Abstract
Background and objectives
MicroRNAs (miRs) are noncoding RNAs that regulate protein translation and melanoma progression. Changes in plasma miR expression following surgical resection of metastatic melanoma are under-investigated. We hypothesize differences in miR expression exist following complete surgical resection of metastatic melanoma.
Methods
Blood collection pre- and post-surgical resection was performed in 6 individuals with solitary melanoma metastases. miR expression in extracted RNA was quantified using the NanoString nCounter Digital analyzer.
Results
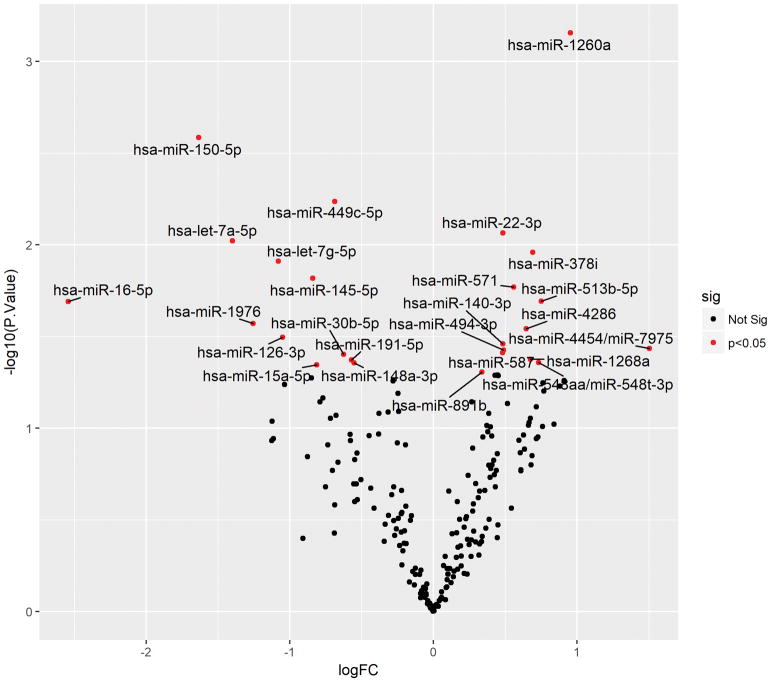
Pre- and post-surgical plasma samples contained 216 miRs with expression above baseline. Comparison of post-surgical to pre-resection samples revealed differential expression of 25 miRs: miR-let-7a, miR-let7g, miR-15a, miR-16, miR-22, miR-30b, miR-126, miR-140, miR-145, miR-148a, miR-150-5p, miR-191, miR-378i, miR-449c, miR-494, miR-513b, miR-548aa, miR-571, miR-587, miR-891b, miR-1260a, miR 1268a, miR-1976, miR-4268, miR-4454 (P<0.05). Utilizing P<0.0046 as a cutoff to control for 1 false positive among the 216 miRs revealed that post-surgical melanoma plasma samples had up-regulation of miR-1260a (P=0.0007) and down-regulation of miR-150-5p (P=0.0026) relative to pre-surgical samples.
Conclusions
Differential expression of miR-150-5p and miR-1260a is present in plasma following surgical resection of metastatic melanoma in this small sample (n=6) of melanoma patients. Therefore, further investigation of these plasma miRs as non-invasive biomarkers for melanoma is warranted.
Keywords: miR-1260a, miR-150-5p, NanoString, miR, miRNA
Synopsis for Table of Contents
Assessment of the plasma miRNA profile in metastatic melanoma patients pre-and post-surgical resection revealed differential expression of miR-150-5p and miR-1260a. These microRNAs may therefore exhibit diagnostic value as noninvasive biomarkers of tumor burden or recurrence in melanoma patients.
Introduction
Melanoma remains the deadliest form of skin cancer. In 2017 an estimated 87,110 patients will be newly diagnosed with melanoma in the United States while 9,730 persons will die of the disease [1]. Surgical resection is the mainstay of therapy [2,3]. Unfortunately, there are many challenges associated with the management of malignant melanoma. For instance, the 5-year recurrence free survival is 76–90% for cutaneous melanoma without lymph node involvement following curative surgical resection [2,4]. Recurrence free survival drops to 35–58% when metastasis to the lymph nodes is present [2,4]. Routine imaging has been assessed as a means of surveillance for earlier detection of recurrent disease. However, imaging is not an economical measure as the median cost to detect a single recurrence in patients with stage II disease is over $28,000 [4,5]. Other groups have investigated circulating biomarkers including the proteins S-100B and lactate dehydrogenase (LDH), but the use of these biomarkers can be associated with low specificity, and thus their incorporation into clinical guidelines has not been universal [6,7]. The management of metastatic melanoma has greatly improved with the recent approval of several immunomodulatory agents, but primary and secondary resistance is still commonplace [3]. These issues have prompted investigation into novel, non-invasive diagnostic assays that can be used in melanoma surveillance, such as levels of microRNAs (miRs) in bodily fluids.
miRs are 19–22 nucleotide RNA inhibitors of protein translation. There are over 800 unique miRs, each with the capacity to inhibit multiple targets. Thus, the regulatory capacity of this RNA type is immense. Importantly, unique miR expression patterns can be discerned for different types of cancer, and even different stages of disease. Investigation into miRs associated with malignant melanoma is ongoing. Several miRs are frequently dysregulated in melanoma cancers, including miR-21, miR-125b, miR-150, miR-155, miR-205, and miR-211[8]. However, many of these analyses were performed on tissue samples. This limits the applicability of this technique of measuring miRs to patients with larger tumors that can be easily biopsied, and examines the miR profile at only a single time point and at a single place within the tumor [9,10]. miRs are stable within the circulation thereby making the analysis of plasma miRs an ideal candidate for development as a minimally invasive diagnostic tool. Repetitive blood sampling is also more feasible than tissue sampling. Notably, some groups have questioned the applicability of tissue-derived miR tumor studies for plasma applications as plasma and tissue expression profiles do not always correlate [11]. These concerns have prompted our further investigation into plasma-based miR studies in the peri-operative setting.
