Abstract
The current reference method in the U.S.A. for measuring in vivo population red blood cell (RBC) kinetics utilizes 51‐chromium (51Cr) RBC labeling for determining red cell volume (RCV), 24-hour post-transfusion RBC recovery (PTR24), and long-term red cell survival (RCS). Here we provide evidence supporting adoption of a method for kinetics that uses the biotin labeled RBC (BioRBC) as a superior, versatile method for both regulatory and investigational purposes. RBC kinetic analysis using biotin-labeled RBC has important methodological, analytical, and safety advantages over 51Cr labeled-RBC. We critically review recent advances in labeling human RBC at multiple and progressively lower biotin label densities for concurrent, accurate, and sensitive determination of both autologous and allogeneic RBC population kinetics. BioRBC methods valid for RBC kinetic studies, including successful variations used by the authors, are presented along with pharmacokinetic (PK) modeling approaches for the accurate determination of RBC PK parameters in health and disease. The advantages and limitations of the BioRBC method—including its capability of determining multiple BioRBC densities simultaneously in the same individual throughout the entire RBC life span—are presented and compared with the 51Cr method. Finally, potential applications and limitations of kinetic BioRBC determinations are discussed.
INTRODUCTION
Overview of RBC kinetics
Advances needed to improve the quality and safety of RBC storage for transfusion and investigation of hematological biology and disease states are dependent upon the safe and accurate in vivo determination of post-transfusion RBC kinetics. Achieving this requires labeling RBC—or the ability to distinguish allogeneic RBC populations. In 1919 Winfred Ashby used an method based on red cell antigens to accurately measure red cell lifespan for the first time.1 In a subsequent series of classical investigations beginning in the 1950s, Patrick Mollison advanced the isotopic labeling of autologous and allogeneic RBC for RBC kinetic studies.2
The two approaches to determine in vivo RBC kinetics use either a population or a cohort label.3 In the former, a representative population of autologous or allogeneic blood containing RBC of all ages is labeled with biotin or isotopes of chromium, phosphorus, or technetium. Following labeled RBC transfusion, red cell survival (RCS) is determined on the basis of the temporal decline in labeled RBC. 51Cr RBC labeling has an extensive track record and is the FDA’s current RCS regulatory reference method for RCS.4–6
The cohort labeling approach is characteristically based on in vivo hemoglobin labeling of an age-defined RBC population. Labeling occurs predominantly in reticulocytes in the peripheral blood and bone marrow following either oral or intravenous administration of isotopic (stable or radioactive) heme precursors of iron or glycine (i.e., using nitrogen or carbon).7–10
This review focuses on RBC population labeling with biotin for use in kinetic studies. Here we use kinetics to encompass red cell volume (RCV), short-term 24-hr post-transfusion RBC recovery (PTR24), and long-term RCS including RBC lifespan. RBC kinetic studies are critical in understanding of anemia attributable to inadequate RBC production, shortened RBC lifespan, or blood loss attributable to hemorrhage or phlebotomy, or a combination of these.
Comparison of biotin and 51Cr labeled RBC for kinetic studies
Comparison of 51Cr and biotin RBC labeling methods for RBC kinetic purposes reveals important differences in their methodological features (Table 1). Of particular importance is their different susceptibility to hemo-concentration and hemo-dilution artifact. Because 51Cr labeled RBC are measured as the concentration of radioactivity in whole blood, 51Cr is susceptible to such artifacts. In marked contrast, BioRBC are measured as the proportion of labeled RBC relative to total RBC and thus not susceptible to hemo-concentration and hemo-dilution error. This feature of biotin labeling permits accurate measurement of BioRBC equally well from capillary, venous, or arterial blood draws.11
Table 1.
Comparison of differences in 51Cr and biotin RBC labeling for use in RBC population kinetic studies
| Methodological features | 51Cr | Biotin | Comments |
|---|---|---|---|
| 1) Artifact from hemo-concentration or hemo-dilution | Yes | No | Both capillary and venous (or arterial) blood can be used with BioRBC54 |
| 2) Duration of accurate measurement of labeled RBC after reinfusion | 30 d | >120+ d | Problem of variable 51Cr elution4 |
| 3) Ability to use multicolor flow cytometry to track transfused BioRBC changes by recovering these for ex vivo analysis | No | Yes | Useful for investigation of RBC senescence and age-dependent changes of normal and pathological RBC3 |
| 4) Ability to study >1 RBC population concurrently | No | Yes | Direct comparison to eliminate subject-to-subject variability |
| 5) Use in vulnerable patient populations | No | Yes | Ethical considerations prohibit radiation exposure for research in vulnerable populations, e.g., fetuses, infants, children and pregnant women55 |
| 6) Volume of blood required by assay (mL) | 0.1-1.0 | 0.01 | BioRBC are suitable for fetuses and infants53 |
| 7) Radioactive waste disposal | Yes | No | 51Cr disposal is a hazard and expense |
| 8) Complexity of labeling procedure | 1 wash | 4-6 washes | 4 h for biotin versus 1.5 h for 51Cr |
| 9) Development of antibodies | No | Infrequent | There is no evidence of harm from anti-BioRBC antibodies25,56 |
| 10) Requirement for FDA IND (in U.S.A.) | No | Yes | Immunogenicity testing (https://www.federalregister.gov/documents/2016/04/25/2016-09449/assay-development-and-validation-for-immunogenicity-testing-of-therapeutic-protein-products-revised) |
A second crucially important advantage of biotin RBC labeling is that it permits accurate measurement of RCS for the entire RBC life span, instead of the 28-day limit for 51Cr labeling.4 This short time period for 51Cr measurement is required because of its elution of 0.70 to 1.55% per day from intra-erythrocytic hemoglobin molecules to which it binds.12 In contrast, biotinylation results in the covalent amido link attachment of biotin to lysine residues of outward facing RBC membrane proteins.13 Because biotin binds permanently to the RBC surface, the total number of circulating RBC can easily be determined throughout the entire RBC life span of up to 120 d or more.14,15 In addition, if magnetic beads or flow cytometric cell sorting methodologies are applied, BioRBC can be isolated to characterize potentially age-dependent RBC changes for investigating mechanisms underlying RBC senescence and injury.16,17
Other unique advantages of biotin RBC labeling include its ability to label multiple populations of RBC concurrently when assayed by flow cytometry along with its ethical advantages in kinetic studies of vulnerable populations, e.g., fetuses, infants, children, and pregnant women, which are not possible with 51Cr RBC labeling. The ability to follow multiple BioRBC populations concurrently—rather than sequentially as is required with 51Cr labeling—offers important advantages such as reducing time, effort, cost, and statistical demands. For example, autologous or allogeneic long-term RCS can be simultaneously determined for one or more RBC storage media or for pathogen reduction treatments.
Flow cytometry detection of BioRBC is another advantage as it permits highly reproducible and accurate measurement using 10 μL or less of sample. This feature—along with biotin being non-radioactive—is advantageous in studies of fetuses, infants, and toddlers. With its greater blood volume requirement and use of radioactivity, 51Cr RBC labeling carries ethical—and expensive hazardous radioactive waste disposal—considerations prohibiting its research use in vulnerable populations.
The advantages of 51Cr RBC labeling over biotin labeling are few. For one, 51Cr-RBC labeling requires a single wash step, making 51Cr labeling simpler and less time consuming. Unlike biotin’s labeling of RBC surface proteins,11 51Cr labeling of intra-erythrocytic hemoglobin does not carry the possibility of eliciting an antibody response. It is noteworthy that human BioRBC studies in the U.S.A. require a FDA IND application. Because blood transfusion in Europe is not considered a drug administration, BioRBC studies in humans are not regulated by the European Medical Agency; only ethical approval is required.
BioRBC Method: Consensus and Variations
The RBC biotinylation and detection procedures described below represent a consensus developed jointly by the Iowa, Arkansas, Cincinnati, and Amsterdam groups based on published evidence and our unpublished experience.14,18–20 Where variations occur among sites (e.g., wash buffer composition, choice of fluorescent dye), assessments of these effects—if any—are presented along with recommended methods our laboratories have used successfully.
Biotinylation of RBC for in vivo RCS studies
Autologous and allogeneic RBC are biotinylated at room temperature using optimal sterile techniques. Labeling should be conducted as quickly as feasible, and the Bio-RBC (or mixture of BioRBC populations; see below) should be transfused as soon as possible. The goal is that the total time during which the BioRBC are at room temperature (i.e., during biotinylation and the transfusion procedure) does not exceed 4 h. If immediate transfusion is not possible, the BioRBC should be kept in a monitored refrigerator set at 4°C (range 1° to 6°). For research studies, this period can be extended, provided sterility concerns can be justified and explained as part of informed consent (see “Pathogen contamination” section below). In doing so, bacterial cultures of the BioRBC transfusion product may be performed during initial adoption of the BioRBC method to document sterility and checked intermittently thereafter for quality control.
The anticoagulant used for collection and storage of RBC prior to biotinylation (e.g., EDTA, heparin, or RBC storage media) has no detectable effect on BioRBC product quality, presumably because biotinylation is performed on washed RBC (see below). Similarly, irradiation of BioRBC infusates prepared from RBC concentrates stored under blood bank conditions for up to 35 d demonstrates no in vitro qualitative effect on density of BioRBC labeling.21 Whether irradiation induces additional damage to BioRBC that reduces RCS in vivo has yet to be determined.22,23
We recommend removing plasma by washing RBC two (U Iowa) or three (U Cincinnati) times in freshly prepared phosphate wash buffer pH 7.4; the V/V ratio of wash buffer to packed RBC is 4:1 (see wash buffers section below). Most human BioRBC studies have used sulfo-NHS biotinylating reagent (N-Hydroxysulfosuccinimidobiotin [biotin 3-sulfo-NHS]; EZ-Link Sulfo-NHS-Biotin No-Weigh Format Product #21326, ThermoFisher Scientific; https://www.thermofisher.com/order/catalog/product/21326).11 GMP sulfo-NHS biotinylating reagent has recently become available from Agro-Bio/Groupe-STAGO (2 allée de la Chavannerie, 45240 La Ferté Saint Aubin, France). We recommend this GMP product as the biotinylating regent of choice. We recommend preparing sulfo-NHS biotinylating reagent within 20 minutes prior to use to avoid inactivating hydrolysis of the biotinylating reagent (see below). Inactivation can be minimized by preparing the biotinylation reagent in wash buffer adjusted to pH 5.0 with HCl in which the buffer capacity of this solution is insufficient to alter the reaction mixture pH.24 Sulfo-NHS-biotin (1 mg) is dissolved in a volume of wash buffer suitable for the desired ratio of the mass of biotinylating reagent to the volume of packed RBC (see discussion below); then, the solution is filter sterilized using a 0.22 μm, PVDF syringe filter unit (EMD Millipore).
