Abstract
Purpose
Pseudomonas aeruginosa infections in hospitals constitute an important problem due to the increasing multidrug resistance (MDR) and carbapenems resistance. The knowledge of resistance mechanisms in Pseudomonas strains is an important issue for an adequate antimicrobial treatment. Therefore, the objective was to investigate other antimicrobial resistance mechanisms in MDR P. aeruginosa strains carrying blaIMP, make a partial plasmids characterization, and determine if modifications in oprD gene affect the expression of the OprD protein.
Methodology
Susceptibility testing was performed by Kirby Baüer and by Minimum Inhibitory Concentration (presence/absence of efflux pump inhibitor); molecular typing by Pulsed-field gel electrophoresis (PFGE), resistance genotyping and integrons by PCR and sequencing; OprD expression by Western blot; plasmid characterization by MOB Typing Technique, molecular size by PFGE-S1; and blaIMP location by Southern blot.
Results
Among the 59 studied P. aeruginosa isolates, 41 multidrug resistance and carbapenems resistance isolates were detected and classified in 38 different PFGE patterns. Thirteen strains carried blaIMP; 16 blaGES and four carried both genes. This study centered on the 17 strains har-boring blaIMP. New variants of β-lactamases were identified (blaGES-32, blaIMP-56, blaIMP-62) inside of new arrangements of class 1 integrons. The presence of blaIMP gene was detected in two plasmids in the same strain. The participation of the OprD protein and efflux pumps in the resistance to carbapenems and quinolones is shown. No expression of the porin OprD due to stop codon or IS in the gene was found.
Conclusions
This study shows the participation of different resistance mechanisms, which are reflected in the levels of MIC to carbapenems. This is the first report of the presence of three new variants of β-lactamases inside of new arrangements of class 1 integrons, as well as the presence of two plasmids carrying blaIMP in the same P. aeruginosa strain isolated in a Mexican hospital.
Keywords: Pseudomonas aeruginosa, resistance mechanisms, plasmid, integrons
Introduction
Pseudomonas aeruginosa is a well-known cause of severe and potentially life-threatening nosocomial infections, which include pulmonary disease, urinary tract infections and bacteremia. P. aeruginosa has an extraordinary capacity to develop antimicrobial resistance due to chromosomal mutations and the acquisition of resistance genes encoding β-lactamases, particularly carbapenemases, and combined with the presence of other resistance mechanisms, has generated an increase of multidrug resistance (MDR) strains.1,2 Previous studies have demonstrated that metallo-β-lactamases (MBLs) represent an important antimicrobial resistance determinant among P. aeruginosa strains causing nosocomial infections.3 The first MBL, variant blaIMP, was described in Japan,4 and to date 64 variants of blaIMP3 have been reported which are available in GenBank. The blaIMP genes are usually located in integrons, which are prevalent in gram-negative bacteria, and can be located in plasmids and transposons, facilitating their dissemination.5 P. aeruginosa strains harboring integrons with blaIMP genes cassettes have been reported in Europe, Asia, and South America.6–8 However, its presence has been recently found in Mexico, reports include the presence of carbapenem-resistant P. aeruginosa strains containing either blaIMP-15 or blaIMP-18 genes, both inside integrons class 1.9–11
In addition, a number of studies describe the presence of blaIMP-1, blaIMP-9, blaIMP-15, blaIMP-29, blaIMP-45, in conjugative plasmids of narrow and broad-host range belonging to incompatibility groups IncP-2 and IncP-9 in some cases.4,10,12–14 In P. aeruginosa, at least 13 incompatibility groups have been identified: IncP-1 to IncP-7 and IncP-9 to IncP-14. The plasmids belonging to the groups IncP-2, IncP-5, IncP-7, IncP-10, IncP-12 and IncP-13 are defined as plasmids of host narrow range. The plasmids with IncP-1, IncP-4 and IncP-6 are broad-range host plasmids.13,15,16 Due to this diversity in the Inc groups, Alvarado et al17 designed degenerate oligonucleotides that allow the identification of transmissible plasmids based on the sequence of relaxases, which are multidomain proteins present in mobilizable and conjugative plasmids.
Other mechanisms such as alterations or lack of porin OprD, and overexpression of efflux pumps have been associated with resistance to drugs in strains isolated from hospitalized patients. Loss of porin OprD decreases the susceptibility to carbapenems.18 Previous studies have shown that mutations in oprD gene results in the absence corresponding to the OprD porin band in SDS-PAGE gels.19,20
On the other hand, in P. aeruginosa, four clinically relevant RND efflux pumps: MexAB-OprM,21 MexCD-OprJ22 MexEF-OprN23 and MexXY-OprM24 have been identified, and the overexpression of these pumps, has been associated with increases of β-lactam, aminoglycosides and fluoroquinolone resistance.25 Also, it has been reported that the pump inhibitor phenylalanine arginyl β-naphthylamide (EPI), allows the participation of these pumps in the resistance to meropenem and fluoroquinolones, through a mechanism of competitive inhibition, this is because the pumps recognize EPI as a substrate instead of the target antibiotics.25,26
According to the information about the participation of different resistant mechanisms developed by P. aeruginosa, this work was carried out to investigate other antimicrobial resistance mechanisms in MDR P. aeruginosa strains arrying blaIMP, as well as to make a partial characterization of plasmids and determine if mutations in the oprD gene could have an effect on the expression of the OprD protein.
Material and methods
Bacterial strains and culture conditions
Fifty-nine P. aeruginosa clinical isolates were obtained from 57 patients with nosocomial infection (two patients with two different strains) hospitalized in Hospital Regional del ISSSTE of Puebla, Mexico, during the period April 2013 to July 2015. The isolates were identified by conventional biochemical tests and VITEK-2 system.
Antimicrobial susceptibility testing
Susceptibility to 13 anti-pseudomonal agents (piperacillin, ticarcillin, piperacillin/tazobactam, ceftazidime, cefepime, aztreonam, imipenem, meropenem, gentamicin, tobramycin, amikacin, ciprofloxacin, and norfloxacin) was performed by the disc-diffusion method. Minimum Inhibitory Concentration (MIC) of CAZ, FEP, IPM, MEM, CIP, AN, and GM was determined in the strains carrying blaIMP by agar dilution method using the breakpoints recommended by the Clinical and Laboratory Standard Institute.27 The MDR strains and carbapenem resistance (RC) were selected to continue the study. MDR was defined according to Magiorakos’ proposal as: non-susceptible to ≥1 agent in ≥3 antimicrobial categories.28
Molecular typing of P. aeruginosa isolates
The clonal relationship was performed by Pulsed-field gel electrophoresis (PFGE). Genomic DNA was digested using SpeI restriction enzyme (Thermo Fisher Scientific, Waltham, MA, USA) and macrorestriction fragments were separated as previously described29 on a CHEF-DR II (BioRad Laboratories Inc., Hercules, CA, USA). Banding patterns were visualized by ethidium bromide staining and photographed. Dendrogram and cluster analysis were performed with NTSYSpc2.2. software, using the Unweighted Pair Group Method with Arithmetic Averages (UPGMA) and based on the criteria of Tenover et al.30 The sequence type (ST) was determined in the three strains with new variants of β-lactamases by the multilocus sequence typing (MLST) technique as previously described by Curran et al31 Allelic profile and a sequence type (ST) were assigned using the Pseudomonas aeruginosa MLST website.32
Detection of resistance genes
The presence of different resistance genes was studied by PCR using specific primers for resistance to: β-lactams (blaIMP, blaVIM, blaGIM, blaSPM, blaNDM, blaGES, blaKPC, blaVEB, blaPSE, blaCMY, blaPER, blaOXA-2, blaOXA-40, blaOXA-50), aminoglycosides (aac(3′)-IIa, aacA4), quinolones (qnrA, qnrB, qnrS, aac(6′)Ib-cr) and colistin (mcr-1). Primers used for amplification and for sequencing are listed in Table S1. The amplicons obtained were sequenced. The nucleotide and deduced protein sequences were analyzed and compared with those available at GenBank. The strains carrying blaIMP were selected and used for the assays that are described below.