Only a few analyses of plasma miR levels in melanoma patients have been attempted [12–14]. Armand-Labat et al. found a combination of miR-185 and miR-1246 to be associated with the diagnosis of metastatic melanoma compared to healthy subjects[13]. However, a major limitation of the aforementioned studies is the controls utilized, which often consist of independent groups (i.e., a separate group of non-diseased individuals). This approach may accentuate inter-individual variability, which could confound the results. A method to control for this variability includes the analysis of the same patient before and after surgery [15]. Indeed, this scheme has been used in the setting of other neoplasms including lung, breast, and esophageal cancer [16]. However, to our knowledge, these studies have not been previously performed in melanoma patients. In order to determine levels of plasma miRs in patients with malignant melanoma while controlling for inter-individual variability, we sought to investigate the miR plasma expression profile in patients undergoing resection of malignant melanoma.
Materials and Methods
Patient Samples
Between 2013–2015, six individuals at The Ohio State University Wexner Medical Center with clinically evident locoregional and distant metastatic melanoma undergoing surgical therapy were selected for participation. This work was conducted under the auspices of an IRB approved protocol (No. 1999C0348).
Blood Collection
In all cases, the pre-operative samples were harvested using sterile peripheral venipuncture prior to surgical incision. Post-surgery samples were obtained at the time of a post-operative clinic visit. Peripheral venous blood was collected in EDTA containing blood collection tubes following standard venipuncture. Hemolysis was minimized by adherence to sound blood collection techniques. Blood collection was performed pre-operatively and at about 3 weeks following surgical therapy. Peripheral whole blood was centrifuged at 1700 rpm for 11 minutes. The resultant plasma supernatant was isolated, aliquoted within one hour of collection, and stored at −80°C until RNA isolation.
Isolation of RNA
200 μL of thawed plasma from each pre-operative and post-operative sample was used for RNA isolation with miRNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacture’s recommendations. 3 miR spike-ins with known concentrations were used as loading controls: ath-miR-159a, cel-miR-248 and osa-miR-414. Additional RNA purification was performed using Amicon® 0.5 mL 3kDa columns as per the manufacturer’s recommendations (Millipore, Billerica, MA). Isolated RNA was stored at −80°C until NanoString analysis was performed.
NanoString
Isolated and purified RNA (3 μL) was loaded onto the NanoString nCounter (NanoString Technologies, Seattle, WA) platform and miR expression quantification was carried out as previously described[17]. Normalization of melting temperatures and miR identification was facilitated by ligation of individual miRs within the sample to DNA tags. Excess tags were washed away. miRs within the plasma sample were hybridized to capture reporter probes at 64°C for 18 hours. The reporter probes contain unique fluorescent signals thereby permitting downstream identification of individual miRs. Excess probes were removed using two-step magnetic bead-based purification, target/probe complexes were immobilized onto a streptavidin coated cartridge via nCounter Prep Station (NanoString Technologies, Seattle, WA) and the florescence of each hybridized miR was analyzed by an nCounter Digital analyzer (NanoString Technologies, Seattle, WA). A high-density scan containing 600 fields of view was performed. The investigation included 5 positive, 5 negative, and 5 housekeeping genes (beta-actin, beta-2-microglobulin, glyceraldehyde-3-phosphate dehydrogenase, ribosomal protein L19, and ribosomal protein large subunit P0).
Data Analysis
Statistical analysis was performed using a linear mixed effect model. The technical normalized data was first filtered using the negative controls (cutoff=mean of negative control expression+2SD). Individual miRs were removed from further analysis if 90% of the samples had expression levels lower than the cutoff. The data was then quantile normalized and analyzed with linear mixed effects models to take account of the correlation among observations from the same patient. Controlling for 1 false positive among the tested miRs, a p-value <0.0046 can be considered significant for differential expression status between pre- and post-resection plasma samples. The ggplot2 R package was used to create the volcano plot.
Results
Clinical Data
Six patients with metastatic melanoma with a median age of 63 (range 49–77 years old) were included in the analysis (3 male, 3 female). Four patients had stage III disease (axillary nodes n=1, inguinal nodes n=2, in-transit metastasis of the leg n=1) while 2 patients had stage IV disease (both had an isolated splenic metastasis). All patients had post-operative blood samples harvested within 5 months of surgery while most were drawn within approximately 3 weeks (Table 1). The average length of follow-up was 22.3 months and 4 patients are currently alive without any evidence of disease. Despite adjuvant therapy, 2 patients recurred following metastasectomy at 13 and 14 months post-surgery and subsequently died from their illness at 17 and 21 months, respectively.
Table I. Clinical Patient Characteristics.
