Abstract
Advances in bioinformatics allow identification of single nucleotide polymorphisms (variants) from RNA sequence data. In an allograft biopsy, two genomes contribute to the RNA pool, one from the donor organ and the other from infiltrating recipient’s cells. We hypothesize that imbalances in genetic variants of RNA sequence data of kidney allograft biopsies provide an objective measure of cellular infiltration of the allograft. We performed mRNA sequencing of 40 kidney allograft biopsies, selected to represent a comprehensive range of diagnostic categories. We analyzed the sequencing reads of these biopsies and of 462 lymphoblastoid cell lines from the 1000 Genomes Project, for RNA variants. The ratio of heterozygous to non-reference genome homozygous variants (Het/Hom ratio) on all autosomes was determined for each sample, and the ESTIMATE score was computed as a complementary estimate of the degree of cellular infiltration into biopsies. The Het/Hom ratios (P=0.02) and the ESTIMATE scores (P<0.001) were associated with the biopsy diagnosis. Both measures correlated significantly (r=0.67, P<0.0001), even though the Het/Hom ratio is based on mRNA sequence variation while the ESTIMATE score uses mRNA expression. Het/Hom ratio and the ESTIMATE score may offer unbiased and quantitative parameters for characterizing cellular traffic into human kidney allografts.
1. Introduction
Kidney transplantation has moved from a high-risk experimental procedure to a safe lifesaving therapy. Besides the clinical application of potent immunosuppressive drugs and effective infectious prophylaxis, refinements in diagnostics including standardized readings of kidney allograft biopsy specimens with the use of the Banff classification schema has contributed to the current success rates (1–3). Multiple challenges however still remain. An often-repeated concern is that long-term survival rates of kidney allografts have not improved pari-pasu with improvements in the short-term outcomes (4). There are concerns related to inter-observer variability regarding interpretation of biopsy findings (5, 6). In addition, tubulitis in the kidney allograft, an essential criterion for histological diagnosis of Banff acute T cell mediated rejection, is often missed in the absence of immunostaining of the allograft biopsy for T cell CD3 protein (7). An additional issue is that the current Banff scoring system is semi-quantitative (2, 3). Thus, development of precise quantitative measures to monitor allograft status might be of value.
Transplantation is a unique situation from a genetic perspective. Two diploid genomes, the genome of the recipient and the genome of the donor, are brought together by a clinical intervention. An allograft with graft infiltrating cells therefore contains genomes from two distinct sources; the donor genome from the graft parenchymal cells and the recipient genome from graft infiltrating cells. Quantification of the relative contribution of recipient’s genome and donor genome within an allograft biopsy may hence offer an objective readout for the degree of allograft invasion by recipient’s cells. Advances in genomics can be innovatively applied leveraged to carry out such quantification.
RNA sequencing has enabled genome-wide transcriptome profiling at an unprecedented level of precision (8–11), and is being increasingly applied to clinical investigation, including kidney allograft pathologies (12–14). Recent advances in computational methods have enabled the extraction of additional genomic information from the RNA sequence data (15–19). Variant calling of single nucleotide polymorphisms (SNPs), hitherto a domain of DNA sequencing, is now possible using mRNA sequencing data and novel bioinformatics tools (20–22). In this approach, the ratio of heterozygous variants to non-reference homozygous variants (Het/Hom ratio) is generally used as quality control measure in DNA sequencing (23, 24), as it offers accurate resolution of genome admixture, often due to experimental errors, such as sample cross-contamination. Because the kidney allograft infiltrated by recipient’s cells is a unique situation of admixture of two genomes, we reasoned that variant calling on RNA sequencing of kidney allograft biopsy specimens and therefrom derived Het/Hom ratios could function as a quantitative measure for the invasion of the transplant organ by recipient’s cells. We further reasoned that a method used in cancer studies to estimate purity scores for the degree of stromal and immune cell invasion of tumor tissue (called ESTIMATE) may also be of use to gauge graft infiltration, as this method is based on cell-type specific gene expression levels (25).
In this first-in-its-kind study, we report that kidney allograft biopsies manifesting cellular invasion have higher Het/Hom ratios and ESTIMATE scores compared to pristine biopsies from patients without manifest allograft rejection. We observe significant relationships between Banff biopsy classification and Het/Hom ratios and between Banff biopsy classification and the ESTIMATE scores. Moreover, we find a robust correlation between the Het/Hom ratios and ESTIMATE scores. This is of interest as the two methods are based on different readouts of the mRNA sequencing data: The Het/Hom ratio is derived from genetic variance while the ESTIMATE score is based on cell-type specific gene expression. In order to establish robust Het/Hom ratios of single genomes using RNA sequencing data, we leveraged a publicly available database of mRNA and small RNA sequencing data from 462 lymphoblastoid cell lines (LCLs) from five distinct populations included in the 1000 Genomes Project (26).
