsite –1, respectively. Additionally, His151 is suitably placed to interact with the terminal GlcNAc unit (Fig 5B). The C-terminus (394–408) of the experimental structure does not show interpretable electron density, probably due to flexibility, and was therefore not modeled.
Analysis of oligomeric state
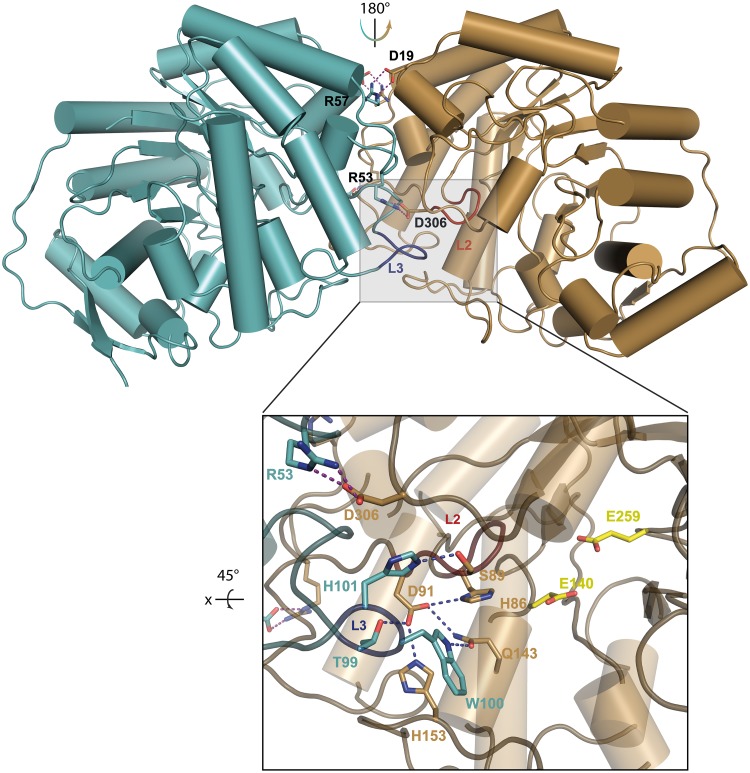
To determine the oligomeric state of the CaMan5_18 in vitro, we applied several techniques: (i) analysis of crystal contacts between CaMan5_18 monomers, (ii) SEC analysis, and (iii) inspection of the protein unfolding profile as a function of temperature. The enzyme is present as a two-fold symmetrical homodimer in the crystal (Fig 6), featuring an extensive dimer interface generated by five loop regions: L1 (residues 46–57), L2 (residues 89–92), L3 (residues 98–102), L4 (residues 152–156), and L5 (residues 299–314), where especially L2 and L3 shape and stabilize the active site (Fig 6, inset). Ion links can be established between Asp19 and Arg57 as well as between Arg53 and Asp306 from both monomers across the two-fold-symmetrical dimer interface.
Fig 6. Dimerization of CaMan5_18.
The dimer interface between subunit A (cyan) and subunit B (brown) with the side chains highlighted that stabilize the dimeric state through intersubunit salt links (Asp19-Arg57 and Arg53-Asp306). Loops L2 (red; residues 89–92) and L3 (blue; residues 92–102) participate in dimer formation, as well as in forming the blockage of the active site required for exo-mode activity. Inset: Interactions formed by loops L2 and L3 in the active site. The steric blockage is mainly provided by L2, supported by L3 and L5 (residues 299–314). The catalytic residues Glu140 and Glu259 are colored yellow.
Analysis of the dimer interface using PISA (Proteins, Interfaces, Structures and Assemblies; http://www.ebi.ac.uk/pdbe/pisa/) [55], supports a stable dimeric state with a buried surface area of 3270 Å2 and a complex-formation significance score of 1.0. An estimated solvation free energy gain (ΔGint), i.e., the gain in solvation free energy upon dimer formation calculated as the difference in solvation energy between free monomers and the dimer state, of –20.8 kcal/mol; and a free energy of assembly dissociation (ΔGdiss), i.e., the free-energy difference between dissociated and associated states, of 21.3 kcal/mol.
In agreement with a dimeric protein, the unfolding profiles observed for CaMan5_18 during thermofluor denaturation experiments are biphasic, as exemplified by two unfolding transitions at pH 7.5 corresponding to Tm values of 48.8°C and 58.0°C (Parts A and B in S5 Fig). Further evidence for a natural dimer in solution was provided by the result from SEC analysis. Whereas the molecular weight of monomeric CaMan5_18 is 45.7 kDa, the protein elutes as a 98-kDa species on a Superdex 200 column (Part C in S5 Fig).
Substrate specificity
Initially, the activity of CaMan5_18 was investigated by screening different pNP-glycosides as substrates. Of the pNP sugars, only pNP-βMan served as substrate for CaMan5_18 with a specific activity of 2.6 U (Table 2). Possible endo activity was tested on relevant complex substrates, e.g., mannan and glucomannan, however, no hydrolysis was observed, which indicated that CaMan5_18 is an exo-β-mannosidase.
Table 2. Substrate screening.
| Substrate | Specific activity (U) |
|---|---|
| pNP-β Man | 2.6 ± 0.13 |
| pNP-β Xyl | ND |
| pNP-β Fuc | ND |
| pNP-α Fuc | ND |
| pNP-β Gal | ND |
| pNP-β Glc | ND |
| pNP-β cellobiose | ND |
| Mannan | ND |
| Glucomannan | ND |
| Xylan | ND |
| Barley β-glucan | ND |
| CMC | ND |
| Cellobiose | ND |
ND, not detected.
Furthermore, the kinetic parameters (Vmax, KM, kcat and kcat/KM) were determined for CaMan5_18 with pNP-βMan as substrate under the conditions of pHopt and Topt for in-vitro activity (see below). For pNP-βMan, a Michaelis constant, KM, of 10.4 mM was obtained, and a turnover number, kcat, of 7.76 s-1, corresponding to a catalytic efficiency of 0.746 s-1 mM-1. Since the only purpose of the mutant was to attempt trapping of a protein-substrate co-crystal complex, no further biochemical or kinetic characterization was performed.