Clinical characteristics of six patients undergoing plasma microRNA analysis before and after surgical resection of metastatic melanoma are listed including site of disease involvement and timing of post-operative blood draw. Abbreviations: NU (non-ulcerated), U (ulcerated), POD (post-operative day), WLE (wide local excision), SLNB (sentinel lymph node biopsy) expressed as ratio of number of lymph nodes containing microscopic disease relative to the total number of lymph nodes biopsied, LND (lymph node dissection) expressed as a ratio of number of lymph nodes containing microscopic disease to the total number of lymph nodes resected, mo (month), NAT(no adjuvant treatment), XRT (radiation therapy), CY (cyclophosphamide), VI (vincristine), DA (dacarbazine), IPI (ipilimumab).
| Patient | Age | Sex | Primary Site | Stage | Site of Disease | Procedure | Post-op blood draw | Post-op recurrence | Time to recurrence | Other therapy |
|---|---|---|---|---|---|---|---|---|---|---|
| A | 49 | F | Right hip 0.78 mm NU 1 mitotic figure No SLNB |
IIIC | Groin | Superficial and deep inguinal LND 9/22 (superficial 7/13, deep 2/9) |
POD 20 | Yes | 13 mo. | XRT to R groin and CY, VI, DA post op |
| B | 57 | F | Left ankle 1.87 mm NU No mitotic figures SLNB (0/2) |
IIIC | Groin | Superficial and deep inguinal LND | POD 20 | No | N/A | IPI high dose |
| C | 57 | M | Left back 4.5 mm NU 5 mitotic figures, SLNB (0/4) |
IV | Spleen | Splenectomy | POD 11 | Yes | 14 mo. | NAT |
| D | 77 | F | Left leg 0.7 mm NU Unknown mitotic figures No SLNB |
IIIB | Lower Extremity | WLE of in-transit metastasis with SLNB (0/1) | POD 25 | No | N/A | NAT |
| E | 71 | F | Left flank 0.98 mm NU No mitotic figures SLNB (0/X) |
IIIC | Left axilla | Axillary LND (2/35) with in-transit metastases | POD 129 | No | N/A | XRT to axillae |
| F | 68 | M | Left neck 3.0 mm U >10 mitotic figures |
IV | Spleen | Splenectomy | POD 18 | No | N/A Alive (15 mo.) |
Neo-adjuvant therapy (IPI) |
Positive and Negative Controls
Both positive and negative controls were utilized. Spike-in loading controls consisted of three different small RNA sequences that were added to the sample preparation during RNA isolation: ath-miR-159a, cel-miR-248 and osa-miR-414. Each of the positive control molecules for NanoString were present in the CodeSet at known concentrations (0.125 fM, 0.5 fM, 2 fM, 8 fM, 32 fM, 128 fM) and were hybridized in the same manner as endogenous miR sample. This allowed for assessment of variation in sample preparation and chip hybridization. Under ideal conditions for NanoString, the positive controls should show a strong linear trend when plotting the observed versus the expected log transformed expression values. Conversely, the negative controls consist of probes without target transcripts, allowing for assessment of the non-specific background within the system. The negative controls should be tightly clustered under the positive control line. Both of these patterns were observed for the 12 plasma samples in this study, suggesting an adequately controlled system with efficient sample preparation and hybridization and minimal non-specific background.
microRNA Expression following Surgical Resection
An analysis of miR expression profiles using the NanoString platform on 6 individuals with malignant melanoma revealed 216 miRs that had a change in expression from baseline levels. These 216 miRs were thus used as the total set of miRs for subsequent analysis. A comparison of plasma samples following surgical resection to pre-resection samples revealed 25 miRs within this group that had differential expression without adjustment for multiple comparisons. The 25 miRs included the following: miR-let-7a, miR-let7g, miR-15a, miR-16, miR-22, miR-30b, miR-126, miR-140, miR-145, miR-148a, miR-150, miR-191, miR-378i, miR-449e, miR-494, miR-513b, miR548aa, miR-571, mir-587, miR-891b, miR-1260a, miR-1268a, miR-1976, miR-4268, and miR-4454 (P<0.05, shown in Table II). However, use of a P value of <0.05 allowed for increased false positives among the 216 miRs with expression above baseline without adjustment for multiple comparisons. A stringent cutoff P value of P<0.0046 was then applied, which controlled for one false positive among the 216 tested miRs. With examination of the 216 miRs using this stricter cutoff, post-surgical melanoma plasma samples demonstrated significant up-regulation of miR-1260a (1.935 fold increase, P=0.0007) and down-regulation of miR-150-5p (3.12 fold decrease, P=0.0026) relative to pre-surgical samples, shown in Figure 1. Therefore, differential expression of miR 150-5p and miR-1260a is present in plasma following surgical resection of metastatic melanoma.
Table II. Dysregulated microRNAs following surgical resection of metastatic melanoma with top target pathway.
