Abstract
Sterol regulator element binding proteins (SREBPs) are a family of transcription factors involved in the biogenesis of cholesterol, fatty acids and triglycerides. They also regulate physiological functions of many organs, such as thyroid, brain, heart, pancreas and hormone synthesis. Beside the physiological effects, SREBPs participate in some pathological processes, diabetes, endoplasmic reticulum stress, atherosclerosis and chronic kidney disease associated with SREBP expression changes. In the liver, SREBPs are involved in the pathogenesis of nonalcoholic fatty liver disease, nonalcoholic steatohepatitis, hepatitis and hepatic cancer. There are several SREBP inhibitors that have potential for treating obesity, diabetes and cancer. This review assesses the recent findings about the roles of SREBPs in the physiology of organs’ function and pathogenesis of liver diseases.
Keywords: SREBP, Liver, Lipid
Introduction
The sterol regulator element binding proteins (SREBPs) are a family of transcription factors. These proteins are synthesized as 110-amino acid inactive precursors; then, they are inserted into the endoplasmic reticulum (ER) membrane.1 In the ER, SREBPs interact with a sterol sensor, the SREBP-cleavage activating protein (Scap).2 The SREBP/Scap complex moves to the Golgi apparatus, where the mature or nuclear forms of SREBP are generated by two proteases, the site 1 protease and the site 2 protease, and an anchoring protein. The insulin-induced gene (Insig)-1/2 also contributes.3 Then, the nuclear SREBPs translocate to the nucleus and bind to the target gene promoters, such as those of lipid metabolism-related genes.4 The expressions of these transcriptional genes regulated by feed-forward and feedback mechanisms (i.e. increased cholesterol in the cells) inhibit the proteolytic activation of SREBPs and decrease expression of SREBP target genes.5
The SREBP family consists of three members: SREBP-1a; SREBP-1c, from the SREBF-1 protein coding gene; and, SREBP-2, from the SREBF-2 protein coding gene.6,7 SREBP-1c is mainly expressed in the liver, white adipose tissue, adrenal gland, skeletal muscle and brain of mice and humans;7 but SREBP-1a is expressed in cell lines, spleen and intestinal tissues.8
Physiological function and regulation of SREBPs
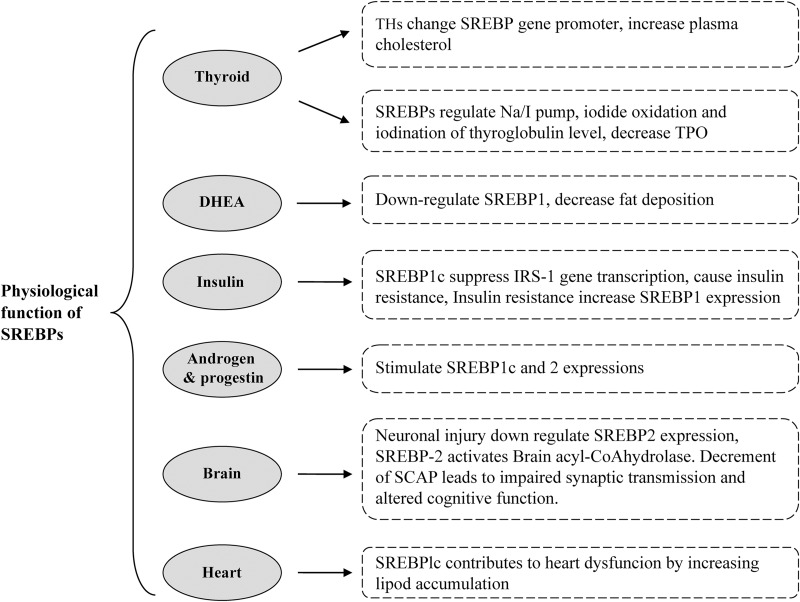
The SREBP family plays a key role in lipid homeostasis (cholesterol and triglyceride). Moreover, SREBPs are also involved in the normal functions of some organs. One of these organs is the thyroid. Thyroid hormones (THs) change the SREBP-2 gene promoter; then, the low-density lipoprotein (LDL) receptor expression deceases and plasma cholesterol level increases.9,10 SREBPs are also known as regulators of the Na/I pump, iodide oxidation and iodination of thyroglobulin in TH synthesis.11 Another study by Rauer et al.12 showed 25-hydroxycholesterol, as a SREBP-1c inhibitor, decreased the mRNA levels of thyroid peroxidase by 50%. Rochira and colleagues13 have also revealed that in HepG2 cells, 3,5,3’-triiodo-l-thyronine regulates SREBP-1 expression in a dose-/time-dependent manner. Their results also indicated that the AKT/PI3K signaling pathway may be involved in this process. The 3,5-diiodo-l-thyronine, unlike 3,5,3’-triiodo-l-thyronine, blocks SREBP-1c activation in HepG2 cells, activates β-oxidation and reduces lipogenic factor expression via nongenomic mechanisms.14 In this way, the findings show that THs can effect SREBP expression, while, on the other hand, SREBP-1 can affect the thyroid gland and decrease thyroid peroxidase levels. Dehydroepiandrosterone is one of the adrenal hormones; the reduction of which is associated with poor sexual function. Dehydroepiandrosterone stimulates intracellular activity of the cyclic adenosine 3’,5’-monophosphate and cyclic adenosine 3’,5’-monophosphate-dependent protein kinase A that leads to down-regulation of SREBP-1 and PPARα, and inactivation of carnitinepalmitoyltransferase, therefore decreasing fat deposition.15 Androgens and progesterone also stimulate SREBP-1c and 2 expressions in normal cell lines, increase mRNA and protein levels of fatty acid synthase (FAS), acetyl-CoA-carboxylase, acetyl-CoA-synthase and HMG-CoA-reductase.16 In this context, another organ affected by SREBPs is the pancreas, and its hormone, insulin. In insulin signaling, it has been demonstrated that SREBP-1c binding to the Irs-1 promoter region suppresses Irs-1 gene transcription that leads to insulin resistance in the skeletal muscle.17,18 Insulin resistance is also associated with increased AMP-activated protein kinase (AMPK)α phosphorylation, FAS, ChREBP and SREBP-1 mRNA expression.19 Activation of both PKβ/Akt and PKCk pathways lead to SREBP-1c expression increase.20
Indeed, the activated liver X receptor (LXR)α-C/EBPβ complex binds to the SREBP-1c promoter in sites that are required for insulin induction,21 and LXRα loss leads to decrement of SREBP-1c, FAS and lipogenic factors and improvement of insulin sensitivity in LXRαβ-deficient-ob/ob mice.22 But, Eberlé et al.1,21 did not find any effect of SREBP-1c stimulation, and determined that these effects were exclusive to SREBP-1a and SREBP-2. In another attempt, Cagen et al.21 have shown that insulin activates LXR, specificity protein 1 and nuclear factor-Y that are required for the full action of SREBP-1c. Taken together, the role of SREBPs in control of insulin signaling and secretion is important and inhibition of SREBP pathways may be a potential treatment in the future, possibly for obesity with type 2 diabetes.23,24
Brain is the most cholesterol-rich organ, and mostly de novo pathways synthesize cholesterol. Studies have shown SREBP-2 expression in the normal hippocampus, cortex and striatum;25 it has also been shown that neuronal injury induced by kainite results in a down-regulation of SREBP-2 expression in lesion areas of the brain.26 Other documents have reported that SREBP-2 expression is decreased in streptozotocin-induced mice and in Alzheimer’s disease.27 Reduction of the cholesterol sensor Scap in the brains of mice causes impaired synaptic transmission and altered cognitive function.28,29
24 S-hydroxycholesterol is an important metabolic product of cholesterol in the brain.37 Wang et al.30 showed that under cholesterol excess 24S-hydroxycholesterol is increased, acting as a sensor and reducing cholesterol synthesis enzymes through SREBP-2 down-regulation. Brain acyl-CoA hydrolase is responsible for hydrolyzing the long-chain acyl-CoA among neurons; it has been demonstrated that SREBP-2 binds to the sterol regulatory element motif, activating brain acyl-CoA hydrolase enzyme and thereby the conversion of long-chain acyl-CoAs to fatty acids and CoA-SH.31 In total, these research findings have provided insights into the interaction between SREBP-2 and cholesterol metabolisms in neurons.