Detection and characterization of the genetic structures of integrons
The presence of type 1 and 2 integrases genes (intl1 and intl2), as well as the 3′-conserved segment of class 1 integrons (qacEΔ1+ sul1) were studied by PCR in all blaIMP-harboring strains. The characterization of class 1 integron variable regions was determined by PCR and sequencing (Table S1).33
Mechanisms of resistance to quinolones
Amino acid changes in GyrA and ParC proteins were studied in the P. aeruginosa strains. For this purpose, gyrA and parC genes were amplified by PCR, amplified fragments were purified and sequenced.34 Deduced amino acid sequences were compared with those previously reported for GyrA and ParC in P. aeruginosa PAO1 (GenBank accession number for GyrA/ParC: NP_251858.1 and NP_253651.1 respectively).
Modifications of porin OprD
The oprD gene was amplified by PCR and sequenced in all blaIMP-harboring strains. The mutations were determined by comparison with the sequence of the P. aeruginosa PAO1 strain (GeneID: 881970).35,36
Expression of protein OprD
The strains were cultured in BHI medium for 20 hours; the cells were collected and lysed with RIPA buffer (Sigma Aldrich, St. Louis, MO, USA). The total protein extract was precipitated with acetone, recovered by centrifugation and proteins were separated by means of 12% SDS-PAGE gel. Proteins were separate in duplicate to obtain two gels, one was stained with Coomassie brilliant blue20 and the other was used for Western blot.
Western blot
Gel destined for Western blot was transferred to a nitrocellulose membrane (Hybond-ECL; Amersham Pharmacia Biotech, Amersham, UK) for detection of OprD protein. The membrane was blocked using 5% non-fat skim milk overnight at 4°C. Then was washed with PBS and incubated with rabbit polyclonal antisera (1:10000) against OprD (obtained in this laboratory) followed by incubation with a second antibody phosphatase alkaline-conjugated goat anti-mouse IgG (1:5000) (Zymed Laboratories, Thermo Fisher Scientific, Waltham, MA, USA) and visualized with 5-bromo-4-chloro-3-indolyl phosphate/p-nitro blue tetrazolium system (BCIP/NBT) (Sigma Aldrich). The total extract protein of P. aeruginosa PAO1 and the OprD recombinant protein (obtained in this laboratory) were used as positive controls.
Influence of efflux pumps inhibitor on MIC to meropenem and ciprofloxacin
The efflux pumps inhibitor EPI (L-phenylalanine-argininenaphthylamide, PAβN, Sigma-Aldrich) was used to determine the involvement of the RND pumps in the resistance to MEM and CIP. The MIC of MEM and CIP was performed by the agar diffusion method without and with EPI at 25 mg/L. A decrease of two-fold or higher in MIC values in presence of EPI was considered as a participation of RND-type efflux pumps in the resistance to these two antibiotics.37
Extraction and size determination of plasmids
Plasmid extraction was performed using the Quick Prep method,38 and the size and number of plasmids were determined by PFGE-S1. For this method, the bacterial DNA was embedded in agarose plugs as described by Kaufmann.39 Plasmid digestions were done with nuclease S1 (8 U per plug) (New England Biolabs, Ipswich, MA, USA). The PFGE conditions were used as follows: pulse time ranging from 1 s to 30 s for 22 hours, the gel was run at 6 V cm and at 14°C. Two strains with plasmids of known size were used for size determination: E. coli NCTC 50192 with four plasmids (154, 66, 48 and 7 kb) and Rhizobium etli CFN42Δe with six plasmids (624, 371, 250, 194, 184 and 60.5 kb).
Plasmid typing
The types of relaxases were determined in the strains carrying plasmids by the Degenerate Primer MOB Typing (DPMT) method,17 and the incompatibility groups as was reported by Caratolli et al.40
Location of blaIMP gene in plasmids by southern blot
The PFGE conditions used were as follows: pulse time ranging from 5 s to 45 s for 20 hours at 6 V/cm and at 14°C after nuclease S1 (180 U/µL, Takara Bio Inc., Kusatsu, Shiga Prefecture, Japan) digestion. Southern blot was performed using the DIG HIGH Prime DNA Labeling and Detection Starter Kit II. The hybridization was performed overnight at 40°C with the blaIMP gene as probe; detection was made as is described by the manufacturer (Hoffmann-La Roche, Basel, Switzerland).
Ethics
All isolates were collected during routine sampling, and patient’s data were maintained under anonymity. The protocol for this study was approved by the Ethical Committee of Hospital Regional ISSSTE, Puebla (Registry number 188.2018)
Results
The 59 isolates were recovered from 57 patients hospitalized in different units (Internal Medicine, 51%; Intensive Care Unit, 17%; Urology, 7%; Surgery, 9%; and other units, 16%) and from different types of samples (bronchial secretions, 36%; urine culture, 32%; wounds, 8%; blood culture, 8%; catheter, 7%; and others, 9%).
Antimicrobial susceptibility
Forty-one of the 59 P. aeruginosa isolates showed MDR-RC (69%). The following percentages of resistance were detected: ticarcillin 74%, imipenem 69%, ciprofloxacin 69%, norfloxacin 69%, meropenem 68%, gentamicin 68%, tobramycin 66%, amikacin 63%, cefepime 61%, ceftazidime 59%, piperacillin 47%, aztreonam 41% and piperacillin/tazobactam 24%. All isolates were susceptible to colistin.
Clonal relationship of the isolates
A total of 38 different PFGE patterns were detected among the 41 MDR-RC P. aeruginosa isolates. Dendrogram shows that six strains were grouped into three clusters (A to C). Cluster A included strains PE09 and PE16, with an 84% of similarity, these strains were collected from the same service (Internal Medicine); cluster B included the strains PE54 and PE55 presenting a similarity of 95%, these strains were obtained from different services (Urology and Internal Medicine, respectively); and cluster C included strains PE57 and PE58 with 87% of similarity, these strains were also obtained from different services (Neurology and Internal Medicine, respectively). The strains from the same patient PE06 and PE52 (isolated from urine) and PE26 and PE30 (from expectoration and urine respectively) were non-related (Figure S1). MLST was performed on strains PE06, PE21, and PE52 that carried new genetic variants; results showed that these belong to lineages ST308, ST167, and ST385, respectively.
Detection of resistance genes
Among the 41 MDR P. aeruginosa strains, the following resistance genes were detected (% strains) to β-lactams: blaIMP (41%), blaGES (49%), blaOXA-2 (85%) and blaOXA-50 (100%); and to aminoglycosides: aacA4 (70%) and aac(3′)-IIa (7%). None of the strains harbored the genes: blaVIM, blaGIM, blaSPM, blaNDM, blaVEB, blaPSE, blaKPC, blaCMY, blaPER, blaOXA-40, qnrA, qnrB, qnrS or aac(6′)Ib-cr
The following variants were detected among the 16 blaGES-positive strains: blaGES-2 (n=6 strains), blaGES-19 (n=4), blaGES-20 (n=1) and blaGES-26 (n=5); and the variants among the 17 blaIMP-positive strains were as follows: blaIMP-18 (n=10 strains), blaIMP-18+ blaIMP-56 (n=2), blaIMP-56 (n=1), and blaIMP-62 (n=4). The two new variants, blaIMP-56 and blaIMP-62, were deposited in the GenBank database with the accession number KU351745.1 for blaIMP-56 sequence, and accession number KX753224.1 for blaIMP-62 sequence (Table 1). Association of blaIMP and blaGES genes was detected in four isolates: blaIMP-62 +blaGES32 in PE21; blaIMP-62+ blaGES-2 in PE23; blaIMP-18+ blaGES-1 in PE63; and blaIMP-62+ blaGES-26 in PE83 (Table 1). The sequence of the new variant blaGES-32 was deposited in the GenBank database (accession number KX753225).
Table 1.