Reaction pH, kinetic stability and thermal stability
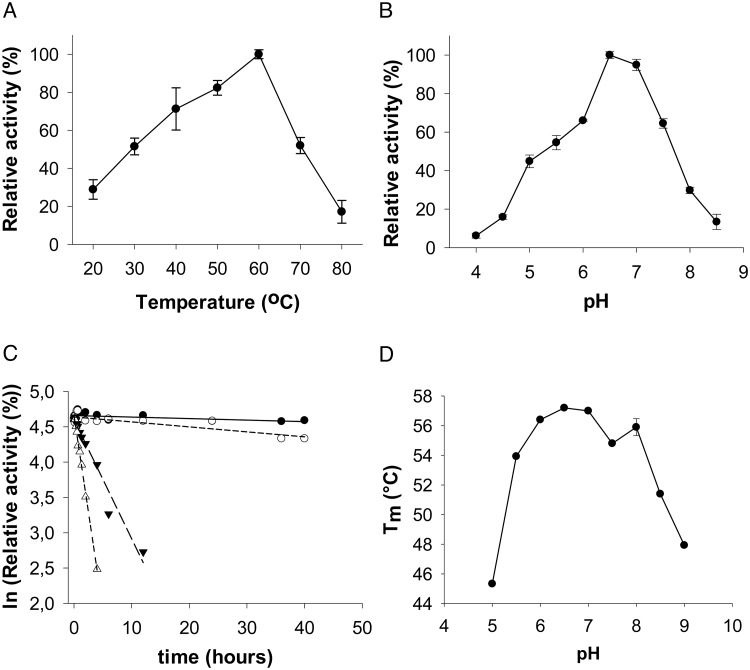
Optimum temperature and pH for pNP-βMan hydrolysis by CaMan5_18 was determined to be 60°C (Fig 7A), and 6.5 (Fig 7B), respectively. The activity decreases by 50% at temperatures above 70°C, and declines rapidly below 60% at pH values near the narrow pHopt. Analysis of the kinetic stability shows that the activity remains stable at lower temperatures, 40°C and 50°C, over an extended period of time with half-life time values of 322 h and 98 h, respectively. At higher temperatures, 60°C and 70°C, the kinetic stability is reduced, as evidenced by half-life time values of 4 h and 1.2 h, respectively (Fig 7C). ThermoFluor analysis of the thermal stability to unfolding as a function of pH showed that the enzyme’s structural integrity remains intact within the pH range 6.5 to 8.0, and that Tm decreases rapidly at higher pH values (Fig 7D).
Fig 7. CaMan5_18 activity on pNP-βMan.
Dependency of CaMan5_18 activity with pNP-βMan as substrate on (A) temperature, and on (B) pH. (C) Kinetic stability of CaMan5_18 hydrolysis of pNP-βMan. Symbols: filled circles, 40°C; empty circles, 50°C; filled triangles, 60°C; empty triangles, 70°C. (D) ThermoFluor analysis of thermal stability to unfolding as a function of pH.
Product analysis
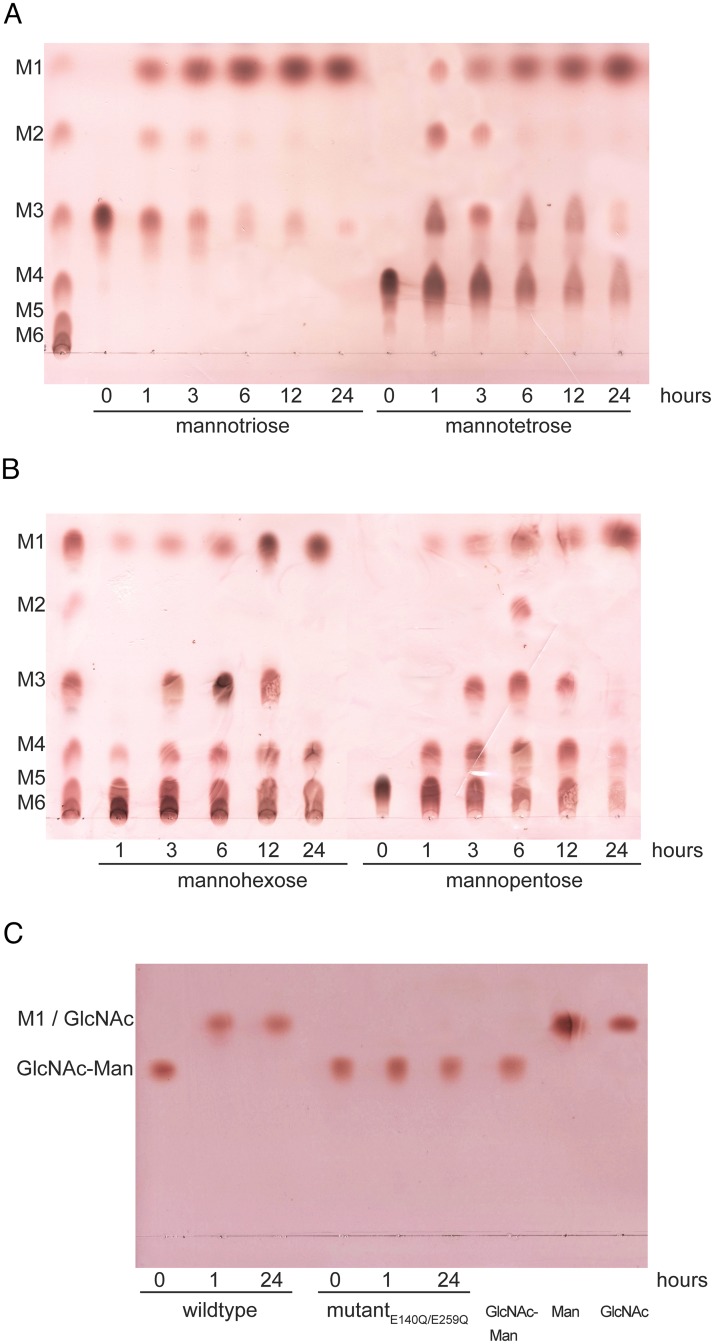
Released monosaccharides and oligosaccharides upon enzyme treatment were identified using TLC and MALDI-TOF mass spectrometry. As stated above, the initial substrate screening of pNP sugars indicated hydrolytic activity only for β-1,4-linked mannose. Time-dependent TLC analyses of hydrolysis products from β-linked mannooligosaccharides of degree of polymerization (DP) of 3 to 6 revealed that M1 (mannose) is accumulating slowly with time for all substrates tested (Fig 8).
Fig 8. TLC analysis of product formation.
Separation of the hydrolysis products from time-dependent hydrolysis of mannooligosaccharides and Man-GlcNAc by wild-type CaMan5_18. (A) M3 and M4; (B) M6 and M5 with the mannooligosaccharide standards M1-M6 at the left; (C) Man-GlcNAc hydrolysis by CaMan5_18 wild type and the E140Q/E259Q double mutant.
For the mannotriose (M3) as substrate (Fig 8A), the products M1 and M2 appear early, after 1 h. Accumulation of M2 occurs at 1–3 h, and is depleted at 3–6 h. After 24 h, most M3 substrate has been converted to M1. For mannotetraose (M4) as substrate (Fig 8A), the products M1, M2 and M4 appear after 1 h. As for the M3 substrate, accumulation of M2 takes place at 1–3 h, and is depleted at 3–6 h. After 24 h, most of the M4 substrate has been converted to M1 and some residual M3, but even at 24 h, residual M4 substrate remains.