In healthy people, lipid storage is minimal in the heart, but Marfella et al.32 reported a significant correlation between increase of SREBP-1c levels and increase of cardiomyocyte triglyceride accumulation among metabolic syndrome patients, which is associated with ejection fraction lowering and cardiac dysfunction. In addition, SREBP-1c gene silencing reduces triglyceride sand very low-density lipoprotein (VLDL) content in bovine hepatocytes33 (Fig. 1).
Fig. 1. Schematic diagram of the physiologic function of SREBP in several organs.
Interaction between SREBPs and other transcription factors
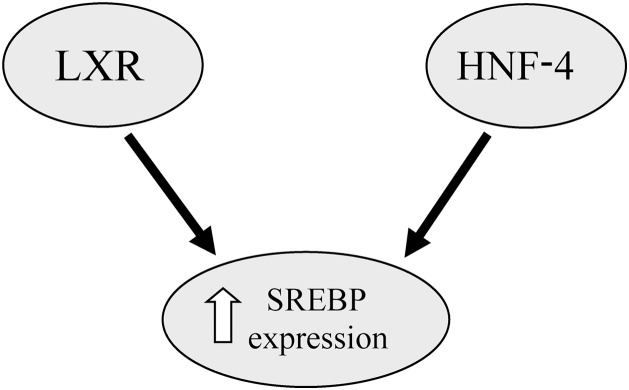
The LXRs, LXRα and LXRβ, are ligand-activated transcription factors and members of the DNA-binding transcription factors. Their functions are related to retinoid X receptors.34 The LXRs play an important role in cholesterol homeostasis and hepatic lipogenesis.35 LXRα allows for SREBP-1c induction.11 Studies have shown that activation of LXRα increases SREBP1c expression, which leads to hepatic lipogenesis and hypertriglyceridemia.36 In addition, insulin stimulates SREBP-1c expression through the nuclear receptor of LXR.37The hepatocyte nuclear factor-4 alpha (HNF-4α) is another nuclear receptor protein involved in hepatic lipid homeostasis through regulation of VLDL and apolipoprotein B function.37 In this context, Misawa et al.38,39 reported that overexpression of HNF-4 in HEK293 cells augmented the expression of SREBP-responsive genes and that it seemedlikeHNF-4 potentiates the SREBP functions and stimulates the expression of SREBP-responsive genes in the enterohepatic cells. Another study showed activation of SREBP2 blocking HNF-4α expression in the mouse liver.40 (Fig. 2)
Fig. 2. Diagram of the interaction between SREBP and transcription factors.
SREBPs in liver disease
Role of SREBPs in nonalcoholic fatty liver
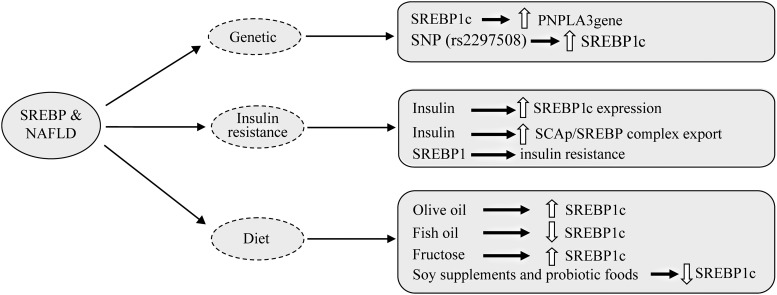
SREBPs are involved in some metabolic disorders, including obesity, type 2 diabetes, dyslipidemia, atherosclerosis, etc.41,42 Nonalcoholic fatty liver disease (NAFLD), the most common liver disease, is simple hepatic steatosis, and nonalcoholic steatohepatitis (NASH) is a developed form of NAFLD that is associated with hepatic inflammation and fibrosis. NASH could lead to cirrhosis and hepatocarcinoma.35 Genetic background, obesity and insulin resistance are considered contributing factors to the pathophysiology of NASH.43 Studies on single nucleotide polymorphisms (i.e. rs2297508) and SREBP-1 gene variations have shown a positive relation with increased risk of NAFLD development.37 Other documents have confirmed that patatin-like phospholipase3 (PNPLA3) plays a key role in NASH development as well44,45 and that PNPLA3gene polymorphisms are strongly associated with severity of NAFLD.46 SREBP-1c binds to the PNPLA3gene and activates its expression; then, PNPLA3 stimulates lipid accumulation, as shown in mouse hepatocytes.46,47
The stimulatory effects of insulin on lipogenesis in the liver and the adipose tissue are well known, but the cellular mechanisms remain unclear. To our knowledge, insulin effects on hepatic lipogenesis are partly mediated by SREB-1c, from SREBP-1c gene expression until entry to the cell nucleus.48 Indeed, insulin stimulates SREBP-1c gene expression, enhancing SCAP/SREBP complex export to the Golgi and proteolytic processing on the nascent SREBP-1c by reducing levels of Insig-1/2.49 Furthermore, we know that AMPK, an energy sensor for cellular energy of homeostasis, inhibits cleavage and transcriptional activation of SREBP via phosphorylation. In this regard, Li and colleagues41 reported that metformin stimulated AMPK activity; it also suppressed SREBP-1c cleavage and nuclear translocation via Ser372 phosphorylation, leading to liver steatosis attenuation in diet-induced insulin-resistant LDL receptor-deficient mice. On the other hand, there are many documents that provided evidence of SREBP-1c inducing lipogenic enzymes and causing lipid deposition associated with insulin resistance.50 Sun et al.51 found that early insulin therapy in type 2 diabetic rats leads to down-regulation of SREBP1 in hepatic tissue and fat shifting from liver to the adipose tissue.