Phenotype and genotype of resistance of P. aeruginosa clinical isolates carrying blaIMP gene
| Strain | PFGE type | Phenotype of resistance |
aMIC (μg/mL)
|
Genotype | Amino acid changes
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CAZ | FEP | MEM (+ bEPI) | IMP | CIP (+ bEPI) | AN | GM | GyrA | ParC | ||||
| PE06 | 15 | PIP, TIC, TZP, CAZ, FEP, ATM, IPM, MEM, GM, NN, AN, CIP, NOR | >512 | >512 | >512 (>512) | >512 | >512 (>512) | >512 | 32 | blaIMP-18, blaIMP-56, blaOXA-2, blaOXA-50, aadA1, aacA6 | T83I | S87L |
| PE09 | 6a | TIC, CAZ, FEP, IPM, MEM, GM, NN, AN, CIP, NOR | >512 | 512 | >512 (32) | 64 | 64 (32) | 256 | >512 | blaIMP-18, blaOXA-2, blaOXA-50, aadA4 | T83I | S87L |
| PE10 | 8 | PIP, TIC, CAZ, FEP, ATM, IPM, MEM, GM, NN, AN, CIP, NOR | 512 | 256 | 64 (64) | 64 | 32 (16) | 64 | >512 | blaIMP-18, blaOXA-2, blaOXA-50, aadA4 | T83I | S87L |
| PE15 | 7 | TIC, CAZ, FEP, IPM, MEM, GM, NN, AN, CIP, NOR | 512 | 256 | 64 (64) | >512 | 32 (16) | 64 | >512 | blaIMP-18, blaOXA-2, blaOXA-50, aadA4 | T83I | S87L |
| PE16 | 6b | TIC, CAZ, FEP, IPM, MEM, GM, NN, AN, CIP, NOR | 512 | 256 | 64 (64) | >512 | 32 (16) | 64 | >512 | blaIMP-18, blaOXA-2, blaOXA-50, aadA4 | T83I | S87L |
| PE20 | 1 | PIP, TIC, CAZ, FEP, IPM, MEM, GM, NN, AN, CIP, NOR | >512 | >512 | >512 (128) | >512 | 128 (64) | >512 | >512 | blaIMP-18, blaOXA-2, blaOXA-50, aadA4 | T83I | S87L |
| PE21 | 28 | TIC, CAZ, FEP, IPM, MEM, GM, NN, AN, CIP, NOR | >512 | 512 | >512 (>512) | 128 | 128 (64) | >512 | >512 | blaIMP-62, blaGES-32, blaOXA-2, blaOXA-50, aacA33, aadA1, aadA6, aacA4, aacA7 | T83I | S87L |
| PE22 | 12 | TIC, CAZ, FEP, IPM, MEM, GM, NN, AN, CIP, NOR | 512 | 256 | 16 (16) | 64 | 128 (64) | 128 | >512 | blaIMP-18, blaOXA-2, blaOXA-50, aac(3′)-IIa, aadA4 | T83I | S87L |
| PE23 | 10 | TIC, FEP, IPM, MEM, GM, NN, AN, CIP, NOR | 16 | 32 | 32 (32) | 64 | 32 (16) | 64 | >512 | blaIMP-62, blaGES-2, blaOXA-2, blaOXA-50, aacA4, aacA29b | T83I | S87L |
| PE27 | 25 | TIC, CAZ, FEP, IPM, MEM, GM, NN, AN, CIP, NOR | 512 | 256 | 64 (64) | >512 | 64 (32) | 128 | 512 | blaIMP-62, blaGES-2, blaOXA-2, blaOXA-50, aacA4 | T83I | S87L |
| PE46 | 32 | TIC, CAZ, FEP, IPM, MEM, GM, NN, AN, CIP, NOR | >512 | 512 | >512 (32) | 64 | 64 (32) | 512 | >512 | blaIMP-18, blaGES-2, blaOXA-2, blaOXA-50, aacA4 | T83I | S87L |
| PE52 | 16 | PIP, TIC, TZP, CAZ, FEP, ATM, IPM, MEM, GM, AN, CIP, NOR | >512 | >512 | >512 (>512) | >512 | >512 (512) | >512 | 32 | blaIMP-18, blaIMP-56, blaOXA-2, blaOXA-50, aadA1, aacA6 | T83I | wild type |
| PE62 | 20 | PIP, TIC, CAZ, FEP, ATM, IPM, MEM, GM, NN, AN, CIP, NOR | >512 | >512 | >512 (>512) | >512 | >512 (512) | >512 | 16 | blaIMP-18, blaOXA-2, blaOXA-50, | T83I | S87L |
| PE63 | 38 | PIP, TIC, TZP, CAZ, FEP, ATM, IPM, MEM, GM, NN, AN, CIP, NOR | 256 | 232 | 512 (512) | 512 | 128 (64) | 512 | >512 | blaIMP-18, blaGES-1, blaOXA-2, blaOXA-50, aacA4, aadA1 | wild | P105T |
| PE64 | 21 | PIP, TIC, CAZ, FEP, ATM, IPM, MEM, GM, NN, AN, CIP, NOR | 256 | 512 | >512 (512) | >512 | 512 (256) | >512 | 32 | blaIMP-18, blaOXA-2, blaOXA-50 | T83I | S87L |
| PE73 | 26 | PIP, TIC, CAZ, FEP, ATM, IPM, MEM, GM, NN, AN, CIP, NOR | >512 | >512 | >512 (>512) | >512 | >512 (512) | >512 | 32 | blaIMP-56, blaOXA-2, blaOXA-50 | T83I | S87L |
| PE83 | 13 | TIC, CAZ, FEP, IPM, MEM, GM, NN, AN, CIP, NOR | >512 | >512 | >512 (>512) | 64 | 256 (128) | >512 | >512 | blaIMP-62, blaGES-26, blaOXA-2, blaOXA-50, aac(3′)-IIa, aacA4, aadA6, aacA7 | T83I | S87L |
Notes:
Minimum Inhibitory Concentration;
PAβN (L-phenylalanine-arginine-β-naphthylamide).
Abbreviations: PFGE, pulsed-field gel electrophoresis; PIP, piperacillin; TIC, ticarcillin; TZP, tazobactam; CAZ, ceftazidime; FEP, cefepime; IMP, imipenem; MER, meropenem; GM, gentamicin; NN, tobramycin; AN, amikacin; CIP, ciprofloxacin; NOR, norfloxacin; ATM, aztreonam.
Nine blaIMP-positive strains also carried the aacA4 gene (amikacin MIC 64 to >512 µg/mL), and two strains contained aac(3′)-IIa+ aacA4 (amikacin MIC 128 to >512 µg/mL and gentamicin MIC >512 µg/mL). None of the acetylases studied were amplified in three strains resistant to amikacin MIC >512 µg/mL and gentamicin MIC 16–32 µg/mL (Table 1).
Detection and characterization of the genetic structures of integrons
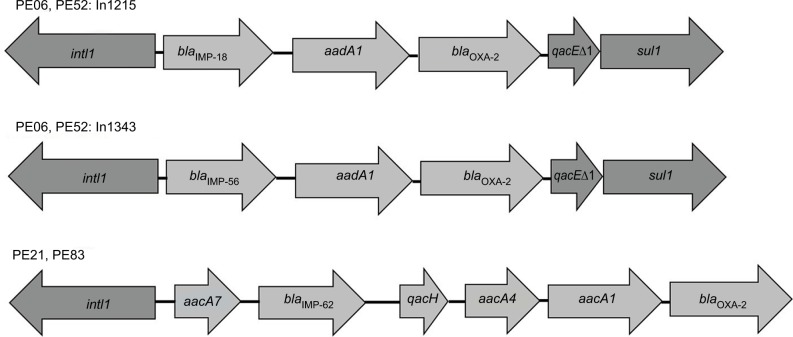
To determine the genetic context of blaIMP, class 1 integrons were detected in all strains, but not class 2 integrons. Two new class 1 integron arrangements were identified in two strains obtained from the same patient (PE06 and PE52): blaIMP-18+ aadA1+blaOXA-2 and blaIMP-56+ aadA1+blaOXA-2. These new arrangements were deposited in Integrall and GenBank database with the accession numbers KY646160 and KY646161 for the In1215/In1343 sequence of PE52 respectively.
The strains PE21 and PE83 showed an identical class 1 integron array without 3′-CS: aacA7+blaIMP-62+ qacH + aacA4+ aadA1+ blaOXA-2 (Figure 1). These strains were unrelated according to PFGE and they were obtained from different patients. By analyzing the variable regions of class 1 integrons, we identified some other resistance genes in these strains, such as aacA33, aadA1, aadA6 and aacA7.
Figure 1.
Diversity of class 1 integron arrangements detected in blaIMP-carrying P. aeruginosa clinical isolates.
Notes: The In1215 containing blaIMP-18 with the accession number KY646160 and the In1343 containing blaIMP-56 with the accession number KY646161. Arrows indicate the open reading frames.