The biotinylation reaction is initiated with addition of one volume of sulfo-NHS-biotin solution to a 1:10 volumes of RBC adjusted to 25% HCT with wash buffer such that the HCT of final reaction mixture is 22.7%. After inverting approximately 10 times, the mixture is incubated at room temperature for 30 min. The reaction is stopped by sedimentation of RBC at 1000 RCF followed by one (U Iowa) or two (U Cincinnati) 4:1 V/V buffer washes, sedimentation, and resuspension. To remove RBC microaggregates in clinical studies, we recommend that BioRBC for clinical studies be resuspended in wash buffer to a packed BioRBC HCT of 50%. This packed BioRBC suspension is passed through an 18‐micron filter (Hemo-Nate Utah Medical Products Inc., Midvale, UT) and transfused as 100% BioRBC, still at a HCT 50%.
Because the number of biotin labels per RBC increases linearly with the ratio of the mass (μg) of biotinylating reagent to the volume (mL) of packed RBC being biotinylated,11 individual RBC populations can be reproducibly labeled at biotin densities that produce discrete histogram peaks for counting (Figure 1). The BioRBC density populations are designated as BioRBC-N, where N represents the ratio of mass of biotinylating reagent to volume of packed RBC used in the labeling reaction. For human multidensity BioRBC studies, we recommend a minimum of 3-fold serial dilutions of the biotinylating reagent in wash buffer. Each BioRBC-N is enumerated by flow cytometry as a percentage of total RBC in the blood sample. Independent tracking of multiple populations of BioRBC concurrently in the same individual eliminates confounding subject-to-subject variation.11 Studies in humans have used BioRBC-2 to BioRBC-162.11 At biotinylation densities ≤BioRBC-2, adequate resolution from the unlabeled RBC is not yet consistently possible (unpublished). At BioRBC≥162, mean corpuscular volume is increased, in vitro RBC morphology is altered (swelling and spicule formation), and long-term RCS is significantly shortened.15,25
Figure 1.
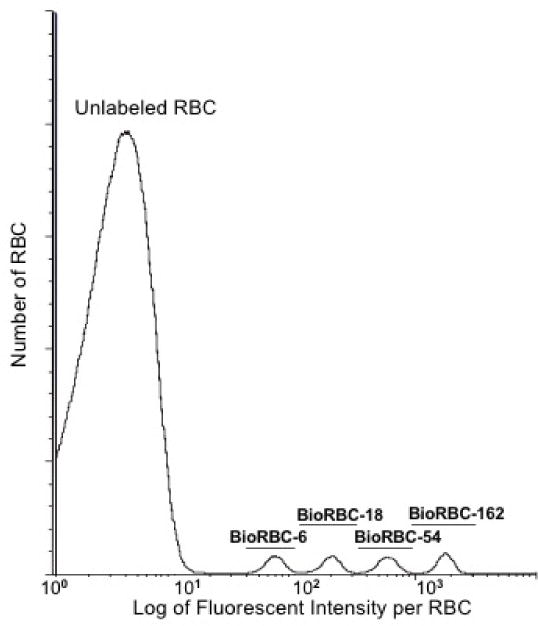
Flow cytometric histogram of the number of RBC versus fluorescent intensity per RBC for RBC labeled with biotin at four densities. Shown are the results of a single in vivo sample from a healthy adult following transfusion with a mixture of BioRBC-6, -18, -54, and -162. Discrete separation of the four BioRBC-N populations were achieved with Alexa Fluor 488 streptavidin.
Successful wash and biotinylation buffers
Use of RBC wash solution is employed in three critically important biotinylation steps: pre-biotinylation washing; during biotinylation; and post-biotinylation. Initial pre-biotinylation washing of whole blood removes plasma proteins and proteins loosely adherent to RBC surfaces. Failure to adequately wash whole blood substantially and unpredictably decreases the number of biotin labels per RBC. The resultant BioRBC product may be unsuitable for determining RBC survival.26,27 The final post-biotinylation wash step removes potentially toxic unreacted sulfo-NHS-biotin, reaction byproducts, and hydrolysis products from the final BioRBC suspension (see Safety Issues for BioRBC below).
Published studies of in vivo human BioRBC kinetics have primarily used two wash solutions: 1) Dulbecco’s Phosphate Buffered Saline (PBS) (Invitrogen, Carlsbad, CA); and 2) “Iowa Wash Buffer.”11 Saline-adenine-glucose-mannitol (SAGM; Fresenius Kabi, The Netherlands) is approved for RBC storage media in Europe and has been used successfully in biotinylating RBC.21,28 Although not yet reported as successful in RBC biotinylation in clinical studies, Plasma-Lyte A, USP is likely a suitable biotinylation buffer based on composition and in vitro studies (see below). Plasma-Lyte A is available commercially in both Europe and the USA, and is FDA approved for use with RBC.
The compositions of various BioRBC wash solutions have similarities with one another (Table 2). Dulbecco’s PBS, Iowa Wash Buffer, and Plasma-Lyte A all have similar pH; SAGM has a lower pH and includes adenine and mannitol. Dulbecco’s PBS and Plasma-Lyte A are isotonic with plasma and have similar ionic strength; Iowa Wash Buffer and SAGM have higher osmolalities due to inclusion of dextrose as an RBC energy substrate. Iowa Wash Buffer has two limitations: 1) unlike Dulbecco’s PBS and Plasma-Lyte A, preparation requires multiple sterile solution transfers that increase chances of bacterial contamination; and 2) Iowa Wash Buffer components are occasionally unavailable because of manufacturer shortages.
Table 2.
Composition of wash buffers.
| Dulbecco’s PBS | Iowa Wash Buffer | Plasma-Lyte A | SAGM | |
|---|---|---|---|---|
| pH | 7.0 - 7.2 | 7.4 | 7.4 | 5-6 |
| Sodium Chloride (mM) | 137.9 | 149.6 | 89.99 | 150 |
| Potassium Chloride (mM) | 2.67 | — | 4.96 | |
| Sodium Phosphate Dibasic (mM) | 8.06 | 0.972 | — | |
| Sodium Phosphate Monobasic (mM) | — | 1.94 m/M | — | |
| Potassium Phosphate Monobasic (mM) | 1.47 | — | — | |
| Glucose (mM) | — | 21.6 | — | 45 |
| Sodium Bicarbonate (mM) | — | 19.4 | — | |
| Magnesium Chloride (mM) | — | — | 3.15 | |
| Sodium Gluconate (mM) | — | — | 23.01 | |
| Sodium Acetate Trihydrate (mM) | — | — | 27.04 | |
| Adenine (mM) | 1.25 | |||
| Mannitol (mM) | 30 | |||
| Osmolality (mOsmol) | 270 - 300 | 374 | 294 | 375 |
The in vitro RBC biotinylation characteristics for Iowa Wash buffer, Dulbecco PBS and Plasma-Lyte A buffer were compared; isotonic 0.9% sodium chloride injection, USP, served as control (Figure 2, unpublished data). For each of the three buffers and control, all the BioRBC-N peaks (6, 18, 54, and 162) exhibited nearly identical fluorescent intensity per RBC, thus indicating equivalent biotin label densities. In addition, enumeration of BioRBC-N in each histogram peak agreed within experimental error (±3%) among the four. Fluorescence intensity per RBC was linear with the mass of sulfo-NHS-biotin (Figure 3). Finally, in two separate groups of healthy adult subjects (one at Iowa, the other at Cincinnati), RCS was compared for RBC labeled in Dulbecco’s buffer or Iowa Wash Buffer. Despite different study populations, sites, and small differences in concentration of biotinylation reagent, RCS was not significantly different between the two groups. (Figure 4).
Figure 2.
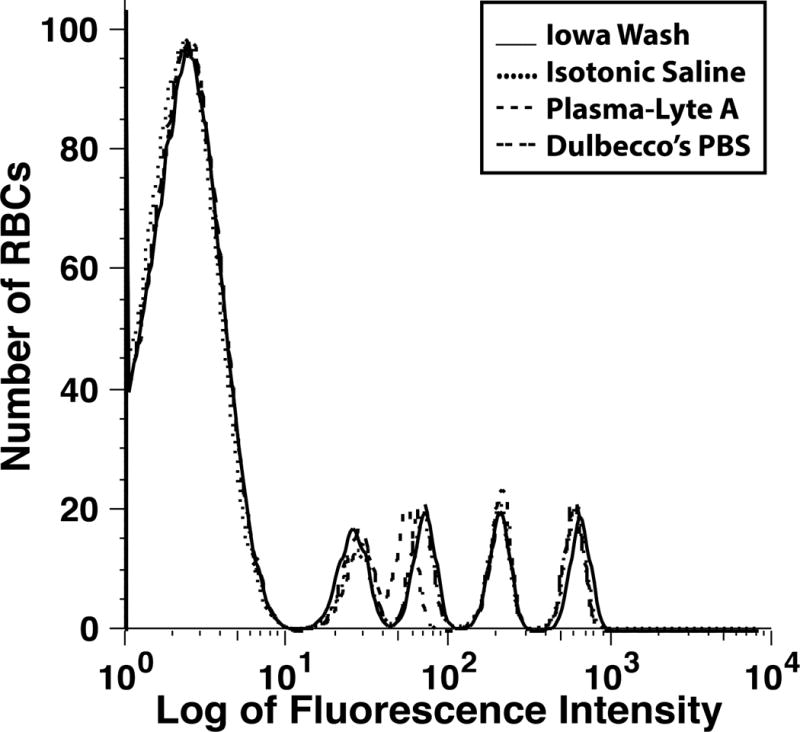
Four histograms of a mixture of unlabeled and BioRBC-6, -18, -54, and -162 prepared in vitro using three wash buffers (Table 2) and isotonic saline.