Ding and colleagues52 have reported that curcumin inhibits SREBP expression, and improves serum lipid levels and insulin sensitivity in high-fat diet-induced obese mice. The current findings may consider two opposite roles for insulin in relation to SREBP1 that dependent on the tissue types, obese or nonobese, etc.8 In rats, injection of leptin leads to fat deposition with up-regulation of SREBP-1 though the JAK2-STAT3/PI3K signalingpathway.53
Obesity is another feature of NAFLD pathogenesis. Western diets and modern diets are factors causing obesity and fatty liver diseases. High-fat diet, especially with different fat sources, usually induces obesity and lipid accumulation in the hepatocytes and adipocytes.54 In this context, Ronis et al.55 reported that olive oil increased SREBP1c expression greater than corn oil or echium oil in overfed male rats. Another study showed that docosahexaenoic acid/eicosapentaenoic acid at a 1:2 ratio decreased serum triglycerides, total cholesterol, and LDL-cholesterol levels, lowered SREBP-1C and FAS mRNA expression and alleviated liver damage in mice. It seems that this process is probably mediated by both activation of AMPK and inhibition of mechanistic target of rapamycin complex 1.56,57 Moreover, it has been reported that dietary consumption of fish oil as a source of n-3 polyunsaturated fatty acids, down-regulates SREBP1c mRNA expression but probably does not influence SREBP-2 expression.58 Obesity can also cause insulin resistance, increased leptin levels and, importantly, stimulation of SREBP1c expression.59
Today, fructose has markedly increased in our diet and overconsumption of fructose has also been shown to stimulate SREBP1c expression and lead to hepatic lipid accumulation.60 In contrast, some foods such as soy supplements and probiotic foods are considered to decease insulin secretion, and to suppress SREBP1c expression and enzymes involving lipid synthesis61 (Fig. 3).
Fig. 3. Diagram of the involvement of SREBPs in nonalcoholic fatty liver disease.
Role of SREBPs in ER stress
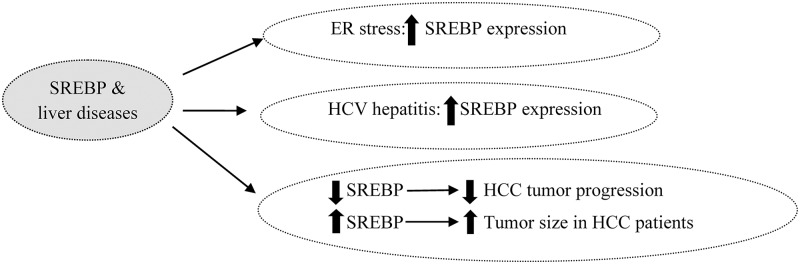
ER, a critical membranous organelle, plays a key role in lipid synthesis, nascent protein folding and Ca+2 ion storage.62 In special circumstances, such as pharmacological stimuli, oxidative stress, viral infections and dietary demands, ER homeostasis can disrupt and create an ER stress phenomenon63 that causes abnormalities in insulin action, inflammatory responses, lipoprotein B100 degradation and hepatic lipogenesis.64 Three ER transmembrane sensors, inositol-requiring protein 1, protein kinase-like ER kinase and activating transcription factor 6, are activated by the glucose-regulated protein 78 required for folding of proteins in the ER. Tunicamycin, cow milk casein or oxidative stress induce acute ER stress,65,66 while overfeeding of fatty acids, cholesterol and fructose, due to the produced obesity and insulin resistance, induce chronic ER stress that is not fully restored.67 Many studies have shown that ER stress and glucose-regulated protein 78 activation up-regulate SREBPs directly and indirectly.68
Recently, it has been reported that elevated uric acid levels are associated with ER stress induction and hepatic lipid accumulation via SREBP-1c activation.69 One of the ω-3 polyunsaturated fatty acid derivatives and resolving D1, alleviated tunicamycin-induced ER stress and decreased SREBP1c via inhibition of c-Jun N-terminal kinase expression in hepatocytes.20,70 In contrast, AMPK activation down-regulates SREBP 1c expression and alleviates ER stress response through suppression of mechanistic target of rapamycin complex 1 signaling in the ER stress-induced hepatocytes.41 In the study by Sun et al.71, early insulin therapy reduced c-Jun N-terminal kinase and IRS-1 expression; it also improved ER stress and steatosis in the rat liver. Several pharmacologic agents, including rosiglitazone, naltrexone and tauroursodeoxycholic acid, also attenuated ER stress liver injury and down-regulated both SREBP1 and SREBP2 expression.72,73 (Fig. 4).
Fig. 4. Diagram of the role of SREBPs in endoplasmic reticulum stress, hepatitis C virus infection and hepatic cancer.
Role of SREBPs in hepatitis disease
Hepatitis C virus (HCV)-2 infection markedly leads to chronic hepatitis, liver cirrhosis and possibly hepatocellular carcinoma.74 Liver steatosis is frequently found in the patients who are infected with HCV, but the molecular mechanisms of HCV-associated steatosis are not clear. Since SREBPs are the key transcriptional factors in lipogenic gene expression, they are likely important in the HCV-induced liver steatosis.75 Some studies have shown that HCV nonstructural protein 2, HCV nonstructural 4B protein and HCV-3a core protein increase SREBPs expression.76 These results suggest activation of the PI3K and Akt-2 pathway, enhancement of HCV entry, replication and translation of HCV, increased SREBP-1 activityandsteatosis.77
Contrary to these studies, McPherson et al.78 have reported that among HCV patients, there was no significant difference in the hepatic expression of SREBP-1c or FAS mRNA compared with normal subjects. Moreover, a negative relationship was found between hepatic SREBP-1c mRNA expression and grade of steatosis. These findings state that SREBP-1c may play transient and not a prominent role in HCV-related steatosis.20 In other research, Kim et al.79 found that curcumin decreases HCV gene expression via suppression of the Akt-SREBP-1 pathway. Another study indicated that, among patients with HCV infection, although SREBP-2 expression was unchanged, HMG-CoA reductase, HMG-CoA synthase and SREBP-1c expression was up-regulated.80
Subtilisinkexin isozyme-1/sphingosine 1-phosphate is recognized as a novel regulator of the HCV lifecycle. In hepatomacells, subtilisinkexin isozyme-1/sphingosine 1-phosphate-specific protein-based inhibitor blocked HCV from establishing infection and reduced lipid droplets; it could be considered as a therapeutic target against HCV infection and liver steatosis, though more evidence is required to support this hypothesis.81 These studies indicate that more evidence is also required to clarify the role of SREBP-1 in patients with HCV. Silent information regulator 1 (SIRT1) is another regulator of hepatic lipid metabolism. In 2013, Sun and colleagues82 found that HCV replication inHuh-7.5 cells decreased SIRT1, up-regulated SREBP-1c, FAS, ACC and SREBP-2, and increased lipid profile. Recently, it has been reported that retinoid-interferon-induced mortality 19 reduces HCV-infected Huh7 cells and that activation of retinoid-interferon-induced mortality 19 attenuates intracellular lipid droplets through a decrease of SREBP-1c83 (Fig. 4).
