SUMMARY
Intracellular bacterial pathogens secrete a repertoire of effector proteins into host cells that are required to hijack cellular pathways and cause disease. Despite decades of research, however, the molecular functions of most bacterial effectors remain unclear. To address this gap, we generated quantitative genetic interaction profiles between 36 validated and putative effectors from three evolutionarily divergent human bacterial pathogens and 4,190 yeast deletion strains. Correlating effector-generated profiles with those of yeast mutants, we recapitulated known biology for several effectors with remarkable specificity and predicted previously unknown functions for others. Biochemical and functional validation in human cells revealed a role for an uncharacterized component of the Salmonella SPI-2 translocon, SseC, in regulating maintenance of the Salmonella vacuole through interactions with components of the host retromer complex. These results exhibit the power of genetic interaction profiling to discover and dissect complex biology at the host-pathogen interface.
eTOC
Despite playing a crucial role in virulence, the molecular targets of most bacterial effector proteins remain completely unknown. Using budding yeast as a heterologous host, we generated genetic interaction profiles for 36 bacterial effector proteins and validated target pathways/complexes predicted by our yeast screen with experiments in human cells. We report that the Salmonella protein SseC binds to and modulates the human retromer complex, which is required for maintenance of the Salmonella-containing vacuole.

INTRODUCTION
Intracellular bacterial pathogens have evolved a variety of specific adaptations to survive and replicate within their hosts. Central to the pathogenesis of many bacteria are sophisticated secretion systems that translocate proteins and nucleic acids from the cytosol of bacteria into target eukaryotic cells. These translocated proteins, deemed effectors, are essential for invading bacteria to establish a replicative niche and cause disease by modulating a diverse set of host pathways including cytoskeletal dynamics, gene expression, inflammation, and antimicrobial defenses. However, assigning precise molecular mechanisms of action to effectors has been thwarted by genetic intractability of certain bacteria, functional redundancy, interdependency between effector proteins, lack of sequence similarity between effectors and annotated protein domains, and the spatiotemporal complexity of their expression during infection. In this study, we examined effector proteins from three divergent human and animal pathogens: Salmonella enterica serovar Typhimurium, Coxiella burnetii, and Brucella melitensis. While these bacterial pathogens differ significantly in their intracellular lifestyles and their modes of pathogenesis, the virulence of each of these species is centered on its ability to translocate effectors into host cells.
S. Typhimurium, a common cause of food poisoning and a major cause of diarrheal disease worldwide, encodes two type III protein secretion systems (T3SS). Salmonella pathogenicity islands 1 and 2 (SPI-1/SPI-2) are required for reprogramming the actin cytoskeleton to promote bacterial entry into non-phagocytic cells and for maintaining and renovating the Salmonella-containing vacuole (SCV), respectively (Kimbrough and Miller, 2002; Que et al., 2013; Waterman and Holden, 2003). Coxiella burnetii is a zoonotic pathogen and select agent that causes Q-fever, a flu-like disease that can lead to life-threatening endocarditis. The ability of C. burnetii, which utilizes a type IVB (T4BSS) or Dot/Icm secretion system to survive and replicate in a low pH, phagolysosome-like compartment (Coxiella-containing vacuole (CCV)), depends on the activity of many of its 143 annotated effector proteins (Larson et al., 2016). Brucella melitensis is a zoonotic select agent associated with spontaneous abortion in animals and both acute (febrile) and chronic (skeletal, cardiac, neurological complications) infection in humans. Although the full repertoire of Brucella T4ASS effectors remains poorly characterized, a number of the 15 known Brucella spp. effectors have been implicated in establishing a Brucella-containing vacuole (BCV) decorated with markers of the host endoplasmic reticulum (ER), proper trafficking of the BCV, and virulence in macrophages (de Figueiredo et al., 2015; Delrue et al., 2001; Pei et al., 2008).
Because many effectors target conserved eukaryotic pathways, yeast provides a convenient and appropriate surrogate host for heterologous expression of bacterial proteins (Lesser and Miller, 2001; Popa et al., 2016). Indeed, studies of bacterial effectors in yeast have provided insights into the functions of several effectors from diverse intracellular bacterial species (Curak et al., 2009; Popa et al., 2016; Siggers and Lesser, 2008). In this study, we build upon the success of focused bacterial effector yeast genetic screens (Burnaevskiy et al., 2013) and take advantage of epistasis mini-array profile (E-MAP) methodology, a type of synthetic genetic array analysis (SGA), which has been employed widely to identify functional interplay between yeast genes (Braberg et al., 2013; Collins et al., 2007; Patrick et al., 2015; Schuldiner et al., 2005), as well as heterologously expressed genes from pathogenic yeast species (Brown and Madhani, 2012) and human neurodegenerative proteins (Sun et al., 2011). An E-MAP improves upon traditional suppressor/enhancer screens by being quantitative, not requiring a baseline mutant growth defect, and by generating functional profiles for each query gene, together allowing a more robust prediction of gene function. These genetic interaction profiles, by virtue of containing 4,000+ pairwise genetic interaction data points, serve as rich functional signatures revealing nuanced ways in which an effector protein interacts with all pathways in a yeast cell.
In our application of the E-MAP, we constructed a set of 36 bacterial effector protein-expressing query strains that included both well-characterized effectors as well as putative effectors with no known molecular function. The resulting effector-host genetic interaction profiles were informative in predicting genes, complexes, and/or pathways potentially modulated by each bacterial effector. In addition to recapitulating known biology for several well-characterized effectors, the E-MAP identified a previously unappreciated role for the Salmonella protein SseC in maintaining the integrity of the SCV through interaction with the retromer complex (Burd and Cullen, 2014). We find that SseC physically interacts with components of the retromer complex and that depletion of the cellular retromer complex disrupts the integrity of the SCV, allowing for Salmonella to replicate in the cytosol of host cells. These data illustrate the utility of unbiased high-throughput yeast genetic interaction profiling over traditional yeast genetic screens to predict the molecular functions of bacterial effector proteins.
RESULTS
A high-throughput yeast genetic interaction screen generates functional signatures for a diverse set of bacterial effector proteins
We began by selecting a set of effector proteins to query from S. Typhimurium, C. burnetii and B. melitensis, three bacterial species with vastly different intracellular lifestyles (Figure 1A) and secretion systems (Figure 1B). Our criteria for selecting S. Typhimurium and C. burnetii effectors were that they had been previously validated substrates of the T3SS or T4BSS, respectively, and their ablation had been shown to confer a replication defect either in a cell line or mouse model of infection (Table 1). To demonstrate the ability of the E-MAP to recapitulate known biology, we included a handful of effectors with experimentally validated target pathways (e.g. ST SseG (Salcedo and Holden, 2003), ST SptP (Fu and Galán, 1999), ST GtgE (Spanò and Galán, 2012; Spanò et al., 2016; 2011), and CBU1314 (Weber et al., 2016)). The remainder of the effectors screened had little to no previous molecular characterization (Table 1). In the case of B. melitensis, because bona fide secreted effector proteins have not yet been described, we leveraged sequence homology to include orthologs of three validated B. abortus effectors (BME0390/VceA; BME0736/RicA; BME1111/VceC). The remaining B. melitensis query genes were selected using a recently developed bioinformatics tool called S4TE (searching algorithm for type-IV secretion system effectors) that identifies putative type IV effectors based on various protein features (e.g. homology to known effectors, presence of protein domains, features at the C-terminus, etc.) (Meyer et al., 2013) (Figure S1A). These putative B. melitensis effectors are of interest not only because they may provide insights into mechanisms of Brucella pathogenesis, but also because they allow us to test the utility of the EMAP as a way to identify and prioritize putative bacterial effectors that are most likely to interface with host biology.
Figure 1. Yeast E-MAP reveals the molecular function of bacterial effector proteins from three distinct human pathogens.
(A) Internalization and trafficking of Salmonella enterica serovar Typhimurium, Brucella melitensis and Coxiella burnetii. (B) Secretion systems utilized by S. Typhimurium, C. burnetii, and B. melitensis. (C) Modeling genetic interactions in yeast. Combining the expression of a bacterial effector protein with a cellular deletion can lead to nonlinear effects on cell growth, i.e. a genetic interaction. (D) Genetic interactions are quantified with the S-score (Collins et al., 2006). (E) Genetic interaction profiles for a given query gene across a library of cellular gene deletions reveal similarities in gene function when compared to one another. (F) Schematic representation of the E-MAP screen. Query strains express bacterial effectors from a low-copy CEN6/ARSH4 plasmid under the control of the GALS promoter. Query strains were mated with the S. cerevisiae non-essential gene deletion library with bacterial effector protein expression induced by plating yeast on media containing 2% galactose. (G) Heatmap of pairwise genetic interactions (S-scores) between effector-expressing query strains (y-axis) and cellular gene mutants (x-axis).
Table 1. Bacterial effectors included in E-MAP screen.
When known, the virulence phenotype of an effector mutant (in mice or in mammalian cells ex vivo) is listed.