Figure 3.
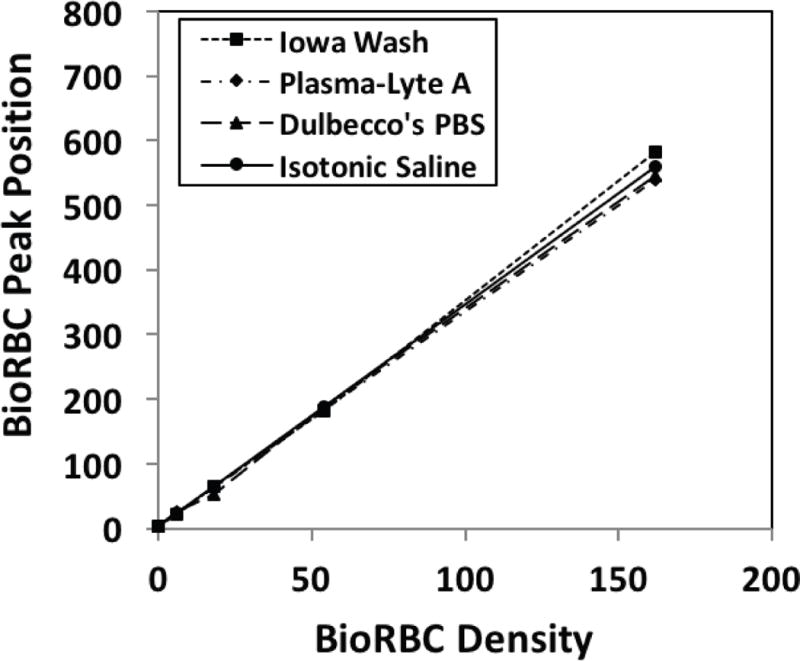
Fluorescent intensity per RBC versus mass of sulfo-NHS-biotin per mL of RBC for BioRBC-6, -18, -54, and -162. The least square regression plots for these in vitro mixtures of BioRBC prepared using the three wash buffers and isotonic saline are nearly superimposable. Fluorescein-conjugated avidin was used as the fluorescent stain (ImmunoPure® Avidin, Fluorescein Conjugated, cat#21221, Thermo Scientific, Rockford, IL). Data from Figure 2.
Figure 4.
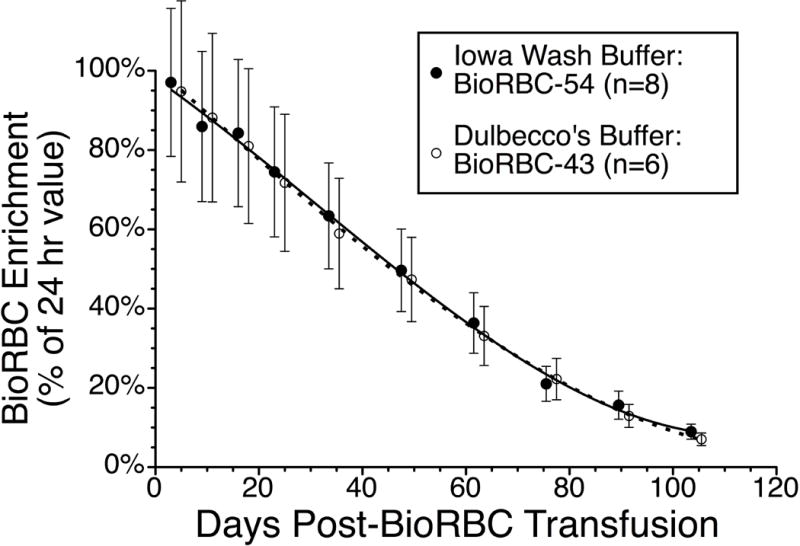
Comparison of autologous in vivo BioRBC survival in two groups of normal adults at two study sites using two different biotinylation buffers. Shown are the mean ± 95% CI for eight Iowa adults whose BioRBC‐54 were prepared using Iowa Wash Buffer18 (filled circles), and six Cincinnati adults whose BioRBC‐54 were prepared using Dulbecco’s PBS buffer (open circles).52 RCS of the two were compared using by linear mixed model analysis as described previously.53 The relative difference between the two buffers did not change over time, i.e., buffer*days interaction P=0.886, with no overall difference between the two buffers (p=0.490).
Infusion of one or a mixture of BioRBC-N
Following biotinylation, aliquots of one BioRBC-N or a mixture of several BioRBC-N (final HCT of about 50%) are prepared in a single syringe. For studies employing transfusion of more than one RBC population, red cell counts are determined before mixing the individual densities in order to estimate the volume (by syringe mass difference) of each population to include in the final mixture. The BioRBC suspension is passed through an 18 micron Hemo-Nate blood filter (Utah Medical Products Inc., Midvale, Utah) into a second syringe, which is used for administering the BioRBC transfusion. Just prior to transfusion, aliquots are removed for bacterial pathogen testing and RBC flow cytometry to determine the relative percentage of RBC in each BioRBC-N) and enumeration of total RBC per fL. Finally, to determine total RBC number infused, mass infused is determined by mass difference of the syringe (weights to the nearest milligram) and converted to volume infused using 1.06 for the specific gravity of the infusate. The BioRBC are transfused over 5 to 10 min.
Safety issues for BioRBC
Toxicity of biotinylation reaction byproducts
Cytotoxicity was assessed using an ultrasensitive and reliable, ATP bioluminescence hematopoietic stem cell cytotoxicity assay (HALO-Tox HT; HemoGenix, Colorado Springs, Colorado; http://hemogenix.com). Aliquots of supernatant wash solutions from a biotinylation reaction used to produce BioRBC-9 and BioRBC-243 densities were tested. No cytotoxicity was observed for these washes.18 Thus, BioRBC labeling does not expose subjects to detectable cytotoxicity byproducts.
Pathogen contamination
For allogeneic BioRBC studies, the donor blood should be obtained from a unit that has had all routine blood bank infectious disease testing. Post-biotinylation bacterial contamination is minimized by following established good laboratory practices, including strict aseptic technique. Biotinylation should be performed under a sterile Class 2 laminar flow hood. Contamination testing should include a 0.5 mL aliquot of the BioRBC infusate for endotoxin determination (e.g., Pyrosate® Kit, #PSD125, 0.125 EU/mL, Associates of Cape Cod, Inc., East Falmouth MA). Because endotoxin test results are available within 30 to 60 min, administration of endotoxin positive infusates can be avoided. For post-transfusion quality control purposes, >0.5 mL volumes of BioRBC infusates can be tested for bacterial contamination by incubation at 37°C for five days (e.g., BACTEC™ system, BD, Franklin Lakes NJ). The anticipated rare, positive bacterial cultures should be interpreted in context of the subject’s clinical condition at that time. For some study populations in whom autologous BioRBC are being studied (e.g., fetuses and neonates), there may be insufficient BioRBC infusate volumes for endotoxin and/or bacterial testing. In the future, molecular detection using 0.1 to 1.0 mL of BioRBC infusate for bacterial and fungal DNA may prove superior and require less blood than bacterial culture.29 Finally, the recent report that RBC biotinylation can be performed in two hours in a closed system offers promise that bacterial contamination during the biotinylation procedure can be further reduced.21
Anti-BioRBC antibody surveillance
We recommend monitoring for the few patients in whom anti-BioRBC antibodies are induced by BioRBC transfusion. Using an IgG gel card assay method,25 we have reported that transient anti-BioRBC antibody responses are induced in approximately 10% of adults.25 Of note, when BioRBC label densities ≤BioRBC-18 are used, the development of anti-BioRBC antibodies has not been detected.25 We hypothesize that infusing fewer RBC with lower biotin label densities (i.e., fewer total biotin labels) decreases the likelihood of antibody induction. Although neither clinical abnormalities nor reduced BioRBC survival have been detected following initial induction of antibodies,25,30 this is a potential safety issue.
We observed accelerated removal of BioRBC in a BioRBC alloimmunized adult when the subject was re-challenged five years later with a mixture of BioRBC-6, -18, -54, and -162. This re-challenge caused a brisk, substantial anamnestic increase in anti-BioRBC antibody titer and dramatically shortened BioRBC survival.25 We infer that accelerated removal was caused by coating of circulating BioRBC-N with anti-BioRBC antibody. This phenomenon renders such subjects unsuitable for subsequent BioRBC kinetic studies. Because <2% of circulating RBC are biotin-labeled in such studies, rapid removal of nearly all BioRBC did not result in hematological laboratory abnormalities; biotin nutritional status was also not affected.25
Induction of anti-BioRBC antibodies should be monitored in individuals prior to and following BioRBC transfusion. For human studies, monitoring typically requires a FDA IND. We recently published a sensitive and specific IgG gel card assay for detecting anti-BioRBC antibodies.25 This assay detects both induced and pre-existing (naturally occurring) anti-BioRBC antibodies. Pre-existing antibodies are present in about 0.5% of naïve adults. We recommend that blood for monitoring be obtained before and every four weeks for 20 weeks after BioRBC administration. Neither blood sample anticoagulant nor multiple freeze thaw cycles affect anti-BioRBC antibody detection or titer (unpublished).