For mannopentaose (M5) as substrate (Fig 8B), small amounts of M1 and some M4, but no M2 or M3, are formed early, after 1 h. Concomitantly with conversion of M5 substrate, we observe an accumulation of M1, M3 and M4 at 3–12 h, and small amounts of M2 appearing after 6 h. After 24 h, the product mixture is dominated by M1 and residual M4 and some remaining M5 substrate. For mannohexaose (M6) as substrate (Fig 8B), small amounts of M1, M4 and some M5, but no M2 or M3, are formed early, after 1 h. Concomitantly with conversion of M6 substrate, there is an accumulation of M1, M3, M4 and M5 at 3–12 h, but no M2. After 24 h, the product mixture is dominated by M1 and residual M4, M5 and considerable amounts of M6 substrate. The result of the product analysis of β-1,4-linked mannooligosaccharides is consistent with CaMan5_18 being an exo-β-1,4-mannosidase.
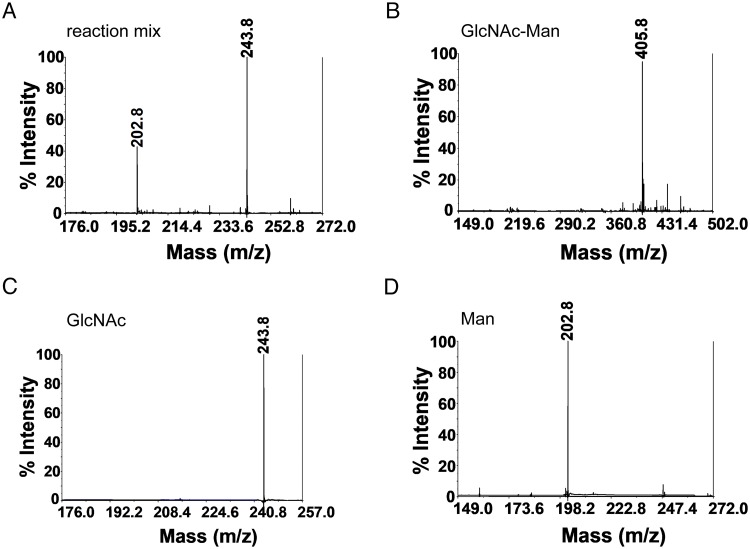
For the Man-GlcNAc substrate that corresponds to the relevant substrate in an N-glycan, full conversion of the substrate to Man and GlcNAc was achieved at 1 h, i.e., the earliest time point tested, and as expected, the CaMan5_18 mutant E140Q/E259Q was unable to hydrolyze the substrate (Fig 8C). Since the products Man and GlcNAc co-migrated on TLC, MALDI-TOF mass spectrometry successfully confirmed the product identities (Fig 9).
Fig 9. MALDI—TOF-MS analysis of Man-GlcNAc hydrolysis.
(A) Enzymatic reaction products using Man-GlcNAc as substrate after 1 h of incubation. Controls: (B) Man-GlcNAc; (C) GlcNAc; and (D) Man.
Discussion
In this work, we propose gene candidates likely to code for enzyme activities involved in a cytoplasmic pathway for degradation of the Man3-GlcNAc2 N-glycan core structure (Fig 3A, Table 3). The gene coding for CaMan5_18 clusters with two genes: AEE72694 (predicted GH38; exo-α-1,3- or exo-α-1,6-mannosidase; EC 3.2.1.24) and AEE72696 (predicted GH20; exo-β-1,4-N-acetylhexoaminidase; EC 3.2.1.52), located immediately upstream and downstream of the CaMan5_18 gene, respectively (Fig 2). We find it legitimate to propose that these three genes, together with nearby genes annotated as sugar transporters and sugar-binding proteins, are part of a distinct locus with relevance to an N-glycan degradation pathway (Figs 2 and 3A). Indeed this enzyme triad would be able to completely convert the Man3-GlcNAc2 core to monosaccharides on the cytoplasmic side.
Table 3. Genes implied in N-glycan degradation (NGD) by C. acnes strain 266.
| Gene accession code | CAZy family | Secretion signal | EC number | Predicted activity | NGD locus | Pathway | Representative homolog with known 3-D structure (sequence identity) [citation] |
|---|---|---|---|---|---|---|---|
| AEE72694 | GH38 | No | 3.2.1.24 | exo-α-1,3- mannosidase, exo-α-1,6-mannosidase | 1 | Cytoplasmic | Ams1, UniProt P22855, PDB 5JM0 (30.1%) [56] SpGH38, UniProt Q99YP5, PDB 2WYH (22.5%) [57] |
| AEE72695 | GH5_18 | No | 3.2.1.25 | exo-β-1,4-mannosidase (confirmed) | 1 | Cytoplasmic | This work, 6GVB; no available structurally characterized homologs with sequence identity > 19% |
| AEE72696 | GH20 | No | 3.2.1.52 | exo-β-1,4-N-acetylhexosaminidase | 1 | Cytoplasmic | SP_2141, UniProt A0A0H2US73, PDB 5AC4 (20.0%) [58] |
| AEE72704 | GH18 | No | 3.2.1.96 | endo-β-N-acetylglucosaminidase | 1 | Cytoplasmic | endoT, UniProt C4RA89, PDB 4AC1 (41.7%) [59] |
| AEE71833 | GH29 | No | 3.2.1.51 | α-L-fucosidase | – | Cytoplasmic | TM_0306, UniProt Q9WYE2, PDB 1HL8 (27.1%) [60] BT2970, UniProt Q8A3I4, PDB 2WVS (24.9%) [61] |
| AEE71274 | GH4 | No | 3.2.1.86 | 6-phospho-glucosidase | 2 | Cytoplasmic | Gan4C, UniProt W8R9V4, PDB 5C3M(31.6%) [N/A] |
| AEE71275 | GH38 | No | 3.2.1.24 | exo-α-1,3- mannosidase, exo-α-1,6-mannosidase | 2 | Cytoplasmic | Ams1, UniProt P22855, PDB 5JM0 (30.0%) [56] SpGH38, UniProt Q99YP5, PDB 2WYH (23.5%) [57] |
| AEE71278 | GH125 | No | 3.2.1.– | exo-α-1,6-mannosidase | 2 | Cytoplasmic | CPE0426/CpGH125, UniProt Q8XNB2, PDB 3QT3 (44.5%) [62] SP_2144/SpGH125, UniProt A0A0H2URZ6, PDB 3QPF (42.6%) [62] |
| AEE71279 | GH20 | No | 3.2.1.52 | exo-β-1,4-N-acetylhexosaminidase | 2 | Cytoplasmic | nahA, UniProt A1RBZ5, PDB 3RCN (36.5%) [N/A] |
| AEE72802 frameshift* | GH33 | Yes | 3.2.1.18 | sialidase | – | Extracellular | nedA/NanH, UniProt Q02834, 1EUS (59.0%) [45] |
| AEE73363 | GH3 | Yes | 3.2.1.52 | β-1,2-N-acetylhexosaminidase | – | Extracellular | nagZ/YbbD, UniProt P40406, 3BMX (34.7%) [48] |
| AEE73032 | GH35 | Yes | 3.2.1.23 | β-1,4-galactosidase | – | Extracellular | N-terminal β-galactosidase (β-Gal) TIM-barrel domain, UniProt Q700S9, 1XC6 residues 41–265 (34.4%) [49] |
*) Full-length representative: ALT42513, C. acnes strain PA_12_1_L1
A similar pathway appears to exist in the human pathogen Streptococcus pyogenes where the exo-α-mannosidase SpGH38 hydrolyzes mannose α-1,3 and α-1,6 linkages in host N-glycans [57]. However, in Streptococcus pneumoniae, the corresponding GH38 is predicted to have exo-α-1,3-mannosidase activity, while a GH125 would provide the complementary exo-α-1,6-mannosidase activity [21]. Interestingly, we have identified another C. acnes locus that contains the GH38/GH125 gene pair in combination with genes coding for GH20 and GH4 members (S6 Fig). This pair is conserved in all C. acnes types except for the type-III strains. While this may indicate that C. acnes uses alternative pathways for hydrolysis of the N-glycan core, this latter locus lacks a candidate gene for the β-mannosidase activity.