| Effector | Virulence phenotype | Molecular function | Reference |
|---|---|---|---|
| ST AvrA | Y (mouse) | SPI-1; inhibits JNK/NFkB signaling; limits host inflammation | Lin, Z. et al. (2016). J Biol Chem. 291(52) Wu, H. et al. (2012). Cell Microbiol. 14(1) |
| ST CigR | Y (BMDM and mouse) | SPI-2; ND | Kidwai, A.S. et al. (2013). PLoS One. 8(8):e70753 Figueira, R. et al. (2013). MBio. 4(2):e00065 Lawley, T.D. et al. (2006). PLoS Pathog. 2(2):e11 |
| ST GtgE | Y (mouse) | SPI-2; Rab32 protease; prevents recruitment of Rab32 to SCV | Núñez-Hernández, C. et al. (2014). Infect Immun. 82(1) Spanò, S. (2016. Cell Host Microbe. 19(2) |
| ST PipB | N (fibroblasts) | SPI-2; ND | Chen, L.M. et al. (1996). Mol Microbiol. 21(5) |
| ST SipD | Y (mouse) | SPI-1; needle “tip” protein; required for invasion into host cells | Lawley, T.D. et al. (2006). PLoS Pathog. 2(2):e11 Kaniga, K. et al. (1995). J Bacteriol. 177(24) |
| ST SopD | Y (mouse) | SPI-1; membrane fission during invasion | Lawley, T.D. et al. (2006). PLoS Pathog. 2(2):e11 Jiang, X. et al. (2004). Mol Microbiol. 54(5) Lara-Tejero, M. and Galán, J.E. (2009) Infect Immun. 77(7) Bakowski, M.A. et al. (2007). Cell Microbiol. 9(12) |
| ST SptP | ND | SPI-1 restores actin cytoskeleton following invasion by acting as a GAP for Rac1 and Cdc42 | Fu, T. and Galán, J.E. (1999). Nature. 401(6750) |
| ST SpvB | Y (mouse) | SPI-2; destabilizes actin cytoskeleton via ADP ribosylation of actin monomers | Lawley, T.D. et al. (2006). PLoS Pathog. 2(2):e11 Lesnick, M.L. et al. (2001). Mol Microbiol. 39(6) |
| ST SseC | Y (RAW264.7 and mouse) | SPI-2; component SPI-2 translocon | Klein, J.R. and Jones, B.D. (2001). Infect Immun. 69(2). Hensel, M. et al. (1998). Mol Microbiol. 30(1) |
| ST SseG | Y (RAW264.7 and mouse) | SPI-2; recruitment/tethering of the SCV to the Golgi apparatus | Hensel, M. et al. (1998). Mol Microbiol. 30(1) Salcedo, S.P. et al. (2003). EMBO J. 22(19) |
| ST SseI | Y (mouse) | SPI-2; localizes to the plasma membrane; interferes with host cell migration in vivo | Lawley, T.D. et al. (2006). PLoS Pathog. 2(2):e11 McLaughlin, L.M. et al. (2009). PLoS Pathog. 5(11):e100671 |
| ST SseK1 | N (RAW264.7 or mouse) | SPI-2; GlcNAcylation of host proteins; inhibition of NFkB | Baison-Olmo, F. et al. (2015). Front Microbiol. 6:396 Gunster, R.A. et al. (2017). Infect Immun. 85(3) |
| ST SseK2 | Y (mouse) | SPI-2; ND | Lawley, T.D. et al. (2006). PLoS Pathog. 2(2):e11 |
| ST SseK3 | ND | SPI-2; GlcNAcylation of host proteins; inhibition of NFkB | Gunster, R.A. et al. (2017). Infect Immun. 85(3) |
| ST SteA | Y (BMDM and mouse) | SPI-2; Sif formation; modulation of SCV membrane dynamics | Lawley, T.D. et al. (2006). PLoS Pathog. 2(2):e11 McQuate, S.E. et al. (2017). Cell Microbiol. 19(1) Domingues, L. et al. (2014). Infect Immun. 82(7) |
| ST SteB | ND | SPI-2; ND | NA |
| CBU CirA | Y (J774.1 and HeLa) | RhoA GAP; disrupts host actin cytoskeleton | Weber, M.M. et al. (2013). J Bacteriol. 195(17) Weber, M.M. et al. (2016). Infect Immun. 84(9) |
| CBU0388 | Y (J774.1 and HeLa) | ND | Weber, M.M. et al. (2013). J Bacteriol. 195(17) |
| CBU CirB | Y (J774.1 and HeLa) | ND | Weber, M.M. et al. (2013). J Bacteriol. 195(17) |
| CBU0794 | N (J774.1 or HeLa) | ND | Weber, M.M. et al. (2013). J Bacteriol. 195(17) |
| CBU CirC | Y (J774.1 and HeLa) | ND | Weber, M.M. et al. (2013). J Bacteriol. 195(17) |
| CBU1198 | Y (J774.1) | ND | Weber, M.M. et al. (2013). J Bacteriol. 195(17) |
| CBU1217 | N (J774.1 or HeLa) | ND | Weber, M.M. et al. (2013). J Bacteriol. 195(17) |
| CBU1314 | Y (J774.1 and HeLa) | Localizes to the host cell nucleus; interacts with chromatin | Weber, M.M. et al. (2013). J Bacteriol. 195(17) Weber, M.M. (2016). Microbes Infect. 18(5) |
| CBU CirD | Y (J774.1 and HeLa) | ND | Weber, M.M. et al. (2013). J Bacteriol. 195(17) |
| CBU CirE | Y (J774.1 and HeLa) | ND | Weber, M.M. et al. (2013). J Bacteriol. 195(17) |
| BME0304 | Y (J774.1) | Putative transcriptional regulatory element | Wu, Q. et al. (2006). BMC Microbiol. 6:102 |
| BME0390 | ND | ND; Homolog of VceA in B. abortus | de Jong, M.F. et al. (2008). Mol Microbiol. 70(6) |
| BME0736 | Y (BMDM) | Homolog of RicA in B. abortus; binds to Rab2, controls kinetics of BCV trafficking | De Barsy, M. et al. (2011). Cell Microbiol. 13(7) |
| BME0948 | ND | ND | NA |
| BME1044 | ND | ND | NA |
| BME1111 | Y (BMDM and mouse) | Homolog of VceC in B. abortus; causes ER stress | De Jong, M.F. et al. (2013). MBio. 4(1) |
| BME1361 | ND | ND | NA |
| BME1482 | ND | ND | NA |
| BME1577 | ND | ND | NA |
| BME1658 | ND | ND | NA |
ND = No data
NA = Not available
The central principle behind the E-MAP is that the combinatorial effect of gene mutants on cellular growth reveals gene function. A genetic interaction exists when a double mutant displays a nonlinear growth defect based on the expected contribution of each single mutant (Figure 1C). In this study, the pair of mutants examined was 1) expression of a bacterial effector protein and 2) a cellular gene deletion. Genetically interacting genes are functionally related, and the magnitude and sign of the genetic interaction is quantified with the S-score (Figure 1D) (Collins et al., 2006). Positive genetic interactions between an effector and a cellular mutant indicate that expression of the effector protein caused the deletion strain to grow better than the average growth of effector-expressing mutant strains. Negative genetic interactions represent situations in which expression of the effector protein caused the deletion strain to grow worse than average (Figure 1D). The compilation of these effector-host genetic interactions for each bacterial effector constitutes a genetic interaction profile, i.e. a rich functional ‘signature’, reflecting the genetic interplay between the effector protein and host genes across biological processes. Genetic interaction profiles that show high similarity tend to be functionally similar, belonging to the same protein complex and/or cellular pathway (Figure 1E).
To interrogate the genetic interaction landscape of bacterial effector proteins from these three pathogens in S. cerevisiae, we cloned 43 effectors into a stable, low copy number, autonomously replicating (CEN6/ARSH4) plasmid under the control of a truncated galactose-inducible promoter (GALS) (Figure 1F). The GALS promoter, which lacks one and a half of the 3UAS elements required for full induction by galactose, was chosen to avoid toxicity associated with overexpression of exogenous proteins (induction is about 10–80x less in GALS vs. various GAL1 promoters) (Mumberg et al., 1994). Our use of the GALS promoter was strategic; unlike traditional suppressor screens, the E-MAP requires queries to be viable in order to generate a reproducible, quantifiable set of mutant yeast that express effector proteins. Because the E-MAP does not require effector expression to induce a discernable growth defect in wild-type yeast, the only pre-requisite for inclusion in the screen is that effector-expressing query strains are viable when grown on galactose. While the vast majority of effectors induced little to no growth defect when expressed by the GALS promoter, several of the effectors did cause slow growth on galactose (SseG, SopD, and CBU0388) but were healthy enough to survive the screen (Figure S1B). Several additional Salmonella effectors (PipA, SseF, SseJ, SopB, SifB, SopE, SopE2) were cloned but could not be included in the screen due to severe sickness/lethality on galactose (Figure S1C). Although no C. burnetii or B. melitensis effectors cloned were found to be lethal when expressed by the GALS promoter, previous results have shown that high expression of some C. burnetii effectors (e.g. CBU CirA and CirD) is indeed lethal to S. cerevisiae, highlighting the utility of the GALS promoter in the context of this screen (Weber et al., 2013). Using the E-MAP methodology in S. cerevisiae (Collins et al., 2006), we generated 36 bacterial effector genetic interaction profiles from a mutant library containing 4,465 non-essential host gene deletions—over 160,000 pairwise genetic interaction measurements (Figure 1F–G, Table S1).
Genetic interaction profiling reveals conserved eukaryotic cellular pathway targets for effectors from S. Typhimurium, C. burnetii and B. melitensis
Because previous iterations of the E-MAP have demonstrated that strong correlation of genetic interaction profiles between host genes indicates participation in related cellular pathways, we surmised that correlation between host profiles and effector profiles would similarly predict that an effector interfaces with a particular biological process. To begin to test this, we correlated our effector genetic interaction profiles against a previously generated collection of host genetic interaction profiles (Costanzo et al., 2016) with the Pearson’s correlation coefficient (PCC) followed by a z-score transformation (Table S1). Because it is conceivable that deletion of some yeast genes will generate pleiotropic effects and thus phenotypically resemble a number of bacterial effector-expressing query strains non-specifically, we imposed a strict filter for specificity, whereby a yeast mutant could not share a z-score greater than 5 with more than one effector to be included in our network analysis (Figure 2).
Figure 2. Correlated genetic interaction profiles predict interplay between bacterial effectors and diverse host pathways.
Yeast gene mutants that specifically correlated with a single effector, i.e. did not correlate with more than one effector (z-score > 5) are shown. Blue lines indicate effector-host correlations and grey lines indicate host-host protein-protein interactions as annotated by BIOGRID. Functional pathway “clouds” were manually annotated based on enriched GO term analysis (Table S2).
To guide the functional classification of the putative effector targets, we calculated enriched cellular pathways (Table S2) and manually curated genes into the broad categories of ribosome biology, ER/Golgi biology, actin cytoskeleton and nuclear biology to visualize functional similarity between effectors from each of the three bacterial pathogens (Figure 2). From this analysis, interplay between effectors and a number of infection-relevant host pathways began to emerge (Figure 2). Several effectors show strong enrichment with genes involved in trafficking through the endocytic pathway, particularly between the ER and Golgi apparatus (ST GtgE, ST SseG, ST SseC, BME0304, ST GtgE, and CBU CirD). Such interplay with vesicle trafficking is consistent with the role many effectors play in subversion of the canonical endocytic pathway. We also observed profile correlation between effectors and host genes involved in actin cytoskeleton organization (ST SptP, ST SseK2, BME0304), consistent with host cytoskeleton remodeling that occurs during internalization and trafficking of these intracellular bacteria pathogens (Colonne et al., 2016; Van Nhieu and Romero, 2017). Putative targets also extend into the nucleus with several effectors showing similar genetic interaction profiles with yeast mutants in kinetochore/microtubule biology (CBU0388), chromatin remodeling (CBU0794, CBU1314, BME1111), pre-mRNA splicing (CBU1314), and cell cycle regulation (BME1111 and CBU0388). Collectively, this analysis demonstrates that E-MAP profiling of bacterial effectors can predict putative target complexes that are specific for individual effectors and relate to a number of conserved eukaryotic biological pathways.
We next hypothesized that these same effector phenotypic signatures could be correlated with previously generated profiles from chemical-genetics screen. To this end, we compiled drug-host genetic interaction profiles for more than 13,000 compounds (Nelson et al., 2018). Consistent with our E-MAP-derived cellular pathway designations, the profile of CBU0388 correlated with compounds associated with microtubule processes and chromosome segregation, ST SseG and ST SseC with drugs associated with vesicle-mediated transport, and ST SptP with actin polymerase inhibitors like Latrunculin B. Intriguingly, correlation of effector profiles with drug profiles may not only provide insights into effector mechanisms of action but also identify compounds with anti-bacterial activity.
Lastly, we sought to determine whether genetic interaction profiles could predict functional similarities between bacterial effectors within and across the three pathogens queried. To this end, we measured the correlation of the genetic interaction profiles of all pairwise combinations of the 36 validated and putative effector proteins. Interestingly, we observed several highly correlated clusters of effectors, suggesting shared biology and/or common molecular targets. For example, strong correlations emerged among CBU0388, CBU1314, ST SopD, and ST SptP (Figure S2, cluster 2), consistent with their enrichment for correlation with genes involved in the actin cytoskeleton (Figure 2). Another cluster predicted similarity between ST SseG and ST SseC, suggesting a role for ST SseC similar to that described for ST SseG in modulating SCV trafficking in host cells (Salcedo and Holden, 2005) (Figure S2, cluster 3).