2. Materials and Methods
2.1. Study Groups and Allograft Biopsies
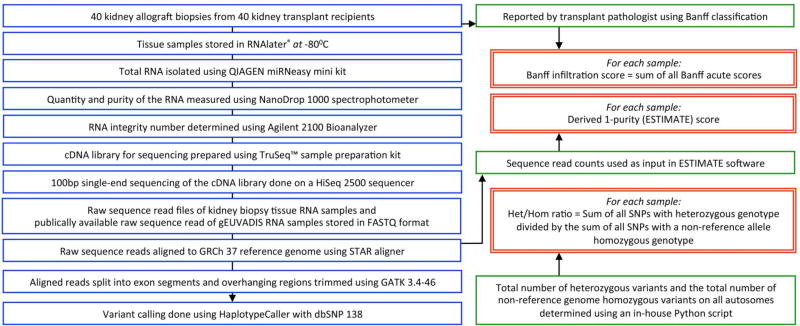
We studied 40 adult recipients of human kidney allografts, a subset of kidney allograft recipients transplanted and followed at our center, the New York Presbyterian-Weill Cornell Medicine. All recipients provided written informed consent to participate in the study, and our Institutional Review Board approved the study. The clinical and research activities that we report here are consistent with the principles of the ‘Declaration of Istanbul on Organ Trafficking and Transplant Tourism’. The overall workflow used in this investigation is shown in Figure 1.
Figure 1. Workflow of the study.
Forty allograft biopsy specimens obtained from 40 unique kidney allograft recipients were RNA sequenced. For each sample: (i) Biopsy infiltration score was derived as the sum of Banff acute scores, reported by the transport pathologist; (ii) A purity score was computed using the aligned RNA sequencing reads as input in ESTIMATE software; and (iii) Het/Hom ratio was computed after variant calling was done from the RNA sequencing data. Throughout the manuscript, the ratio of heterozygous to non-reference genome homozygous variants on all autosomes, for each sample, is called as the Het/Hom ratio, and the ESTIMATE score and 1-Purity score are used interchangeably.
Each study participant underwent ultrasound guided percutaneous core needle biopsy of the allografts and 40 unique kidney allograft biopsy specimens were obtained. Among the 40 biopsies, 35 biopsies were performed for clinical reasons (for-cause biopsies) in patients with graft dysfunction and the remaining 5 biopsies were protocol (surveillance) biopsies in clinically stable patients. Biopsy tissue sections were stained with hematoxylin and eosin, periodic acid Schiff, and Masson trichrome, as well as for polyomavirus, for complement factor 4 degradation product d (C4d), and for CD3, C20 and CD68. The biopsies were categorized by our pathologists blinded to RNA sequencing data and with the use of Banff 2017 update of the Banff ‘97 classification of allograft pathology (1, 3). We selected these 40 unique biopsies for inclusion in this study since the allograft biopsy diagnosis represented major Banff diagnostic categories; acute T-cell mediated rejection (acute TCMR, N=11), active antibody mediated rejection (active ABMR, N=7), chronic active antibody mediated rejection (chronic active ABMR, N=11), interstitial fibrosis and tubular atrophy (IFTA, N=6) or Normal biopsies (N=5). Each patient provided a single biopsy sample to this study.
2.2. RNA Sequencing of Allograft Biopsies
At the time of allograft biopsy, a portion of the biopsy tissue was immediately submerged in RNAlater® RNA stabilization solution (Life technologies, Grand Island, NY) and stored at −80°C. We isolated total RNA from the stored biopsy tissues using miRNeasy mini kit (Qiagen, Inc., Valencia, CA). The quantity and purity of the RNA were measured using NanoDrop 1000 spectrophotometer (Thermo Scientific, Wilmington, DE) and the RNA integrity number using Agilent 2100 Bioanalyzer (Agilent Technologies Inc., Santa Clara, CA). For mRNA sequencing we used 400ng of total RNA obtained from each biopsy sample. We used the TruSeq™ sample preparation kit v2 (Illumina, Inc., San Diego, CA) to prepare individual cDNA libraries. Briefly, this consists of poly-A selection of mRNA and conversion to single-stranded cDNA using random hexamer primer followed by second strand generation to create double stranded cDNA. Sequencing adapters were then ligated to the fragmented cDNA. This was followed by PCR amplification and pooling of the libraries. Six cDNA samples were pooled per lane of the flow cell for 100bp single-end sequencing on a HiSeq 2500 sequencer (Illumina, Inc., San Diego, CA). The raw sequencing data was stored in FASTQ format.