As a first step towards experimentally confirming our proposed cytoplasmic N-glycan-degradation pathway, we chose to characterize the GH5_18 gene product that was predicted to code for a putative β-mannosidase. The rationale for this choice was that, unlike the other proposed enzyme activities, GH5 enzymes have not previously been linked to N-glycan degradation. The amino-acid sequences of exo-acting β-mannosidases (EC 3.2.1.25) are found in any of the three GH families 1, 2 and 5 [30], which belong to the GH-A clan that features the (β/α)8 TIM-barrel fold. These enzymes follow a retaining reaction mechanism according to the classical Koshland double-displacement mechanism [63] where two conserved glutamate residues act as acid/base catalyst and nucleophile [64]. Of the 31 β-mannosidase sequences that are currently classified in the CAZy database, eight belong to GH1, 21 to GH2, and only two are GH5 members, namely, the exo-β-mannosidase CmMan5A from Cellvibrio mixtus (UniProt Q6QT42; PDB code 1UUQ [65]) and the exo-β-mannosidase TthMan5 from Pseudothermotoga (Thermotoga) thermarum (UniProt F7YX66) [52]. While the majority of GH5 enzymes are endo-acting, exo-acting enzymes dominate the GH1 and GH2 families.
The proposed model of N-glycan metabolism in S. pneumoniae lacks a candidate for cleaving the β-1,4 linkage in Man-GlcNAc, and in the N-glycan-processing pathways of Xanthomonas campestris [66] and the human colonic bacterium Bacteroides thetaiotaomicron this β-mannosidase activity is provided by a GH2 enzyme [67]. In B. thetaiotaomicron, the gene coding for the β-mannosidase is located in a polysaccharide-utilization locus (PUL), which also contains three GH20 genes, a GH2 gene and a putative GH33 sialidase gene. A related GH2 enzyme in Bacteroides fragilis predicted to degrade the N-glycan core displays β-mannosidase activity, although activity on Man-GlcNAc or Man-GlcNAc2 has not been demonstrated [68].
Of all aryl-glycosides and polysaccharide substrates tested, CaMan5_18 showed activity only for pNP-βMan (Table 4). Activity on aryl-mannosides such as pNP-βMan is the generally accepted way to distinguish exo-acting mannosidases from endo-acting mannanases [65]. The exo-β-mannosidases BtMan2A [67] and TthMan5 [52] perform well with pNP-βMan as substrate whereas the exo-β-mannosidase CmMan5A does not (Table 4). Similar to CmMan5A, CaMan5_18 displays low but convincing activity on pNP-βMan. However, product analysis by TLC reveals a product pattern consistent with an exo-mannosidase (Fig 8). Although CaMan5_18 can accept mannooligosaccharides (DP 3–6) as substrates, and given enough time, convert most of these to free mannose, the efficiency declines as a function of increasing DP (Fig 8). Furthermore, using M5 and M6 as substrates, neither M2 nor M3 is generated early, i.e., after 1 h (Fig 8B), which is supportive of an exo-mode activity on β-1,4-linked mannooligosaccharides.
Table 4. Kinetic parameters for CaMan5_18 and related enzymes on pNP-βMan.
In contrast to the mannooligosaccharides, wild-type CaMan5_18 hydrolyzes Man-GlcNAc efficiently to mannose and GlcNAc in 1 h (Fig 8C), while no reaction products were observed when incubating the double mutant E140Q/E259Q with this substrate. This confirms the importance of the proposed catalytic amino acids, as well as the relevance of Man-GlcNAc as substrate. Since both hydrolysis products, Man and GlcNAc, are co-migrating on TLC, further analysis using MALDI-TOF mass spectrometry was needed to confirm the product identities. The results from MALDI-TOF mass spectrometry showed clear separation of Man and GlcNAc, which provided positive confirmation of the hydrolysis products (Fig 9).
Several observations based on the crystal structure also support that Man-GlcNAc2 is a more preferred substrate for CaMan5_18 compared with longer mannose-containing substrates: (i) loop 2 (chain A, residues 89–92), loop 3 (chain B, residues 98–102) and loop 5 (chain A, residues 299–314) together form a steric blockage which precludes binding sites at the minus end beyond –1, analogous to the steric barrier observed in CmMan5A (residues 378–412); (ii) the substrate-binding region encompasses three glycosyl-binding subsites, i.e., subsite –1 and two product-binding sites +1 and +2. The residue His151 is suitably positioned to interact with one or both of the GlcNAc units, predicted to be located in subsites +1 and +2, to facilitate positioning of the substrate. In agreement with this function, His151 is conserved in Cutibacterium GH5_18 sequences as well as in other enzymes belonging to Propionibacteriaceae. We have used several experimental methods to show that CaMan5_18 exists as a functional dimer in solution, a feature that appears to be uncommon for GH5 enzymes. In the crystal structure, important residues involved in dimer stabilization (Asp19, Asn52, Arg53, Arg57, Ser89, Asp91, Thr99, Trp100, His101, Gln143, His153 and Asp306) are conserved in all or most GH5_18 sequences (S4 Fig), suggesting that GH5_18 enzymes in general are likely to form functional dimers.
Prior to the intracellular deconstruction of the Man3-GlcNAc2 core, the full-length N-glycan antenna has to be trimmed on the extracellular side by secreted GH enzymes, and the resulting Man3-GlcNAc2 oligosaccharide must be transported across the plasma membrane. We could identify possible candidate genes for the extracellular pathway, but in contrast to the cytoplasmic pathway, the secreted GH genes were dispersed and did not map to any distinct gene cluster that could be assigned as a carbohydrate-processing locus. Gene candidates containing signal-peptide sequences were readily identified (Fig 3B): a β-1,2-N-acetylhexoaminidase gene (GH3; AEE73363), a β-1,4-galactosidase gene (GH35, AEE73032), and a potential sialidase gene (GH33; AEE72802).
The only gene that we have so far not been able to identify in the C. acnes genome is that coding for peptide-N-glycosidase F activity (PNGase F), i.e., the amidase responsible for hydrolyzing the amide bond to the host protein asparagine. As an alternative to PNGase-F activity, an endo-β-N-acetylglucosaminidase (GH18; AEE72208) would be able to cleave within the chitobiose core of N-linked glycans to release the glycan from the host protein as is the case for S. pyogenes, which produces two endoglycosidases, EndoS and EndoS2 that selectively hydrolyze Fc-bound N-glycans on therapeutic antibodies [14,69]. Another possibility is that another symbiotic bacterial skin colonizer provides the PNGase-F enzyme.