Plasma microRNA analysis via NanoString was performed in 6 individuals with solitary melanoma metastasis before and after complete surgical resection. Differentially expressed miRs (P<0.05) are listed with their respective targets. Top target pathways were identified using ToppFunn functional enrichment analysis.
| miRNA | Mean Fold Change | Top Pathway Enriched |
|---|---|---|
| miR-let-7a | 0.36 | Cyclins and cell cycle regulation |
| miR-let7g | 0.47 | P53 signaling pathway |
| miR-15a | 0.48 | Pathways in cancer |
| miR-16 | 0.31 | Adaptive Immune System |
| miR-22 | 1.20 | Regulation of nuclear SMAD2/3 signaling |
| miR-30b | 0.54 | Pre-NOTCH expression and processing |
| miR-126 | 0.48 | mTOR signaling pathway |
| miR-140 | 1.00 | Pathways in cancer |
| miR-145 | 0.45 | TGF-beta signaling pathway |
| miR-148a | 0.61 | Post-transcriptional silencing by small RNAs |
| miR-150-5p | 0.26 | Pre-NOTCH transcription and translation |
| miR-191 | 0.59 | Coregulation of androgen receptor activity |
| miR-378i | 1.10 | - |
| miR-449c | 0.54 | Telomeres, telomerase, cellular aging, and immortality |
| miR-494 | 0.66 | Insulin/IGF pathway-protein kinase B signaling cascade |
| miR-513b | 1.15 | - |
| miR-548aa | 1.13 | - |
| miR-571 | 1.08 | P53 pathway |
| miR-587 | 1.02 | - |
| miR-891b | 0.94 | Ca2+ pathway |
| miR-1260a | 1.77 | - |
| miR-1268a | 1.24 | Transcription factor CREB and its extracellular signals |
| miR-1976 | 0.42 | - |
| miR-4286 | 1.27 | - |
Figure 1. Volcano plot showing the fold change versus negative log of the p-value of differentially expressed miRNAs.
The false discovery rate (FDR) cutoff imposed to define the statistical significance threshold of p<0.0046 (at y=2.337) indicates statistical significance of two miRs that passed this criterion, miR-1260a and miR-150-5p.
Pathway Analysis
A list of experimentally-validated gene targets was collected for miR-150-5p and miR-1260a using the miRTarBase database [18]. Computationally predicted target genes were also identified for miR-150-5p and miR-1260a using the TargetScan database. Gene targets with a cumulative weighted context++ score less than zero were identified for each miR, determined by TargetScan (329 gene targets for hsa-miR-150-5p, and 4067 gene targets for hsa-miR-1260a). The cumulative weighted context++ score is a measure of the total repression expected from multiple sites of the same miRNA, for each predicted mRNA target [19]. The gene targets identified by TargetScan were then assessed for overlap with the miRTarBase identified gene target lists, reducing the number of genes for analysis to 32 gene targets for miR-150-5p, and 120 gene targets for miR-1260a [20].
A pathway enrichment analysis was then performed on each list of gene targets using the ToppFunn functional enrichment tool. This revealed significant interaction between miR-150-5p gene targets and NOTCH signaling, transcription, and translation, IL-4 mediated signaling, oncogene induced senescence, and p53-associated pathways (Bonferroni q-value < 0.05). Pathway enrichment analysis of miR-1260a gene targets did not reveal any pathway interactions that met a significance q-value <0.05. However, significant co-expression of miR-1260a gene targets with miR-506 and miR-124 (q-value = 0.001) was indicated by the enrichment. The top pathway enriched for each differentially expressed miR by ToppFunn analysis is listed in Table II.
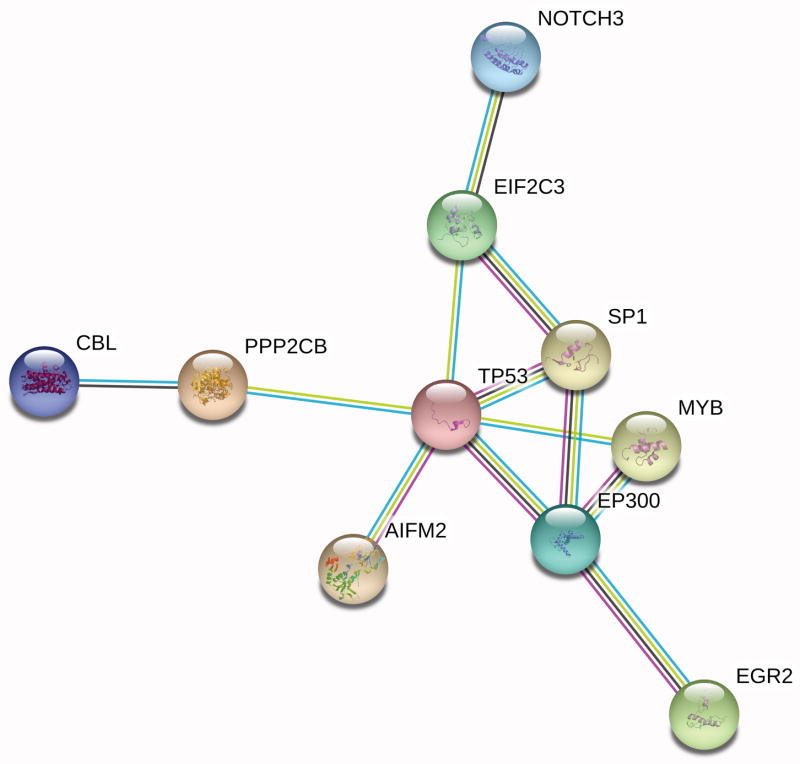
The common gene targets determined for miR-150-5p from miRTarBase and TargetScan were also used to create a target protein-protein interaction (PPI) network using the String database (Fig 2). The String network is generated using experimental data, as well as textmining, coexpression, and data from curated databases to identify protein-protein interactions associated with the target genes of miR-150-5p. Using this approach protein-protein interactions were detected between 10 gene targets of miR-150-5p via String network generation, including NOTCH3, TP53, MYB, and EGR2.