SREBP inhibitor agents
Nowadays, research on SREBP inhibitory agents is being conducted. Inhibitors of SREBP processing can be helpful in reducing the risk of atherosclerosis, metabolic syndrome and obesity.84 There are several SREBP inhibitors, such as 24-HC, 25-HC, 27-HC, etc. But since LXR up-regulates SREBP expression, it’s important that inhibitors of SREBP do not activate LXR expression.57,58 Botulin, as a specific inhibitor of SREBP, suppresses SREBP maturation and decreases cholesterol and fatty acid synthesis.59 In this context, Quan and colleagues85 reported that betulinic acid reduced hepatic steatosis and levels of SREBP1 in HepG2 cells and in livers of mice that were fed a high-fat diet. They also showed that this function is mediated by the Ca(+2)-calmodulin dependent protein kinase-AMPK-SREBP1 signaling pathway.
Another agent is fatostatin, that interacts with Scap, blocks ER-Golgi translocation of SREBP and decreases blood glucose and liver steatosis in obese ob/ob mice.86 Fatostatin also has antitumor properties and inhibits cell growth.87 Other natural compounds, including and rographolide and anhydroicaritin, ameliorate obesity, insulin resistance, liver steatosis and hyperlipemia via suppression of SREBP activation.88 Emodin from the Rheum palmatum herb has anti-inflammatory and anticancer effects, and attenuates obesity but decreases insulin sensitivity through regulation of the SREBP pathway.89
However, it has been revealed that antipsychotic drugs, such as clozapine, olanzapine and haloperidol, increase lipogenesis gene expression via SREBP activation; although, there are some differences in rates of SREBP activation among these drugs.90
Role of SREBPs in cancer
Cholesterol and lipid are requirements for new membrane building and for maintaining active signaling in developing cancer cells;91 therefore, lipid metabolism-related transcriptional genes and their enzymes change in carcinoma cells. Hepatocellular carcinoma (HCC) is one of the most common liver malignancies in the world.84 Many research studies have demonstrated that SREBP up-regulation, and FAS and LDL receptor overexpression occur in prostate, breast and glioblastoma.92,93 Previously, Li et al.94 demonstrated that overexpression of SREBP-1 is associated with large tumor size, high histological grade and advanced tumor-node-metastasis stage in HCC patients, and that SREBP-1 down-regulation suppressed cell proliferation and apoptosis in both HepG2 and MHCC97L cells.94
One of the effects of SREBP-2 in cell proliferation is mediated by regulation of farnesyldiphosphate synthase gene transcription.95 It has also been shown that tocotrienol (a minor form of vitamin E) reduced SREBP-2 activity and improved cell viability in prostate cancer cells.96 Obesity, fatty liver and hepatitis have potential roles in pathogenesis of HCC.97 In this regard, Zhang et al.98 have indicated that miR-449 inhibits SIRT1, and decreases SREBP1c expression and that of downstream target genes, including fatty acid synthase and 3-hydroxy-3-methylglutaryl CoA reductase. Also, miR-449 can repress DNA synthesis and proliferation, both in HepG2 and Huh7 cells.98
Overexpression of NS5ABP37protein, a HCC oncogenomic screen and hepatitis C virus nonstructural protein 5A‐associated binding protein, decreased intracellular triglyceride and total cholesterol contents, down-regulated SREBP1c and SREBP2 expression and inhibited cancer cell proliferation in human hepatoma cells.99 Moreover, SREBP pathway blocking by L-Scap-/- and L-gp78-/- mice led to reduced SREBP1c, SREBP1a and SREBP2 expression and to decrease in related enzymes, such as fatty acid synthase, ACC, LDLR and HMGCs, improving HCC tumor progression.100 In human NAFLD-associated HCC, SREBP-1 up-regulated HDAC8 and suppression of the histone deacetylase HDAC8, and decreased insulin resistance and NAFLD-associated HCC.101
Among SREBP inhibitor agents, it has been shown that fatostatin, andrographolides and silibinin have anticancer effects. Li et al.87 and other researchers have reported that fatostatin blocks cell proliferation through lipid-independent and Scap-independent mechanisms, causing G2-M cell-cycle arrest and inducing apoptosis. Silibinin also has been shown to induce apoptosis in prostatic cancer cells.102 (Fig. 4)
Conclusions
SREBPs (SREBP1c, SREBP1a and SREBP2) are found in several organs and participate in physiologic and pathologic functions of the body. SREBPs are involved in lipid and glucose homeostasis as well as hormones synthesis. SREBPs also have a key role in the pathogenesis of NASH, obesity and cancers. Moreover, they cause lipid-regulated cellular disorders in hepatocytes that can lead to steatosis and liver injury. Several SREBP inhibitors, including natural and synthetic agents, have potential treatment effects in obesity, hepatic steatosis and even tumor cells.
Abbreviations
- AMPK
AMP-activated protein kinase
- ER
endoplasmic reticulum
- FAS
fatty acid synthase
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- HNF-4α
hepatocyte nuclear factor-4 alpha
- Insig
insulin-induced gene
- LDL
low-density lipoprotein
- LXR
liver X receptor
- NAFLD
nonalcoholic fatty liver disease
- NASH
nonalcoholic steatohepatitis
- PNPLA2
patatin-like phospholipase3
- S1-p
sphingosine1-phosphate
- Scap
SREBP-cleavage activating protein
- SIRT1
silent information regulator 1
- SREBP
sterol regulator element binding proteins
- TH
thyroid hormone
- VLDL
very low-density lipoprotein
References
- 1.Eberlé D, Hegarty B, Bossard P, Ferré P, Foufelle F. SREBP transcription factors: master regulators of lipid homeostasis. Biochimie. 2004;86:839–848. doi: 10.1016/j.biochi.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 2.Edwards PA, Tabor D, Kast HR, Venkateswaran A. Regulation of gene expression by SREBP and SCAP. Biochim Biophys Acta. 2000;1529:103–113. doi: 10.1016/S1388-1981(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 3.Gong Y, Lee JN, Lee PC, Goldstein JL, Brown MS, Ye J. Sterol-regulated ubiquitination and degradation of Insig-1 creates a convergent mechanism for feedback control of cholesterol synthesis and uptake. Cell Metab. 2006;3:15–24. doi: 10.1016/j.cmet.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 4.Bengoechea-Alonso MT, Ericsson J. SREBP in signal transduction: cholesterol metabolism and beyond. Curr Opin Cell Biol. 2007;19:215–222. doi: 10.1016/j.ceb.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shimano H. SREBPs: physiology and pathophysiology of the SREBP family. FEBS J. 2009;276:616–621. doi: 10.1111/j.1742-4658.2008.06806.x. [DOI] [PubMed] [Google Scholar]