Although the phylotype 1A1 C. acnes strain 266 was isolated from a pleuropulmonary infection rather than skin [26], its NGD locus 1 (Fig 2) is representative for the clade-IA phylotype. It is interesting to note that the proposed NGD pathway is present only in IA strains, while appearing in a truncated version in the IB, IC, II and III phylotypes (S1 Fig). Phylotype IA1 strains dominate in follicles of Caucasian patients with acne, whereas IA1, IA2, IB and II display heterogeneous distributions on healthy skin [70]. The skin in patients with severe acne and healthy subjects show markedly different distributions of C. acnes phylotypes [71].
In acne sufferers, a homogeneous distribution of C. acnes phylotypes is observed, with 1A1 as the dominating phylotype, as well as an overall loss of phylotype diversity and absence of phylotype II [71]. Healthy subjects however, carried a more diverse composition of phylotypes with roughly equal distribution of 1A1 and II. In the same study, the distribution of clonal complexes (CC) was analyzed, and revealed that the CC18 subgroup was more prevalent in acne patients, whereas CC53 (phylotype II and K based on the SLST classification) dominated in healthy subjects.
The intact NGD locus 1 would enable IA strains to efficiently depolymerize and utilize all types of host N-glycans for nutrition. This is further supported by the presence of associated genes coding for ABC transporters. The ability to process host N-glycans is likely to present a selective advantage for IA strains over non-IA strains under conditions of limiting nutrient supply. The presence of a GH33 sialidase (AEE72802) indicates that the extracellular NGD pathway in C. acnes strain 266 may also contribute to virulence. The homologous GH33 enzyme produced by the C. acnes strain KPA171202 (96% sequence identity to AEE72802, was experimentally confirmed to be an exo-α-sialidase that cleaves off sialoglycoconjugates in human sebocyte cultures [72]. Nakatsuji and co-workers showed that the C. acnes KPA171202 sialidase increased host susceptibility to C. acnes cytotoxicity and adhesion, and furthermore, that mice immunized with the sialidase acquired protective immunity against C. acnes [72]. Beyond confirming a role of this sialidase in the degradation of sialoglycoconjugates, the study highlights the strategic importance of C. acnes enzymes that degrade host glycans for new treatment of acne vulgaris.
Conclusions
The commensal bacterium Cutibacterium acnes is a natural habitant of the human skin microbiota, but under certain circumstances, C. acnes can turn pathogenic and cause acne vulgaris. We have mined the genome of C. acnes and identified a set of genes that are likely candidates for degradation of host N-glycans. We have also presented the biochemical and structural characterization of one enzyme in the proposed pathway, namely the exo-β-1,4-mannosidase that is able to catalyze hydrolysis of the β-1,4-glycosidic bond between the second GlcNAc and the first mannose residue in the canonical eukaryotic N-glycan core. This is the first reported enzymatic activity of subfamily GH5_18, and the first characterized GH5 enzyme likely to be involved in N-glycan degradation. Our results provide a valuable platform for further efforts to elucidate the complete set of activities of the proposed C. acnes N-glycan degradation pathway.
Based on our structural and functional characterization of CaMan5_18, and whole-genome analysis, we propose a pathway for complete N-glycan degradation and utilization by C. acnes strain 266, results that offer new insights into the host-microbe interaction of this bacterium. We propose that this pathway allows the C. acnes to use host N-glycans as a means of nutrition, and possibly, also to contribute to virulence in susceptible individuals. The ability to degrade N-glycans would complement the demonstrated ability of C. acnes to disassemble O-glycans, which makes this bacterium well adapted to utilize a wide range of host glycans.
Supporting information
Genomic comparison of the N-glycan processing locus 1 in C. acnes using the comparative genomics platform Sybil (http://sybil.sourceforge.net). The C. acnes 266 strain was used as reference genome (region 1545000–1564000). A subset of genomes was selected to represent the different C. acnes phylogenetic groups (IA1, IA2, IB, II and III). The GH genes of each locus are highlighted. Predicted function of other genes is given in Fig 2.
(PDF)
The tree was generated using PhyML and was based on 55 GH5_18 sequences. Numbers at the nodes are derived from the Approximate Likelihood-Ratio Test. Sequences from the Bifidobacterium, Streptomyces and Cutibacterium are indicated.
(PDF)
Sequence alignment using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo) highlighting the difference in amino-acid sequence of the different C. acnes phylotypes IA, IB, II and III strains. CaMan5_18 (AEE72695) originates from the type-IA1 C. acnes strain 266. The figure was prepared with ESPript3 (http://espript.ibcp.fr/ESPript/ESPript/).
(PDF)
Sequence alignment using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo). Strictly conserved and highly conserved residues are highlighted with red boxes and red lettering, respectively. Secondary-structure elements are shown on top as spirals and arrows for α-helices and β-strands, respectively. Solid stars indicate the active-site residues that are conserved. The GenBank accession numbers are as follows: CaMan5_18 (AEE72695); CaGH5_18 from Cutibacterium avidum 44067 (AGJ77370); KfGH5_18 from Kribbella flavida DSM 17836 (ADB34475); BlGH5_18 from Bifidobacterium longum subsp. infantis 157F (BAJ71452); SkGH5_18 from Sanguibacter keddieii DSM 10542 (ACZ21418); ScGH5_18 from Streptomyces coelicolor A3(2) (CAB61915) and AhGH5_18 from Arcanobacterium haemolyticum DSM 20595 ADH91800). The figure was prepared with ESPript3 (http://espript.ibcp.fr/ESPript/ESPript/).
(PDF)
Analysis of oligomeric state. (A) ThermoFluor-derived melting curve for CaMan_18 in 50 mM sodium phosphate (pH 7.5), and in the presence of the dye SYPRO Orange. Relative fluorescence units (RFU) were plotted against temperature. The protein unfolding curve is biphasic, which is consistent with two unfolding transitions. (B) Representation of the data in A by plotting the first derivative (dRFU)/dT of the raw data against temperature. Two Tm values were derived from this curve, 48.8°C and 58.0°C. (C) Overlay of the SEC chromatograms for the standard proteins (black lines) and of CaMan5_18 (red line). Inset: Standard curve used to extrapolate the molecular weight (R2 = 0.9974). CaMan5_18 elutes at a volume corresponding to a molecular weight of 98 kDa.
(PDF)
Gene organization of a second putative N-glycan processing locus in the genome of C. acnes 266. GH genes predicted as mannosidases are colored green, and GH genes with predicted N-acetylhexosaminidase activity are blue. The GH4 gene with unknown activity is colored dark gray. Other associated genes are colored light gray and include: predicted sugar ABC transporter permease (PERM), transcriptional-regulator gene (REG), sugar ABC-transporter substrate-binding protein (SBP), hypothetical proteins (HP), HAD-family phosphatase involved in carbohydrate transport (PHO), and sugar kinase (ROK). Accession numbers (GenBank, or RefSeq when GenBank was not available) are shown below each gene.