Figure 2. Gene interaction network generated using miR-150-5p experimentally validated gene targets.
Only interactions with a high confidence interaction score are shown (0.9 or higher, scale of 0.1 to 1.0). Edges represent protein-protein interactions (PPI). PPI enrichment p-value = 0.00371. (Color legend: from curated databases = cyan blue, experimentally determined = magenta, textmining = green, coexpression = black).
Discussion
The present analysis of plasma miR expression in patients with malignant melanoma before and after surgical resection suggests the existence of distinct differences in miR expression related to the presence of melanoma tumors. In particular, 25 miRs had differential expression above baseline, while the application of a stringent cutoff (P<0.0046) revealed that just two miRs exhibited the greatest difference following surgery. MiR-150-5p was downregulated (P=0.0026) in post-surgical plasma samples relative to pre-operative samples while levels of miR-1260a were up-regulated following resection (P=0.0007). This is the first approach involving the utilization of paired plasma melanoma samples which may account, in part, for differences relative to other plasma-based melanoma miR studies [12–14,21].
Expression of miR-150-5p, the mature 5′ strand of the miR-150 stem-loop precursor, was downregulated in our samples following resection of melanoma tumors. MiR-150 expression has previously been observed in malignant melanoma tissue samples relative to benign nevi[8]. miR-150 has several relevant targets including the v-myb avian myeloblastosis viral oncogene homolog (MYB) as well as neurogenic locus notch homolog protein 3 (NOTCH 3), and early growth response 2 (EGR2) [22]. It has been observed that loss of c-MYB function can play a role in the progression of malignant melanoma [23]. These effects may come about in part through c-MYB regulation of apoptosis [24]. The role of target gene early growth response protein 2 (EGR2) in PTEN-induced apoptosis is also supportive of a potential action by miR-150-5p in mediating gene suppression in cancer.
The String network in Figure 2 was generated to demonstrate protein-protein interactions between gene targets identified for miR-150-5p. Designation as an interaction in the network is experimentally determined or identified based on co-expression, data from curated databases, or textmining. The String network of protein-protein interactions determined from the gene targets of miR-150-5p includes the protein known as eukaryotic translation initiation factor 2C, 3 (EIF2C3) or Argonaute 3, a catalytic component of the RNA induced silencing complex (RISC) that cleaves mRNA in microRNA-mediated RNA interference. The Argonaute 3 protein is shown to interact with TP53 based on data from textmining and curated databases in the String network. Support for this protein-protein interaction has been documented in non-small cell lung carcinoma (NSCLC), in which suppression of p53 expression by miR-150-5p has been observed in vitro. Additionally, miR-150 has been identified as a member of a regulatory network in NSCLC tumorigenesis via regulation of p53 expression [25].
Interestingly, this suggested role of miR-150-5p in the promotion of tumorigenesis is contrasted by demonstration of an interaction between Argonaute 3 with NOTCH3 in the presence of miR-150-5p in the String network based on co-expression, data from curated databases, and textmining. This interaction is suggestive of inhibition of NOTCH3 expression by Argonaute 3 in the presence of high miR-150-5p. Therefore, the NOTCH3-Argonaute 3 interaction indicated by the String network suggests downregulation of NOTCH3 in melanoma with high miR-150-5p expression, which contradicts patterns of NOTCH3 expression in cancer and melanoma reported historically. High NOTCH3 expression has been observed in multiple cancer types, and has been previously associated with malignant progression, abnormal differentiation, metastasis, tumor angiogenesis, and a poor prognosis [26]. Upregulation of NOTCH3 expression has also been demonstrated in melanoma-endothelial cell co-cultures. Additionally, NOTCH3 overexpression has been documented in malignant melanoma, and is associated with enhanced migration by melanoma cells in vitro [27]. Since microRNAs function to ultimately inhibit protein expression, the interaction of Argonaute 3 with NOTCH3 may indicate a protective function of miR-150-5p expression against melanoma progression. However, without co-evaluation of plasma miR-150-5p and tissue NOTCH expression, the relation between these two molecules and the significance of their interaction in the setting of melanoma remains to be determined.
A third protein-protein interaction of interest is indicated by the generated String network between Argonaute 3 and SP1, in the presence of miR-150-5p, supported experimental data, co-expression data, data from curated databases, and textmining. SP1 has been previously identified as a core transcriptional regulator in metastatic melanoma, associated with regulation of 24 differentially expressed hub genes involved in the immune response, tumor cell development, and tumorigenesis in melanoma [28]. This suggests a potential regulatory role for miR-150-5p on differentially expressed genes in the setting of metastatic melanoma through interaction with SP1 via Argonaute 3.
These protein interactions suggest miR-150-5p-mediated regulation of these genes or their associated protein expression may impact melanoma progression. Down-regulation of miR-150-5p expression following surgical resection of melanoma in these samples may indicate the reduction of tumor burden, and by extension, may potentially serve as a candidate biomarker of tumor recurrence for future studies. However, without additional information regarding downstream protein expression of these targets, elaboration on the cause of miR-150-5p upregulation in the presence of melanoma, the events leading to reduction of miR-150 expression post-surgery, and its effect on gene targets in the tumor environment is needed.