- 7.Deng X, Cagen LM, Wilcox HG, Park EA, Raghow R, Elam MB. Regulation of the rat SREBP-1c promoter in primary rat hepatocytes. Biochem Biophys Res Commun. 2002;290:256–262. doi: 10.1006/bbrc.2001.6148. [DOI] [PubMed] [Google Scholar]
- 8.Ferré P, Foufelle F. SREBP-1c transcription factor and lipid homeostasis: clinical perspective. Horm Res. 2007;68:72–82. doi: 10.1159/000100426. [DOI] [PubMed] [Google Scholar]
- 9.Lopez D, Abisambra Socarrás JF, Bedi M, Ness GC. Activation of the hepatic LDL receptor promoter by thyroid hormone. Biochim Biophys Acta. 2007;1771:1216–1225. doi: 10.1016/j.bbalip.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 10.Shin DJ, Osborne TF. Thyroid hormone regulation and cholesterol metabolism are connected through Sterol Regulatory Element-Binding Protein-2 (SREBP-2) J Biol Chem. 2003;278:34114–34118. doi: 10.1074/jbc.M305417200. [DOI] [PubMed] [Google Scholar]
- 11.Ringseis R, Rauer C, Rothe S, Gessner DK, Schütz LM, Luci S, et al. Sterol regulatory element-binding proteins are regulators of the NIS gene in thyroid cells. Mol Endocrinol. 2013;27:781–800. doi: 10.1210/me.2012-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rauer C, Ringseis R, Rothe S, Wen G, Eder K. Sterol regulatory element-binding proteins are regulators of the rat thyroid peroxidase gene in thyroid cells. PLoS One. 2014;9:e91265. doi: 10.1371/journal.pone.0091265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gnoni GV, Rochira A, Leone A, Damiano F, Marsigliante S, Siculella L. 3,5,3’triiodo-L-thyronine induces SREBP-1 expression by non-genomic actions in human HEP G2 cells. J Cell Physiol. 2012;227:2388–2397. doi: 10.1002/jcp.22974. [DOI] [PubMed] [Google Scholar]
- 14.Rochira A, Damiano F, Marsigliante S, Gnoni GV, Siculella L. 3,5-Diiodo-l-thyronine induces SREBP-1 proteolytic cleavage block and apoptosis in human hepatoma (Hepg2) cells. Biochim Biophys Acta. 2013;1831:1679–1689. doi: 10.1016/j.bbalip.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 15.Tang X, Ma H, Shen Z, Zou S, Xu X, Lin C. Dehydroepiandrosterone activates cyclic adenosine 3’,5’-monophosphate/protein kinase A signalling and suppresses sterol regulatory element-binding protein-1 expression in cultured primary chicken hepatocytes. Br J Nutr. 2009;102:680–686. doi: 10.1017/S0007114509289021. [DOI] [PubMed] [Google Scholar]
- 16.Heemers H, Maes B, Foufelle F, Heyns W, Verhoeven G, Swinnen JV. Androgens stimulate lipogenic gene expression in prostate cancer cells by activation of the sterol regulatory element-binding protein cleavage activating protein/sterol regulatory element-binding protein pathway. Mol Endocrinol. 2001;15:1817–1828. doi: 10.1210/mend.15.10.0703. [DOI] [PubMed] [Google Scholar]
- 17.Ide T, Shimano H, Yahagi N, Matsuzaka T, Nakakuki M, Yamamoto T, et al. SREBPs suppress IRS-2-mediated insulin signalling in the liver. Nat Cell Biol. 2004;6:351–357. doi: 10.1038/ncb1111. [DOI] [PubMed] [Google Scholar]
- 18.Repa JJ, Liang G, Ou J, Bashmakov Y, Lobaccaro JM, Shimomura I, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ben Djoudi Ouadda A, Levy E, Ziv E, Lalonde G, Sané AT, Delvin E, et al. Increased hepatic lipogenesis in insulin resistance and Type 2 diabetes is associated with AMPK signalling pathway up-regulation in Psammomys obesus. Biosci Rep. 2009;29:283–292. doi: 10.1042/BSR20080141. [DOI] [PubMed] [Google Scholar]
- 20.Ferré P, Foufelle F. Hepatic steatosis: a role for de novo lipogenesis and the transcription factor SREBP-1c. Diabetes Obes Metab. 2010;12(Suppl 2):83–92. doi: 10.1111/j.1463-1326.2010.01275.x. [DOI] [PubMed] [Google Scholar]
- 21.Cagen LM, Deng X, Wilcox HG, Park EA, Raghow R, Elam MB. Insulin activates the rat sterol-regulatory-element-binding protein 1c (SREBP-1c) promoter through the combinatorial actions of SREBP, LXR, Sp-1 and NF-Y cis-acting elements. Biochem J. 2005;385:207–216. doi: 10.1042/BJ20040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beaven SW, Matveyenko A, Wroblewski K, Chao L, Wilpitz D, Hsu TW, et al. Reciprocal regulation of hepatic and adipose lipogenesis by liver X receptors in obesity and insulin resistance. Cell Metab. 2013;18:106–117. doi: 10.1016/j.cmet.2013.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kahn BB, Flier JS. Obesity and insulin resistance. J Clin Invest. 2000;106:473–481. doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saltiel AR, Kahn CR. Insulin signalling and the regulation of glucose and lipid metabolism. Nature. 2001;414:799–806. doi: 10.1038/414799a. [DOI] [PubMed] [Google Scholar]
- 25.Ong WY, Hu CY, Soh YP, Lim TM, Pentchev PG, Patel SC. Neuronal localization of sterol regulatory element binding protein-1 in the rodent and primate brain: a light and electron microscopic immunocytochemical study. Neuroscience. 2000;97:143–153. doi: 10.1016/S0306-4522(00)00031-2. [DOI] [PubMed] [Google Scholar]
- 26.Kim JH, Ong WY. Localization of the transcription factor, sterol regulatory element binding protein-2 (SREBP-2) in the normal rat brain and changes after kainate-induced excitotoxic injury. J Chem Neuroanat. 2009;37:71–77. doi: 10.1016/j.jchemneu.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Barbero-Camps E, Fernández A, Martínez L, Fernández-Checa JC, Colell A. APP/PS1 mice overexpressing SREBP-2 exhibit combined Aβ accumulation and tau pathology underlying Alzheimer’s disease. Hum Mol Genet. 2013;22:3460–3476. doi: 10.1093/hmg/ddt201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki R, Ferris HA, Chee MJ, Maratos-Flier E, Kahn CR. Reduction of the cholesterol sensor SCAP in the brains of mice causes impaired synaptic transmission and altered cognitive function. PLoS Biol. 2013;11:e1001532. doi: 10.1371/journal.pbio.1001532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stoeckel LE, Arvanitakis Z, Gandy S, Small D, Kahn CR, Pascual-Leone A, et al. Complex mechanisms linking neurocognitive dysfunction to insulin resistance and other metabolic dysfunction. F1000Res. 2016;5:353. doi: 10.12688/f1000research.8300.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y, Muneton S, Sjövall J, Jovanovic JN, Griffiths WJ. The effect of 24S-hydroxycholesterol on cholesterol homeostasis in neurons: quantitative changes to the cortical neuron proteome. J Proteome Res. 2008;7:1606–1614. doi: 10.1021/pr7006076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takagi M, Suto F, Suga T, Yamada J. Sterol Regulatory Element-Binding Protein-2 modulates human brain acyl-CoA hydrolase gene transcription. Mol Cell Biochem. 2005;275:199–206. doi: 10.1007/s11010-005-1990-y. [DOI] [PubMed] [Google Scholar]