(PDF)
Acknowledgments
We thank the beamline staff scientists for support during data collection at PROXIMA 1 beamline at SOLEIL synchrotron (France). Part of this work was facilitated by the Protein Science Facility at Karolinska Institutet/SciLifeLab (http://psf.ki.se). The research leading to these results has received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under BioStruct-X (grant agreement no 283570). Parts of this project received support from iNEXT (PID 1678) for data collection at SOLEIL project No. 20151194 (France).
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was funded by the Swedish Research Council VR (vr.se) grant numbers 2013-5717 to C.D., and by the Swedish Research Council Formas (www.formas.se) grant number 2013-1741 to C.D. and grant numbers 2012-1513 and 2014-176 to H.A. For synchrotron data collection, C.D. also received funding from the European Community’s Seventh Framework Programme (FP7/2007–2013) under BioStruct-X (grant agreement no 283570), and iNEXT (PID 1678) for data collection at SOLEIL project No. 20151194 (France).
References
- 1.Apweiler R, Hermjakob H, Sharon N. On the frequency of protein glycosylation, as deduced from analysis of the SWISS-PROT database. Biochim Biophys Acta. 1999;1473: 4–8. [DOI] [PubMed] [Google Scholar]
- 2.Walsh CT, Garneau-Tsodikova S, Gatto GJ Jr. Protein posttranslational modifications: The chemistry of proteome diversifications. Angew Chem Int Ed. 2005;44: 7342–7372. [DOI] [PubMed] [Google Scholar]
- 3.Moreman KW, Tiemeyer M, Nairn AV. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev. 2012;13: 448–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aebi M. N-linked protein glycosylation in the ER. Biochim Biophys Acta. 2013;1833: 2430–2437. 10.1016/j.bbamcr.2013.04.001 [DOI] [PubMed] [Google Scholar]
- 5.Jung D, Alt FW. Unraveling V(D)J recombination: Insights into gene regulation. Cell. 2004;116: 299–311. [DOI] [PubMed] [Google Scholar]
- 6.Schroeder HW, Cavacini L. Structure and function of immunoglobulins. J Allergy Clin Immunol. 2010;125: 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breitling J, Aebi M. N-Linked protein glycosylation in the endoplasmic reticulum. Cold Spring Harb Perspect Biol. 2013;5: a013359 10.1101/cshperspect.a013359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirschberg CB, Snider MD. Topography of glycosylation in the rough endoplasmic reticulum and Golgi apparatus. Annu Rev Biochem. 1987;56: 63–87. 10.1146/annurev.bi.56.070187.000431 [DOI] [PubMed] [Google Scholar]
- 9.Struwe WB, Reinhold VN. The conserved oligomeric Golgi complex is required for fucosylation of N-glycans in Caenorhabditis elegans. Glycobiology. 2012;22: 863–875. 10.1093/glycob/cws053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang P, Wang H, Gai J, Zhang X, Lv Y, Jian Y. Evolution of protein N-glycosylation process in Golgi apparatus which shapes diversity of protein N-glycan structures in plants, animals and fungi. Sci Rep. 2017;7: 40301–40314. 10.1038/srep40301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stanley P, Taniguchi N, Aebi M. N-Glycans, in Essentials of Glycobiology, Cold Spring Harbor (NY) p. 99–111; 2015. [Google Scholar]
- 12.Marcobal A, Southwick AM, Earle KA, Sonnenburg JL. A refined palate: Bacterial consumption of host glycans in the gut. Glycobiology. 2013;23: 1038–1046. 10.1093/glycob/cwt040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garbe J, Collin M. Bacterial hydrolysis of host glycoproteins—powerful protein modification and efficient nutrient acquisition. J Innate Immun. 2012; 4: 121–131. 10.1159/000334775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collin M, Olsén A. Effect of SpeB and EndoS from Streptococcus pyogenes on human immunoglobulins. Infect Immun 2001;69: 7187–7189. 10.1128/IAI.69.11.7187-7189.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Collin M, Fischetti VA. A novel secreted endoglycosidase from Enterococcus faecalis with activity on human immunoglobulin G and ribonuclease B. J Biol Chem. 2004;279: 22558–22570. 10.1074/jbc.M402156200 [DOI] [PubMed] [Google Scholar]
- 16.Byers HL, Tarelli E, Homer KA, Beighton D. Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human alpha1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology. 1999;9: 469–479. [DOI] [PubMed] [Google Scholar]
- 17.Burnaugh AM, Frantz LJ, King SJ. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J Bacteriol. 2008;190: 221–230. 10.1128/JB.01251-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manfredi P, Renzi F, Mally M, Sauteur L, Schmaler M, Moes S et al. The genome and surface proteome of Capnocytophaga canimorsus reveal a key role of glycan foraging systems in host glycoproteins deglycosylation. Mol Microbiol. 2011;81: 1050–1060. 10.1111/j.1365-2958.2011.07750.x [DOI] [PubMed] [Google Scholar]
- 19.Renzi F, Manfredi P, Mally M, Moes S, Jenö P, Cornelis GR. The N-glycan glycoprotein deglycosylation complex (Gpd) from Capnocytophaga canimorsus deglycosylates human IgG. PLoS Pathog. 2011;7: e1002118 10.1371/journal.ppat.1002118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cao Y, Rocha ER, Smith CJ. Efficient utilization of complex N-linked glycans is a selective advantage for Bacteroides fragilis in extraintestinal infections. Proc Natl Acad Sci USA. 2014;111: 12901–12906. 10.1073/pnas.1407344111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robb M, Hobbs JK, Woodiga SA, Shapiro-Ward S, Suits MDL, McGregor N et al. Molecular characterization of N-glycan degradation and transport in Streptococcus pneumoniae and its contribution to virulence. PLoS Pathog. 2017;13: e1006090 10.1371/journal.ppat.1006090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lichtenberger R, Simpson MA, Barker J, Navarini AA. Genetic architecture of acne vulgaris. J European Acad Dermatol Venerol. 2017;31: 1978–1990. [DOI] [PubMed] [Google Scholar]
- 23.Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9: 244–253. 10.1038/nrmicro2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Achermann Y, Goldstein EJ, Coenye T, Shirtliff ME. Propionibacterium acnes: from commensal to opportunistic biofilm-associated implant pathogen. Clin Microbiol Rev. 2014;27: 419–440. 10.1128/CMR.00092-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koutsioulis D, Landry D, Guthrie EP. Novel endo-alpha-N-acetylgalactosaminidases with broader substrate specificity. Glycobiology. 2008;18: 799–805. 10.1093/glycob/cwn069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland C, Mak TN, Zimny-Arndt U, Schmid M, Meyer TF, Jungblut PR et al. Proteomic identification of secreted proteins of Propionibacterium acnes. BMC Microbiol. 2010;10: 230 10.1186/1471-2180-10-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bek-Thomsen M, Lomholt HB, Scavenius C, Enghild JJ, Brüggemann H. Proteome analysis of human sebaceous follicle infundibula extracted from healthy and acne-affected skin. PLoS One. 2014;9: e107908 10.1371/journal.pone.0107908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brüggemann H, Henne A, Hoster F, Liesegang H, Wiezer A, Strittmatter A et al. The complete genome sequence of Propionibacterium acnes, a commensal of human skin. Science. 2004;305: 671–673. 10.1126/science.1100330 [DOI] [PubMed] [Google Scholar]