The potential utility of circulating miR-150 may extend beyond that of a tumor biomarker. Liu et al. demonstrated that miR-150-treated tumor associated macrophages (TAMs) have dramatically increased expression of VEGF. They also showed that intravascular administration of miR-150 by a microvesicle delivery system resulted in increased serum VEGF levels and promoted tumor development in vivo using an S-180 xenograft sarcoma mouse model. Furthermore, attenuation of miR-150 by intravascular microvesicle administration of anti-miR-150 RNA resulted in smaller tumors and reduced VEGF protein expression. These findings suggest that miR-150 may act as an oncomir via enhanced TAM activation and angiogenesis in the tumor microenvironment. Thus, miR-150 is a potential therapeutic target [29]. Therefore, further exploration of the interaction between miR-150, TAMs, and VEGF in melanoma is warranted.
Recently, the role and implications of miR-150-5p expression specifically in melanoma patients has been debated. Some reports indicate an oncogenic role for miR-150-5p based on the presence of high levels of circulating miR-150 in melanoma patients with high risk of recurrence [30]. This finding was supported by the upregulation of miR-150-5p in the plasma of melanoma patients observed by Fogli et al. concurrently with miR-15b-5p and mir-149-3p, suggestive of a “triple classifier” of miRs for identification of melanoma-baring patients when compared to miR expression in healthy donors [31]. Segura et al. found similar miR-150 overexpression in formalin fixed, paraffin embedded melanoma metastases and primary melanoma samples relative to benign nevi[32]. However, these findings are opposed by the observation of reduced miR-150-5p expression in tissue and serum of melanoma patients with a poor prognosis. Tembe et al. found that miR-150-5p expression was significantly lower in the tissue of BRAF mutant melanomas. Additionally, higher miR-150-5p expression in serum of patients with melanoma was associated with better survival, with subsequent loss of its expression portending a poor prognosis. Since miR-150-5p is associated with multiple oncogene targets and oncogenic pathways, Tembe et al. suggested that reduced miR-150-5p expression in serum and tissue in the presence of melanoma may be associated with an autoregulatory tissue reaction to tumor progression, rather than acting as a driver of tumor proliferation [33]. Given these discrepancies, further studies investigating the role of miR-150-5p in the progression of melanoma, its association with mutation status, clinical characteristics, and potential implications of its presence in circulation are necessary.
miR-1260a was found to be up-regulated in patients following surgical resection. However, while miR-1260a has experimentally-validated gene targets, no significant pathway enrichment was associated with these gene targets. Scarce evidence exists for a role of miR-1260 in melanoma. Sand et al. found a 2.14 over-expression of miR-1260 in cutaneous malignant melanoma[34]. In contrast, miR-1260 downregulation has been found in other neoplastic settings, including follicular lymphoma [35]. In a recent study, miR-1260 expression was increased in asthma patients undergoing longer treatment duration compared to short treatment courses. However, the implication of these findings in the neoplastic setting is uncertain [36]. Further functional assays will be needed to elucidate the role of miR-1260a in the context of melanomas.
Studies investigating plasma miRs in patients with melanoma using NanoString have highlighted other potential miRs of interest including over-expression of miR-17, miR-19a, miR-21, miR-126 and miR-149 [14]. However, this sample was compared against an independent control group of normal individuals. Since plasma is an indirect analysis of the solid tumor milieu, plasma miR profiles have the potential to simultaneously reflect both ongoing neoplastic and non-neoplastic processes. Indeed, the authors contend the miRs of interest also have overlap with patterns seen in non-neoplastic inflammatory conditions [14]. Armand-Labat et al. compared plasma miR expression in metastatic melanoma to healthy subjects and found decreased expression of miR-185 and increased expression of miR-1246 associated with malignant melanoma [13]. However, sizeable differences in age groups were evident between malignant and control groups which may account for some of the difference. Differences in miR profiles are associated with non-neoplastic entities that are more common in elderly individuals such as cardiovascular disease and Alzheimer’s disease [37]. Thus, the need exists for carefully selected controls to account for potential confounding variables. Utilization of paired samples from individual patients may help to minimize any potential biases.
A major limitation to this study is the small sample size utilized. This reflects the inherent constraints of NanoString platform which limit the analysis to a maximum of twelve samples. However, NanoString has been previously shown to be a reproducible method of analyzing a high number of targets with low input of starting RNA [38]. It also provides greater sensitivity with lower relative expenses than other high throughput platforms such as RNA-seq [39]. These attributes justify the use of the NanoString platform in this setting. Therefore, future studies to expand the patient sample size are needed to confirm these findings. An increased sample size would allow for substantial strengthening of the associations observed in this study, thus improving the potential utility of these miRs as biomarkers in the plasma of patients with metastatic melanoma. Overall, this study highlights potential candidate miRs of interest that deserve focused attention during the design of subsequent validation studies using independent samples.
A second limitation to this study is apparent in the approach used for pathway analysis. The absence of significant pathway enrichment using gene targets of miR-1260a, but presence of significant co-expression of miR-506 and miR-124 may indicate that the list of gene targets generated by miRTarBase and TargetScan is not sensitive to miR-1260a. This indicates that use of gene targets as the source of pathway identification for enrichment may limit the assessment of miR function and interactions.
The use of a String network for assessment of protein-protein interactions also carries the inherent limitation of revealing interactions between proteins without directionality to indicate upstream or downstream effects of one protein on the other. Therefore, the interpretation and potential implications of the indicated interactions must be considered in light of this limitation.