- 32.Coppola A, Marfella R, Coppola L, Tagliamonte E, Fontana D, Liguori E, et al. Effect of weight loss on coronary circulation and adiponectin levels in obese women. Int J Cardiol. 2009;134:414–416. doi: 10.1016/j.ijcard.2007.12.087. [DOI] [PubMed] [Google Scholar]
- 33.Deng Q, Li X, Fu S, Yin L, Zhang Y, Wang T, et al. SREBP-1c gene silencing can decrease lipid deposits in bovine hepatocytes cultured in vitro. Cell Physiol Biochem. 2014;33:1568–1578. doi: 10.1159/000358720. [DOI] [PubMed] [Google Scholar]
- 34.Schulman IG. Liver X receptors link lipid metabolism and inflammation. FEBS Lett. 2017;591:2978–2991. doi: 10.1002/1873-3468.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma Z, Deng C, Hu W, Zhou J, Fan C, Di S, et al. Liver X receptors and their agonists: targeting for cholesterol homeostasis and cardiovascular diseases. Curr Issues Mol Biol. 2017;22:41–64. doi: 10.21775/cimb.022.041. [DOI] [PubMed] [Google Scholar]
- 36.Faulds MH, Zhao C, Dahlman-Wright K. Molecular biology and functional genomics of liver X receptors (LXR) in relationship to metabolic diseases. Curr Opin Pharmacol. 2010;10:692–697. doi: 10.1016/j.coph.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Ponugoti B, Fang S, Kemper JK. Functional interaction of hepatic nuclear factor-4 and peroxisome proliferator-activated receptor-gamma coactivator 1alpha in CYP7A1 regulation is inhibited by a key lipogenic activator, sterol regulatory element-binding protein-1c. Mol Endocrinol. 2007;21:2698–2712. doi: 10.1210/me.2007-0196. [DOI] [PubMed] [Google Scholar]
- 38.Misawa K, Horiba T, Arimura N, Hirano Y, Inoue J, Emoto N, et al. Sterol regulatory element-binding protein-2 interacts with hepatocyte nuclear factor-4 to enhance sterol isomerase gene expression in hepatocytes. J Biol Chem. 2003;278:36176–36182. doi: 10.1074/jbc.M302387200. [DOI] [PubMed] [Google Scholar]
- 39.Tarling E, Salter A, Bennett A. Transcriptional regulation of human SREBP-1c (sterol-regulatory-element-binding protein-1c): a key regulator of lipogenesis. Biochem Soc Trans. 2004;32:107–109. doi: 10.1042/bst0320107. [DOI] [PubMed] [Google Scholar]
- 40.Yin L, Ma H, Ge X, Edwards PA, Zhang Y. Hepatic hepatocyte nuclear factor 4α is essential for maintaining triglyceride and cholesterol homeostasis. Arterioscler Thromb Vasc Biol. 2011;31:328–336. doi: 10.1161/ATVBAHA.110.217828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eberlé D, Clément K, Meyre D, Sahbatou M, Vaxillaire M, Le Gall A, et al. SREBF-1 gene polymorphisms are associated with obesity and type 2 diabetes in French obese and diabetic cohorts. Diabetes. 2004;53:2153–2157. doi: 10.2337/diabetes.53.8.2153. [DOI] [PubMed] [Google Scholar]
- 43.Luyckx FH, Lefebvre PJ, Scheen AJ. Non-alcoholic steatohepatitis: association with obesity and insulin resistance, and influence of weight loss. Diabetes Metab. 2000;26:98–106. [PubMed] [Google Scholar]
- 44.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dongiovanni P, Donati B, Fares R, Lombardi R, Mancina RM, Romeo S, et al. PNPLA3 I148M polymorphism and progressive liver disease. World J Gastroenterol. 2013;19:6969–6978. doi: 10.3748/wjg.v19.i41.6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 47.Qiao A, Liang J, Ke Y, Li C, Cui Y, Shen L, et al. Mouse patatin-like phospholipase domain-containing 3 influences systemic lipid and glucose homeostasis. Hepatology. 2011;54:509–521. doi: 10.1002/hep.24402. [DOI] [PubMed] [Google Scholar]
- 48.Foretz M, Guichard C, Ferré P, Foufelle F. Sterol regulatory element binding protein-1c is a major mediator of insulin action on the hepatic expression of glucokinase and lipogenesis-related genes. Proc Natl Acad Sci U S A. 1999;96:12737–12742. doi: 10.1073/pnas.96.22.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yellaturu CR, Deng X, Park EA, Raghow R, Elam MB. Insulin enhances the biogenesis of nuclear sterol regulatory element-binding protein (SREBP)-1c by posttranscriptional down-regulation of Insig-2A and its dissociation from SREBP cleavage-activating protein (SCAP).SREBP-1c complex. J Biol Chem. 2009;284:31726–31734. doi: 10.1074/jbc.M109.050914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shimomura I, Hammer RE, Richardson JA, Ikemoto S, Bashmakov Y, Goldstein JL, et al. Insulin resistance and diabetes mellitus in transgenic mice expressing nuclear SREBP-1c in adipose tissue: model for congenital generalized lipodystrophy. Genes Dev. 1998;12:3182–3194. doi: 10.1101/gad.12.20.3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun WP, Bi Y, Liang H, Cai MY, Chen X, Zhu YH, et al. Effects of early insulin therapy on sterol regulatory element binding protein 1 pathway and lipid accumulation in liver of type 2 diabetic rats. Zhonghua Yi Xue Za Zhi. 2011;91:1809–1812. [PubMed] [Google Scholar]
- 52.Ding L, Li J, Song B, Xiao X, Zhang B, Qi M, et al. Curcumin rescues high fat diet-induced obesity and insulin sensitivity in mice through regulating SREBP pathway. Toxicol Appl Pharmacol. 2016;304:99–109. doi: 10.1016/j.taap.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 53.Wu L, Chen G, Liu W, Yang X, Gao J, Huang L, et al. Intramuscular injection of exogenous leptin induces adiposity, glucose intolerance and fatty liver by repressing the JAK2-STAT3/PI3K pathway in a rat model. Gen Comp Endocrinol. 2017;252:88–96. doi: 10.1016/j.ygcen.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 54.Meng X, Zou D, Shi Z, Duan Z, Mao Z. Dietary diacylglycerol prevents high-fat diet-induced lipid accumulation in rat liver and abdominal adipose tissue. Lipids. 2004;39:37–41. doi: 10.1007/s11745-004-1199-1. [DOI] [PubMed] [Google Scholar]