The gel card assay quantifies the interaction of the antigen (i.e., biotin bound to the surface of reagent BioRBC-N) with plasma anti-BioRBC antibodies.25 Quantification is performed by visual assessment of agglutinated BioRBC-N retained in a gel matrix containing immobilized anti-human IgG (Coombs reagent). Positive control plasma is useful but is not commercially available; plasma from antibody-reactive, BioRBC-exposed individuals can be used and is available by contacting JAW. We employ plasma with a high titer anti-BioRBC antibody; when diluted 1:100 in Diluent 2 from the gel card manufacturer, this plasma produces a strong agglutination reaction with reagent BioRBC-54. To rule out non-specific reactivity, plasma to be tested should be assayed with reagent BioRBC and simultaneously with unlabeled RBC from the same type O donor. The gel card assay can be performed with either fresh or glycerolized reagent BioRBC stored frozen at −70°C.31 Reagent BioRBC are used after thawing and deglycerolizing. For maximum sensitivity, but less specificity, reagent BioRBC-256 should be used for both pre-BioRBC transfusion screening and post-infusion monitoring for anti-BioRBC antibodies.25
Flow cytometric detection and enumeration of multiple BioRBC populations
Multiple BioRBC populations can be prepared for separate single BioRBC-N infusion (e.g., to repetitively determine RCV during active hemorrhage) or simultaneous infusion of a mixture of BioRBC-N (e.g., to evaluate pathogen reduction treatment). The number of BioRBC populations that can be used simultaneously is limited by these factors: 1) the BioRBC-N populations in the histogram must not overlap, allowing accurate enumeration of each population (e.g., Figure 1); 2) the least heavily biotinylated population of BioRBC must not overlap the unlabeled RBC; and 3) the RCS parameter(s) of primary interest must not be artifactually shortened by biotinylation.
Staining protocol
We recommend that washed RBC from a 10 μL sample of venous or capillary whole blood be stained with streptavidin conjugated with phycoerythrin (SA-PE; BD Biosciences, San Jose, CA) as follows. The blood sample is mixed with 500 μL of wash buffer and gently vortexed. These RBC are sedimented by centrifuging at 400 RCF for 2.5 min; the supernate is aspirated and discarded and the wash steps are repeated twice more. The final washed RBC pellet is resuspended in 100 μL of wash buffer and the staining reaction is initiated by adding 10 μL of wash buffer producing containing SA-PE that results in a final SA-PE concentration of 12.6 nM. After a 30-min room temperature incubation in the dark, the binding (“staining”) reaction is stopped by adding 500 μL of wash buffer containing 1.63 μM biotin (Sigma, St. Louis, MO). The addition of wash buffer containing free biotin prevents cross-linking of the BioRBC by the tetravalent (strept)avidin; crosslinking is particularly problematic in the pure BioRBC samples used for initial assessment of adequate peak separation (see below). To remove free SA-PE and minimize nonspecific binding, four post-staining 500 μL washes are performed. The third and fourth washes should also contain 2% BSA to reduce nonspecific RBC aggregation. The final RBC pellet is resuspended in 500 μL of wash buffer containing biotin and 2% BSA to produce a RBC suspension suitable for flow cytometry.
Some investigators prefer to inspect the flow cytometric histograms for BioRBC fluorescent intensities prior to BioRBC infusion to confirm peak spacing of unlabeled and BioRBC prior to administration. This can be accomplished without adding significant time prior to BioRBC infusion by rapidly staining a small, nonsterile sample of the BioRBC.
Flow cytometric aspects
Prior to enumeration, a gate is applied to the side scatter versus forward scatter plot (Figure 5). During the first two weeks following BioRBC transfusion, about one third of the biotin labels per RBC are lost as assessed by 125I-avidin binding32 and as assessed by the relative shift of median fluorescent intensity of two BioRBC-N peaks and the leading edge of the unlabeled RBC peak (unpublished observations DMM, JAW, DN, SVK, NIM). This loss is likely attributable to in vivo cleavage of the biotin label by hydrolysis mechanisms that are both enzymatic and non-enzymatic.13 One of the advantages of the biotin RBC labeling method is that this loss of biotin label does not interfere with tracking of the BioRBC-N, provided the shift in fluorescent intensity of histogram peaks does not result in BioRBC peak overlap with unlabeled RBC (Table 1).32 In studies employing multiple BioRBC-N, the leftward peak migrations occur in parallel such that the peaks do not merge. Thus, loss of label does not limit the accuracy of RCV14,20,33 or RCS determination under conditions of either normal32 or accelerated removal.19
Figure 5.
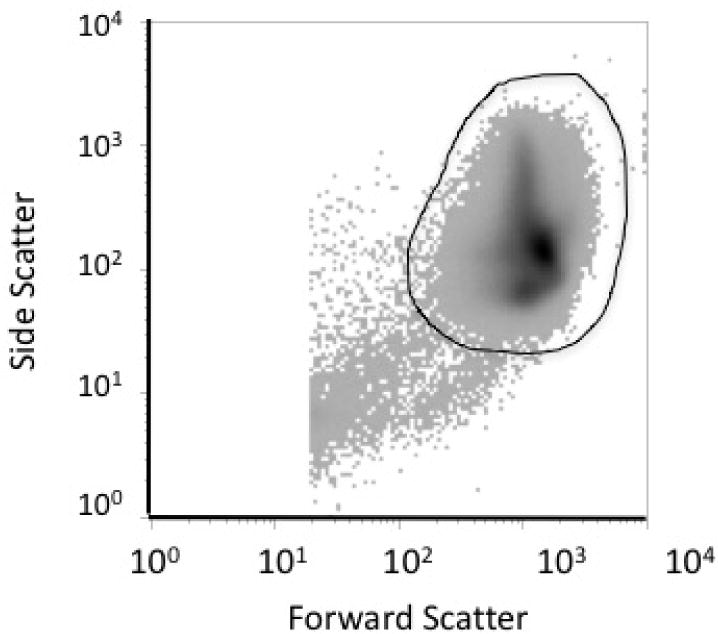
Example of RBC gate of dot matrix density plot of log plot side scatter versus log forward scatter. Dark line depicts manually drawn gate for RBC to exclude confounding hemolyzed RBC, white cells, and platelets; gate typically include >99% of flow cytometric events.
In studies of individual premature infants, we occasionally observe overlap of the lowest density BioRBC peak with the unlabeled RBC peak or with adjacent BioRBC peaks. This has been observed using Alexa Fluor 488 SA stain: 1) when low density adult allogeneic BioRBC peaks (e.g., BioRBC-6) overlap with unlabeled infant RBC; or 2) when adjacent density peaks of adult allogeneic BioRBC and premature infant BioRBC are administered (unpublished observations). Whether density of biotin label per RBC depends on the developmental stage of the donor (i.e., fetus to adult) remains unknown. If encountered, this may be remedied by increasing biotinylating reagent concentration for the lowest BioRBC population (mass/mL RBC) or by increasing the difference in biotinylating reagent concentration between the BioRBC densities, respectively.
As long as the BioRBC-N peaks do not overlap with each other or the unlabeled RBC, the proportion of BioRBC in each population can be accurately determined at enrichments down to the lower limit of BioRBC quantitation (LLOQ). We have empirically defined the analytical reproducibility to be about 3%; LLOQ is defined as 4-fold greater than the BioRBC enrichment at which the CVs of replicates for a given BioRBC-N does not increase to >5%.15 Defined in this way, LLOQ depends on total RBC counted. For example, LLOQ is ~0.06% for 1×106 RBC counted and minimum BioRBC-N enumerated would be 600 cells. At progressively lower BioRBC enrichments, statistical counting error becomes the major contributor to analytical error (e.g., √N/N = 24/600 = 4%). Thus, the choice of total RBC counted is a balance between charges for flow cytometer time and these counting statistics.
Performance of (strept)avidin conjugated fluorescent labels
For BioRBC flow cytometric studies, fluorescein or fluorescein analogues conjugated to avidin or streptavidin generate the primary fluorescence signal by covalent binding to the RBC surface proteins. For the unlabeled RBC, fluorescence arises primarily from hemoglobin. In our hands, difference in the fluorescence intensity was maximized empirically using a Fortessa flow cytometer (BD Biosciences, San Jose, CA) with 488 nm laser at a voltage of 681with a 515/20 band-pass filter. This optical set up produced the largest separations of the individual BioRBC histogram peaks, e.g., BioRBC-6, -18, -54, and -162, from each other and the largest separation from the unlabeled RBC peak. Peak separation is improved using SA-PE rather than fluorescein-conjugated avidin or Alexa Fluor 488 streptavidin. This conclusion is supported by an in vivo RCS study in which BioRBC-2 demonstrated substantial overlap with the leading edge of the unlabeled RBC peak when stained with Alexa Fluor 488, but there was complete peak resolution for the same sample when stained with SA-PE (Figure 6). Accurate enumeration of BioRBC-2 was possible with SA-PE, but not with Alexa Fluor 488. Concentration of SA-PE in the staining reaction was 12.6 nM; optical choices were 561 nm laser at voltage = 660 with a 582/15 band-pass filter. The staining concentration of SA-PE was optimized empirically by examining a range of concentrations from 0.3 to 126 nM.
Figure 6.