Correlations between bacterial effector genetic interaction profiles and yeast mutants identify known targets of characterized effectors
SptP’s genetic interaction profile specifically correlates with yeast mutants related to its target, Cdc42
To validate the extent to which genetic interaction profiling recapitulates known biology, we first selected ST SptP, a S. Typhimurium effector with well-known molecular functions, for further analysis. ST SptP, a SPI-1 T3SS substrate, is a tyrosine phosphatase and a GTPase-activating protein (GAP) for the mammalian Rho GTPases Rac-1 and Cdc42, which are important for regulating actin dynamics as well as growth and budding in yeast cells (Fu and Galán, 1998; 1999). By mimicking the action of a cellular GAP, SptP modulates Cdc42 by converting Cdc42-GTP to Cdc42-GDP (Figure 3C). These activities are required for ST SptP to reverse SopE-dependent changes to the host actin cytoskeleton and promote host-cell recovery after bacterial invasion (Fu and Galán, 1998). Previous work has shown that ST SptP targets Cdc42 in yeast and can act as an inhibitor of Cdc42-dependent budding and mating pathways (Rodríguez-Pachón et al., 2002).
Figure 3. Genetic interaction profiling recapitulates actin and filamentous growth phenotypes associated with ST SptP expression in S. cerevisiae.
(A) Enriched GO terms forcorrelating genetic interaction profiles for ST SptP. (B) Genes whose genetic interaction profile correlated (z-score >5) with only ST SptP (C) Schematic representation of CDC42-GTP to GDP conversion. Mutants of CDC24 and CDC28 whose profiles correlated with that of ST SptP are indicated (along with each of their z-scores) (D) Correlation of ST SptP and CDC genes, with black brackets indicating CDC24 and CDC28 mutants
Pathway and gene level analyses validated the actin cytoskeleton as being the predominant target of ST SptP. The Gene Ontology (GO) terms ‘actin filament bundle’, ‘actin binding’, and ‘cytoskeleton’ were strongly enriched as well as the ‘cell bud growth’ and ‘mating projection tip’ pathways (Figure 3A), the latter of which corroborate ST SptP’s ability to influence growth downstream of Cdc42-dependent Slt2 MAPK signaling (Rodríguez-Pachón et al., 2002). Yeast mutants with strongly correlating genetic interaction profiles specific to ST SptP reveal a number of actin cytoskeleton genes, including direct regulators of Cdc42 such as Bem4 and Cdc24 (Figure 3B). The lack of correlation between ST SptP and certain cdc24 mutants (cdc24-2, -3, and -4) is consistent with these mutants having different genetic interaction profiles than cdc24-1, -5, -11, and -H, as evidenced by hierarchical clustering (Figure S3). We also observed positive correlations between ST SptP and mutants of Cdc28 (Cyclin dependent kinase 1), an activator of Cdc24 (Figure 3D). Together, these results confirm that yeast genetic profiling can predict pathways and molecules targeted by characterized bacterial effectors.
Subcellular localization of bacterial effectors is predicted by genetic interaction profile correlations
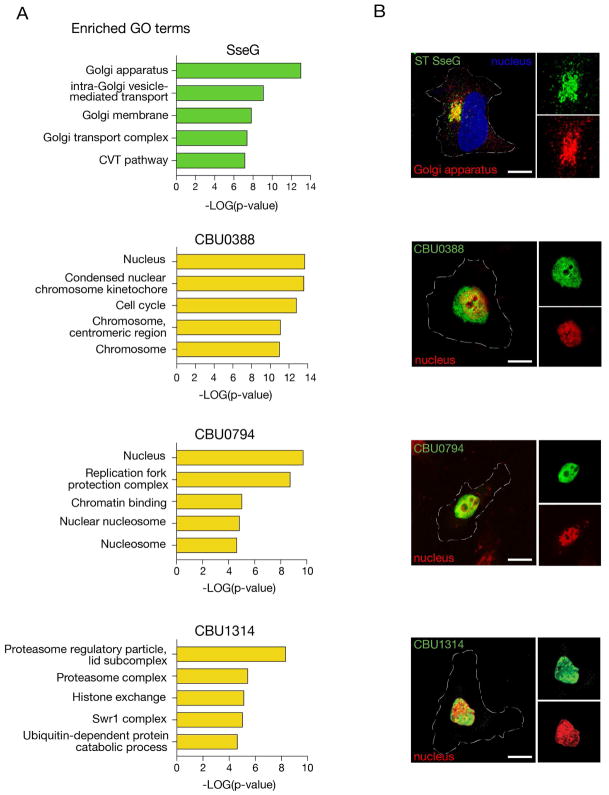
We next wanted to determine whether correlation of genetic interaction profiles could predict subcellular compartments, and specific host cell pathways contained therein, targeted by different effectors. Based on the cohort of host profiles that strongly correlated with that of each effector (Figure 2), we identified several effectors (ST SseG, CBU0388, CBU0794, CBU1314) for which the E-MAP predicted a likely subcellular compartment—either the Golgi apparatus (ST SseG) or the nucleus (CBU0388, CBU0794, and CBU1314). To confirm these predictions, we transfected tagged constructs into HeLa cells and the subcellular localization of each ectopically expressed effector was followed by immunofluorescence microscopy. Previous work has shown that ST SseG is involved in targeting the SCV to the Golgi (Salcedo and Holden, 2005). Our E-MAP analysis uncovered strong enrichment of Golgi-related pathways among correlating yeast mutants and we saw striking localization of ST SseG to the Golgi in HeLa cells (Figure 4A–B). Likewise, we observed significant enrichment for correlations with genes involved in nuclear biology with the three C. burnetii effectors (Figure 4A). Within these broad categories, enrichment with particular nuclear genes/pathways was evident: CBU0388 predominantly interfaces with the centromere and kinetochore, and CBU0794 correlates with mutants involved in chromatin/histone biology. In the case of CBU1314, we saw GO term enrichment driven by genes in both the SWR1 histone exchange complex and the proteasome, perhaps suggesting a ubiquitin-dependent mechanism through which CBU1314 affects chromatin remodeling (Figure 4A). Consistent with these predictions, we observed nuclear localization of GFP-tagged versions of each of these effectors when exogenously expressed in HeLa cells (Figure 4B). Together, these data reveal the utility of using GO term analysis based on highly correlating E-MAP z-scores to predict subcellular localization and putative molecular function of bacterial effectors.
Figure 4. Subcellular localization of bacterial effectors is predicted by E-MAP GO term enrichment.
(A) Enriched GO terms for significantly correlating genetic interaction profiles for ST SseG, CBU0388, CBU0794, and CBU1314. (B) HeLa cells transiently transfected to express 3xFLAG-ST SseG or GFP-CBU0388, GFP-CBU0794, GFP-CBU1314; co-stained with an antibody against RCAS1 to visualize the Golgi apparatus or Hoescht to visualize nuclei. Scale bar = 10 μm.
Yeast genetic interaction profiling leads to the identification of the human retromer complex as a target of the S. Typhimurium effector ST SseC
Having shown that the E-MAP could confirm known effector biology at the molecular level, we were interested in whether the E-MAP could predict the molecular target of an uncharacterized effector. One effector that quickly rose to the top of our list was ST SseC. It is currently unknown if SseC interfaces with any host cell biology beyond its role as a component of the SPI-2 translocon pore (Nikolaus et al., 2001). Examination of the top specific z-score correlations indicated obvious connections between ST SseC and the retromer complex (Figure 5A). The retromer is an evolutionarily conserved protein complex that plays a key role in trafficking endosomal cargo away from the lysosome degradative pathway (Burd and Cullen, 2014). All five components of the retromer complex (GO:0030904) had z-scores >5 that were specific for ST SseC: PEP8, VPS29, VPS17, VPS5, VPS35) (Figure 2 and 5A). Because the enrichment of genetic interaction profile correlations between ST SseC and the retromer was so prominent, we were curious to determine whether individual genetic interactions predicted a connection between ST SseC and the retromer. Interestingly, ST SseC expression did not generate significant positive or negative pairwise genetic interactions with retromer genes (Figure 5B). This is in contrast with the high degree of genetic interaction profile similarity between ST SseC and retromer mutants (Figure 5B). Because of the striking correlations of genetic interaction profiles with retromer mutants and ST SseC, and because very little is known about how the retromer influences Salmonella biology, we focused on the potential role of SseC in mediating retromer function during Salmonella infection.
Figure 5. ST SseC interacts with the retromer complex in vivo and in vitro.
(A) Highest ranking genes that specifically correlate with ST SseC, with gene #19 included to depict all five components of the retromer complex (underlined). (B) Distribution of all ST SseC S-scores and z-scores with the five retromer mutants represented as red circles. (C) Immunoblot analysis of interaction between 3xFLAG-ST SseC and VPS35 (HA-tagged and endogenous), HA-VPS26A, HA-TBC1D5. (D) In vitro pull-down assays between ST SseC and single retromer components (VPS26A, VPS29, and VPS35) (left panel) or combinations of retromer components (right panel). SscA, a chaperone for SseC (Cooper et al., 2013), was included in all reactions. CTL (control) (E) Equilibrium binding of ST SseC with combination of retromer components by SPR. The dissociation constant (Kd) was derived by fitting of the equilibrium binding data to a one site binding model (right panel).
Previous iterations of the E-MAP have convincingly shown that strong genetic interaction profile similarity is often observed between components of protein complexes (Collins et al., 2006). Therefore, we next sought to determine whether ST SseC physically interacted with the retromer in human cells. By both co-immunoprecipitation (Figure 5C) and affinity purification mass spectrometry (Figure S4A), we observed that ST SseC specifically interacted with several members of the core mammalian retromer complex including the vacuolar protein sorting-associated protein 35 (hVPS35; yeast VPS35) and vacuolar protein sorting-associated protein 26, subunits A and B (hVPS26A/B; yeast Pep8). Our AP-MS analysis also detected an interaction with TBC1 domain family member 5 (TBC1D5), a Rab7 GAP and likely regulator of the retromer, which we confirmed by co-immunoprecipitation (Figure 5C, S5A)(Seaman et al., 2009). Importantly, we did not detect an interaction of the retromer complex with ST SteB (by AP-MS) or ST SseG (by co-IP or AP-MS) despite having somewhat similar genetic interaction profiles with ST SseC (PCC=0.4 and 0.7, respectively), underscoring the specificity of the ST SseC-retromer physical interaction. Together with our genetic profiling data, both co-immunoprecipitation and mass spectrometry experiments suggest that ST SseC forms a complex with components of the retromer in vivo.
To further interrogate the precise nature of the interaction between ST SseC and retromer components, we individually expressed and purified human VPS35, VPS26A, VPS29 as well as ST SseC (co-expressed with its chaperone SscA to promote protein stability) and conducted an in vitro pull-down assay. Interactions were tested between ST SseC and each retromer component individually, as well as all combinations of the components. Remarkably, we found that both VPS35 and VPS26A were required for their interaction with ST SseC (Figure 5D, right blot, lanes 3 and 7) as no binding is observed between ST SseC and single retromer components (Figure 5D, left blot, lanes 1, 3, 5) or other combinations (Figure 5D, right blot, lanes 1, 5; inputs in Figure S4B). Next, to determine the binding affinity and kinetic properties of retromer binding by ST SseC, we performed surface plasmon resonance (SPR). Again, ST SseC binding was only observed in the presence of both VPS35 and VPS26A (Figure 5E). No binding was observed with the chaperone SscA alone (Figure S4C). Subsequent equilibrium binding studies revealed that ST SseC has a strong affinity for the VPS35/VPS26A subcomplex (380nM) (Figure 5E, right panel). Based on our genetic and biochemical studies, we hypothesize that ST SseC has evolved to interact with a specific surface of the retromer (VPS35/VPS26A) in order to remodel the cytosolic surface of the SCV and ensure proper trafficking and survival of intracellular S. Typhimurium.