Modifications and expression of porin OprD
The oprD gene was analyzed in the 17 blaIMP-carrying strains (Table 2). In two strains the oprD gene was not amplified and by immunoblot the protein could not be detected. Four strains showed deletions of two amino acids: 378 and 379 (called loop L7-short), which generated an OprD protein of 441 amino acids, and the protein expression was confirmed by immunoblot, showing a positive signal of approximately 48 kDa. The OprD porin was not detected by immunoblot in nine strains with premature stop codon (aminoacid 295, 417, 422 and 426), and in the PE20 strain with an insertion sequence (IS1394) in the amino acid 232. The PE63 strain showed the oprD gene wild type and the expression of protein was detected (Table 2, Figure 2).
Table 2.
Changes in OprD porin in clinical isolates of P. aeruginosa carrying blaIMP
| Strain |
aMIC (μg/mL)
|
OprD size (Number of amino acids) | Amino acid changes in OprD sequence | Insertion/deletion | |
|---|---|---|---|---|---|
| IMP | MEM | ||||
| PE06 | >512 | >512 | 426 | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S, A281G, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, ∆378, ∆379, K380Y, N381A, Y382G, G383L E426STOP | Stop codon |
| PE09 | 64 | >512 | 441 | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, A267S, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, ∆378, ∆379, K380Y, N381A, Y382G, G383L | Loop L7-short |
| PE10 | 64 | 64 | 441 | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S, A281G, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, ∆378, ∆379, K380Y, N381A, Y382G, G383L | Loop L7-short |
| PE15 | >512 | 64 | bNA | bNA | bNA |
| PE16 | >512 | 64 | 422 | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S, A281G, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, ∆378, ∆379, K380Y, N381A, Y382G, G383L A422STOP | Stop codon |
| PE20 | >512 | >512 | 232 | D43N, S57E, S59R, E202Q, I210A, E230K, I232M, Y233A, R234A, IS1394 | Truncated by IS1394 |
| PE21 | 128 | >512 | 417 | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S A281G, K296Q, Q301E R310G V359L, M372V, S373D, D374S, N375S, N376S, V377S, ∆378, ∆379, K380Y, N381A, Y382G, G383L W417STOP | Stop codon |
| PE22 | 64 | 16 | 441 | D43N, S57E, S59R, E202Q, I210A, L224F, E230K, S240T, N262T, A267S A281G, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, ∆378, ∆379, K380Y, N381A, Y382G, G383L | Loop L7-short |
| PE23 | 64 | 32 | 417 | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S A281G, K296Q, Q301E, L307M, R310G, A316G, M372V, S373D, D374S, N375S, N376S, V377S, ∆378, ∆379, K380Y, N381A, Y382G, G383L W417STOP | Stop codon |
| PE27 | >512 | 64 | bNA | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S | bNA |
| PE46 | 64 | >512 | 441 | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S, W277R, A281G, A282G, Y283S, T284G, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, ∆378, ∆379, K380Y, N381A, Y382G, G383L | Loop L7-short |
| PE52 | >512 | >512 | 426 | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S, A281G, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, ∆378, ∆379, K380Y, N381A, Y382G, G383L, E426STOP | Stop codon |
| PE62 | >512 | >512 | 426 | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S, A280G, A281G, Y283S, T284A, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, ∆378, ∆379 K380Y, N381A, Y382G, G383L, E426STOP | Stop codon |
| PE63 | 512 | 512 | 443 | Wild type | - |
| PE64 | >512 | >512 | 295 | D43N, S57E, S59R E202Q, I210A, E230K, S240T, N262T, A267S, A281G, K295STOP | Stop codon |
| PE73 | >512 | >512 | 426 | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S, A281G, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, ∆378, ∆379, K380Y, N381A, Y382G, G383L, E426STOP | Stop codon |
| PE83 | 64 | >512 | 417 | D43N, S57E, S59R, E202Q, I210A, E230K, S240T, N262T, A267S A281G, K296Q, Q301E, R310G, V359L, M372V, S373D, D374S, N375S, N376S, V377S, ∆378, ∆379, K380Y, N381A, Y382G, G383L W417STOP | Stop codon |
Notes:
Minimum Inhibitory Concentration;
Not amplified or partially amplified; IMP, imipenem; MEM, meropenem.
Figure 2.
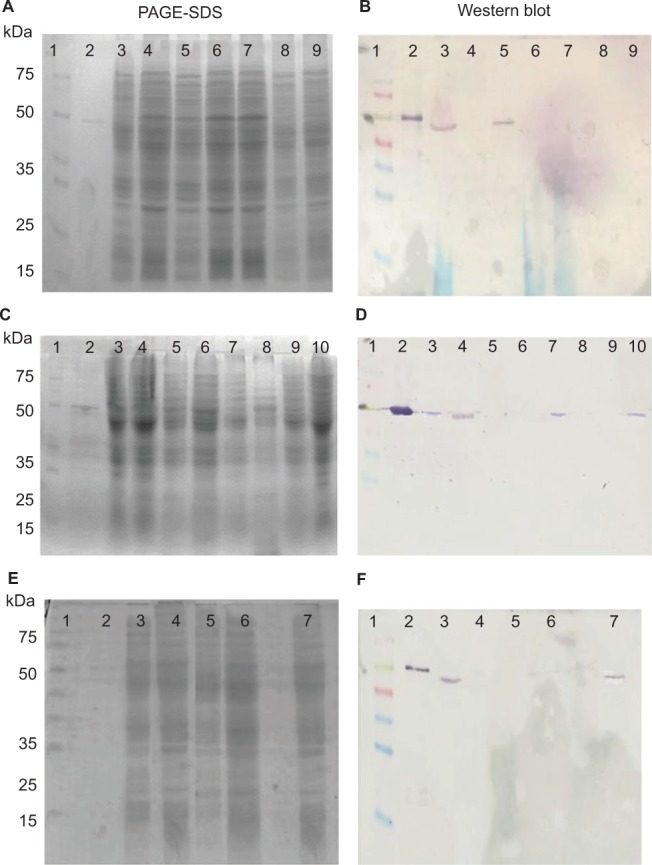
Detection of the OprD protein in 17 clinical blaIMP-carrying P. aeruginosa. (A, C, E) PAGE-SDS of extract total protein, (B, D, F) Western blot of extract total protein with polyclonal antisera anti-oprD.
Notes: For each lane, 30 μg of proteins were loaded. P. aeruginosa PAO1 and OprD protein recombinant were used as positive controls. (A, B) Line 1: Molecular weight marker (Spectratm); Line 2: recombinant OprD protein; Line 3: P. aeruginosa PAO1; Line 4: PE06 (stop codon); Line 5: PE09 (L7-short); Line 6: PE15 (not amplified); Line 7: PE16 (stop codon), Line 8: PE20 (IS1394); Line 9: PE21 (stop codon). (C, D) Line 1: Size marker; Line 2: recombinant OprD protein; Line 3: P. aeruginosa PAO1; Line 4: PE22 (L7-short); Line 5: PE23 (stop codon); Line 6: PE27 (partially amplified), Line 7: PE46 (L7-short); Line 8: PE52 (stop codon); Line 9: PE62 (stop codon); Line 10: PE63 (wild type). (E, F) Line 1: Size marker; Line 2: recombinant OprD protein; Line 3 P. aeruginosa PAO1; Line 4: PE64 (stop codon); Line 5: PE73 (stop codon); Line 6: PE83 (stop codon) and Line 7: PE10 (L7-short). The experiment was made in duplicate.
Influence of efflux pumps inhibitor on MIC to meropenem and ciprofloxacin
In order to investigate if RND-superfamily efflux pumps were involved in resistance to meropenem and ciprofloxacin, strains carrying blaIMP, were exposed with EPI. In the strains PE09 and PE46 there was a decrease in meropenem MIC value ≥16-fold (≥512–32 µg/mL), in the strain PE20 there was a 4-fold decrease (>512–128 µg/mL) and in the strain PE64 there was a decrease of ≥2 fold (>512–512 µg/mL). In the other strains, no change was detected in meropenem MIC value by EPI presence. In the 17 strains, the ciprofloxa-cin MIC value without EPI (32 to >512 µg/mL) showed a decrease in the order of ≥2 fold (16–512 µg/mL) (Table 1). In the case of the strains with ciprofloxacin or meropenem MIC values of >512 µg/mL and that decreased to 512 µg/mL in presence of EPI, additional experiments were performed using concentrations of 1024 µg/mL of the antibiotic to corroborate this decrease (data not showed).