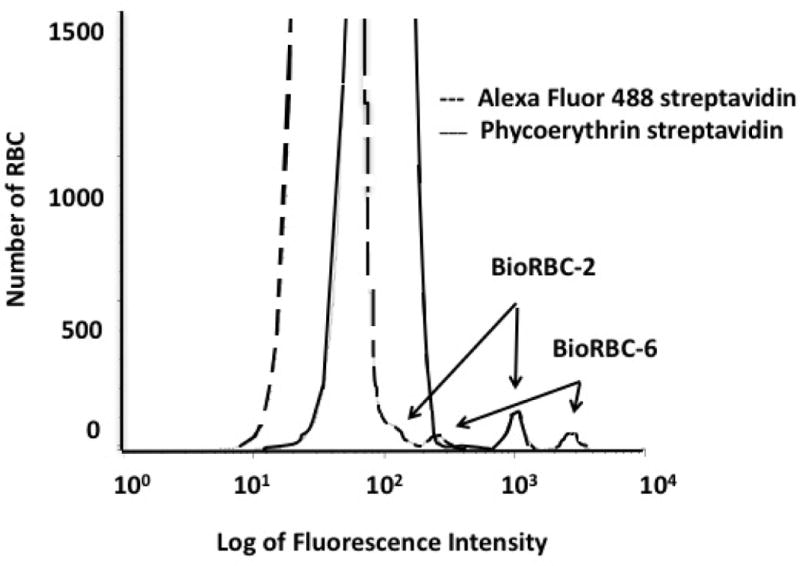
Peak resolution for a mixture of BioRBC-2, BioRBC-6, and unlabeled RBC: Comparison of phycoerythrin streptavidin (___) to Alexa Fluor 488 streptavidin (---). Blood sample was obtained from an RCS study in an adult.
Quantitation of multiple BioRBC-N populations with overlap
In the ideal case in which individual BioRBC-N histograms are completely discrete, the counting windows required are readily identified by visual inspection, and enumeration of the peaks in the windows provides values directly applicable to determining RCS kinetics. However, when the initial labeling produces minimal inter-peak separation or when a portion of the biotin label is lost over time in circulation, overlap of histograms for the individual BioRBC-N may occur and simple “drop line” analysis produces inaccurate enumeration. Figure 7 depicts histogram from the seventh day of an RCS study that utilized autologous BioRBC-2 and BioRBC-18; BioRBC-2 has substantial overlap with the leading edge of unlabeled RBC. To avoid overlap error, a mixed effect analysis using mixture modeling was successfully applied to estimate the number of RBCs in each BioRBC-N population. The mixture modeling approach involves multiple probability density functions; the overall total histogram is constructed by the superposition of the individually scaled and positioned density functions representing the individual BioRBC populations together with the unlabeled RBCs. Multiple overlapping populations can be accurately determined using this approach.34
Figure 7.
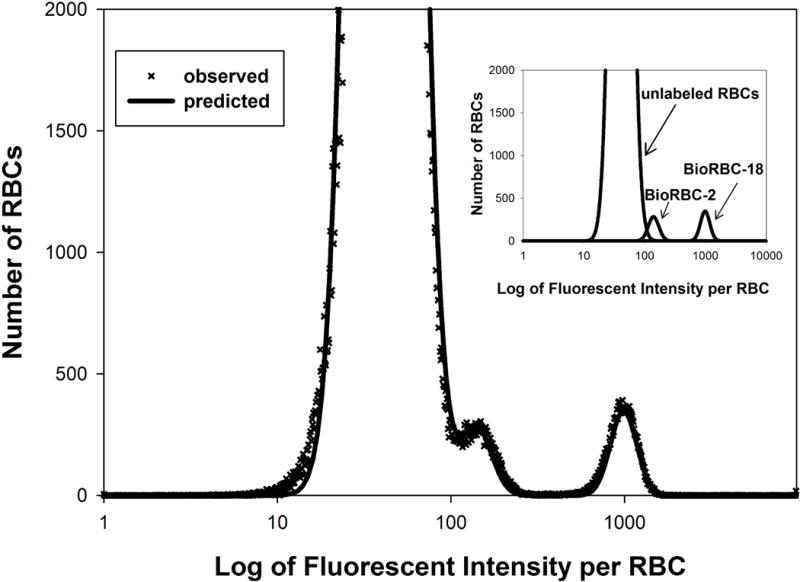
Representative flow cytometric histogram of blood sample obtained after BioRBC transfusion. The number of RBCs enumerated is plotted versus the log of fluorescent intensity per RBC. Peaks are evident for unlabeled RBC, BioRBC-2, and BioRBC-18. The unlabeled RBC peak overlaps the BioRBC-2 peak. The inset illustrates peak quantitation adjusting for peak overlap using mixed effect analysis using mixture modeling.
Interlaboratory comparison of BioRBC enumeration
Three of our sites compared the resolution of BioRBC-N populations and quantitation of BioRBC-N populations: 1) Arkansas; 2) Iowa; and 3) Cincinnati. To avoid site specific differences in biotin labeling (e.g., effects due to shipping), a single source of frozen glycerolized unlabeled RBC, BioRBC-2, -6, -18, and -54 and a mixture of these BioRBC-N were analyzed. Three healthy adults donated RBC. These four BioRBC densities were prepared from each donor and glycerolized along with unlabeled RBC; a mixture of these BioRBC-N and unlabeled RBC were prepared with enrichments of each BioRBC-N that resemble proportions commonly used in in vivo RCS studies, (e.g., ~1% of total RBC).
Arkansas optical settings were as described above. Iowa optical settings were BD LSR-Violet, 561 nm excitation laser at 20 mW, and 582/15 band-pass filter. Cincinnati optical settings were BD LSR-Fortessa, 561 nm excitation laser at 20 mW, and 586/15 band-pass filter. The stain manufacturer and the staining method was the same for all centers. Qualitative examination of histograms demonstrated similar peak positions. Comparison by linear mixed modeling for this limited number of samples was performed on the data set of mean of replicates (n=2 for Arkansas and Iowa; n=1 for Cincinnati for each BioRBC-N from each of the three adult participants. For each site, all peaks were discretely resolved without overlap. Despite analyzing the same samples, a small but statistically significant difference between the three sites in the percentage of RBC in each peak was detected (P<0.0008). Among the three sites, differences ranged from −21.5% to +41.6% with a mean of 4.0% and SD of 11%. We speculate that differences among sites could have arisen from staining or from flow cytometric differences. Although such differences are small, this limited experiment indicates that all time points in given RCS curve should be assayed at the same site. Such transportation is feasible; in vivo blood samples can be stored in Iowa wash buffer at 4°C for at least 10 days.
Analysis of BioRBC kinetic data
Quantification of 24-h post-transfusion RBC recovery (PTR24) short-term survival
Multiple determinations of PTR24 can be made based on the ratio of BioRBC-N enrichment at 24 h relative to that immediately following the BioRBC transfusion. In all studies using fresh autologous or stored donor RBC in naïve subjects, PTR24 values for the BioRBC-N have consistently agreed within analytical error, providing evidence that there is negligible early cell loss of BioRBC-N.14,18,20,35 The FDA relies exclusively on 51Cr-labeled RBC for determination of PTR24 for regulatory review of safety and efficacy (e.g., licensing of RBC storage media).36 Although the FDA has yet to sanction BioRBC PTR24 determinations for this purpose, available results document that concurrent determination of PTR24 using biotin- or 51Cr-labeled RBC yields indistinguishable results.14,32,33
Quantification of PK parameters for long-term RBC survival
Non-steady state changes in the total number of circulating RBC (e.g., due to intervening transfusion of unlabeled blood or with somatic growth) require correction using mathematical modeling to accurately determine RCS from in vivo survival curves.11 This is done by modeling the temporal change in the fraction of BioRBC in circulation following a rapid infusion; loss in the first 24 h is routinely attributed to damage of RBC (e.g., handling, storage) and not included in such PK models. The quantification of the survival function is based on the principle of proportionality of AUC of the BioRBC fluorescent counting histograms obtained over the experimental time period. Accordingly, the ratio between the AUC at time t and the initial AUC denoted as AUC(t)/AUC(0) is the fraction F(t) of BioRBC remaining at time t. In the determination of long-term RCS, adjustment is made for the fraction of BioRBC that is quickly removed from the circulation, i.e., by 24 h as assessed by PTR24. This loss is accounted for by delaying the beginning of the long-term RCS until 24. Accordingly for long-term RCS, F(t) = AUC(t)/AUC(24), t >24 h.
BioRBC survival parameters obtained from the survival curve
In contrast to removal of drugs from the circulation by processes such as chemical degradation, metabolism, and receptor binding, the kinetics of RBC removal is lifespan based (i.e., RBC age-based). Accordingly, mathematical models derived for such substances (e.g., drugs) are not appropriate for determination of RCS, and interpretation and use of some common parameters such as half-life are misleading and should be avoided. There are three main lifespan-based parameters: 1) mean lifespan: the average time a RBC stays in circulation measured from the time the newly created RBC is released into the circulation; 2) the mean remaining lifespan: the average time RBC currently in circulation subsequently stays in the circulation; and 3) the mean RBC age: the average time that the current population of RBC has stayed in circulation.
Mathematical modeling of long-term RBC survival
a. The simplest point distribution model
Since RBC cannot be followed individually, a mathematical lifespan-based model may be applied in determining RCS. The simplest such model is the steady-state, point distribution model; steady-state point distribution model assumes that all RBC are produced at steady-state and are “born” with the exactly same survival properties, and hence have the same lifespan. A population of circulating RBC will have a uniform distribution of remaining lifespan, leading to a linear survival curve, such that the time-axis intercept equals the RBC lifespan (also referred to traditionally as mean potential lifespan). In this scenario, the mean remaining lifespan and the mean age are both equal to half the lifespan (determined from the intercept).
b. The simple lifespan distribution model
Even under steady-state conditions, RBC released into the circulation on the same day do not all have the same survival; instead of having a “point distribution,” survival of even this narrow cohort follows some distribution. Accordingly, the lifespan distribution of a narrow cohort has a lower limit (LL) for about 95% of the RBC. A simple model that considers this complexity is the steady-state model with lifespan distribution. This model leads to a survival curve model that declines linearly until time t = LL and then exhibits a curved decline. Due to either assay limitations (see discussion of lower limit of quantitation above) or logistical reasons in human studies, following the survival curve beyond 5% remaining may not be practical. Accordingly, long-term survival parameters are commonly defined for only the major portion of the BioRBC and do not include the 5% RBC with the longest survival. For this model, the mean lifespan of the major fraction of RBC will be F(LL), and the mean remaining lifespan and the mean age of the circulating RBC are both equal to half this lifespan. In cases where the survival curves cannot be followed to lowest 5% of remaining RBC, the linear portion of the survival curve can be extrapolated to the time axis to estimate mean RBC lifespan.