2.3. Lymphoblastoid Cell Line (LCL) RNA Sequencing Data
We downloaded FASTQ files of the gEUVADIS (Genetic European Variation in Health and Disease) RNA sequencing project (26). This is a European medical sequencing consortium with a publicly available database of mRNA and small RNA sequencing from 462 LCLs from five distinct populations included in the 1000 Genomes Project: residents from Utah, USA with Northern and Western European ancestry (CEU), Finnish in Finland (FIN), British in England and Scotland (GBR), Tuscany in Italy (TSI), and Yoruba in Ibadan, Nigeria (YRI).
2.4. RNA Sequence Alignment and Variant Calling
RNA sequence reads from the kidney allograft biopsy specimens and the RNA sequence reads downloaded from gEUVADIS were processed using Genome Reference Consortium human genome reference build 37 (GRCh37) as the reference genome. We used the ‘Spliced Transcripts Alignment to a Reference’ (STAR) aligner together with the iGenome’s (Illumina®) Human Ensemble gene annotation to process the data following Genome Analysis ToolKit (GATK) best practices for RNA sequence variant calling (27–31). Briefly, we used Picard (1.107) to sort, add read groups and remove duplicates in the aligned Binary Alignment/Map (BAM) files as generated by STAR aligner (32). We then used GATK (version 3.4–46) to split reads into exon segments and trim overhang regions with mismatches before calling variants for each sample individually using HaplotypeCaller with ‘Single Nucleotide Polymorphism Database build 138’ (dbSNP 138) (33). The variant calls were filtered with settings ‘Quality by Depth’ (QD) <2.0 and ‘Fisher Strand value’ (FS) >30.0. Insertions/Deletions were removed from the analysis. These preprocessing and filtering steps using GATK improve the alignment quality and reduces the rate of false positive variants calls.
2.5. Determination of Het/Hom Ratio
We calculated the number of heterozygous variants and the number of non-reference genome homozygous variants on all autosomes using an in-house Python script and determined the Het/Hom ratio of an individual sample as the number of all SNPs with heterozygous genotype dived by the number of all SNPs with a non-reference allele homozygous genotype (23).
2.6. Determination of ESTIMATE Score
We used the raw sequencing read counts (as generated by STAR aligner in quant mode) as input for the ESTIMATE software (25), a tool for predicting tumor purity and the presence of infiltrating stromal/immune cells in tumor tissues using gene expression data. The ESTIMATE algorithm combines gene expression of 141 stromal and 141 immune genes and performs single-sample gene set-enrichment analysis for each set of selected genes, calculate a stromal score and an immune score to predict the level of infiltrating stromal and immune cells, respectively, and combines these individual scores to provide a final score called the ESTIMATE score which is converted to a tumor purity score, ranging from 0 to 1, with 1 denoting a highly pure sample. For simplicity, we represent ESTIAMTE score as 1-Purity score, so that a higher score represents increasing degrees of immune cell infiltration in the biopsy specimens.
2.7. Statistical Analysis
We used the non-parametric Kruskal-Wallis test to compare Het/Hom ratios and ESTIMATE scores among the different diagnostic categories followed by the Dunn’s test for pair-wise comparisons. We used Spearman rank-order correlation test for assessing associations. We used Prism 6.07 software (GraphPad Software, La Jolla, CA, USA) for statistical tests and generating graphs.
3. Results
3.1. Clinical and Biopsy Characteristics
Table 1 is a summary of the clinical characteristics of the 40 adult recipients of kidney allografts, stratified by Banff kidney allograft biopsy diagnosis. The selection based on biopsy diagnosis resulted in clinical heterogeneity such as time from transplant to biopsy, steroid maintenance therapy but all received calcineurin inhibitor as the primary immunosuppressive drug. A detailed assessment of the biopsy phenotype is shown in Figure 2. Immunostaining of all rejection biopsies showed that the CD3+ T cells were the most abundant cell type in the biopsies classified as acute TCMR and also in biopsies classified as active or chronic active ABMR. CD20+ B cells were the least frequent cell type including those classified as active ABMR or chronic active ABMR (Table 2). Biopsies categorized as active antibody mediated rejection and chronic active antibody mediated rejection fulfilled all the three criteria as per the revised Banff 2017 classification. All 7 patients with biopsies categorized as active ABMR were positive at the time of biopsy for donor HLA-specific IgG antibodies detected using single HLA coated beads and the Luminex platform. The mean fluorescence intensity (MFI) of the highest ranked donor specific bead was 17201 (median, IQR: 13307-22585). All 11 patients with biopsies categorized as chronic active ABMR antibody were also positive at the time of biopsy for donor HLA-specific IgG antibodies. The mean fluorescence intensity of the highest ranked donor specific bead was 4072 (2796-12900).