Mechanisms of resistance to quinolones
The changes in the QRDR region of the GyrA and ParC proteins were determined. A single amino acid substitution in GyrA protein (Thr-83→Ile) was detected in all the strains. In ParC protein, 15 strains showed the substitution Ser-87→Leu and one strain (PE63) the change Pro-105→Thr. ParC protein in the strain PE52 was wild type. The ciprofloxacin MIC showed different values in the 17 strains: four showed 32 µg/mL, three strains 64 µg/mL, four strains 128 µg/mL and six strains 256–>512 µg/mL (Table 1).
Partial characterization of plasmids
Eight strains showed plasmids with different size as was determined by PFGE-S1 method. We confirm the presence of plasmids by Quick Prep method, however, in the strain PE63 was not possible to detect any plasmid (Figure S2). The relaxase MOBP11 was detected in the strains PE06, PE21, PE52, PE62, PE64, PE73 and PE83, and a second relaxase MOBH2 was amplified in the PE21 and PE83 strains. The relaxase MOBP14 was detected in the strain PE63 (Table 3, Figure 3).
Table 3.
Plasmid characterization of 17 clinical blaIMP-carrying P. aeruginosa isolates
| Strain gene | b-lactamase | Plasmids size (kb) | aRelaxase MOB subfamily |
|---|---|---|---|
| PE06 | blaIMP-18, blaIMP-56 | 2(approx 224, approx 188) | MOBP11 |
| PE09 | blaIMP-18 | Absent | Absent |
| PE10 | blaIMP-18 | Absent | Absent |
| PE15 | blaIMP-18 | Absent | Absent |
| PE16 | blaIMP-18 | Absent | Absent |
| PE20 | blaIMP-18 | Absent | Absent |
| PE21 | blaIMP-62, blaGES-32 | 1(approx 368) | MOBP11 MOBH2 |
| PE22 | blaIMP-18 | Absent | Absent |
| PE23 | blaIMP-62, blaGES-2 | Absent | Absent |
| PE27 | blaIMP-62 | Absent | Absent |
| PE46 | blaIMP-18 | Absent | Absent |
| PE52 | blaIMP-18, blaIMP-56 | 2(approx 242, approx 194) | MOBP11 |
| PE62 | blaIMP-18 | 2(approx 248, approx 200) | MOBP11 |
| PE63 | blaIMP-18, blaGES-1 | 3(approx 230, approx 200, approx 182) | MOBP14 |
| PE64 | blaIMP-18 | 1(approx 146) | MOBP11 |
| PE73 | blaIMP-18 | 1(approx 266) | MOBP11 |
| PE83 | blaIMP-62, blaGES-26 | 1(approx 350) | MOBP11 MOBH2 |
Notes:
Detection of relaxases was performed by Degenerate Primer MOB Typing (DPMT).17
Figure 3.
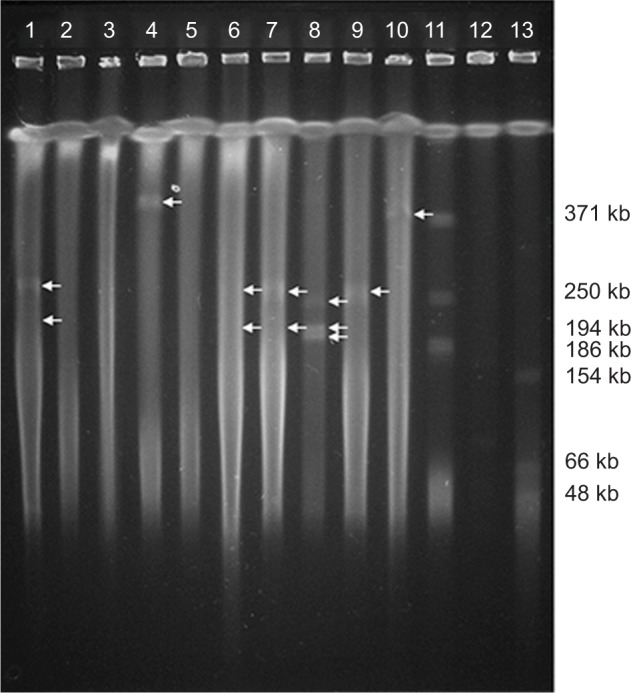
PFGE-S1 plasmids of 10 clinical isolates of P. aeruginosa carrying blaIMP.
Notes: The arrows indicate the plasmids. Lane 1: PE06, Lane 2: PE09, Lane 3: PE20, Lane 4: PE21, Lane 5: PE46, Lane 6: PE52, Lane 7: PE62, Lane 8: PE63, Lane 9: PE73, Lane 10: PE83, Lane 11: Control strain Rhizobium etli CFN42 ∆e, Lane 12: Negative control PAO1, Lane 13: Control strain E. coli NCTC 50192 with four plasmids: 154 kb, 66 kb, 48 kb and 7 kb. The experiment was performed twice.
Abbreviation: PFGE-S1, Pulsed-field gel electrophoresis with nuclease S1.
Location of the blaIMP gene by southern blot
Results obtained showed that blaIMP gene was detected in only four of the eight strains carrying plasmids. The PE06 and PE52 strains, that carried two blaIMP variants (blaIMP-18 and blaIMP-56), and the PE62 and PE63 strains that carried blaIMP-18, each showed two positive hybridization signals in two plasmids (Figure 4). No hybridization signals were detected in the remaining strains (data not shown)
Figure 4.
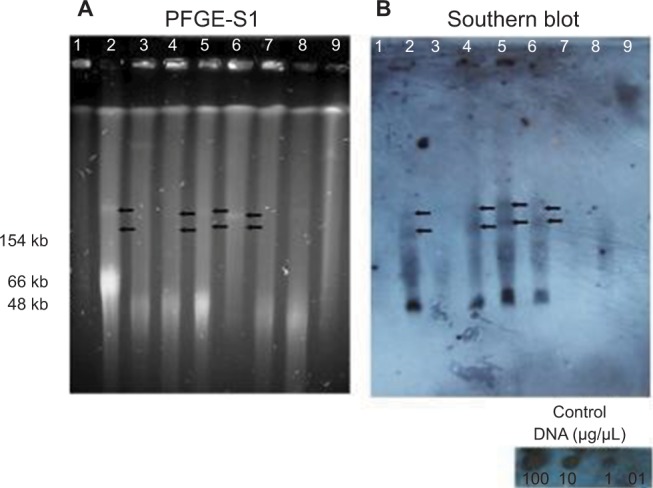
Detection of blaIMP in plasmids of eight P. aeruginosa strains by Southern blot.
Notes: (A) PFGE after digestion with nuclease S1 (180 U/μL, Takara Bio Inc.), (B) Southern hybridization of plasmids (DIG HIGH Prime DNA Labeling and Detection Starter Kit II) using blaIMP as a probe. Line 1: Control strain E. coli NCTC 50192 with four plasmids: 154 Kb, 66 Kb, 48 Kb and 7 Kb, Line 2: PE06; Line 3: PE21; Line 4: PE52; Line 5: PE62; Line 6: PE63; Line 7: PE64; Line 8: PE73 and Line 9: PE83. The black arrows indicate the positive signal corresponding to the hybridization with blaIMP in plasmids. The experiment was done three times.
Abbreviation: PFGE-S1, Pulsed-field gel electrophoresis with nuclease S1.
Discussion
It has been reported that most of the nosocomial infections caused by P. aeruginosa most frequently affect patients with pneumonia associated with mechanical ventilation and this is the main cause of chronic respiratory infection in patients with cystic fibrosis, bronchiectasis or COPD.41 In the present study, isolates were obtained from bronchial secretions (36%). On the other hand, in the last few years an increase of strains with carbapenems’ resistance has been reported all over the world.3,25 In this study we detected that 69% of the strains were resistant to carbapenems, and resistant to nine or more of the 13 antibiotics tested, indicating the dissemination of MDR and CR strains, and an emerging problem in this hospital.
Of 41 MDR-CR strains, 24 were harboring β-lactamase with carbapenemase activity; six strains were carrying blaGES-2, one strain had blaGES-20 and 17 strains were carrying different variants of blaIMP. Twelve strains were carrying blaIMP-18, which is one of the most frequent variants detected in Mexico,9,11 USA42 and Latin America.8 Two new variants, blaIMP-56 and blaIMP-62 were detected in seven strains, alone or with blaIMP-18, these new variants derived from blaIMP-18 and blaIMP-15, respectively and showed the same change (S214G) in the amino acid sequence. Coexistence of blaIMP and blaGES genes in the same strain has not been reported in Mexico, and there is only one report of P. aeruginosa carrying blaGES-5 and blaVIM.43 In this study we found a new variant blaGES-32, this variant had 96% of similarity with blaGES-1, blaGES-5, blaGES-6 and blaGES-7.