c. The non-steady-state lifespan distribution model
This model is the same as just described as the simple lifespan distribution model except the population of circulating RBC are not assumed to have been produced at a steady state. Accordingly, the portion of the survival curve earlier than t = LL deviates from linearity. An exact quantification of the mean RBC lifespan requires a model for the non-steady-state production of the RBC.37,38
There are two methods of quantification leading to approximations of the mean lifespan. The simplest method uses suitable, empirical nonlinear curve fitting to derive the mean lifespan from the majority of the RCS curve (95%). The second uses a model-based, non-steady state analysis of the survival curve. Extensive simulation studies would be required to determine which of these two methods provides the most accurate approximation of mean lifespan. Fortunately, determination of the mean remaining lifespan is not confounded by non-steady-state production of the RBC. This is because mean remaining lifespan can be accurately evaluated from the AUC irrespective of the shape of the survival curve. Nonetheless, due to the deviation of the survival from a straight line, following survival for a sufficient time to ensure accurate representation of the survival curve is necessary. Accordingly, an accurate evaluation of the mean remaining lifespan of the major (95%) portion of the circulating RBC is obtained by integrating the survival curve until no more than 5% of BioRBC remain. In contrast, the determination of the mean age of RBC in circulation is confounded by non-steady-state conditions; an expedient approximation can be obtained by equating mean age to the mean remaining lifespan.
d. Models for RBC lifespan distribution
Estimating the lifespan distribution is difficult because this requires accurate and frequent data in the trailing end of the survival curve and because the definition of the tail end is often confounded by analytic limitations under non-steady-state RBC production conditions. Attempts have been made to address this challenging goal. This area of investigation remains largely hypothesis driven and has been reported in detail previously.39
Relative importance of RBC survival parameters
In the context of transfusion medicine, the mean remaining lifespan is among the most relevant parameters. This is because mean remaining lifespan reflects the duration of peripheral tissue oxygen delivery by the remaining transfused RBC. The more frequently reported mean lifespan parameter does not have a simple relationship to tissue oxygen delivery. The more common mean lifespan parameter simply represents a hypothetical population of all freshly produced RBC, which is never transfused. Because mean RBC age does not include information about how long the RBC will remain in circulation, this parameter is problematic as an estimate of the quality of transfused blood.40
Sampling frequency for parameter estimation
RCV
Because the RCV is calculated using the dilution principle, the method assumes that a negligible number of BioRBC are removed from circulation before the first blood sample is obtained post-BioRBC transfusion.14,18,20,41 Thus, after the circulatory mixing time (about 2 minutes), only a single sample is needed. In human studies, the proportions of BioRBC remain stable and within analytical error from 5 to 20 min.14,15
RCS
The frequency and temporal spacing of blood sampling needed in defining the RCS curve are based on several factors: 1) the survival parameter(s) of primary interest; 2) the required accuracy of the survival parameter(s) being determined; 3) anticipated kinetic conditions (e.g., steady-state or non-steady-state); and 4) subject to subject variability in the biological system being investigated. These considerations may differ substantially with different types of investigation. Within the constraints of assay sensitivity, ethical considerations, and feasibility inherent to human studies, following RCS as long as possible is generally advisable. To obtain a comprehensive representation of the major fraction of the BioRBC, the survival should ideally be followed until only 5% remains. Due to 51Cr elution from RBC, this is not possible with 51Cr-labeled RBC. The sampling scheme depends on the complexity and reliability of the survival model applied; the simplest models require the fewest sampling points. For example, 12 to 16 samples equally spaced over the 95% reduction time interval will work well in most situations for adults. However, RCS studies in fetuses and newborn infants can take advantage of frequent blood sampling due to the more complex physiological conditions (e.g., non-steady-state erythropoiesis, laboratory phlebotomy loss, RBC transfusion to treat anemia, and accelerated erythropoiesis due to somatic growth). For RBC population studies in which the sole interest is the variability of RBC lifespan,39 infrequent monthly sampling for the first three months followed by more frequent sampling, e.g., twice weekly, once the limit of RBC survival is approached, is desirable.
FUTURE RESEARCH DIRECTIONS
Despite the methodological development of the BioRBC method to date, additional methodological refinements are needed for investigation of important clinical and scientific questions. Although biotin labeling properties are the same for allogeneic and autologous RBC, we indicate below whether allogeneic RBC, autologous RBC, or both likely to be used.
Regulatory studies in blood banking and transfusion medicine
A multicenter, randomized comparison of long-term RCS of BioRBC with 51Cr-labeled RBC has not been reported. Such a definitive kinetic study would likely be done using autologous BioRBC and should promote acceptance by the FDA of the biotin method over the 51Cr RBC labeling method. If so, this would pave the way for more accurate, comprehensive BioRBC studies impacting important regulatory issues such as evaluation of improved RBC storage media and pathogen reduction procedures for allogeneic RBC.
Investigation of mechanisms of RBC senescence
BioRBC transfusion allows isolation of progressively older allogeneic and autologous RBC using flow cytometric or magnetic targeted separation of BioRBC. Over time, the age distribution of the isolated BioRBC becomes narrower.16 Physical cell sorting may be avoided by using multicolor flow analysis to characterize age-dependent changes in BioRBC properties such as hemoglobin composition, membrane protein expression, phosphatidylserine externalization, and complement deposition. In concurrent comparison of the survival of injured or stored RBC with fresh RBC, mechanisms of “storage lesion” injury and improvements in storage media can be rapidly and comprehensively examined with less cost and effort.42 Additional potentially promising research areas include: 1) investigation of mechanisms of RBC removal under normal and pathological conditions, e.g., hemolytic anemia by examining the properties of senescing RBC; and 2) investigation of the pre-clinical and clinical efficacy and toxicity of therapeutic approaches targeting shared “eat-me” signals43,44 and immune-mediated checkpoints targeting tumor cell evasion.45,46
Circulating RCV measured repeatedly on a real time, emergent basis
For repeated RCV determination in the same patient (e.g., acute gastrointestinal bleeding or heart failure),47 repeated administration of mini-doses of allogeneic BioRBC (e.g., 5-10 mL of packed BioRBC-N) could provide such information. A potential allogeneic donor source would be Group O RBC that are Rh(D) negative or Rh(D) compatible. The feasibility of doing this has been recently enhanced by the report of successful biotinylation of fresh RBC in a closed system.21 This approach increases safety and flexibility of such studies, particularly in Europe where freshly biotinylated RBC may be stored up to one week. Alternatively, frozen, glycerolized, universal donor BioRBC could be employed in a similar fashion—but with pre-transfusion storage for months to years instead of only a week. The safety and efficacy of the glycerolized BioRBC approach seems feasible because frozen-thawed glycerolized RBC are commonly used clinically in treating hemolytic disorders, e.g., sickle cell disease and beta thalassemia.
This same mini-dose approach might also be applied in regulatory or research studies determining PTR24. While similar studies have been performed using glycerolized 51Cr labeled RBC, use of BioRBC would permit concurrent, multi-BioRBC-N density PTR24 studies to examine the effect of duration of BioRBC storage at 4°C or −20°C (when glycerolized). Such studies have potentially important clinical and research applications.47–50
Emergent RCS determinations
BioRBC might also be used in predicting the efficacy of therapeutic allogeneic transfusions in critically ill, high-risk patients who have antibodies to multiple RBC antigens, e.g., sickle cell disease and thalassemia. Donor-recipient specific testing could include administering mini-doses of low density BioRBC-N to assess BioRBC survival after 12 to 72 h in circulation. These mini-dose RCS determinations would provide predictive information about long-term RCS of specific donor units. Only RBC units with near normal BioRBC survival would be transfused. In subsequent crisis hemolytic or aplastic situations, allogeneic BioRBC prepared in a closed system and frozen could be used immediately and more effectively with savings of time, effort, and cost.21
Enhancement of fluorescent probes
Extrapolating from ongoing improvements in flow cytometry instrumentation and fluorescent probes, future methodological refinements in BioRBC detection will continue. Amplification of the biotin signal without increasing background noise (non-specific binding to unlabeled RBC) has potential for further reducing the BioRBC “dose” administered. To investigate this, we (DM, SK, NM, JW) investigated whether the fluorescence per BioRBC can be increased by employing quantum dots coated with SA-containing fluor molecules. The separation of unlabeled RBC and BioRBC-6 in vitro using QDot605-SA, QDot655-SA, and QDot800-SA (Invitrogen, Carlsbad, CA) was quantitated. We observed that enumeration of BioRBC by either QDot605-SA or QDot655-SA agreed with the AlexaFluor488 results but reproducibility was inferior to Alexa Fluor 488 SA and SA-PE (unpublished). Clarification of the value of quantum dots or similar signal enhancing methodologies awaits further investigation.
Characterization of anti-BioRBC antibody induction and functional implications
Additional studies are needed to elucidate the stimuli that induce anti-BioRBC antibody production in relevant patient populations and disease states such as sickle cell anemia.11 Additionally, the clinical consequences of such antibodies (if any) will likely emerge as greater numbers of subjects are studied. Despite the fact that the only detectable anti-BioRBC antibody effect reported thus far is accelerated BioRBC removal,51 laboratory and clinical investigations exploring meaningful functional consequences of anti-BioRBC antibodies are important.
The topics raised in this review provide a firm basis for future investigations of BioRBC methodologies. The content of this review may be of particular help to investigators in the U.S.A. where human BioRBC studies require FDA approval. While much has been learned about this novel and important methodology, more remains to be explored. We speculate that BioRBC methodology will find important applications in research fields yet to be anticipated. Hence, the future focus, scope, and clinical impact of BioRBC studies seem promising.