The retromer regulates the functional integrity of the SCV
To further validate our in vivo and in vitro biochemistry results demonstrating a physical interaction between ST SseC and the retromer, we examined where retromer components localize during S. Typhimurium infection and whether their localization was dependent on SseC. Briefly, we infected HeLa cells with WT or ΔsseC S. Typhimurium and measured recruitment of endogenous retromer (VPS35) to the SCV over a time-course of infection using immunofluorescence microscopy and a native VPS35 antibody. We detected robust accumulation of VPS35 that associated with invading Salmonella and also localized to the plasma membrane ruffles at very early time points following S. Typhimurium invasion (15 min. post-infection) (Figure 6A), supporting a role for the retromer in sorting the SCV immediately upon pathogen uptake. Following this population of VPS35-positive SCVs over time, we observed that wild-type SCVs no longer co-localized with the retromer starting at 2 hours post-infection. In stark contrast, ΔsseC SCVs remained retromer-positive at the 2 hour time-point (Figure 6A–C). Importantly, retromer dissociation from the SCV is concomitant with a dramatic increase in SseC expression (Figure 6D), as has been previously reported for SPI-2 upregulation (Hautefort et al., 2008). These data are consistent with a role for SseC in promoting remodeling or releasing the retromer from the SCV at 2 hours post infection.
Figure 6. The retromer complex dynamically associates with the SCV and is required for S. Typhimurium intracellular survival in macrophages.
(A) Co-localization of VPS35 and the SCV in HeLa cells infected with wild-type S. Typhimurium at MOI=30. (B) Infection as in (A) but alongside ΔsseC S. Typhimurium. (C) Quantitation of (B). (D) SseC expression over a time-course of early infection. ΔsseC and WT ST labels indicate SseC expression in starting inoculum. SseC expression is shown relative to 16S rRNA expression. Error bars represent SEM of 6 technical replicates. (E) shRNA knockdown of VPS35 in human THP-1 macrophages. (F) CFU recovery of S. Typhimurium-infected THP-1 cells (MOI=30) expressing SCR or VPS35 shRNA. A late time point (18h post infection) was taken as part of a separate experiment. (G) shRNA knockdown of VPS35 in HeLa cells. (H) CFU recovery of wild-type S. Typhimurium (MOI = 30) from wild-type and VPS35 knockdown HeLa cells at 4h and 8h post-infection, relative to 1h. (I) CFU recovery of total (black) and cytosolic (red) S. Typhimurium from wild-type and VPS35 knockdown HeLa cells. (J) Digitonin permeabilization assay to detect cytosolic (green) versus total intracellular bacteria (red). (K) Quantitation of percent cytosolic bacteria at 1 and 2 hours post-infection. (L) Potential model for retromer/SCV interactions. Upon internalization in wild-type cells, (1) SPI-1 T3SS releases SopB, which generates PI(3)P. Sorting nexins 1&3 bind to PI(3)P at the nascent SCV. VPS35 binds to SNX1 and/or 3. Retromer association with the SCV promotes trafficking to the Golgi apparatus. (2) Subsequent acidification of the vacuole promotes SseC/SPI-2 T3SS expression. SseC interacts with VPS35/VPS26a. (3) SseC (and/or other SPI-2 effectors) promote release of the retromer complex from the SCV, likely through recruitment of the Rab7 GAP TBC1D5 and (4) the integrity of the SCV is maintained and the SCV is tethered in the vicinity of the Golgi apparatus. In the absence of SseC (ΔsseC), VPS35 is retained at the retromer past 2h post-infection, perhaps leading to changes in SCV localization and/or maintenance of the vacuole at later time points. In the absence of VPS35 (VPS35KD), a population of SCVs does not maintain its integrity and a higher percentage of Salmonella are exposed to the cytosol, leading to hyper-replication. *p<0.05 by two-tailed student’s t-test compared to wild-type. Scale bar = 10μm. Unless otherwise stated, error bars represent STDEV of 3 different biological samples.
Previous work has shown that sorting nexins (SNX) 1 and 3, components of the retromer cargo recognition complex, are recruited to the sites of Salmonella uptake in a manner that relies on generation of phosphatidylinositol 3-phosphate [PI(3)P] by the SPI-1 effector SopB (Bujny et al. 2008; Braun et al. 2010). Because recruitment of the downstream retromer vacuolar sorting complex (e.g. VPS35) also required SopB (Figure S5A–B), we propose that the retromer (first the sorting nexins and then the sorting complex) is recruited to the SCV concomitant with S. Typhimurium uptake following SopB-dependent generation of PI(3)P.
To more precisely define a functional role for the retromer during S. Typhimurium infection, we next evaluated the fate of invading bacteria in cells lacking VPS35. We generated THP-1 human macrophages that stably express an shRNA targeting VPS35 (Figure 6E). Following infection of VPS35KD macrophages with wild-type S. Typhimurium, we observed a steady decrease in S. Typhimurium replication over time (Figure 6F–G). Because cytosolic S. Typhimurium are rapidly degraded in THP-1 cells (Beuzón et al., 2002), we hypothesized that S. Typhimurium may hijack the retromer to maintain the integrity of SCV. Therefore, the loss of retromer could cause S. Typhimurium to be released into the cytosol, where in the case of phagocytic cells it is destroyed. To test this hypothesis, we took advantage of non-immune HeLa cells, which cannot control cytoplasmic replication of S. Typhimurium (Beuzón et al., 2002; Brumell et al., 2002). In support of our hypothesis, we observed an increase in S. Typhimurium replication in HeLa cells stably expressing an shRNA against VPS35 (Figure 6G–H). To differentiate whether we were detecting vacuolar or cytosolic bacteria, we employed a chloroquine protection assay, which selectively kills vacuolar bacteria. In the VPS35 knockdown cells, we noticed a striking increase in the number of cytosolic bacteria at both 4 and 8 hours post-infection, concomitant with an increase in total bacteria (Figure 6I), further supporting our hypothesis that VPS35 is required for maintenance of the SCV.
These data suggest that in the absence of VPS35 there is an increase in the initial number Salmonella that escape the nascent SCV and go on to hyperreplicate within the cytosol of epithelial cells. To more definitively determine the number of cytosolic bacteria in infected HeLa cells, especially at early time points, we utilized a digitonin permeabilization assay to selectively permeabilize the plasma membrane and deliver anti-LPS antibodies directly to the cytosol of Salmonella-infected epithelial cells. Subsequent saponin permeabilization, which breaks down the SCV, and a second anti-LPS staining allows for enumeration of total intracellular bacteria. To ensure that our digitonin treatment was selectively permeabilizing the plasma membrane, and not intracellular vacuolar compartments, we performed a control experiment using an antibody against luminal Lamp-1, wherein the lack of Lamp-1 staining signals that intracellular membranes are intact (Figure S5E). Using this optimized assay we quantified cytosolic versus total bacteria at 1 and 2 hours post infection in both control and VPS35 knockdown HeLa cells. We found that there was significantly more cytosolic S. Typhimurium in VPS35 knockdown cells compared to control cells (~5% versus 20%, respectively) (Figure 6J–K). Using this assay we were also able to define cells with hyperreplicating cytosolic bacteria (Figure S5C–D). Corroborating the CFU data in Figure 6I, we found that ~2 fold more cells harbored >50 cytosolic bacteria in the VPS35 knockdown versus control cells. Collectively these data strongly suggests that the retromer plays a role in maintenance of the SCV.
DISCUSSION
While no single molecular approach can capture all relevant effector-host interactions, the results from this study strongly argue that genetic interaction profiling in yeast is a powerful platform for uncovering intracellular pathogen effector functions. By using a low copy plasmid, a weak GALS promoter, and applying stringent specificity filters, our screen was designed to generate high-confidence target predictions while minimizing artifacts stemming from exogenous effector overexpression. Although screening effector function in yeast has certain limitations stemming from the evolutionary distance between yeast and humans, most notably the lack of innate immune molecules in yeast, the genetic interaction profiling approach presented here can be adapted for effector screens in human cells and the development of such technologies is ongoing.
S. Typhimurium and the retromer complex
Genetic interaction profiling coupled with in vivo and in vitro biochemistry allowed us to uncover the retromer complex as the specific molecular target of ST SseC. Subversion of retrograde transport is emerging as an important adaptation for intracellular pathogen survival, with the retromer being a common target of translocated pathogen effectors (Personnic et al., 2016). Recent work has shown that Chlamydia trachomatis lncE subverts host restriction by binding to the Sorting Nexin 5 (SNX5) protein and disrupting retromer trafficking (Elwell et al., 2017; Mirrashidi et al., 2015). Similarly, L. pneumophila RidL binds the VPS29 retromer subunit to alter retrograde trafficking and support intracellular bacterial growth (Finsel et al., 2013). Retromer is also required for C. burnetii replication and survival in host cells, although the effectors that mediate this interplay remain unknown (McDonough et al., 2013).
To date, the VPS26-VPS29-VPS35 retromer sorting complex has not been implicated in Salmonella infection. Interestingly, while both SNX1 and 3 have been shown to be important for the formation of Salmonella vacuole-associated tubules, only SNX1 has been found on the SCV itself, with kinetics consistent with what we observed for VPS35, whereby co-localization is concomitant with uptake but drops off ~3 hours post-infection (Bujny et al., 2008). Together, our data suggests a model whereby SPI-1 secretion of SopB generates PI(3)P, which is recognized by SNX1 and/or 3, promoting recruitment of the VPS26A-VPS29-VPS35 retromer sorting complex, which is then likely remodeled or released by ST SseC (Figure 6L). Based on our observation that ST SseC and the Rab7 GAP/negative regulator of retromer TBC1D5 also interact, we propose that ST SseC engages the retromer at the VPS26A-VPS35 interface, promoting recruitment of TBC1D5, which then stimulates the GTPase activity of Rab7 and drives retromer release from the SCV. While additional experimentation is needed to fully elucidate the consequence of prolonged SCV-retromer association, we hypothesize it will lead to mistrafficking of the SCV, perhaps interfering with SseF/G-dependent tethering of the SCV at the Golgi apparatus (Figure 6L). This idea of dynamic, spatiotemporally regulated interactions between effectors and host molecules is a theme in Salmonella pathogenesis: just as SopE/SopE2 and SptP have opposing effects on polarization of the actin cytoskeleton, so too may SopB and SseC regulate recruitment and release of the retromer, respectively.