On the other hand, 17 MDR-RC strains did not carry any β-lactamase genes related with carbapenems’ resistance. Previous studies have reported the absence of these genes in CR isolates, suggesting the existence of other mechanisms responsible for the resistance.19,25
Integrons are ubiquitous platforms of great importance that participate in the capture of resistance genes, which are carried by mobile elements, plasmids, and transposons. These elements allow bacteria to acquire the capacity to adapt to environmental changes. Class 1 integrons are the most frequently reported in P. aeruginosa related to resistance to β-lactams and aminoglycosides.44 In this study, the PE06 and PE52 strains harbored two new class 1 integrons: In1245 and In1343, and the strains PE21 and PE83 carried an integron with the identical partial arrangement in the variable region: aacA7+ blaIMP-62+qacH+aacA4+ aadA1+blaOXA-2. These results suggest the existence of a dissemination of integrons in unrelated strains. Other strains with blaIMP were found in class 1 integrons, but the genetic array in the variable region could not be determined.
Three main high-risk clones ST111, ST175 and ST235 have been reported.45 In our study, strain PE21 corresponds to a ST167; when performing the analysis of eBURST V3, we find that it derives from group O where high-risk ST111 is found, but they only share two alleles, so we think that they are not related. The other two, ST308 and ST385, were not found to be related to the high risk clones reported. In the case of high risk clones, only ST235 has been reported in Mexico.45 It is important to note that our strains carry resistance mechanisms different from those reported in high risk clones.19 The ST found in this study has already been reported in different parts of the world45 and the ST308 in Mexico.46 So this is the first report that shows the presence of ST167 and ST385 in Mexico.
Since this study was focused on the strains carrying blaIMP and they showed different levels of resistance to carbapenems, it was interesting to analyze other mechanisms that could be contributing to the resistance such as mutations of the oprD gene and its expression, and the participation of the efflux pumps.
Four groups were formed according to MIC values for carbapenems. In the first group, eight strains were grouped with high MIC values for imipenem and meropenem (128 to >512 µg/mL), in all of them, we detected one or two carbapenemases and they presented a premature stop codon or SI in the oprD gene. Previous studies have shown that strains with a premature stop codon in the middle of the protein OprD sequence exhibited a loss in protein expression19,47; in the analyzed strains, the premature stop codon was located at the end of the sequence, however, the protein was not expressed. The insertion sequence (IS1394) found in the PE20 strain, correspond to a 1,100 bp sequence, coding for a transposase. The first report of an inactivation of OprD porin in clinical isolates by insertion sequences that lead to carbapenems resistance in P. aeruginosa occurred in 2004.36 At present there are several reports that show the inactivation of OprD porin by insertion sequences.18,19,48 In regard to IS1394 inserted into oprD gene in P. aeruginosa, the first report appears in Puerto Rico.49 It is important to mention that this is the first report that describes the presence of IS1394 in a strain isolated in Mexico. We suggest that the OprD porin, and the presence of MBL contributed synergistically to the acquisition of a higher level of MIC to carbapenems, except for the PE63 strain, which showed a wild type porin. Another mechanism that may be contributing to the bacteria resistance is the efflux pumps, as is suggested by the results observed in the strains PE20 and PE64, which showed a decrease of CMI for MEM in presence of EPI, as has been reported in other studies.26 These results also suggest that other resistance mechanisms, eg, the role of AmpC,50 could be involved.
The second group included three strains (PE15, PE16 and PE27), these strains showed high MIC values for imipenem (>512 µg/mL) and low levels for meropenem (64 µg/mL). These strains exhibited a premature stop codon and did not express OprD porin. In these strains we did not observe the contribution of efflux pumps suggesting that another mechanism might participate in carbapenem resistance, as has been reported in other works.1,2
The third group with three strains, showed low MIC values for IMP (64 µg/mL) and high values for meropenem (>512 µg/mL). Two of these strains (PE09, PE46) showed a Loop L7-short; therefore, the porin OprD probably is not participating in the carbapenems’ resistance. It is important to highlight that the strains under study showed a high polymorphism in the oprD gene, which may be involved in the variable susceptibility presented.19 These strains in the presence of EPI, showed a decrease of MIC value for meropenem, suggesting that efflux pump is participating in the meropenem resistance. In PE83 strain, a premature stop codon was detected, and the porin was not expressed, suggesting that the OprD porin and the two new blaIMP-62 and blaGES-26 variants detected in this isolate, could be related in the resistance to carbapenems.
The fourth group involved three strains with low MIC values for IMP (64 µg/mL) and low values for MEM (16–64 µg/mL). Imipenem resistance in PE10 y PE22 strains is probably due only to the presence of blaIMP-18. These strains showed a Loop L7-short, so that we suggested that porin is perhaps involved with the low resistance to meropenem.51 Results obtained in the PE23 strain suggest that the presence of the two carbapenemases, and the porin oprD are participating in carbapenems’ resistance, because EPI did not modify MIC values for meropenem.
The presence of aminoglycoside-modifying enzymes was detected in 14 strains, all of them showed high MIC values for aminoglycosides (gentamicin and amikacin). Therefore, there is no doubt that in addition to the participation of these enzymes, there are other aminoglycoside resistance mechanisms involved.52
The mutations detected in gyrA and parC genes are not enough to explain the high ciprofloxacin MIC values shown by 10 strains (128 to >512 µg/mL), therefore, it was necessary to identify other mechanisms involved. The 17 strains showed a decrease of ciprofloxacin MIC value in the presence of EPI, suggesting that efflux pumps also are participating in the resistance to ciprofloxacin since it has been reported that PAβN is a broad-spectrum efflux pump inhibitor (EPI), which is capable of significantly reducing fluoroquinolone resistance in P. aeruginosa.53
In eight strains carrying blaIMP we detected 1–3 plasmids of different sizes, but only the blaIMP gene was detected in two plasmids of four strains. Strain PE06 and PE52 (blaIMP-18 and blaIMP-56) have a relaxase MOBP11, and this relaxase has been found in conjugative plasmids of a broad host range belonging to the complex IncP1.
On the other hand, blaIMP-18 was detected in PE62 and PE63 strains, in two plasmids in each strain. It is important to mention that blaIMP-18 has not been reported in plasmids,8,9,11 therefore, in Mexico this is the first report of plasmids that carry blaIMP-18. In the PE62 strain the relaxase MOBP11 was detected and in the strain PE63 the relaxase MOBP14 was detected, the latter one is present in mobilizable plasmids with a broad range belonging to the incompatibility groups IncP6 and IncQ2.17,54
It has been reported that IncP1 in Pseudomonas corresponds to the IncP of E. coli, a complex formed by six subgroups (α, β, γ, ε, δ and ζ), with the difference between them the protein TrfA, a protein that initiates the replication of plasmids (determined by phylogenetic studies and amino acid sequencing).55 In these strains we performed a search for IncP1, but it was not possible to amplify them. Probably other types of Inc belonging to the IncP complex could be present.56
Results obtained in this work suggest the existence of horizontal gene transference between bacteria within this hospital. It is important to underline that the knowledge of the mechanisms, mainly of carbapenems’ resistance in clinical isolates of P. aeruginosa will help to decide the best therapeutic strategy. This information is of great value in taking measures to avoid the dissemination of emergent mechanisms of resistance in the hospital setting.
Conclusion
This study highlights the participation of various resistance mechanisms that are reflected in the levels of MIC to car-bapenems shown by the studied strains. This is the first report in Mexico that describes the presence of three new variants of β-lactamases (blaGES-32, blaIMP-56, blaIMP-62) inside of new arrangements of class 1 integrons and the presence of two plasmids carrying blaIMP in the same strain isolated in a Mexican hospital. Non-expression of the OprD protein is shown when it presents premature stop codons at the end of the sequence.
Supplementary materials
Dendrogram of 41 multidrug-resistant P. aeruginosa clinical isolates obtained in ISSSTE Regional Hospital of Puebla, Mexico.
Notes: The isolates showing a Dice coefficient ≥80% were considered as genetically related for this study. PFGE type (PT), PFGE clusters (A, B and C).
Abbreviation: PFGE, pulsed-field gel electrophoresis.
Plasmid extraction by the Quick Prep method of 17 clinical blaIMP-carrying P. aeruginosa isolates.