Acknowledgments
The authors acknowledge helpful comments and critiques from flow cytometry experts Andrea Harris (UAMS) and Justin Fishbaugh (University of Iowa) as well as and the technical contributions of Sarah Hill, B.S. and Monica DeLay, M.S., to optimize the Cincinnati flow cytometry analysis method. M. Bridget Zimmerman, Ph.D. provided statistical expertise and input. Mark Hart provided valuable editorial and secretarial assistance.
Support: The sponsors of this work had no role in this work including: study design; the collection, analysis, and interpretation of data; the writing of the report; and the decision to submit the manuscript for publication. The sponsors of this work include: The United States Public Health Service National Institutes of Health Grant P01 HL046925; The Thrasher Research Fund 0285-3; The National Center for Research Resources, a part of the National Institutes of Health (NIH) CTSA Grants U54TR001356 (U Iowa) and UL1TR000039 (UAMS) and 1S10 RR027219. Support was also provided (D.M.M.) by the NHLBI Summer Undergraduate Research Program to Increase Diversity in Research (SURP grant) R25 HL108825 (UAMS). Some of the flow cytometry data were obtained at the Flow Cytometry Facility, which is a Carver College of Medicine/Holden Comprehensive Cancer Center core research facility at the University of Iowa. The facility is funded through user fees and the generous financial support of the Carver College of Medicine, Holden Comprehensive Cancer Center, and Iowa City Veteran’s Administration Medical Center.
ABBREVIATIONS
- BioRBC
biotin-labeled red blood cell(s)
- BV
blood volume
- HCT
hematocrit
- RCV
circulating red blood cell volume
- RCS
red blood cell survival
- RBC
red blood cell(s)
- SA-PE
streptavidin conjugated with phycoerythrin
Footnotes
Conflicts of Interest: The authors have the following potential conflicts of interest items to disclose:
Donald M. Mock is a consultant for MedDay Pharmaceuticals, Paris, France. There are no conflicts of interest for the remaining authors.
AUTHOR CONTRIBUTIONS
Multiple authors (DMM, DN, JW, GA, JAC, and PVP) contributed to the writing of the initial and all subsequent drafts. SVK, NIM, JAC, and RLS contributed to conception, acquisition, analysis, and interpretation of several aspects of the paper and critically reviewed the paper. RSF and RGS contributed to the content of the paper and critically reviewed the paper. DDK contributed to the content of the paper and critically reviewed the paper. RVB contributed to the content of the paper and critically reviewed the paper. APJV contributed to the content of the paper and critically reviewed the paper.
None of the coauthors received an honorarium for writing or submitting this manuscript.
References
- 1.Ashby W. The determination of the length of life of transfused blood corpuscles in man. J Exp Med. 1919;29:267–81. doi: 10.1084/jem.29.3.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garratty G. Patrick Loudon Mollison. Transfusion. 2012;52:684–5. doi: 10.1111/j.1537-2995.2012.03586.x. [DOI] [PubMed] [Google Scholar]
- 3.Franco RS. The measurement and importance of red cell survival. Am J Hematol. 2009;84:109–14. doi: 10.1002/ajh.21298. [DOI] [PubMed] [Google Scholar]
- 4.International Committee for Standardization in Haematology. Recommended methods for radioisotope red-cell survival studies. Br J Haematol. 1980;45:659–66. doi: 10.1111/j.1365-2141.1980.tb07189.x. [DOI] [PubMed] [Google Scholar]
- 5.International Committee for Standardization in Haematology. Recommended methods for measurement of red-cell and plasma volume. J Nucl Med. 1980;21:793–800. [PubMed] [Google Scholar]
- 6.Dumont LJ, AuBuchon JP. Evaluation of proposed FDA criteria for the evaluation of radiolabeled red cell recovery trials. Transfusion. 2008;48:1053–60. doi: 10.1111/j.1537-2995.2008.01642.x. [DOI] [PubMed] [Google Scholar]
- 7.Shemin D, Rittenberg D. The life span of the human red blood cell. J Biol Chem. 1946;166:627–36. [PubMed] [Google Scholar]
- 8.Khera PK, Smith EP, Lindsell CJ, Rogge MC, Haggerty S, Wagner DA, Palascak MB, Mehta S, Hibbert JM, Joiner CH, Franco RS, Cohen RM. Use of an oral stable isotope label to confirm variation in red blood cell mean age that influences HbA1c interpretation. Am J Hematol. 2015;90:50–5. doi: 10.1002/ajh.23866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinn CT, Smith EP, Arbabi S, Khera PK, Lindsell CJ, Niss O, Joiner CH, Franco RS, Cohen RM. Biochemical surrogate markers of hemolysis do not correlate with directly measured erythrocyte survival in sickle cell anemia. Am J Hematol. 2016;91:1195–201. doi: 10.1002/ajh.24562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ricketts C, Cavill I. Ferrokinetics: methods and interpretation. Clin Nucl Med. 1978;3:159–64. doi: 10.1097/00003072-197804000-00014. [DOI] [PubMed] [Google Scholar]
- 11.Mock DM, Widness JA, Veng-Pedersen P, Strauss RG, Cancelas JA, Cohen RM, Lindsell CJ, Franco RS. Measurement of posttransfusion red cell survival with the biotin label. Transfus Med Rev. 2014;28:114–25. doi: 10.1016/j.tmrv.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein HG, Anstee DJ. The transfusion of red cells. In: Klein HG, Anstee DJ, editors. Mollison’s blood transfusion in clinical medicine. Chichester, West Sussex, UK: John Wiley and Sons, Inc; 2014. p. 362. [Google Scholar]
- 13.Bogusiewicz A, Mock NI, Mock DM. Instability of the biotin-protein bond in human plasma. Anal Biochem. 2004;327:156–61. doi: 10.1016/j.ab.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 14.Mock DM, Lankford GL, Widness JA, Burmeister LF, Kahn D, Strauss RG. Measurement of circulating red blood cell volume using biotin labeled red cells: Validation against 51Cr labeled red cells. Transfusion. 1999;39:149–55. doi: 10.1046/j.1537-2995.1999.39299154728.x. [DOI] [PubMed] [Google Scholar]
- 15.Mock DM, Matthews NI, Zhu S, Strauss RG, Schmidt RL, Nalbant D, Cress GA, Widness JA. Red blood cell (RBC) survival determined in humans using RBCs labeled at multiple biotin densities. Transfusion. 2011;51:1047–57. doi: 10.1111/j.1537-2995.2010.02926.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franco RS, Puchulu-Campanella ME, Barber LA, Palascak MB, Joiner CH, Low PS, Cohen RM. Changes in the properties of normal human red blood cells during in vivo aging. Am J Hematol. 2013;88:44–51. doi: 10.1002/ajh.23344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen RM, Lindsell CJ. When the blood glucose and the HbA(1c) don’t match: turning uncertainty into opportunity. Diabetes Care. 2012;35:2421–3. doi: 10.2337/dc12-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mock D, Matthews N, Zhu S, Burmeister L, Zimmerman M, Strauss R, Schmidt R, Nalbant D, Cress G, Widness J. Red blood cell (RBC) volume can be independently determined in vivo in humans using RBCs labeled at different densities of biotin. Transfusion. 2011;51:148–57. doi: 10.1111/j.1537-2995.2010.02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mock DM, Lankford GL, Widness JA, Burmeister LF, Kahn D, Strauss RG. Accelerated removal of antibody-coated red blood cells from the circulation is accurately tracked by a biotin label. Transfusion. 2012;52:1097–105. doi: 10.1111/j.1537-2995.2011.03397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nalbant D, Bhandary P, Matthews NI, Schmidt RL, Bogusiewicz A, Cress GA, Zimmerman MB, Strauss RG, Mock DM, Widness JA. Comparison of multiple red cell volume methods performed concurrently in premature infants following allogeneic transfusion. Pediatr Res. 2013;74:592–600. doi: 10.1038/pr.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Back DZ, Vlaar R, Beuger B, Daal B, Lagerberg J, Vlaar APJ, de Korte D, van Kraaij M, van Bruggen R. A method for red blood cell biotinylation in a closed system. Transfusion. 2018 doi: 10.1111/trf.14535. In press. [DOI] [PubMed] [Google Scholar]
- 22.Davey RJ, McCoy NC, Yu M, Sullivan JA, Spiegel DM, Leitman SF. The effect of prestorage irradiation on posttransfusion red cell survival. Transfusion. 1992;32:525–8. doi: 10.1046/j.1537-2995.1992.32692367195.x. [DOI] [PubMed] [Google Scholar]
- 23.Moroff G, Holme S, AuBuchon JP, Heaton WA, Sweeney JD, Friedman LI. Viability and in vitro properties of AS-1 red cells after gamma irradiation. Transfusion. 1999;39:128–34. doi: 10.1046/j.1537-2995.1999.39299154725.x. [DOI] [PubMed] [Google Scholar]
- 24.Mock DM, Matthews NI, Strauss RG, Burmeister LF, Schmidt R, Widness JA. Red blood cell volume can be independently determined in vitro using sheep and human red cells labeled at different densities of biotin. Transfusion. 2009;49:1178–85. doi: 10.1111/j.1537-2995.2009.02095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmidt RL, Mock DM, Franco RS, Cohen RM, North AK, Cancelas JA, Geisen C, Strauss RG, Vlaar AP, Nalbant D, Widness JA. Antibodies to biotinylated red blood cells in adults and infants: improved detection, partial characterization, and dependence on red blood cell-biotin dose. Transfusion. 2017;57:1488–96. doi: 10.1111/trf.14075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cavill I, Trevett D, Fisher J, Hoy T. The measurement of the total volume of red cells in man: A non-radioactive approach using biotin. Br J Haematol. 1988;70:491–3. doi: 10.1111/j.1365-2141.1988.tb02522.x. [DOI] [PubMed] [Google Scholar]