Table 1.
Characteristics of kidney transplant recipients and RNA samples
| Kidney Allograft Biopsies (N=40) | |||||
|---|---|---|---|---|---|
| Characteristics | Acute T Cell Mediated Rejection |
Active Antibody Mediated Rejection |
Chronic Active Antibody Mediated Rejection |
Interstitial Fibrosis/ Tubular Atrophy |
Normal |
| N=11 | N=7 | N=11 | N=6 | N=5 | |
|
| |||||
| At the time of transplant | |||||
| Recipient information | |||||
| Age, years, mean (SD) | 51 (14) | 43 (15) | 51 (13) | 47 (11) | 45 (12) |
| Women, n (%) | 5 (45) | 4 (57) | 6 (55) | 2 (33) | 3 (60) |
| Racial categories, Black, n (%) | 3 (27) | 3 (43) | 2 (18) | 0 (0) | 0 (0) |
| Previous transplants, n (%) | 1 (9) | 0 (0) | 1 (10) | 0 (0) | 0 (0) |
| Donor information | |||||
| Age (years), mean (SD) | 44 (17) | 40 (18) | 44 (13) | 50 (7) | 40 (17) |
| Women, n (%) | 9 (82) | 3 (43) | 5 (46) | 2 (33) | 4 (80) |
| Racial categories, Black, n (%) | 1 (9) | 1 (14) | 1 (9) | 0 (0) | 0 (0) |
| Deceased donor, n (%) | 4 (36) | 2 (29) | 4 (36) | 5 (83) | 2 (40) |
| HLA-ABDR mismatch, median (IQR) | 4 (2.5–5) | 4.5 (3–5) | 4 (3–5) | 3 (3–4.5) | 5 (2–6) |
| Induction therapy, n (%) | 11 (100) | 7 (100) | 10 (91) | 3 (50) | 5 (100) |
| Antithymocyte globulin, n (%) | 8 (73) | 6 (86) | 10 (91) | 3 (100) | 5 (100) |
| After transplant and before the index biopsy | |||||
| Calcineurin inhibitors, n (%) | 11 (100) | 7 (100) | 11 (100) | 6 (100) | 5 (100) |
| Corticosteroids maintenance, n (%) | 3 (27) | 1 (14) | 9 (82) | 4 (67) | 0 (0) |
| At the time of index allograft biopsy | |||||
| Reason for biopsy, clinically indicated: surveillance, n | 11:0 | 7:0 | 11:0 | 6:0 | 0:5 |
| Time from transplantation to biopsy, months, median (IQR) | 13.5 (4.4–53.6) | 0.9 (0.4–15.3) | 62 (25–76) | 78 (43–108) | 6.3 (6.2–6.4) |
| Biopsy within 12 months of transplantation, n (%) | 4 (36) | 4 (57) | 1 (9) | 0 (0) | 5 (100) |
| Serum creatinine, mg/dl, median (IQR) | 2.10 (1.71–3.06) | 3.36 (2.51–3.70) | 1.71 (1.45–2.51) | 3.95 (3.00–5.09) | 1.20 (1.00–1.50) |
| RNA | |||||
| Quantity of total RNA from biopsy tissue, µg, median (IQR) | 2.45 (1.64–2.77) | 1.77 (1.27–1.95) | 1.95 (1.44–2.15) | 1.21 (0.90–1.48) | 1.18 (0.89–1.35) |
| Purity of RNA, OD260/OD280 ratio, median (IQR) | 2.11 (2.07–2.19) | 2.03 (2.02–2.36) | 2.03 (1.97–2.11) | 1.96 (1.88–1.99) | 2.02 (1.96–2.02) |
| RNA integrity number, median (IQR) | 7.2 (7.0–7.9) | 7.7 (7.3–7.8) | 7.9 (7.7–8.1) | 7.1 (6.7–7.9) | 7.2 (6.6–7.3) |
| Quantity of total RNA used for mRNA sequencing, ng | 400 | 400 | 400 | 400 | 400 |
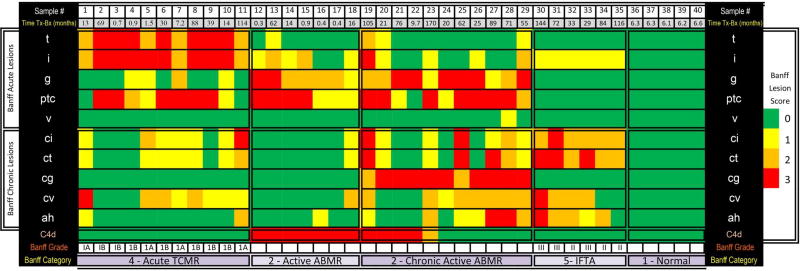
Figure 2. Histopathological characteristics of the 40 kidney allograft biopsies.