Little is known about how components of T3SS translocon pores interface with host cell molecules. The Shigella flexerni translocon pore component IpaC has been shown to interact with intermediate filaments (Russo et al., 2016). Similar data has been shown for the Salmonella SPI-1 translocon pore component SipC, domains of which are presumed to interface with the host to promote actin bundling and nucleate actin polymerization (Hayward and Koronakis, 1999; Myeni and Zhou, 2010). Our finding that the Salmonella SPI-2 translocon pore component SseC (Hensel et al., 1998; Nikolaus et al., 2001) directly interacts with the retromer complex was unexpected. The idea that ST SseC evolved to bind a specific interface between VPS35/VPS26A has exciting implications for future structure-function analysis of this and other pathogen secretion systems and highlights how the study of host-pathogen interactions can illuminate basic host cell biology. Because genetically ablating SseC disrupts the translocation of other effectors (Klein and Jones, 2001; Nikolaus et al., 2001), structural studies of the Salmonella SPI-2 complex or SseC-retromer structure are needed so that we may generate mutants that uncouple SseC’s role in regulation of retromer dynamics from its role in secretion of SPI-2 effectors. Currently, we can only speculate that a portion of ST SseC is accessible to the cytosolic face of the SCV such that it can interact with the retromer.
With hundreds of bacterial effector proteins from medically important pathogens awaiting characterization, there is a critical need to interrogate how these molecules interface with host cells using unbiased, high-throughput approaches. Our application of the E-MAP platform in yeast identified conserved eukaryotic targets of effectors from evolutionarily distant bacterial pathogens. The degree to which these highly-specialized bacterial effector proteins recapitulated precise biochemical activities in the budding yeast speaks to an incredible degree of conservation between yeast and mammals and argues strongly for the continued use of heterologous model systems to ascribe functions to proteins of unknown function.
STAR Methods
KEY RESOURCES TABLE
The table highlights the genetically modified organisms and strains, cell lines, reagents, software, and source data essential to reproduce results presented in the manuscript. Depending on the nature of the study, this may include standard laboratory materials (i.e., food chow for metabolism studies), but the Table is not meant to be comprehensive list of all materials and resources used (e.g., essential chemicals such as SDS, sucrose, or standard culture media don’t need to be listed in the Table). Items in the Table must also be reported in the Method Details section within the context of their use. The number of primers and RNA sequences that may be listed in the Table is restricted to no more than ten each. If there are more than ten primers or RNA sequences to report, please provide this information as a supplementary document and reference this file (e.g., See Table S1 for XX) in the Key Resources Table.
Please note that ALL references cited in the Key Resources Table must be included in the References list. Please report the information as follows:
REAGENT or RESOURCE: Provide full descriptive name of the item so that it can be identified and linked with its description in the manuscript (e.g., provide version number for software, host source for antibody, strain name). In the Experimental Models section, please include all models used in the paper and describe each line/strain as: model organism: name used for strain/line in paper: genotype. (i.e., Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J). In the Biological Samples section, please list all samples obtained from commercial sources or biological repositories. Please note that software mentioned in the Methods Details or Data and Software Availability section needs to be also included in the table. See the sample Table at the end of this document for examples of how to report reagents.
SOURCE: Report the company, manufacturer, or individual that provided the item or where the item can obtained (e.g., stock center or repository). For materials distributed by Addgene, please cite the article describing the plasmid and include “Addgene” as part of the identifier. If an item is from another lab, please include the name of the principal investigator and a citation if it has been previously published. If the material is being reported for the first time in the current paper, please indicate as “this paper.” For software, please provide the company name if it is commercially available or cite the paper in which it has been initially described.
-
IDENTIFIER: Include catalog numbers (entered in the column as “Cat#” followed by the number, e.g., Cat#3879S). Where available, please include unique entities such as RRIDs, Model Organism Database numbers, accession numbers, and PDB or CAS IDs. For antibodies, if applicable and available, please also include the lot number or clone identity. For software or data resources, please include the URL where the resource can be downloaded. Please ensure accuracy of the identifiers, as they are essential for generation of hyperlinks to external sources when available. Please see the Elsevier list of Data Repositories with automated bidirectional linking for details. When listing more than one identifier for the same item, use semicolons to separate them (e.g. Cat#3879S; RRID: AB_2255011). If an identifier is not available, please enter “N/A” in the column.
A NOTE ABOUT RRIDs: We highly recommend using RRIDs as the identifier (in particular for antibodies and organisms, but also for software tools and databases). For more details on how to obtain or generate an RRID for existing or newly generated resources, please visit the RII or search for RRIDs.
Please use the empty table that follows to organize the information in the sections defined by the subheading, skipping sections not relevant to your study. Please do not add subheadings. To add a row, place the cursor at the end of the row above where you would like to add the row, just outside the right border of the table. Then press the ENTER key to add the row. Please delete empty rows. Each entry must be on a separate row; do not list multiple items in a single table cell. Please see the sample table at the end of this document for examples of how reagents should be cited.
TABLE FOR AUTHOR TO COMPLETE
Please upload the completed table as a separate document. Please do not add subheadings to the Key Resources Table. If you wish to make an entry that does not fall into one of the subheadings below, please contact your handling editor. (NOTE: For authors publishing in Current Biology, please note that references within the KRT should be in numbered style, rather than Harvard.)
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-VPS35 | Santa Cruz Biotechnology | Sc-374372 |
| Anti-HA High Affinity from rat IgG1 | Sigma-Aldrich | 3F10 |
| Monoclonal ANTI-FLAG M2 antibody produced in mouse | Sigma-Aldrich | F3165 SIGMA |
| Salmonella O Group B antiserum | Becton, Dickson and Company | L006761 (0800) |
| Anti-Lamp-1 | Developmental Studies Hybridoma Bank (University of Iowa) | G1/139/5 |
| Bacterial and Virus Strains | ||
| Salmonella enterica serovar Typhimurium | Helene Andrews-Polymenis, TAMHSC | SL1344 |
| Escherichia coli strain BL21(DE3) | Novagen | Cat.# 69450 |
| Escherichia coli strain STBL3 | ThermoFisher | C737303 |
| Biological Samples | ||
| none | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| FLAG peptide | Sigma-Aldrich | F4799 Sigma |
| Biotin | SIGMA-ALDRICH | Lot#SLBS8478 |
| Streptavidin Agarose beads | EMD Millipore Corp. | Lot#2975711A |
| Yeast extract | BD Biosciences | 212720 |
| agar | BD Biosciences | 214030 |
| Peptone | BD Biosciences | 211820 |
| Yeast nitrogen base w/o amino acids | BD Biosciences | 291920 |
| Yeast nitrogen base | BD Biosciences | 233520 |
| Dextrose | Fisher | D16-3 |
| Galactose | Fisher | BP656-500 |
| Nat | WernerBioAgentsG mbH | 96736-11-7 |
| G418 | Gibco | 11811-098 |
| Canavanine | Sigma | C9758 |
| S-AEC | Sigma | A2636 |
| raffinose | Sigma | R0250 |
| potassium acetate | Sigma | P5708 |
| Adenine hemisulfate | Sigma | A9126 |
| Alanine | Sigma | A7627 |
| Asparagine | Sigma | A7094 |
| Aspartic acid | Sigma | A9256 |
| Cysteine | Sigma | C7352 |
| Glutamine | Sigma | G3126 |
| Glutamic Acid | Sigma | G1626 |
| Glycine | Sigma | G7126 |
| Inositol | Sigma | I7508 |
| Isoleucine | Sigma | I2752 |
| Leucine | Sigma | L8000 |
| Methionine | Sigma | M9625 |
| Para-aminobenzoic acid | Sigma | A0254 |
| Phenylalanine | Sigma | P2126 |
| Proline | Sigma | P0380 |
| Serine | Sigma | S4500 |
| Threonine | Sigma | T8625 |
| Tryptophan | Sigma | T0254 |
| Tyrosine | Sigma | T3754 |
| Uracil | Sigma | U0750 |
| Valine | Sigma | V0500 |
| Critical Commercial Assays | ||
| Biotin CAPture Kit | GE Healthcare | product# 28920233 |
| DirectZol RNA miniprep kit | Zymo Research | R2052 |
| Deposited Data | ||
| Experimental Models: Cell Lines | ||
| THP-1 | ATCC | TIB-202 |
| HeLa | ATCC | CCL-2 |
| MATα query strain | PMID: 17101447 | can1Δ::STE2pr- SpHIS5 lyp1Δ::STE3pr- LEU2 his3Δ1 leu2Δ0 MET15+ ura3Δ0 |
| MATa nonessential deletion library (BY4741) | Open Biosystems | YSC1053 |
| Experimental Models: Organisms/Strains | ||
| THP-1 VPS35 shRNA knockdown | This paper | N/A |
| Hela VPS35 shRNA knockdown | This paper | N/A |
| Oligonucleotides | ||
| CGG GGA GGA AGG TGT TGT G | This paper | ST_16S_rRNA_F |
| GAG CCC GGG GAT TTC ACA TC | This paper | ST_16S_rRNA_R |
| CAGGAGCAGATCCAGAAAGC | This paper | ST_SseC_F |
| GCCGGTAATCCAGTCAAAAA | This paper | ST_SseC_R |
| GCTGTGAAGGTCCAGTCATT | This paper | hVPS35_KD_F |
| CCGGAGTTCACCAAGCATATTA | This paper | hVPS35_KD_R |
| CTGGACTTCGAGCAAGAGATG | This paper | hACT_F (human actin) |
| GAGTTGAAGGTAGTTTCGTGGA | This paper | hACT_R (human actin) |
| TggtgaactccggacttctaTTCAAGAGATAGAtagaagtccggagttcaccTTTTTTC | IDT | pSICO_vps35_KD #4_F (targeting sequence in lowercase) |
| TCGAGAAAAAAggtgaactccggacttctaTCTCTTGAAtagaagtccggagttcaccA | IDT | pSICO_vps35_KD #4_R |
| TcctaggttaagtcgccctTTCAAGAGATAGAagggcgacttaacctaggTTTTTTC | IDT | pSICO_SCR_F |
| TCGAGAAAAAAcctaggttaagtcgccctTCTCTTGAAagggcgacttaacctaggA | IDT | pSICO_SCR_R |
| Recombinant DNA | ||
| pDest-3xFLAG-VPS35 | This paper | |
| pDest-3xFLAG-VSP26A | This paper | |
| pDest-3xFLAG-VSP29 | This paper | |
| pDest-3xFLAG-GFP | This paper | |
| pDest-3xFLAG-GFP | This paper | |
| pET22b(+) vector | Novagen | Cat.#69744 |
| SscA gene | Genscript | Gene ID: 1252917 |
| pET28a-SUMO | Chang Shu et al. 2012 | |
| Software and Algorithms | ||
| Biacore X100 Evaluation software version 2.0 (GE Healthcare) | GE Healthcare | https://www.biacore.com/lifesciences/service/downloads/downloads/index.html |
| E-MAP analysis software | PMID: 16859555 | https://sourceforge.net/projects/emap-toolbox/ |
| Matlab | Mathworks | version 7.13 (R2011b) |
| Colony measure | na | https://sourceforge.net/projects/ht-col-measurer/ |
| R | version 3.1.3 | |
| Other | ||
| 1× HBS-EP+ buffer | GE Healthcare | BR-1008-26 |
| HTP agar plates | Singer Instruments | PP-200 |
| HTP 1536-pin yeast pads | Singer Instruments | RP-MP-1586 |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Robert O. Watson (robert.watson@medicine.tamhsc.edu)
EXPERIMENTAL MODEL AND SUBJECT DETAILS
HeLa and THP-1 cell lines were maintained in DMEM + high glucose + 10% FBS + 10mM HEPES buffer. Cells were grown to approximately 80% confluency before each passage. Passages were kept to a minimum (<20). For selection, cells were grown with 100ug/ml hygromycin B.