- 27.Hudson IRB, Cavill IAJ, Cooke A, Holland BM, Hoy TG, Trevett D, Turner TL, Wardrop CAJ. Biotin labeling of red cells in the measurement of red cell volume in preterm infants. Pediatr Res. 1990;28:199–202. doi: 10.1203/00006450-199009000-00006. [DOI] [PubMed] [Google Scholar]
- 28.Veale MF, Healey G, Sparrow RL. Effect of additive solutions on red blood cell (RBC) membrane properties of stored RBCs prepared from whole blood held for 24 hours at room temperature. Transfusion. 2011;51(Suppl 1):25S–33S. doi: 10.1111/j.1537-2995.2010.02960.x. [DOI] [PubMed] [Google Scholar]
- 29.Liesenfeld O, Lehman L, Hunfeld KP, Kost G. Molecular diagnosis of sepsis: New aspects and recent developments. Eur J Microbiol Immunol (Bp) 2014;4:1–25. doi: 10.1556/EuJMI.4.2014.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cordle DG, Strauss RG, Lankford GL, Mock DM. Antibodies provoked by the transfusion of biotin-labeled red cells. Transfusion. 1999;39:1065–9. doi: 10.1046/j.1537-2995.1999.39101065.x. [DOI] [PubMed] [Google Scholar]
- 31.Schmid P, Huvard MJ, Lee-Stroka AH, Lee JY, Byrne KM, Flegel WA. Red blood cell preservation by droplet freezing with polyvinylpyrrolidone or sucrose-dextrose and by bulk freezing with glycerol. Transfusion. 2011;51:2703–8. doi: 10.1111/j.1537-2995.2011.03258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mock DM, Lankford GL, Widness JA, Burmeister LF, Kahn D, Strauss RG. Measurement of red cell survival using biotin labeled red cells: Validation against 51Cr labeled red cells. Transfusion. 1999;39:156–62. doi: 10.1046/j.1537-2995.1999.39299154729.x. [DOI] [PubMed] [Google Scholar]
- 33.Mock DM, Lankford GL, Widness JA, Burmeister LF, Kahn D, Strauss RG. RBCs labeled at two biotin densities permit simultaneous and repeated measurements of circulating RBC volume. Transfusion. 2004;44:431–7. doi: 10.1111/j.1537-2995.2004.00654.x. [DOI] [PubMed] [Google Scholar]
- 34.Mock D, Matthews N, Zhu S, Strauss R, Schmidt R, Zimmerman M, Nalbant D, Freise K, Saleh M, Veng-Pedersen P, Widness J. Comparison of red blood cell survival in sheep determined using red blood cells labeled either with biotin at multiple densities or 14C-cyanate: Validation of a model to study human physiology and disease. Transfusion. 2012;52:963–73. doi: 10.1111/j.1537-2995.2011.03512.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nalbant D, Cancelas JA, Mock DM, Kyosseva SV, Schmidt RL, Cress GA, Zimmerman MB, Strauss RG, Widness JA. In premature infants there is no decrease in 24-hour posttransfusion allogeneic red blood cell recovery after 42 days of storage. Transfusion. 2018;58:352–8. doi: 10.1111/trf.14396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hess JR, Biomedical Excellence for Safer Transfusion C Scientific problems in the regulation of red blood cell products. Transfusion. 2012;52:1827–35. doi: 10.1111/j.1537-2995.2011.03511.x. [DOI] [PubMed] [Google Scholar]
- 37.Kuruvilla DJ, Widness JA, Nalbant D, Schmidt RL, Mock DM, Veng-Pedersen P. A method to evaluate fetal erythropoiesis from postnatal survival of fetal RBCs. AAPS J. 2015;17:1246–54. doi: 10.1208/s12248-015-9784-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuruvilla DJ, Widness JA, Nalbant D, Schmidt RL, Mock DM, An G, Veng-Pedersen P. Estimation of adult and neonatal RBC lifespans in anemic neonates using RBCs labeled at several discrete biotin densities. Pediatr Res. 2017;81:905–10. doi: 10.1038/pr.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shrestha RP, Horowitz J, Hollot CV, Germain MJ, Widness JA, Mock DM, Veng-Pedersen P, Chait Y. Models for the red blood cell lifespan. J Pharmacokinet Pharmacodyn. 2016;43:259–74. doi: 10.1007/s10928-016-9470-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kuruvilla DJ, Nalbant D, Widness JA, Veng-Pedersen P. Mean remaining life span: a new clinically relevant parameter to assess the quality of transfused red blood cells. Transfusion. 2014;54:2724–9. doi: 10.1111/trf.12562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mock DM, Mock NI, Lankford GL, Burmeister LF, Strauss RG, Widness JA. Red cell volume can be accurately determined in sheep using a nonradioactive biotin label. Pediatr Res. 2008;64:528–32. doi: 10.1203/PDR.0b013e318183f119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peters AL, Beuger B, Mock DM, Widness JA, de Korte D, Juffermans NP, Vlaar AP, van Bruggen R. Clearance of stored red blood cells is not increased compared with fresh red blood cells in a human endotoxemia model. Transfusion. 2016;56:1362–9. doi: 10.1111/trf.13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Wang L, Zhao F, Tseng S, Narayanan C, Shura L, Willingham S, Howard M, Prohaska S, Volkmer J, Chao M, Weissman IL, Majeti R. Pre-clinical development of a humanized anti-CD47 antibody with anti-cancer therapeutic potential. PLoS One. 2015;10:e0137345. doi: 10.1371/journal.pone.0137345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petrova PS, Viller NN, Wong M, Pang X, Lin GH, Dodge K, Chai V, Chen H, Lee V, House V, Vigo NT, Jin D, Mutukura T, Charbonneau M, Truong T, Viau S, Johnson LD, Linderoth E, Sievers EL, Maleki Vareki S, Figueredo R, Pampillo M, Koropatnick J, Trudel S, Mbong N, Jin L, Wang JC, Uger RA. TTI-621 (SIRPalphaFc): A CD47-blocking innate immune checkpoint inhibitor with broad antitumor activity and minimal erythrocyte binding. Clin Cancer Res. 2017;23:1068–79. doi: 10.1158/1078-0432.CCR-16-1700. [DOI] [PubMed] [Google Scholar]
- 45.Kong BY, Micklethwaite KP, Swaminathan S, Kefford RF, Carlino MS. Autoimmune hemolytic anemia induced by anti-PD-1 therapy in metastatic melanoma. Melanoma Res. 2016;26:202–4. doi: 10.1097/CMR.0000000000000232. [DOI] [PubMed] [Google Scholar]
- 46.Simeone E, Grimaldi AM, Esposito A, Curvietto M, Palla M, Paone M, Mozzillo N, Ascierto PA. Serious haematological toxicity during and after ipilimumab treatment: a case series. J Med Case Rep. 2014;8:240. doi: 10.1186/1752-1947-8-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Montero D, Lundby C, Ruschitzka F, Flammer AJ. True anemia-red blood cell volume deficit-in heart failure: A systematic review. Circ Heart Fail. 2017;10 doi: 10.1161/CIRCHEARTFAILURE.116.003610. [DOI] [PubMed] [Google Scholar]
- 48.Cure P, Bembea M, Chou S, Doctor A, Eder A, Hendrickson J, Josephson CD, Mast AE, Savage W, Sola-Visner M, Spinella P, Stanworth S, Steiner M, Mondoro T, Zou S, Levy C, Waclawiw M, El Kassar N, Glynn S, Luban NLC. 2016 proceedings of the National Heart, Lung, and Blood Institute’s scientific priorities in pediatric transfusion medicine. Transfusion. 2017;57:1568–81. doi: 10.1111/trf.14100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spitalnik SL, Triulzi D, Devine DV, Dzik WH, Eder AF, Gernsheimer T, Josephson CD, Kor DJ, Luban NL, Roubinian NH, Mondoro T, Welniak LA, Zou S, Glynn S, State of the Science in Transfusion Medicine Working G 2015 proceedings of the National Heart, Lung, and Blood Institute’s State of the Science in Transfusion Medicine symposium. Transfusion. 2015;55:2282–90. doi: 10.1111/trf.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Doctor A. Influence of transfused RBC physiology upon recipient oxygen delivery homeostasis. Proceedings of the Food and Drug Administration’s Workshop on New Red Blood Cell Product Regulatory Science. 2016;201:354. [Google Scholar]
- 51.Stowell SR, North AK, Franco RS, Cancelas JA, Mock DM, Strauss RG, Geisen C, Schmidt RL, Nalbant D, Widness JA. Shortened survival of biotin labeled RBCs following a second BioRBC transfusion in an adult with a previous BioRBC antibody response. Transfusion. 2014;54:52A. [Google Scholar]
- 52.Cohen RM, Franco RS, Khera PK, Smith EP, Lindsell CJ, Ciraolo PJ, Palascak MB, Joiner CH. Red cell life span heterogeneity in hematologically normal people is sufficient to alter HbA1c. Blood. 2008;112:4284–91. doi: 10.1182/blood-2008-04-154112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Widness JA, Nalbant D, Matthews NI, Strauss RG, Schmidt RL, Cress GA, Zimmerman MB, Mock DM. Tracking donor RBC survival in premature infants: Agreement of multiple populations of biotin labeled RBCs with Kidd antigen mismatched RBCs. Pediatr Res. 2013;74:689–97. doi: 10.1038/pr.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mock DM, Bell EF, Lankford GL, Widness JA. Hematocrit correlates well with circulating red blood cell volume in very low birth weight infants. Pediatr Res. 2001;50:525–31. doi: 10.1203/00006450-200110000-00017. [DOI] [PubMed] [Google Scholar]
- 55.Diekema DS. Conducting ethical research in pediatrics: a brief historical overview and review of pediatric regulations. J Pediatr. 2006;149:S3–11. doi: 10.1016/j.jpeds.2006.04.043. [DOI] [PubMed] [Google Scholar]
- 56.Mock DM, Widness JA, Strauss RG, Franco RS. Posttransfusion red blood cell (RBC) survival determined using biotin-labeled RBCs has distinct advantages over labeling with 51Cr. Transfusion. 2012;52:1596–8. doi: 10.1111/j.1537-2995.2012.03588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]