- 29.Scholz CF, Brüggemann H, Lomholt HB, Tettelin H, Kilian M. Genome stability of Propionibacterium acnes: a comprehensive study of indels and homopolymeric tracts. Sci Rep. 2016;6: 20662 10.1038/srep20662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lombard V, Golaconda RH, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42: 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brzuszkiewicz E, Weiner J, Wollherr A, Thürmer A, Hüpeden J, Lomholt HB et al. Comparative genomics and transcriptomics of Propionibacterium acnes. PLoS One. 2011;6: e21581 10.1371/journal.pone.0021581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Price MN, Huang KH, Alm EJ, Arkin AP. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 2005;33: 880–892. 10.1093/nar/gki232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52: 696–704. [DOI] [PubMed] [Google Scholar]
- 35.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, Chevenet F et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;36: W465–469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nielsen H. Predicting secretory proteins with SignalP. Methods Mol Biol. 2017;1611: 59–73. 10.1007/978-1-4939-7015-5_6 [DOI] [PubMed] [Google Scholar]
- 37.Robert X, Gouet P. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res. 2014;42: W320–W324. 10.1093/nar/gku316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66: 125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364: 215–230. 10.1385/1-59745-266-1:215 [DOI] [PubMed] [Google Scholar]
- 40.Bricogne G, Vonrhein C, Flensburg C, Schiltz M, Paciorek W. Generation, representation and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr D Biol Crystallogr. 2003;59: 2023–2030. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60: 2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 42.Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66: 213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ericsson UB, Hallberg BM, DeTitta GT, Dekker N, Nordlund P. Thermofluor-based high-throughput stability optimization of proteins for structural studies. Anal Biochem. 2006;357: 289–298. 10.1016/j.ab.2006.07.027 [DOI] [PubMed] [Google Scholar]
- 44.Brzuszkiewicz E, Weiner J, Wollherr A, Thürmer A, Hüpeden J, Lomholt HB et al. Comparative genomics and transcriptomics of Propionibacterium acnes. PLoS.One. 2011;6: e21581 10.1371/journal.pone.0021581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gaskell A, Crennell S, Taylor G. The three domains of a bacterial sialidase: a β-propeller, an immunoglobulin module and a galactose-binding jelly-roll. Structure. 1995;3: 1197–1205. [DOI] [PubMed] [Google Scholar]
- 46.Amaya MF, Watts AG, Damager I, Wehenkel A, Nguyen T, Buschiazzo A et al. Structural insights into the catalytic mechanism of Trypanosoma cruzi trans-sialidase. Structure. 2004;12: 775–784. 10.1016/j.str.2004.02.036 [DOI] [PubMed] [Google Scholar]
- 47.Copley RR, Russell RB, Ponting CP. Sialidase-like Asp-boxes: Sequence-similar structures within different protein folds. Protein Sci. 2001;10: 285–292. 10.1110/ps.31901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Litzinger S, Fischer S, Polzer P, Diedeichs K, Welte W, Mayer C. Structural and kinetic analysis of Bacillus subtilis N-acetylglucosaminidase reveals a unique Asp-His dyad mechanism. J Biol Chem. 2010;285: 35675–35684. 10.1074/jbc.M110.131037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rojas AL, Nagem RAP, Neustroev KN, Arand M, Adamska M, Eneyskaya EV et al. Crystal Structures of β-galactosidase from Penicillium sp. and its complex with galactose. J Mol Biol. 2004;343: 1281–1292. 10.1016/j.jmb.2004.09.012 [DOI] [PubMed] [Google Scholar]
- 50.Jeong JK, Kwon O, Lee YM, Oh DB, Lee JM, Kim S et al. Characterization of the Streptococcus pneumoniae BgaC protein as a novel surface β-Galactosidase with specific hydrolysis activity for the Galβ1-3GlcNAc moiety of oligosaccharides. J Bacteriol. 2009;191: 3011–3023. 10.1128/JB.01601-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Aspeborg H, Coutinho PM, Wang Y, Brumer H III, Henrissat B. Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5). BMC Evol Biol. 2012;12: 186–202. 10.1186/1471-2148-12-186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shi H, Huang Y, Zhang Y, Li W, Li X, Wang F. High-level expression of a novel thermostable and mannose-tolerant β-mannosidase from Thermotoga thermarum DSM 5069 in Escherichia coli. BMC Biotechnol. 2013;13: 83 10.1186/1472-6750-13-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leaver-Fay A, Froning FJ, Atwell S, Aldaz H, Pustilnik A, Lu F et al. Computationally designed bispecific antibodies using negative state repertoires. Structure 2016;24: 641–651. 10.1016/j.str.2016.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou P, Liu Y, Yan Q, Chen Z, Qin Z, Jiang Z. Structural insights into the substrate specificity and transglycosylation activity of a fungal glycoside hydrolase family 5 β-mannosidase. Acta Crystallogr D Biol Crystallogr. 2014;70: 2970–2982. 10.1107/S1399004714019762 [DOI] [PubMed] [Google Scholar]
- 55.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372: 774–797. 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 56.Bertipaglia C, Schneider S, Jakobi AJ, Tarafder AK, Bykov YS, Picco A et al. Higher-order assemblies of oligomeric cargo receptor complexes form the membrane scaffold of the Cvt vesicle. EMBO Rep. 2016; 7:1044–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suits MDL, Zhu Y, Taylor EJ, Walton J, Zechel DL, Gilbert HJ et al. Structure and kinetic investigation of Streptococcus pyogenes Family GH38 α-mannosidase. PLoS One. 2010;5: e9006 10.1371/journal.pone.0009006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Robb M, Robb CA, Higgins MA, Hobbs JK, Paton JC, Boraston AB. A second β-hexosaminidase encoded in the Streptococcus neumoniae genome provides an expanded biochemical ability to degrade host glycans. J Biol Chem. 2015; 290:30888–30900. 10.1074/jbc.M115.688630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stals I, Karkehabadi S, Kim S, Ward M, Van Landschoot A, Devreese B et al. High resolution crystal structure of the endo-N-acetyl-β-D-glucosaminidase responsible for the deglycosylation of Hypocrea jecorina cellulases. PLoS One. 2012; 7:e40854 10.1371/journal.pone.0040854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sulzenbacher G, Bignon C, Nishimura T, Tarling CA, Withers SG, Henrissat B et al. Crystal Structure of Thermotoga maritima α-L-Fucosidase. J Biol Chem. 2004; 279: 13119–13128. 10.1074/jbc.M313783200 [DOI] [PubMed] [Google Scholar]