Conclusions
The analysis of plasma miRs before and after surgical resection of malignant melanoma suggests changes in the miR transcriptome. This relatively non-invasive method of miR profiling has the potential for use during patient surveillance. Differential patterns of miR expression in melanoma patients rendered free of disease warrant further investigation, as these factors exhibit potential diagnostic value.
Acknowledgments
Funding Sources: NIH Grants P01 CA095426 (to M.A. Caligiuri), P30 CA16058 (to M.A. Caligiuri), T32 CA090223 (to W.E. Carson), T32 CA009338 (to M.A. Caligiuri), K24 CA093670 (to W.E. Carson), NLM grant T15LM011270 to ZA
Footnotes
Disclosures: The authors have no disclosures.
References
- 1.Jemal A, Siegel R, Xu J, Ward E. Cancer statistics, 2010. CA Cancer J Clin. 2010;60:277–300. doi: 10.3322/caac.20073. [DOI] [PubMed] [Google Scholar]
- 2.Svedman FC, Pillas D, Taylor A, et al. Stage-specific survival and recurrence in patients with cutaneous malignant melanoma in Europe - a systematic review of the literature. Clin Epidemiol. 2016;8:109–122. doi: 10.2147/CLEP.S99021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pavri SN, Clune J, Ariyan S, Narayan D. Malignant Melanoma: Beyond the Basics. Plast Reconstr Surg. 2016;138:330e–340e. doi: 10.1097/PRS.0000000000002367. [DOI] [PubMed] [Google Scholar]
- 4.Rueth NM, Cromwell KD, Cormier JN. Long-term follow-up for melanoma patients: is there any evidence of a benefit? Surg Oncol Clin N Am. 2015;24:359–377. doi: 10.1016/j.soc.2014.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hengge UR, Wallerand A, Stutzki A, Kockel N. Cost-effectiveness of reduced follow-up in malignant melanoma. J Dtsch Dermatol Ges. 2007;5:898–907. doi: 10.1111/j.1610-0387.2007.06454.x. [DOI] [PubMed] [Google Scholar]
- 6.Mrazek AA, Chao C. Surviving cutaneous melanoma: a clinical review of follow-up practices, surveillance, and management of recurrence. Surg Clin North Am. 2014;94:989–1002. vii–viii. doi: 10.1016/j.suc.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gebhardt C, Lichtenberger R, Utikal J. Biomarker value and pitfalls of serum S100B in the follow-up of high-risk melanoma patients. J Dtsch Dermatol Ges. 2016;14:158–164. doi: 10.1111/ddg.12727. [DOI] [PubMed] [Google Scholar]
- 8.Latchana N, Ganju A, Howard JH, Carson WE., 3rd MicroRNA dysregulation in melanoma. Surg Oncol. 2016;25:184–189. doi: 10.1016/j.suronc.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 9.Jiang L, Lv X, Li J, et al. The status of microRNA-21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem. 2012;114:582–588. doi: 10.1016/j.acthis.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Grignol V, Fairchild ET, Zimmerer JM, et al. miR-21 and miR-155 are associated with mitotic activity and lesion depth of borderline melanocytic lesions. Br J Cancer. 2011;105:1023–1029. doi: 10.1038/bjc.2011.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Margue C, Reinsbach S, Philippidou D, et al. Comparison of a healthy miRNome with melanoma patient miRNomes: are microRNAs suitable serum biomarkers for cancer? Oncotarget. 2015;6:12110–12127. doi: 10.18632/oncotarget.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saldanha G, Potter L, Shendge P, et al. Plasma microRNA-21 is associated with tumor burden in cutaneous melanoma. J Invest Dermatol. 2013;133:1381–1384. doi: 10.1038/jid.2012.477. [DOI] [PubMed] [Google Scholar]
- 13.Armand-Labit V, Meyer N, Casanova A, et al. Identification of a Circulating MicroRNA Profile as a Biomarker of Metastatic Cutaneous Melanoma. Acta Derm Venereol. 2016;96:29–34. doi: 10.2340/00015555-2156. [DOI] [PubMed] [Google Scholar]
- 14.Pfeffer SR, Grossmann KF, Cassidy PB, et al. Detection of Exosomal miRNAs in the Plasma of Melanoma Patients. J Clin Med. 2015;4:2012–2027. doi: 10.3390/jcm4121957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aushev VN, Zborovskaya IB, Laktionov KK, et al. Comparisons of microRNA patterns in plasma before and after tumor removal reveal new biomarkers of lung squamous cell carcinoma. PLoS One. 2013;8:e78649. doi: 10.1371/journal.pone.0078649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leidinger P, Keller A, Backes C, et al. MicroRNA expression changes after lung cancer resection: a follow-up study. RNA Biol. 2012;9:900–910. doi: 10.4161/rna.20107. [DOI] [PubMed] [Google Scholar]