Salmonella enterica serovar Typhimurium strain SL1344 was struck onto fresh LB plates from frozen stocks and grown at 37°C overnight. Plates were subsequently stored at 4°C. Fresh streaks were made approximately every week to ensure the viability of bacteria.
METHOD DETAILS
E-MAP genetic interaction profiling
We began by engineering a yeast expression plasmid that would allow for controlled expression of bacterial effector genes. The truncated galactose inducible promoter expression construct was a modified version of p415 GALS (ATCC 87346) in which the LEU2 cassette was replaced with the NATMX6 cassette via Gibson assembly. Bacterial effectors were PCRed using primers with 25nt of sequence that overlapped with the p415 plasmid from species-specific genomic DNA allowing for Gibson assembly of PCR-amplified effectors and PCR-amplified linearized p415-GALS-NAT plasmid. Constructs were transformed into the MATα query strain (can1Δ::STE2pr-SpHIS5 lyp1Δ::STE3pr-LEU2 his3Δ1 leu2Δ0 MET15+ ura3Δ0) (Schuldiner et al., 2006).
MATα query strains were first grown overnight as a lawn on YPAD (120 mg/L adenine, 10 g/L yeast extract, 20 gL peptone, 20 g/L agar, 20 g/L dextrose) + 100 mg/L Nat at 30C for 24 hr and then pinned onto a fresh YPAD+Nat plate using an automated 1536-dense pinning robot (Rotor, Singer Instruments) and incubated for 24 hr at 30C. These colonies were then mated with a library of haploid MATa nonessential deletion strains (BY4741 strain, Open Biosystems) onto drug-free YPAD plates for 24 hr at RT, and then pinned to YPAD+Nat+G418 to select for diploids. Diploids were then sporulated in triplicate onto NGS media (20 g/L agar, 3 g/L potassium acetate, and 0.02% raffinose) for 5 days at RT, between two large trays with a beaker of water in order to maintain a dark humid environment. From this point on, the technical replicates (n=3) were maintained in parallel. Spores were transferred to haploid selection media (20 g/L agar, 20 g/L dextrose, 6.7 g/L yeast nitrogen base w/o amino acids, 2 g/L Drop-out mix (-HIS -LYS -ARG), 50 mg/L canavanine, and 50 mg/L S-AEC) and incubated for 48 hr at 30C. HS colonies were then transferred to SM media (20 g/L agar, 20 g/L galactose, 1.7 g/L yeast nitrogen base w/o amino acids and w/o ammonium sulfate, 2 g/L Drop-out mix (-HIS -LYS -ARG), 1 g/L monosodium glutamic acid, 50 mg/L canavanine, 50 mg/L S-AEC, and 100 mg/L G418) and incubated for 48 hr at 30C. Lastly, colonies were transferred to DM media (SM media + 100 mg/L Nat) and incubated for 48 hr at 30C. This procedure was repeated from an independent colony of query for a total of two biological replicates.
Photos were taken of each plate, colonies sized extracted with automated software, and genetic interactions calculated (S-score)(Collins et al., 2006). S-scores for each genetic interaction pair were averaged across biological replicates. Previously generated host-host genetic interaction profiles were obtained from http://thecellmap.org/costanzo2016/ (Usaj et al., 2017) and drug-host genetic interaction profiles from http://mosaic.cs.umn.edu/ (Nelson et al., 2018; Piotrowski et al., 2017). Nonessential x nonessential and nonessential x essential matrices were merged and oriented such that the original query strain was in the same orientation as the bacterial effector genetic interaction profiles. The Pearson’s correlation coefficient (PCC) of each effector-host profile was measured against each host-host profile in R, and this PCC was then normalized with a z-score based on the median absolute deviation. A z-score >5 was deemed significant. To minimize false positives, we filtered yeast mutants for network visualization and biological follow up so that a given yeast mutant could have a z-score >5 for only one query. Network edges between yeast mutant genes were derived from BioGRID (https://thebiogrid.org), but for clarity were only included among genes for a given effector. Networks were generated with Cytoscape and hierarchical clustering was done in cluster3 (3.0).
Gene ontology (GO) enrichment analysis
GO terms were downloaded from the Saccharomyces genome database (http://www.yeastgenome.org/). Pathway enrichments from yeast mutants with a z-score > 5 in no more than two effectors were calculated for each effector using the Fisher’s Exact test in R, with p<1×10−5 deemed significant. Because some genes were represented from multiple mutants in the host-host genetic interaction profiles, we only considered genes once in the Fisher’s Exact contingency table if at least one (but not necessarily all) mutants from a gene was in the top 1% for a given effector.
Affinity purification mass spectrometry
293T cells were transfected with 1–10 μg of DNA using PolyJet and were harvested in PBS+0.5M EDTA after 48 hours. Cells were lysed in lysis buffer containing 5% 1M Tris at pH 7.4, 3% NaCl, 0.2% 0.5M EDTA, and 0.8% 20% NP40. Flag resin (Sigma: F2426) was washed using buffer containing 5% 1M Tris at pH 7.4, 3% NaCl, and 0.2% 0.5M EDTA. 1000 μl of the cleared lysate was added to the resin and inverted for 2 hours at 4°C. The resin was washed three times with 1000 μl of IP wash containing 5% 1M Tris at pH 7.4, 3% NaCl, 0.2% 0.5M EDTA, and 0.5% 20% NP40. Samples were eluted three times at room temperature for 15 minutes each using 25X 3x FLAG peptide (Sigma) diluted to 5X with lysis buffer without detergent. IP efficiency was verified by immunoblot and silver stain analysis. Immunoblots were performed using mouse monoclonal anti-flag M2 antibody.
Raw MS files were analyzed by MaxQuant (Cox and Mann, 2008) version 1.3.0.3 and MS/MS spectra searched by the Andromeda search engine (Cox et al., 2011) against a database containing reviewed SwissProt human proteins (UniProt Consortium, 2015), the three bacterial effectors (Uniprot IDs, sseG:H9L486, sseC:O84947, and steB:Q8ZPA6) and GFP. Multiplicity was set to 1 (recommended for label free experiment) and a false discovery rate imposed to 0.01 for peptide and protein identification. Normalization of raw peptide intensities and protein level abundance inference were calculated using the linear mixed-effects model built into the MSstats R package version 3.3.10 (Choi et al., 2014). SAINTq (Teo et al., 2016) was used to assign scores to bait–prey interactions against the negative controls (GPP and empty vectors), providing as input the peptide intensities.
Immunofluorescence microscopy
To determine subcellular localization of CBU0388, CBU0794, and CBU1314 HeLa cells were seeded at 1×106 onto coverslips in a 24-well dish. The following day, cells were transfected with indicated plasmids (pEGFPC1 0388, pEGFPC1 0794 and pEGFPC1 1314) using Lipofectamine 3000 according to the manufacturers protocol. Twenty-four hours after transfection, the cells were fixed using 4% Paraformaldyhyde and stained with Hoescht to visualize the nucleus. Coverslips were imaged on the Nikon A1-Confocal microscope at 40x. HeLa cells were transfected as above to visualize ST SseG-Golgi co-localization, using a pDEST-3xFLAG-SseG plasmid. An antibody specific for the RCAS1 (Cell Signaling) was used to visualize the Golgi apparatus.
To visualize retromer during S. Typhimurium infection, HeLa were infected as described below. At the designated time points, cell were washed three times in PBS and fixed in 4% paraformaldehyde (PFA) for 10 min at room temperature (RT). For time courses, cells were infected with an MOI of 30, centrifuged for 5 min at 1000 x g to maximize bacteria-host cell contact, and incubated for an additional 15 min at 37°C 5% CO 2. Wells were washed three times in PBS and either fixed in 4% PFA for a 15 min time point or the media was replaced with DMEM + 10% FBS with gentamicin (100 μg ml−1) to kill the extracellular bacteria and prevent additional bacterial internalization. At later time points cells were washed an additional three times in PBS, and fixed in 4% paraformaldehyde. The fixed cells were washed three times in PBS and permeablized by incubating them in PBS containing 5% non-fat milk and 0.05% saponin (PBS-MS) (Calbiochem). Cover slips were incubated in primary antibody diluted in PBS-MS for 60 min. The cover slips were then washed 3 times in PBS and incubated in secondary antibody. After two washes in PBS and two washes in deionized water, the cover slips were mounted onto glass slides using Prolong Gold antifade reagent (Molecular Probes). Images were acquired on a Nikon A1-Confocal microscope. When needed, inside/out staining was used to differentiate extracellular from intracellular bacteria. Briefly, before permeabilization with saponin, extracellular bacteria were stained with anti-LPS in PBS containing 5% milk followed by Alexaflour 680-conjugated anti-rabbit antibodies (ThermoFisher). Cells were washed three times, permeablized, and the total bacterial population was stained with anti-LPS followed by Alexaflour 488-conjugated anti-rabbit antibodies (ThermoFisher). After two washes in PBS and two washes in deionized water, the cover slips were mounted onto glass slides using Prolong Gold antifade reagent (Molecular Probes) and images were acquired as described above.
To stain cytosolic bacteria, cells were permeabilized with digitonin before staining. At the indicated time points, coverslips were washed three times with warm KHM buffer (110 mM potassium acetate, 20 mM HEPES, 2 mM MgCl2, pH 7.3), incubated in freshly prepared 50μg/ml digitonin (MP Biomedicals) in KHM for exactly 60 seconds, and washed three times with KHM buffer. Coverslips were incubated in primary antibodies diluted in KHM buffer for 12 minutes at 37deg. Both anti-LPS and anti-Lamp1 primary antibodies were diluted 1:100. Coverslips were then washed once in PBS, fixed in 4%PFA, and washed three times in PBS. Coverslips were then incubated in PBS-MS with Alexfluor-488 anti-rabbit and Alexafluor-647 anti-mouse secondary antibodies. To stain the total bacterial population, coverslips were then incubated in PBS-MS with anti-LPS followed by Alexafluor-594 anti- rabbit. Coverslips were washed, mounted, and imaged as described above.
Co-immunoprecipitations
293T cells were split in 6 well plates containing 8 x 105 cells per well. Cells were transfected with 500 ng of the bacterial effector and 500 ng of the host factor Cells were harvested and 3xFLAG-SseC was purified as above. Immunoblots were performed using anti-HA high affinity rat monoclonal antibody (Roche; 3F10), or anti-VPS35 antibody (Santa Cruz Biotechnology) and anti-flag mouse monoclonal M2 880 antibody (Sigma) diluted 1:5000. Goat monoclonal anti-rat (LICOR) and goat monoclonal anti-mouse (LICOR) were used at dilutions of 1:10000.
Protein expression and purification
Constructs of individual VPS35, VPS26A, VPS29 and ST SseC-SscA complex for protein expression were cloned into pET28a-SUMO or pET22b vectors. All the proteins were expressed in Escherichia coli strain BL21(DE3) overnight at 15 °C. The recombina nt proteins were purified by nickel affinity chromatography followed by gel-filtration chromatography. All the proteins were eluted with a buffer containing 20 mM Tris-HCl, 150 mM NaCl at pH 7.50 (buffer A). Purified proteins were flash-frozen in liquid nitrogen after concentration and stored at –80 °C.