- 55.Ronis MJ, Baumgardner JN, Marecki JC, Hennings L, Wu X, Shankar K, et al. Dietary fat source alters hepatic gene expression profile and determines the type of liver pathology in rats overfed via total enteral nutrition. Physiol Genomics. 2012;44:1073–1089. doi: 10.1152/physiolgenomics.00069.2012. [DOI] [PubMed] [Google Scholar]
- 56.Kobatake Y, Kuroda K, Jinnouchi H, Nishide E, Innami S. Differential effects of dietary eicosapentaenoic and docosahexaenoic fatty acids on lowering of triglyceride and cholesterol levels in the serum of rats on hypercholesterolemic diet. J Nutr Sci Vitaminol (Tokyo) 1984;30:357–372. doi: 10.3177/jnsv.30.357. [DOI] [PubMed] [Google Scholar]
- 57.Mori TA, Burke V, Puddey IB, Watts GF, O’Neal DN, Best JD, et al. Purified eicosapentaenoic and docosahexaenoic acids have differential effects on serum lipids and lipoproteins, LDL particle size, glucose, and insulin in mildly hyperlipidemic men. Am J Clin Nutr. 2000;71:1085–1094. doi: 10.1093/ajcn/71.5.1085. [DOI] [PubMed] [Google Scholar]
- 58.Nakatani T, Kim HJ, Kaburagi Y, Yasuda K, Ezaki O. A low fish oil inhibits SREBP-1 proteolytic cascade, while a high-fish-oil feeding decreases SREBP-1 mRNA in mice liver: relationship to anti-obesity. J Lipid Res. 2003;44:369–379. doi: 10.1194/jlr.M200289-JLR200. [DOI] [PubMed] [Google Scholar]
- 59.Petersen KF, Oral EA, Dufour S, Befroy D, Ariyan C, Yu C, et al. Leptin reverses insulin resistance and hepatic steatosis in patients with severe lipodystrophy. J Clin Invest. 2002;109:1345–1350. doi: 10.1172/JCI15001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang C, Chen X, Zhu RM, Zhang Y, Yu T, Wang H, et al. Endoplasmic reticulum stress is involved in hepatic SREBP-1c activation and lipid accumulation in fructose-fed mice. Toxicol Lett. 2012;212:229–240. doi: 10.1016/j.toxlet.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 61.Shukla A, Brandsch C, Bettzieche A, Hirche F, Stangl GI, Eder K. Isoflavone-poor soy protein alters the lipid metabolism of rats by SREBP-mediated down-regulation of hepatic genes. J Nutr Biochem. 2007;18:313–321. doi: 10.1016/j.jnutbio.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 62.Michalak M, Robert Parker JM, Opas M. Ca2+ signaling and calcium binding chaperones of the endoplasmic reticulum. Cell Calcium. 2002;32:269–278. doi: 10.1016/S0143416002001884. [DOI] [PubMed] [Google Scholar]
- 63.Moslehi A, Nabavizadeh F, Zekri A, Amiri F. Naltrexone changes the expression of lipid metabolism-related proteins in the endoplasmic reticulum stress induced hepatic steatosis in mice. Clin Exp Pharmacol Physiol. 2017;44:207–212. doi: 10.1111/1440-1681.12695. [DOI] [PubMed] [Google Scholar]
- 64.Moslehi A, Farahabadi M, Chavoshzadeh SA, Barati A, Ababzadeh S, Mohammadbeigi A. The effect of amygdalin on endoplasmic reticulum (ER) stress induced hepatic steatosis in mice. Malays J Med Sci. 2018;25:16–23. doi: 10.21315/mjms2018.25.1.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malhotra JD, Kaufman RJ. The endoplasmic reticulum and the unfolded protein response. Semin Cell Dev Biol. 2007;18:716–731. doi: 10.1016/j.semcdb.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lai E, Teodoro T, Volchuk A. Endoplasmic reticulum stress: signaling the unfolded protein response. Physiology (Bethesda) 2007;22:193–201. doi: 10.1152/physiol.00050.2006. [DOI] [PubMed] [Google Scholar]
- 67.Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 68.Kammoun HL, Chabanon H, Hainault I, Luquet S, Magnan C, Koike T, et al. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest. 2009;119:1201–1215. doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Choi YJ, Shin HS, Choi HS, Park JW, Jo I, Oh ES, et al. Uric acid induces fat accumulation via generation of endoplasmic reticulum stress and SREBP-1c activation in hepatocytes. Lab Invest. 2014;94:1114–1125. doi: 10.1038/labinvest.2014.98. [DOI] [PubMed] [Google Scholar]
- 70.Ota T, Gayet C, Ginsberg HN. Inhibition of apolipoprotein B100 secretion by lipid-induced hepatic endoplasmic reticulum stress in rodents. J Clin Invest. 2008;118:316–332. doi: 10.1172/JCI32752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun W, Bi Y, Liang H, Cai M, Chen X, Zhu Y, et al. Inhibition of obesity-induced hepatic ER stress by early insulin therapy in obese diabetic rats. Endocrine. 2011;39:235–241. doi: 10.1007/s12020-010-9429-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Moslehi A, Nabavizadeh F, Dehpour AR, Tavanga SM, Hassanzadeh G, Zekri A, et al. Naltrexone attenuates endoplasmic reticulum stress induced hepatic injury in mice. Acta Physiol Hung. 2014;101:341–352. doi: 10.1556/APhysiol.101.2014.3.9. [DOI] [PubMed] [Google Scholar]
- 73.Basseri S, Austin RC. Endoplasmic reticulum stress and lipid metabolism: mechanisms and therapeutic potential. Biochem Res Int. 2012;2012:841362. doi: 10.1155/2012/841362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lonardo A, Adinolfi LE, Loria P, Carulli N, Ruggiero G, Day CP. Steatosis and hepatitis C virus: mechanisms and significance for hepatic and extrahepatic disease. Gastroenterology. 2004;126:586–597. doi: 10.1053/j.gastro.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 75.Waris G, Felmlee DJ, Negro F, Siddiqui A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J Virol. 2007;81:8122–8130. doi: 10.1128/JVI.00125-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 76.Jackel-Cram C, Qiao L, Xiang Z, Brownlie R, Zhou Y, Babiuk L, et al. Hepatitis C virus genotype-3a core protein enhances sterol regulatory element-binding protein-1 activity through the phosphoinositide 3-kinase-Akt-2 pathway. J Gen Virol. 2010;91:1388–1395. doi: 10.1099/vir.0.017418-0. [DOI] [PubMed] [Google Scholar]
- 77.Shi Q, Hoffman B, Liu Q. PI3K-Akt signaling pathway upregulates hepatitis C virus RNA translation through the activation of SREBPs. Virology. 2016;490:99–108. doi: 10.1016/j.virol.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 78.McPherson S, Jonsson JR, Barrie HD, O’Rourke P, Clouston AD, Powell EE. Investigation of the role of SREBP-1c in the pathogenesis of HCV-related steatosis. J Hepatol. 2008;49:1046–1054. doi: 10.1016/j.jhep.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 79.Kim K, Kim KH, Kim HY, Cho HK, Sakamoto N, Cheong J. Curcumin inhibits hepatitis C virus replication via suppressing the Akt-SREBP-1 pathway. FEBS Lett. 2010;584:707–712. doi: 10.1016/j.febslet.2009.12.019. [DOI] [PubMed] [Google Scholar]