- 17.Alder H, Taccioli C, Chen H, et al. Dysregulation of miR-31 and miR-21 induced by zinc deficiency promotes esophageal cancer. Carcinogenesis. 2012;33:1736–1744. doi: 10.1093/carcin/bgs204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chou CH, Shrestha S, Yang CD, et al. miRTarBase update 2018: a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018;46:D296–D302. doi: 10.1093/nar/gkx1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Riffo-Campos AL, Riquelme I, Brebi-Mieville P. Tools for Sequence-Based miRNA Target Prediction: What to Choose? International Journal of Molecular Sciences. 2016:17. doi: 10.3390/ijms17121987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dweep H, Sticht C, Gretz N. In-Silico Algorithms for the Screening of Possible microRNA Binding Sites and Their Interactions. Curr Genomics. 2013;14:127–136. doi: 10.2174/1389202911314020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Markowitz J, Abrams Z, Jacob NK, et al. MicroRNA profiling of patient plasma for clinical trials using bioinformatics and biostatistical approaches. Onco Targets Ther. 2016;9:5931–5941. doi: 10.2147/OTT.S106288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kunz M. MicroRNAs in melanoma biology. Adv Exp Med Biol. 2013;774:103–120. doi: 10.1007/978-94-007-5590-1_6. [DOI] [PubMed] [Google Scholar]
- 23.Millikin D, Meese E, Vogelstein B, et al. Loss of heterozygosity for loci on the long arm of chromosome 6 in human malignant melanoma. Cancer Res. 1991;51:5449–5453. [PubMed] [Google Scholar]
- 24.Oh IH, Reddy EP. The myb gene family in cell growth, differentiation and apoptosis. Oncogene. 1999;18:3017–3033. doi: 10.1038/sj.onc.1202839. [DOI] [PubMed] [Google Scholar]
- 25.Wang DT, Ma ZL, Li YL, et al. miR-150, p53 protein and relevant miRNAs consist of a regulatory network in NSCLC tumorigenesis. Oncol Rep. 2013;30:492–498. doi: 10.3892/or.2013.2453. [DOI] [PubMed] [Google Scholar]
- 26.Liu L, Yang ZL, Wang C, et al. The Expression of Notch 1 and Notch 3 in Gallbladder Cancer and Their Clinicopathological Significance. Pathol Oncol Res. 2016;22:483–492. doi: 10.1007/s12253-015-0019-4. [DOI] [PubMed] [Google Scholar]
- 27.Howard JD, Moriarty WF, Park J, et al. Notch signaling mediates melanoma-endothelial cell communication and melanoma cell migration. Pigment Cell Melanoma Res. 2013;26:697–707. doi: 10.1111/pcmr.12131. [DOI] [PubMed] [Google Scholar]
- 28.Wang LX, Li Y, Chen GZ. Network-based co-expression analysis for exploring the potential diagnostic biomarkers of metastatic melanoma. PLoS One. 2018;13:e0190447. doi: 10.1371/journal.pone.0190447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu Y, Zhao L, Li D, et al. Microvesicle-delivery miR-150 promotes tumorigenesis by up-regulating VEGF, and the neutralization of miR-150 attenuate tumor development. Protein Cell. 2013;4:932–941. doi: 10.1007/s13238-013-3092-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman EB, Shang S, de Miera EV, et al. Serum microRNAs as biomarkers for recurrence in melanoma. J Transl Med. 2012;10:155. doi: 10.1186/1479-5876-10-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fogli S, Polini B, Carpi S, et al. Identification of plasma microRNAs as new potential biomarkers with high diagnostic power in human cutaneous melanoma. Tumour Biol. 2017;39:1010428317701646. doi: 10.1177/1010428317701646. [DOI] [PubMed] [Google Scholar]
- 32.Segura MF, Belitskaya-Levy I, Rose AE, et al. Melanoma MicroRNA signature predicts post-recurrence survival. Clin Cancer Res. 2010;16:1577–1586. doi: 10.1158/1078-0432.CCR-09-2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tembe V, Schramm SJ, Stark MS, et al. MicroRNA and mRNA expression profiling in metastatic melanoma reveal associations with BRAF mutation and patient prognosis. Pigment Cell Melanoma Res. 2015;28:254–266. doi: 10.1111/pcmr.12343. [DOI] [PubMed] [Google Scholar]
- 34.Sand M, Skrygan M, Sand D, et al. Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell Tissue Res. 2013;351:85–98. doi: 10.1007/s00441-012-1514-5. [DOI] [PubMed] [Google Scholar]
- 35.Wang W, Corrigan-Cummins M, Hudson J, et al. MicroRNA profiling of follicular lymphoma identifies microRNAs related to cell proliferation and tumor response. Haematologica. 2012;97:586–594. doi: 10.3324/haematol.2011.048132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Yang L, Li P, et al. Circulating microRNA Signatures Associated with Childhood Asthma. Clin Lab. 2015;61:467–474. doi: 10.7754/clin.lab.2014.141020. [DOI] [PubMed] [Google Scholar]
- 37.Olivieri F, Rippo MR, Procopio AD, Fazioli F. Circulating inflamma-miRs in aging and age-related diseases. Front Genet. 2013;4:121. doi: 10.3389/fgene.2013.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Veldman-Jones MH, Brant R, Rooney C, et al. Evaluating Robustness and Sensitivity of the NanoString Technologies nCounter Platform to Enable Multiplexed Gene Expression Analysis of Clinical Samples. Cancer Res. 2015;75:2587–2593. doi: 10.1158/0008-5472.CAN-15-0262. [DOI] [PubMed] [Google Scholar]
- 39.Brumbaugh CD, Kim HJ, Giovacchini M, Pourmand N. NanoStriDE: normalization and differential expression analysis of NanoString nCounter data. BMC Bioinformatics. 2011;12:479. doi: 10.1186/1471-2105-12-479. [DOI] [PMC free article] [PubMed] [Google Scholar]