Pull-down assay
Purified biotin-labeled SUMO-fusion ST SseC-SscA (20nmol) was incubated with streptavidin beads in pull-down buffer (20mM Tris-HCl, 150mM NaCl, 5mM DTT, pH 7.5) for 15 min at 4 °C. Individual components of retromer or r etromer complexes (40μmol) were mixed with the beads and incubated on shaker for 30 minutes at 4 °C. Excess proteins were washed off the beads using pull-down buffer. 50 μl of water and 10 μl of 6x SDS loading buffer was added to resin and boiled for 5 minutes, thereafter, the samples were centrifuged briefly. 20μl of supernatant was analyzed by SDS PAGE. The protein bands were visualized by Coomassie blue staining.
SPR binding study
The binding affinities between ST SseC-SscA and the retromer complexes were determined by SPR using a Biacore X100 SPR instrument (GE Healthcare). Biotin-labeled SUMO-fusion sseC-sscA was coupled on the sensor chip using the Biotin CAPture Kit (GE Healthcare). Dilution series of retromer complex VPS35 and VPS26 (31.25, 62.5, 125, 250, 500, 100031.25, 62.5, 125, 250, 500, 2000 nM) in 1× HBS-EP+ buffer (GE Healthcare) were injected over the sensor chip at a flow rate of 30 μL/min. The multi-cycle kinetic/affinity protocol was used in all binding studies. The sensor chip was regenerated with a buffer containing 6 M guanidine hydrochloride and 0.25 M NaOH. The concentration of all controls was set to 2μM. All measurements were duplicated under the same conditions. The equilibrium Kd was determined by fitting the data to a steady-state 1:1 binding model using Biacore X100 Evaluation software version 2.0 (GE Healthcare).
S. Typhimurium infection
Overnight cultures of S. Typhimurium strains were diluted 1:20 in LB broth containing 0.3M NaCl, and grown until they reached an OD600 of 0.9. Unless specified, cell lines at a confluency of 80% were infected with the S. Typhimurium strains at an MOI of 30 for 1 h in Hank’s buffered salt solution (HBSS), and subsequently incubated in DMEM supplemented with gentamicin (100 μg/ml) to kill extracellular bacteria. After 30 min, the concentration of gentamicin was decreased to 10 μg/ml. To measure the number of cytosolic bacteria, infected cells were incubated in the presence of gentimicin with or without 400 uM of chloroquine for one hour prior to harvesting bacteria for CFUs.
SseC expression
HeLa cells expressing the SCR shRNA hairpin were infected with S. Typhimurium of an MOI of 100 as described above. At indicated time points, media was aspirated from cells and replaced with 500ul TRIzol reagent (Invitrogen). RNA was isolated using the DirectZol RNA miniprep kit (Zymo Research). cDNA was synthesized using the iScript cDNA synthesis kit from BioRad. Gene expression in each sample was quantified using the Applied Biosystems StepOne Plus real time PCR machine and primers specific for ST SseC and the ST 16S rRNA.
QUANTIFICATION AND STATISTICAL ANALYSIS
Genetic interactions were quantified with the S-score (Collins et al., 2006) and averaged between replicates. Previously generated host-host genetic interaction profiles were obtained from http://thecellmap.org/costanzo2016/ (Usaj et al., 2017). Nonessential x nonessential and nonessential x essential matrices were merged and oriented such that the original query strain was in the same orientation as the bacterial effector genetic interaction profiles. The Pearson’s correlation coefficient (PCC) of each effector-host profile was measured against each host-host profile in R, and this PCC was then normalized with a z-score based on the median absolute deviation. A z-score >5 was deemed significant. To minimize false positives, we filtered yeast mutants for network visualization and biological follow up so that a given yeast mutant could have a z-score >5 for only one query. Network edges between yeast mutant genes were derived from BioGRID (https://thebiogrid.org), but for clarity were only included among genes for a given effector. Networks were generated with Cytoscape and hierarchical clustering was done in cluster3 (3.0).
For immunofluorescence experiments, cells were plated on three coverslips and >100 bacteria were scored for colocalization with VPS35 or cytosolic staining. Data represents the average number of bacteria across the three coverslips. Data for a single representative experiment is shown but experiment was conducted >3 times. For CFU/enumeration of cytosolic bacteria experiments, each time point represents the average of CFUs from three separate wells. Statistical analysis of data was performed using GraphPad Prism software. Two-tailed unpaired Student’s t tests were used for statistical analyses, and unless otherwise noted, all results are reported as the mean +/− SD or as a percent of the scramble or wild-type control.
DATA AND SOFTWARE AVAILABILITY
Software
Genetic interactions were scored with the E-MAP toolkit (https://sourceforge.net/projects/emap-toolbox/) in Matlab (version 7.13 R2011b). Yeast colony sizes were automatically extracted from photos using the colony measure tool (https://sourceforge.net/projects/ht-col-measurer/).
Data Resources
Genetic interactions scores, z-scores, and all other raw and processed data are containing in Table S1–3.
ADDITIONAL RESOURCES
N/A
Supplementary Material
Lists of mutants in library, S-scores, PCCs, and z-scores.
Only genes with correlation to two or fewer effectors are included. p < 10−5
(Nelson et al. 2017)
HIGHLIGHTS.
Genetic interaction profiling in yeast reveals mechanisms of bacterial effectors
E-MAP functionally classifies virulence factors across diverse bacterial species
Retromer complex binds to the Salmonella effector SseC with high affinity in vitro
Loss of retromer leads to cytosolic hyper-replication of Salmonella in host cells
Acknowledgments
We thank members of the Watson lab for their valuable critiques of the manuscript and members of Christine Guthrie’s lab at UCSF for logistical help with the E-MAP screen. S. Typhimurium mutant strains were graciously shared by Drs. Helene Andrews-Polymenis and Lydia Bogomolnaya at Texas A&M Health Science Center. Mass spectrometry analysis was performed under the supervision of Dr. Larry Dangott at the Protein Chemistry Lab at Texas A&M University. J.A.W is supported by F32 AI116410 (NIH). A.L.C is supported by a CONACYT scholarship (289253). The P.D.F. lab is supported by the Qatar National Research Foundation (NPRP9-001 and NPRP7-1634-2-604), the National Science Foundation (DBI 1532188), the Texas A&M CONACYT program, the NIH CTEHR Program, a CST*R Pilot Grant, and the Texas A&M-CAPES Program. Support to P.L. is provided by the Welsh Foundation (A-1931-20170325) and the Cancer Prevention and Research Institute of Texas (CPRIT) (RP150454). T.A.F. is supported by the NIH (R01HD084339). J.E.S. is supported by NIH/NIAID (R01A1090142) and DTRA/DoD (HDTRA1-12-1-0003). N.J.K. is supported by NCI/NIH (U54CA209891), NIH (R01GM084279), NIH/NIAID (R01AI122747), and NIH/NIAID (R01AI120694). K.L.P. and R.O.W. are supported by NIH/NIAID (R21AI123753) with additional support for R.O.W. from NIH/NIAID (R01AI125512).
Footnotes
DECLARATION OF INTERESTS
The authors report no financial or non-financial competing interests.
AUTHOR CONTRIBUTIONS
Conceptualization, K.L.P., J.A.W., and R.O.W.; Methodology, K.L.P. and J.A.W.; Formal analysis, J.A.W., D.J-M. and M.S.; Investigation, K.L.P., T.J., M.R., S.J.T., P.X., S.L.B., A.L.C., J.X. and R.O.W.; Resources, N.J.K.; Data Curation, J.A.W. and M.S.; Writing – Original Draft, K.L.P. and J.A.W.; Writing – Review and Editing, K.L.P., J.A.W., R.O.W., P.D., S.L.B.; Visualization, K.L.P., J.A.W., M.S., and R.O.W.; Supervision, T.F., P.D., J.E.S., P.L., N.J.K., and R.O.W.; Funding acquisition, T.F., P.D., J.E.S., P.L., N.J.K., and R.O.W.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Beuzón CR, Salcedo SP, Holden DW. Growth and killing of a Salmonella enterica serovar Typhimurium sifA mutant strain in the cytosol of different host cell lines. Microbiology (Reading, Engl) 2002;148:2705–2715. doi: 10.1099/00221287-148-9-2705. [DOI] [PubMed] [Google Scholar]
- Braberg H, Jin H, Moehle EA, Chan YA, Wang S, Shales M, Benschop JJ, Morris JH, Qiu C, Hu F, et al. From structure to systems: high-resolution, quantitative genetic analysis of RNA polymerase II. Cell. 2013;154:775–788. doi: 10.1016/j.cell.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JCS, Madhani HD. Approaching the functional annotation of fungal virulence factors using cross-species genetic interaction profiling. PLoS Genet. 2012;8:e1003168. doi: 10.1371/journal.pgen.1003168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumell JH, Tang P, Zaharik ML, Finlay BB. Disruption of the Salmonella-containing vacuole leads to increased replication of Salmonella enterica serovar typhimurium in the cytosol of epithelial cells. Infect Immun. 2002;70:3264–3270. doi: 10.1128/IAI.70.6.3264-3270.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bujny MV, Ewels PA, Humphrey S, Attar N, Jepson MA, Cullen PJ. Sorting nexin-1 defines an early phase of Salmonella-containing vacuole-remodeling during Salmonella infection. J Cell Sci. 2008;121:2027–2036. doi: 10.1242/jcs.018432. [DOI] [PubMed] [Google Scholar]
- Burd C, Cullen PJ. Retromer: a master conductor of endosome sorting. Cold Spring Harb Perspect Biol. 2014;6:a016774–a016774. doi: 10.1101/cshperspect.a016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnaevskiy N, Fox TG, Plymire DA, Ertelt JM, Weigele BA, Selyunin AS, Way SS, Patrie SM, Alto NM. Proteolytic elimination of N-myristoyl modifications by the Shigella virulence factor IpaJ. Nature. 2013;496:106–109. doi: 10.1038/nature12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M, Chang CY, Clough T, Broudy D, Killeen T, MacLean B, Vitek O. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526. doi: 10.1093/bioinformatics/btu305. [DOI] [PubMed] [Google Scholar]
- Collins SR, Miller KM, Maas NL, Roguev A, Fillingham J, Chu CS, Schuldiner M, Gebbia M, Recht J, Shales M, et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature. 2007;446:806–810. doi: 10.1038/nature05649. [DOI] [PubMed] [Google Scholar]
- Collins SR, Schuldiner M, Krogan NJ, Weissman JS. A strategy for extracting and analyzing large-scale quantitative epistatic interaction data. Genome Biol. 2006;7:R63. doi: 10.1186/gb-2006-7-7-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colonne PM, Winchell CG, Voth DE. Hijacking Host Cell Highways: Manipulation of the Host Actin Cytoskeleton by Obligate Intracellular Bacterial Pathogens. Front Cell Infect Microbiol. 2016;6:107. doi: 10.3389/fcimb.2016.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CA, Mulder DT, Allison SE, Pilar AVC, Coombes BK. The SseC translocon component in Salmonella enterica serovar Typhimurium is chaperoned by SscA. BMC Microbiol. 2013;13:221. doi: 10.1186/1471-2180-13-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, et al. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353:aaf1420–aaf1420. doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p. p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10:1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- Curak J, Rohde J, Stagljar I. Yeast as a tool to study bacterial effectors. Curr Opin Microbiol. 2009;12:18–23. doi: 10.1016/j.mib.2008.11.004. [DOI] [PubMed] [Google Scholar]