Notes: The position of chromosomal DNA is indicated by an arrow at the right. Line 1: Size marker; Line 2: PE06; Line 3: PE09; Line 4: PE15; Line 5: PE16; Line 6: PE20; Line 7: PE21; Line 8: PE22; Line 9: PE23; Line 10: PE27; Line 11: PE46; Line 12: PE52; Line 13: PE62, Line 14: PE63; Line 15: PE64; Line 16: PE73; Line 17: PE10; Line 18: PE83; Line 19: Control E. coli NCTC 50192; Line 20: P. aeruginosa PAO1.
Acknowledgments
This work was supported by the doctoral scholarship from CONACyT granted to Alma López García (78552), and by the projects VIEP-BUAP LOZP-NAT17-I and VIEP/2497/16. The authors wish to express their gratitude to Dr Jesús Silva- Sánchez from the National Institute of Public Health Mexico and Dr Miguel Angel Cevallos from the Center of Genomics Science, UNAM who donated the control strains used in this study.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Meletis G, Exindari M, Vavatsi N, Sofianou D, Diza E. Mechanisms responsible for the emergence of carbapenem resistance in Pseudomonas aeruginosa. Hippokratia. 2012;16(4):303–307. [PMC free article] [PubMed] [Google Scholar]
- 2.Ruppé É, Woerther PL, Barbier F. Mechanisms of antimicrobial resistance in Gram-negative bacilli. Ann Intensive Care. 2015;5(1):61. doi: 10.1186/s13613-015-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hong DJ, Bae IK, Jang IH, et al. Epidemiology and characteristics of metallo-β-lactamase-producing Pseudomonas aeruginosa. Infect Chemother. 2015;47(2):81–97. doi: 10.3947/ic.2015.47.2.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watanabe M, Iyobe S, Inoue M, Mitsuhashi S. Transferable imipenem resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1991;35(1):147–151. doi: 10.1128/aac.35.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao WH, Hu ZQ, Zq H. IMP-type metallo-β-lactamases in Gram-negative bacilli: distribution, phylogeny, and association with integrons. Crit Rev Microbiol. 2011;37(3):214–226. doi: 10.3109/1040841X.2011.559944. [DOI] [PubMed] [Google Scholar]
- 6.Pournaras S, Köck R, Mossialos D, et al. Detection of a phylogenetically distinct IMP-type metallo-β-lactamase, IMP-35, in a CC235 Pseudo-monas aeruginosa from the Dutch-German border region (Euregio) J Antimicrob Chemother. 2013;68(6):1271–1276. doi: 10.1093/jac/dkt004. [DOI] [PubMed] [Google Scholar]
- 7.Mano Y, Saga T, Ishii Y, et al. Molecular analysis of the integrons of metallo-β-lactamase-producing Pseudomonas aeruginosa isolates collected by nationwide surveillance programs across Japan. BMC Microbiol. 2015;15:41. doi: 10.1186/s12866-015-0378-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martínez T, Vázquez GJ, Aquino EE, Goering RV, Robledo IE. Two novel class I integron arrays containing IMP-18 metallo-β-lactamase gene in Pseudomonas aeruginosa clinical isolates from Puerto Rico. Antimicrob Agents Chemother. 2012;56(4):2119–2121. doi: 10.1128/AAC.05758-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sánchez-Martinez G, Garza-Ramos UJ, Reyna-Flores FL, et al. In169, a new class 1 integron that encoded bla(IMP-18) in a multidrug-resistant Pseudomonas aeruginosa isolate from Mexico. Arch Med Res. 2010;41(4):235–239. doi: 10.1016/j.arcmed.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 10.Garza-Ramos U, Morfin-Otero R, Sader HS, et al. Metallo-beta-lactamase gene bla(IMP-15) in a class 1 integron, In95, from Pseudomonas aeruginosa clinical isolates from a hospital in Mexico. Antimicrob Agents Chemother. 2008;52(8):2943–2946. doi: 10.1128/AAC.00679-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Garza-Ramos U, Tinoco P, Silva-Sanchez J, et al. Metallo-beta-lactamase IMP-18 is located in a class 1 integron (In96) in a clinical isolate of Pseudomonas aeruginosa from Mexico. Int J Antimicrob Agents. 2008;31(1):78–80. doi: 10.1016/j.ijantimicag.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 12.Quinones-Falconi F, Galicia-Velasco M, Marchiaro P, et al. Emergence of Pseudomonas aeruginosa strains producing metallo-beta-lactamases of the IMP-15 and VIM-2 types in Mexico. Clin Microbiol Infect. 2010;16(2):126–131. doi: 10.1111/j.1469-0691.2009.02780.x. [DOI] [PubMed] [Google Scholar]
- 13.Xiong J, Hynes MF, Ye H, et al. bla(IMP-9) and its association with large plasmids carried by Pseudomonas aeruginosa isolates from the People’s Republic of China. Antimicrob Agents Chemother. 2006;50(1):355–358. doi: 10.1128/AAC.50.1.355-358.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu J, Yang L, Chen D, et al. Complete sequence of pBM413, a novel multidrug resistance megaplasmid carrying qnrVC6 and blaIMP-45 from Pseudomonas aeruginosa. Int J Antimicrob Agents. 2018;51(1):145–150. doi: 10.1016/j.ijantimicag.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Sagai H, Hasuda K, Iyobe S, et al. Classification of R plasmids by incompatibility in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1976;10(4):573–578. doi: 10.1128/aac.10.4.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boronin AM. Diversity of Pseudomonas plasmids: to what extent? FEMS Microbiol Lett. 1992;100(1-3):461–467. doi: 10.1111/j.1574-6968.1992.tb14077.x. [DOI] [PubMed] [Google Scholar]
- 17.Alvarado A, Garcillán-Barcia MP, de La Cruz F. A degenerate primer MOB typing (DPMT) method to classify gamma-proteobac-terial plasmids in clinical and environmental settings. PLoS One. 2012;7(7):e40438–15. doi: 10.1371/journal.pone.0040438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wolter DJ, Hanson ND, Lister PD. Insertional inactivation of oprD in clinical isolates of Pseudomonas aeruginosa leading to carbapenem resistance. FEMS Microbiol Lett. 2004;236(1):137–143. doi: 10.1016/j.femsle.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 19.Rojo-Bezares B, Estepa V, Cebollada R, et al. Carbapenem-resistant Pseudomonas aeruginosa strains from a Spanish hospital: characterization of metallo-beta-lactamases, porin OprD and integrons. Int J Med Microbiol. 2014;304(3-4):405–414. doi: 10.1016/j.ijmm.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz-Martínez L, López-Jiménez L, D’Ostuni V, et al. A mechanism of carbapenem resistance due to a new insertion element (ISPa133) in Pseudomonas aeruginosa. Int Microbiol. 2011;14(1):51–58. doi: 10.2436/20.1501.01.135. [DOI] [PubMed] [Google Scholar]
- 21.Poole K, Krebes K, Mcnally C, Neshat S. Multiple antibiotic resistance in Pseudomonas aeruginosa: evidence for involvement of an efflux operon. J Bacteriol. 1993;175(22):7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poole K, Gotoh N, Tsujimoto H, et al. Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol. 1996;21(4):713–725. doi: 10.1046/j.1365-2958.1996.281397.x. [DOI] [PubMed] [Google Scholar]
- 23.Köhler T, Michéa-Hamzehpour M, Henze U, et al. Characterization of MexE-MexF-OprN, a positively regulated multidrug efflux system of Pseudomonas aeruginosa. Mol Microbiol. 1997;23(2):345–354. doi: 10.1046/j.1365-2958.1997.2281594.x. [DOI] [PubMed] [Google Scholar]
- 24.Aires JR, Köhler T, Nikaido H, Plésiat P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to amino-glycosides. Antimicrob Agents Chemother. 1999;43(11):2624–2628. doi: 10.1128/aac.43.11.2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chalhoub H, Sáenz Y, Rodríguez-Villalobos H, et al. High-level resistance to meropenem in clinical isolates of Pseudomonas aeruginosa in the absence of carbapenemases: role of active efflux and porin alterations. Int J Antimicrob Agents. 2016;48(6):740–743. doi: 10.1016/j.ijantimicag.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 26.Eshra KA, Shalaby MM. Efflux pump inhibition effect of curcumin and phenylalanine arginyl β- naphthylamide (PAβN) against multidrug resistant Pseudomonas aeruginosa isolated from burn infections in Tanta University Hospitals. Egypt J Med Microbiol. 2017;26(1):113–119. [Google Scholar]