- 61.Lammerts van Bueren A, Ardevol A, Fayers-Kerr J, Luo B, Zhang Y, Sollogoub M et al. Analysis of the reaction coordinate of α-L-fucosidases: A combined structural and quantum mechanical approach. J Am Chem Soc. 2010; 132: 1804–1806. 10.1021/ja908908q [DOI] [PubMed] [Google Scholar]
- 62.Gregg KJ, Zandberg WF, Hehemann JH, Whitworth GE, Deng L, Vocadlo DJ et al. Analysis of a new family of widely distributed metal-independent α-mannosidases provides unique insight into the processing of N-Linked glycans. J Biol Chem. 2011; 286:15586–15596. 10.1074/jbc.M111.223172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koshland D. Stereochemistry and the mechanism of enzymatic reactions. Biol Rev. 1953;28: 416–433. [Google Scholar]
- 64.Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon JP, Davies G. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci. USA 1995;92: 7090–7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dias FM, Vincent F, Pell G, Prates JA, Centeno MS, Tailford LE et al. Insights into the molecular determinants of substrate specificity in glycoside hydrolase family 5 revealed by the crystal structure and kinetics of Cellvibrio mixtus mannosidase 5A. J Biol Chem. 2004;279: 25517–25526. 10.1074/jbc.M401647200 [DOI] [PubMed] [Google Scholar]
- 66.Dupoiron S, Zischek C, Ligat L, Carbonne J, Boulanger A, Dugé de Bernonville T et al. The N-glycan cluster from Xanthomonas campestris pv. campestris: a toolbox for sequential plant N-glycan processing. J Biol Chem. 2015;290: 6022–6036. 10.1074/jbc.M114.624593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tailford LE, Money VA, Smith NL, Dumon C, Davies GJ, Gilbert HJ. Mannose foraging by Bacteroides thetaiotaomicron: structure and specificity of the beta-mannosidase, BtMan2A. J Biol Chem. 2007;282: 11291–11299. 10.1074/jbc.M610964200 [DOI] [PubMed] [Google Scholar]
- 68.Nakayama-Imaohji H, Ichimura M, Iwasa T, Okada N, Ohnishi Y, Kuwahara T. Characterization of a gene cluster for sialoglycoconjugate utilization in Bacteroides fragilis. J Med Invest. 2012;59: 79–94. [DOI] [PubMed] [Google Scholar]
- 69.Sjögren J, Cosgrave EF, Allhorn M, Nordgren M, Björk S, Olsson F et al. EndoS and EndoS2 hydrolyze Fc-glycans on therapeutic antibodies with different glycoform selectivity and can be used for rapid quantification of high-mannose glycans. Glycobiology. 2015;25: 1053–1063. 10.1093/glycob/cwv047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lomholt HB, Scholz CFP, Brüggemann H, Tettelin H, Kilian M. A comparative study of Cutibacterium (Propionibacterium) acnes clones from acne patients and healthy controls. Anaerobe. 2017;47: 57–63. 10.1016/j.anaerobe.2017.04.006 [DOI] [PubMed] [Google Scholar]
- 71.Dagnelie MA, Corvec S, Saint-Jean M, Bourdes V, Nguyen JM, Khammari A et al. Decrease in diversity of Propionibacterium acnes phylotypes in patients with severe acne on the back. Acta Derm Venerol. 2018;98: 262–267. 10.2340/00015555-2847 [DOI] [PubMed] [Google Scholar]
- 72.Nakatsuji T, Liu YT, Huang CP, Zouboulis CC, Gallo RL, Huang CM. Vaccination targeting a surface sialidase of P. acnes: implication for new treatment of acne vulgaris. PLoS One. 2008;3: e1551 10.1371/journal.pone.0001551 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic comparison of the N-glycan processing locus 1 in C. acnes using the comparative genomics platform Sybil (http://sybil.sourceforge.net). The C. acnes 266 strain was used as reference genome (region 1545000–1564000). A subset of genomes was selected to represent the different C. acnes phylogenetic groups (IA1, IA2, IB, II and III). The GH genes of each locus are highlighted. Predicted function of other genes is given in Fig 2.
(PDF)
The tree was generated using PhyML and was based on 55 GH5_18 sequences. Numbers at the nodes are derived from the Approximate Likelihood-Ratio Test. Sequences from the Bifidobacterium, Streptomyces and Cutibacterium are indicated.
(PDF)
Sequence alignment using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo) highlighting the difference in amino-acid sequence of the different C. acnes phylotypes IA, IB, II and III strains. CaMan5_18 (AEE72695) originates from the type-IA1 C. acnes strain 266. The figure was prepared with ESPript3 (http://espript.ibcp.fr/ESPript/ESPript/).
(PDF)
Sequence alignment using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo). Strictly conserved and highly conserved residues are highlighted with red boxes and red lettering, respectively. Secondary-structure elements are shown on top as spirals and arrows for α-helices and β-strands, respectively. Solid stars indicate the active-site residues that are conserved. The GenBank accession numbers are as follows: CaMan5_18 (AEE72695); CaGH5_18 from Cutibacterium avidum 44067 (AGJ77370); KfGH5_18 from Kribbella flavida DSM 17836 (ADB34475); BlGH5_18 from Bifidobacterium longum subsp. infantis 157F (BAJ71452); SkGH5_18 from Sanguibacter keddieii DSM 10542 (ACZ21418); ScGH5_18 from Streptomyces coelicolor A3(2) (CAB61915) and AhGH5_18 from Arcanobacterium haemolyticum DSM 20595 ADH91800). The figure was prepared with ESPript3 (http://espript.ibcp.fr/ESPript/ESPript/).
(PDF)
Analysis of oligomeric state. (A) ThermoFluor-derived melting curve for CaMan_18 in 50 mM sodium phosphate (pH 7.5), and in the presence of the dye SYPRO Orange. Relative fluorescence units (RFU) were plotted against temperature. The protein unfolding curve is biphasic, which is consistent with two unfolding transitions. (B) Representation of the data in A by plotting the first derivative (dRFU)/dT of the raw data against temperature. Two Tm values were derived from this curve, 48.8°C and 58.0°C. (C) Overlay of the SEC chromatograms for the standard proteins (black lines) and of CaMan5_18 (red line). Inset: Standard curve used to extrapolate the molecular weight (R2 = 0.9974). CaMan5_18 elutes at a volume corresponding to a molecular weight of 98 kDa.
(PDF)
Gene organization of a second putative N-glycan processing locus in the genome of C. acnes 266. GH genes predicted as mannosidases are colored green, and GH genes with predicted N-acetylhexosaminidase activity are blue. The GH4 gene with unknown activity is colored dark gray. Other associated genes are colored light gray and include: predicted sugar ABC transporter permease (PERM), transcriptional-regulator gene (REG), sugar ABC-transporter substrate-binding protein (SBP), hypothetical proteins (HP), HAD-family phosphatase involved in carbohydrate transport (PHO), and sugar kinase (ROK). Accession numbers (GenBank, or RefSeq when GenBank was not available) are shown below each gene.
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.