- de Figueiredo P, Ficht TA, Rice-Ficht A, Rossetti CA, Adams LG. Pathogenesis and immunobiology of brucellosis: review of Brucella-host interactions. Am J Pathol. 2015;185:1505–1517. doi: 10.1016/j.ajpath.2015.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delrue RM, Martinez-Lorenzo M, Lestrate P, Danese I, Bielarz V, Mertens P, De Bolle X, Tibor A, Gorvel JP, Letesson JJ. Identification of Brucella spp. genes involved in intracellular trafficking. Cell Microbiol. 2001;3:487–497. doi: 10.1046/j.1462-5822.2001.00131.x. [DOI] [PubMed] [Google Scholar]
- Elwell CA, Czudnochowski N, von Dollen J, Johnson JR, Nakagawa R, Mirrashidi K, Krogan NJ, Engel JN, Rosenberg OS. Chlamydia interfere with an interaction between the mannose-6-phosphate receptor and sorting nexins to counteract host restriction. Elife. 2017;6:213. doi: 10.7554/eLife.22709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finsel I, Ragaz C, Hoffmann C, Harrison CF, Weber S, van Rahden VA, Johannes L, Hilbi H. The Legionella effector RidL inhibits retrograde trafficking to promote intracellular replication. Cell Host Microbe. 2013;14:38–50. doi: 10.1016/j.chom.2013.06.001. [DOI] [PubMed] [Google Scholar]
- Fu Y, Galán JE. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol Microbiol. 1998;27:359–368. doi: 10.1046/j.1365-2958.1998.00684.x. [DOI] [PubMed] [Google Scholar]
- Fu Y, Galán JE. A salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature. 1999;401:293–297. doi: 10.1038/45829. [DOI] [PubMed] [Google Scholar]
- Hautefort I, Thompson A, Eriksson-Ygberg S, Parker ML, Lucchini S, Danino V, Bongaerts RJM, Ahmad N, Rhen M, Hinton JCD. During infection of epithelial cells Salmonella enterica serovar Typhimurium undergoes a time-dependent transcriptional adaptation that results in simultaneous expression of three type 3 secretion systems. Cell Microbiol. 2008;10:958–984. doi: 10.1111/j.1462-5822.2007.01099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward RD, Koronakis V. Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. Embo J. 1999;18:4926–4934. doi: 10.1093/emboj/18.18.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang FC, Holden DW. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Mol Microbiol. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- Kimbrough TG, Miller SI. Assembly of the type III secretion needle complex of Salmonella typhimurium. Microbes Infect. 2002;4:75–82. doi: 10.1016/s1286-4579(01)01512-x. [DOI] [PubMed] [Google Scholar]
- Klein JR, Jones BD. Salmonella pathogenicity island 2-encoded proteins SseC and SseD are essential for virulence and are substrates of the type III secretion system. Infect Immun. 2001;69:737–743. doi: 10.1128/IAI.69.2.737-743.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson CL, Martinez E, Beare PA, Jeffrey B, Heinzen RA, Bonazzi M. Right on Q: genetics begin to unravel Coxiella burnetii host cell interactions. Future Microbiol. 2016;11:919–939. doi: 10.2217/fmb-2016-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser CF, Miller SI. Expression of microbial virulence proteins in Saccharomyces cerevisiae models mammalian infection. Embo J. 2001;20:1840–1849. doi: 10.1093/emboj/20.8.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonough JA, Newton HJ, Klum S, Swiss R, Agaisse H, Roy CR. Host pathways important for Coxiella burnetii infection revealed by genome-wide RNA interference screening. MBio. 2013;4:e00606–e00612. doi: 10.1128/mBio.00606-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer DF, Noroy C, Moumène A, Raffaele S, Albina E, Vachiéry N. Searching algorithm for type IV secretion system effectors 1.0: a tool for predicting type IV effectors and exploring their genomic context. Nucleic Acids Res. 2013;41:9218–9229. doi: 10.1093/nar/gkt718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirrashidi KM, Elwell CA, Verschueren E, Johnson JR, Frando A, von Dollen J, Rosenberg O, Gulbahce N, Jang G, Johnson T, et al. Global Mapping of the Inc-Human Interactome Reveals that Retromer Restricts Chlamydia Infection. Cell Host Microbe. 2015;18:109–121. doi: 10.1016/j.chom.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myeni SK, Zhou D. The C terminus of SipC binds and bundles F-actin to promote Salmonella invasion. J Biol Chem. 2010;285:13357–13363. doi: 10.1074/jbc.M109.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson J, Simpkins SW, Safizadeh H, Li SC, Piotrowski JS, Hirano H, Yashiroda Y, Osada H, Yoshida M, Boone C, et al. MOSAIC: a chemical-genetic interaction data repository and web resource for exploring chemical modes of action. Bioinformatics. 2018;34:1251–1252. doi: 10.1093/bioinformatics/btx732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolaus T, Deiwick J, Rappl C, Freeman JA, Schröder W, Miller SI, Hensel M. SseBCD proteins are secreted by the type III secretion system of Salmonella pathogenicity island 2 and function as a translocon. J Bacteriol. 2001;183:6036–6045. doi: 10.1128/JB.183.20.6036-6045.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick KL, Ryan CJ, Xu J, Lipp JJ, Nissen KE, Roguev A, Shales M, Krogan NJ, Guthrie C. Genetic interaction mapping reveals a role for the SWI/SNF nucleosome remodeler in spliceosome activation in fission yeast. PLoS Genet. 2015;11:e1005074. doi: 10.1371/journal.pgen.1005074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei J, Wu Q, Kahl-McDonagh M, Ficht TA. Cytotoxicity in macrophages infected with rough Brucella mutants is type IV secretion system dependent. Infect Immun. 2008;76:30–37. doi: 10.1128/IAI.00379-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Personnic N, Bärlocher K, Finsel I, Hilbi H. Subversion of Retrograde Trafficking by Translocated Pathogen Effectors. Trends Microbiol. 2016;24:450–462. doi: 10.1016/j.tim.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Piotrowski JS, Li SC, Deshpande R, Simpkins SW, Nelson J, Yashiroda Y, Barber JM, Safizadeh H, Wilson E, Okada H, et al. Functional annotation of chemical libraries across diverse biological processes. Nat Chem Biol. 2017;13:982–993. doi: 10.1038/nchembio.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popa C, Coll NS, Valls M, Sessa G. Yeast as a Heterologous Model System to Uncover Type III Effector Function. PLoS Pathog. 2016;12:e1005360. doi: 10.1371/journal.ppat.1005360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que F, Wu S, Huang R. Salmonella pathogenicity island 1(SPI-1) at work. Curr Microbiol. 2013;66:582–587. doi: 10.1007/s00284-013-0307-8. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Pachón JM, Martín H, North G, Rotger R, Nombela C, Molina M. A novel connection between the yeast Cdc42 GTPase and the Slt2-mediated cell integrity pathway identified through the effect of secreted Salmonella GTPase modulators. J Biol Chem. 2002;277:27094–27102. doi: 10.1074/jbc.M201527200. [DOI] [PubMed] [Google Scholar]
- Russo BC, Stamm LM, Raaben M, Kim CM, Kahoud E, Robinson LR, Bose S, Queiroz AL, Herrera BB, Baxt LA, et al. Intermediate filaments enable pathogen docking to trigger type 3 effector translocation. Nat Microbiol. 2016;1:16025. doi: 10.1038/nmicrobiol.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo SP, Holden DW. SseG, a virulence protein that targets Salmonella to the Golgi network. Embo J. 2003;22:5003–5014. doi: 10.1093/emboj/cdg517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo SP, Holden DW. Bacterial interactions with the eukaryotic secretory pathway. Curr Opin Microbiol. 2005;8:92–98. doi: 10.1016/j.mib.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Weissman JS, Krogan NJ. Quantitative genetic analysis in Saccharomyces cerevisiae using epistatic miniarray profiles (E-MAPs) and its application to chromatin functions. Methods. 2006;40:344–352. doi: 10.1016/j.ymeth.2006.07.034. [DOI] [PubMed] [Google Scholar]
- Schuldiner M, Collins SR, Thompson NJ, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt JF, et al. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 2005;123:507–519. doi: 10.1016/j.cell.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Seaman MNJ, Harbour ME, Tattersall D, Read E, Bright N. Membrane recruitment of the cargo-selective retromer subcomplex is catalysed by the small GTPase Rab7 and inhibited by the Rab-GAP TBC1D5. J Cell Sci. 2009;122:2371–2382. doi: 10.1242/jcs.048686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siggers KA, Lesser CF. The Yeast Saccharomyces cerevisiae: a versatile model system for the identification and characterization of bacterial virulence proteins. Cell Host Microbe. 2008;4:8–15. doi: 10.1016/j.chom.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanò S, Galán JE. A Rab32-dependent pathway contributes to Salmonella typhi host restriction. Science. 2012;338:960–963. doi: 10.1126/science.1229224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanò S, Gao X, Hannemann S, Lara-Tejero M, Galán JE. A Bacterial Pathogen Targets a Host Rab-Family GTPase Defense Pathway with a GAP. Cell Host Microbe. 2016;19:216–226. doi: 10.1016/j.chom.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spanò S, Liu X, Galán JE. Proteolytic targeting of Rab29 by an effector protein distinguishes the intracellular compartments of human-adapted and broad-host Salmonella. Proc Natl Acad Sci USa. 2011;108:18418–18423. doi: 10.1073/pnas.1111959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Z, Diaz Z, Fang X, Hart MP, Chesi A, Shorter J, Gitler AD. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo G, Koh H, Fermin D, Lambert JP, Knight JDR, Gingras AC, Choi H. SAINTq: Scoring protein-protein interactions in affinity purification - mass spectrometry experiments with fragment or peptide intensity data. Proteomics. 2016;16:2238–2245. doi: 10.1002/pmic.201500499. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usaj M, Tan Y, Wang W, VanderSluis B, Zou A, Myers CL, Costanzo M, Andrews B, Boone C. TheCellMap. org: A Web-Accessible Database for Visualizing and Mining the Global Yeast Genetic Interaction Network. G3 (Bethesda) 2017;7:1539–1549. doi: 10.1534/g3.117.040220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nhieu GT, Romero S. Common Themes in Cytoskeletal Remodeling by Intracellular Bacterial Effectors. Handb Exp Pharmacol. 2017;235:207–235. doi: 10.1007/164_2016_42. [DOI] [PubMed] [Google Scholar]
- Waterman SR, Holden DW. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. 2003;5:501–511. doi: 10.1046/j.1462-5822.2003.00294.x. [DOI] [PubMed] [Google Scholar]
- Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, Dealing CM, Roman VA, Banga S, Tan Y, et al. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol. 2013;195:3914–3924. doi: 10.1128/JB.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber MM, Faris R, McLachlan J, Tellez A, Wright WU, Galvan G, Luo ZQ, Samuel JE. Modulation of the host transcriptome by Coxiella burnetii nuclear effector Cbu1314. Microbes Infect. 2016;18:336–345. doi: 10.1016/j.micinf.2016.01.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Lists of mutants in library, S-scores, PCCs, and z-scores.
Only genes with correlation to two or fewer effectors are included. p < 10−5
(Nelson et al. 2017)