- 27.Clinical and Laboratory Standards Institute (CLSI) M100 . Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. USA: Clinical and Laboratory Standards Institute; 2018. [Google Scholar]
- 28.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 29.Rojo-Bezares B, Estepa V, de Toro M, et al. A novel class 1 integron array carrying blaVIM-2 genes and a new insertion sequence in a Pseudomonas aeruginosa strain isolated from a Spanish hospital. J Med Microbiol. 2011;60(Pt 7):1053–1054. doi: 10.1099/jmm.0.030973-0. [DOI] [PubMed] [Google Scholar]
- 30.Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Curran B, Jonas D, Grundmann H, Pitt T, Dowson CG. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J Clin Microbiol. 2004;42(12):5644–5649. doi: 10.1128/JCM.42.12.5644-5649.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jolley KM. Pseudomonas aeruginosa MLST Database, University of Oxford BMC Bioinformatics. [Accessed June 10, 2018]. [updated. 2018 June 4; cited 2010 December 10]. Available from: https://pubmlst.org/paeruginosa/
- 33.Lévesque C, Piché L, Larose C, Roy PH, Ch L. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob Agents Chemother. 1995;39(1):185–191. doi: 10.1128/aac.39.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorgani N, Ahlbrand S, Patterson A, Pourmand N. Detection of point mutations associated with antibiotic resistance in Pseudomonas aeruginosa. Int J Antimicrob Agents. 2009;34(5):414–418. doi: 10.1016/j.ijantimicag.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gutiérrez O, Juan C, Cercenado E, et al. Molecular epidemiology and mechanisms of carbapenem resistance in Pseudomonas aeruginosa isolates from Spanish hospitals. Antimicrob Agents Chemother. 2007;51(12):4329–4335. doi: 10.1128/AAC.00810-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolter DJ, Hanson ND, Lister PD. Insertional inactivation of oprD in clinical isolates of Pseudomonas aeruginosa leading to carbapenem resistance. FEMS Microbiol Lett. 2004;236(1):137–143. doi: 10.1016/j.femsle.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 37.Kumita W, Saito R, Sato K, et al. Molecular characterizations of carbapenem and ciprofloxacin resistance in clinical isolates of Pseudomonas putida. J Infect Chemother. 2009;15(1):6–12. doi: 10.1007/s10156-008-0661-9. [DOI] [PubMed] [Google Scholar]
- 38.Green MR, Sambrook J. Molecular Cloning: A Laboratory Manual. 4th ed. New York: Cold Spring Harbor Laboratory Press; 2012. [Google Scholar]
- 39.Kaufmann ME. Pulsed-field gel electrophoresis. Methods Mol Med. 1998;15:33–50. doi: 10.1385/0-89603-498-4:33. [DOI] [PubMed] [Google Scholar]
- 40.Carattoli A, Bertini A, Villa L, et al. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63(3):219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 41.Oliver A. Epidemiology and carbapenem resistance mechanisms in Pseudomonas aeruginosa: role of high-risk clones in multidrug resistance. Enferm Infecc Microbiol Clin. 2017;35(3):137–138. doi: 10.1016/j.eimc.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Hanson ND, Hossain A, Buck L, Moland ES, Thomson KS. First occurrence of a Pseudomonas aeruginosa isolate in the United States producing an IMP metallo-beta-lactamase, IMP-18. Antimicrob Agents Chemother. 2006;50(6):2272–2273. doi: 10.1128/AAC.01440-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Castillo-Vera J, Ribas-Aparicio RM, Nicolau CJ, et al. Unusual diversity of acquired β-lactamases in multidrug-resistant Pseudomonas aeruginosa isolates in a Mexican hospital. Microb Drug Resist. 2012;18(5):471–478. doi: 10.1089/mdr.2011.0183. [DOI] [PubMed] [Google Scholar]
- 44.Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33(4):757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 45.Oliver A, Mulet X, López-Causapé C, Juan C. The increasing threat of Pseudomonas aeruginosa high-risk clones. Drug Resist Updat. 2015;22:21–22. 41–59. doi: 10.1016/j.drup.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 46.Espinosa-Camacho LF, Delgado G, Soberón-Chávez G, et al. Complete genome sequences of four extensively drug-resistant Pseudomonas aeruginosa strains, isolated from adults with ventilator-associated pneumonia at a tertiary referral hospital in Mexico City. Genome Announc. 2017;5(36):e00925-17-3. doi: 10.1128/genomeA.00925-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ocampo-Sosa AA, Cabot G, Rodríguez C, et al. Alterations of OprD in carbapenem-intermediate and -susceptible strains of Pseudomonas aeruginosa isolated from patients with bacteremia in a Spanish multicenter study. Antimicrob Agents Chemother. 2012;56(4):1703–1713. doi: 10.1128/AAC.05451-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Q, Ba Z, Wu G, et al. Insertion sequence ISRP10 inactivation of the oprD gene in imipenem-resistant Pseudomonas aeruginosa clinical isolates. Int J Antimicrob Agents. 2016;47(5):375–379. doi: 10.1016/j.ijantimicag.2016.02.008. [DOI] [PubMed] [Google Scholar]
- 49.Wolter DJ, Khalaf N, Robledo IE, et al. Surveillance of carbapenem-resistant Pseudomonas aeruginosa isolates from Puerto Rico Medical Center Hospitals: dissemination of KPC and IMP-18 beta-lactamases. Antimicrob Agents Chemother. 2009;53(4):1660–1664. doi: 10.1128/AAC.01172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodríguez-Martínez JM, Poirel L, Nordmann P. Extended-spectrum cephalosporinases in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2009;53(5):1766–1771. doi: 10.1128/AAC.01410-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Epp SF, Köhler T, Plésiat P, et al. C-terminal region of Pseudomonas aeruginosa outer membrane porin OprD modulates susceptibility to meropenem. Antimicrob Agents Chemother. 2001;45(6):1780–1787. doi: 10.1128/AAC.45.6.1780-1787.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.El’garch F, Jeannot K, Hocquet D, Llanes-Barakat C, Plésiat P. Cumulative effects of several nonenzymatic mechanisms on the resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother. 2007;51(3):1016–1021. doi: 10.1128/AAC.00704-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamers RP, Cavallari JF, Burrows LL. The efflux inhibitor phenylalanine-arginine beta-naphthylamide (PAβN) permeabilizes the outer membrane of gram-negative bacteria. PLoS One. 2013;8(3):e60666. doi: 10.1371/journal.pone.0060666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dai X, Zhou D, Xiong W, et al. The IncP-6 plasmid p10265-KPC from Pseudomonas aeruginosa carries a novel ΔISEc33-associated bla KPC-2 gene cluster. Front Microbiol. 2016;7:310. doi: 10.3389/fmicb.2016.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Popowska M, Krawczyk-Balska A. Broad-host-range IncP-1 plasmids and their resistance potential. Front Microbiol. 2013;4:44. doi: 10.3389/fmicb.2013.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bahl MI, Burmølle M, Meisner A, Hansen LH, Sørensen SJ. All IncP-1 plasmid subgroups, including the novel epsilon subgroup, are prevalent in the influent of a Danish wastewater treatment plant. Plasmid. 2009;62(2):134–139. doi: 10.1016/j.plasmid.2009.05.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dendrogram of 41 multidrug-resistant P. aeruginosa clinical isolates obtained in ISSSTE Regional Hospital of Puebla, Mexico.
Notes: The isolates showing a Dice coefficient ≥80% were considered as genetically related for this study. PFGE type (PT), PFGE clusters (A, B and C).
Abbreviation: PFGE, pulsed-field gel electrophoresis.
Plasmid extraction by the Quick Prep method of 17 clinical blaIMP-carrying P. aeruginosa isolates.
Notes: The position of chromosomal DNA is indicated by an arrow at the right. Line 1: Size marker; Line 2: PE06; Line 3: PE09; Line 4: PE15; Line 5: PE16; Line 6: PE20; Line 7: PE21; Line 8: PE22; Line 9: PE23; Line 10: PE27; Line 11: PE46; Line 12: PE52; Line 13: PE62, Line 14: PE63; Line 15: PE64; Line 16: PE73; Line 17: PE10; Line 18: PE83; Line 19: Control E. coli NCTC 50192; Line 20: P. aeruginosa PAO1.