- 80.Nakamuta M, Yada R, Fujino T, Yada M, Higuchi N, Tanaka M, et al. Changes in the expression of cholesterol metabolism-associated genes in HCV-infected liver: a novel target for therapy? Int J Mol Med. 2009;24:825–828. doi: 10.3892/ijmm_00000299. [DOI] [PubMed] [Google Scholar]
- 81.Olmstead AD, Knecht W, Lazarov I, Dixit SB, Jean F. Human subtilase SKI-1/S1P is a master regulator of the HCV Lifecycle and a potential host cell target for developing indirect-acting antiviral agents. PLoS Pathog. 2012;8:e1002468. doi: 10.1371/journal.ppat.1002468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sun LJ, Zhao YH, Li SC, Yu JW, Kang P, Liu W. Inhibition of silent information regulator-1 in hepatocytes induces lipid metabolism disorders and enhances hepatitis C virus replication. Zhonghua Gan Zang Bing Za Zhi. 2013;21:834–839. doi: 10.3760/cma.j.issn.1007-3418.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 83.Kim JH, Sung PS, Lee EB, Hur W, Park DJ, Shin EC, et al. GRIM-19 restricts HCV replication by attenuating intracellular lipid accumulation. Front Microbiol. 2017;8:576. doi: 10.3389/fmicb.2017.00576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Soyal SM, Nofziger C, Dossena S, Paulmichl M, Patsch W. Targeting SREBPs for treatment of the metabolic syndrome. Trends Pharmacol Sci. 2015;36:406–416. doi: 10.1016/j.tips.2015.04.010. [DOI] [PubMed] [Google Scholar]
- 85.Quan HY, Kim DY, Kim SJ, Jo HK, Kim GW, Chung SH. Betulinic acid alleviates non-alcoholic fatty liver by inhibiting SREBP1 activity via the AMPK-mTOR-SREBP signaling pathway. Biochem Pharmacol. 2013;85:1330–1340. doi: 10.1016/j.bcp.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 86.Shao W, Machamer CE, Espenshade PJ. Fatostatin blocks ER exit of SCAP but inhibits cell growth in a SCAP-independent manner. J Lipid Res. 2016;57:1564–1573. doi: 10.1194/jlr.M069583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li X, Chen YT, Hu P, Huang WC. Fatostatin displays high antitumor activity in prostate cancer by blocking SREBP-regulated metabolic pathways and androgen receptor signaling. Mol Cancer Ther. 2014;13:855–866. doi: 10.1158/1535-7163.MCT-13-0797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ding L, Li J, Song B, Xiao X, Huang W, Zhang B, et al. Andrographolide prevents high-fat diet-induced obesity in C57BL/6 mice by suppressing the sterol regulatory element-binding protein pathway. J Pharmacol Exp Ther. 2014;351:474–483. doi: 10.1124/jpet.114.217968. [DOI] [PubMed] [Google Scholar]
- 89.Li J, Ding L, Song B, Xiao X, Qi M, Yang Q, et al. Emodin improves lipid and glucose metabolism in high fat diet-induced obese mice through regulating SREBP pathway. Eur J Pharmacol. 2016;770:99–109. doi: 10.1016/j.ejphar.2015.11.045. [DOI] [PubMed] [Google Scholar]
- 90.Remington G. Understanding antipsychotic “atypicality”: a clinical and pharmacological moving target. J Psychiatry Neurosci. 2003;28:275–284. [PMC free article] [PubMed] [Google Scholar]
- 91.Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J. 2012;279:2610–2623. doi: 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- 92.Guo D, Bell EH, Mischel P, Chakravarti A. Targeting SREBP-1-driven lipid metabolism to treat cancer. Curr Pharm Des. 2014;20:2619–2626. doi: 10.2174/13816128113199990486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lewis CA, Griffiths B, Santos CR, Pende M, Schulze A. Regulation of the SREBP transcription factors by mTORC1. Biochem Soc Trans. 2011;39:495–499. doi: 10.1042/BST0390495. [DOI] [PubMed] [Google Scholar]
- 94.Li C, Yang W, Zhang J, Zheng X, Yao Y, Tu K, et al. SREBP-1 has a prognostic role and contributes to invasion and metastasis in human hepatocellular carcinoma. Int J Mol Sci. 2014;15:7124–7138. doi: 10.3390/ijms15057124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ishimoto K, Tachibana K, Hanano I, Yamasaki D, Nakamura H, Kawai M, et al. Sterol-regulatory-element-binding protein 2 and nuclear factor Y control human farnesyl diphosphate synthase expression and affect cell proliferation in hepatoblastoma cells. Biochem J. 2010;429:347–357. doi: 10.1042/BJ20091511. [DOI] [PubMed] [Google Scholar]
- 96.Huang Y, Wu R, Su ZY, Guo Y, Zheng X, Yang CS, et al. A naturally occurring mixture of tocotrienols inhibits the growth of human prostate tumor, associated with epigenetic modifications of cyclin-dependent kinase inhibitors p21 and p27. J Nutr Biochem. 2017;40:155–163. doi: 10.1016/j.jnutbio.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 97.Singal AG, El-Serag HB. Hepatocellular carcinoma from epidemiology to prevention: translating knowledge into practice. Clin Gastroenterol Hepatol. 2015;13:2140–2151. doi: 10.1016/j.cgh.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang H, Feng Z, Huang R, Xia Z, Xiang G, Zhang J. MicroRNA-449 suppresses proliferation of hepatoma cell lines through blockade lipid metabolic pathway related to SIRT1. Int J Oncol. 2014;45:2143–2152. doi: 10.3892/ijo.2014.2596. [DOI] [PubMed] [Google Scholar]
- 99.Feng S, Han M, Zhou L, Wang Q, Li Z, Li Y, et al. NS5ABP37 inhibits liver cancer by impeding lipogenesis and cholesterogenesis. Cancer Sci. 2017;108:12–22. doi: 10.1111/cas.13117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li N, Zhou ZS, Shen Y, Xu J, Miao HH, Xiong Y, et al. Inhibition of the sterol regulatory element-binding protein pathway suppresses hepatocellular carcinoma by repressing inflammation in mice. Hepatology. 2017;65:1936–1947. doi: 10.1002/hep.29018. [DOI] [PubMed] [Google Scholar]
- 101.Tian Y, To KF, Lai PBS, Cheung YS, Yu J, Wong VWS, et al. Histone deacetylase 8 impairs insulin sensitivity and activates β-catenin signaling in NAFLD-associated hepatocellular carcinoma. Cancer Res. 2015;75:Abstract nr 2073. doi: 10.1158/1538-7445.AM2015-2073. [DOI] [Google Scholar]
- 102.Nambiar DK, Deep G, Singh RP, Agarwal C, Agarwal R. Silibinin inhibits aberrant lipid metabolism, proliferation and emergence of androgen-independence in prostate cancer cells via primarily targeting the sterol response element binding protein 1. Oncotarget. 2014;5:10017–10033. doi: 10.18632/oncotarget.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]