The histopathological characteristics of the 40 kidney allograft biopsies obtained from 40 kidney allograft recipients (“Sample #” 1 through 40) are shown. Biopsy tissue sections were stained with hematoxylin and eosin, periodic acid Schiff, and Masson trichrome for light microscopic evaluation. Staining for polyoma virus was done using affinity-purified and agarose-conjugated IgG2a mouse monoclonal antibody (Calbiochem, San Diego, CA) that recognizes a 94-kDa SV40 large T antigen. Indirect immunofluorescence for complement factor 4 degradation (C4d) product was done on cryosections using a monoclonal anti-C4d antibody (Quidel, Santa Clara, CA). These biopsies were categorized using the Banff 2017 update of the Banff ‘97 classification. The Banff diagnostic categories include acute T-cell mediated rejection (“Acute TCMR, Sample # 1 through 11), active antibody mediated rejection (“Active ABMR, 12 through 18), chronic active antibody mediated rejection (19 through 29), interstitial fibrosis and tubular atrophy (“IFTA”, 30 through 35) and Normal (36 through 40). The median number of glomeruli per biopsy sample was 16 (range 13–24). Colors represent Banff scores 0 to 3. Banff acute scores include the “t” (tubulitis) score, “i” (interstitial inflammation) score, “g” (glomerulitis) score, “ptc” (peritubular capillary inflammation) score and “v” (vascular inflammation). Banff chronic scores include the “ci” (interstitial fibrosis) score, “ct” (tubular atrophy) score, “cg” (chronic glomerulopathy) score, “cv” (chronic vascular lesions) score and “ah” (arteriolar hyaline thickening) score). Also shown in the figure is the staining for complement factor 4d (C4d) in the peritubular capillaries. Sample 13, categorized as active ABMR and samples 20 and 29, categorized as chronic active ABMR, fulfilled the criteria for borderline changes suspicious for acute TCMR as well. Sample 19, categorized as chronic active ABMR, fulfilled the criteria for acute TCMR as well.
Table 2.
Immunostaining of kidney allograft infiltrating cells in the 29 biopsies showing rejection*
| Kidney Allograft Biopsies (N=29) | |||||||
|---|---|---|---|---|---|---|---|
| Immunophenotyping of Infiltrating Cells* |
Acute T Cell Mediated Rejection |
Active Antibody Mediated Rejection |
Chronic Active Antibody Mediated Rejection |
Kruskal- Wallis Test |
Dunn’s Post Test | ||
| N=11 | N=7 | N=11 | Acute T Cell Mediated Rejection vs. Active Antibody Mediated Rejection |
Acute T Cell Mediated Rejection vs. Chronic Active Antibody Mediated Rejection |
Active Antibody Mediated Rejection vs. Chronic Active Antibody Mediated Rejection |
||
|
|
|||||||
| Cells per high power field, median (IQR) | |||||||
|
| |||||||
| CD3+ T cells | 315 (280–438) | 102 (18–221) | 220 (113–270) | 0.001 | <0.01 | <0.05 | >0.05 |
| CD68+ Macrophages | 223 (155–265) | 84 (56–175) | 144 (100–200) | 0.036 | <0.05 | >0.05 | >0.05 |
| CD20+ B cells | 16 (10–23) | 11 (5–16) | 2 (0.5–6) | 0.032 | >0.05 | <0.05 | >0.05 |
Immunohistochemical staining for CD3+ T cells, CD68+ monocytes/macrophages and CD20+ B cells was performed on all kidney allograft biopsies showing acute TCMR, active ABMR or chronic active ABMR using CD3, CD68 and CD20 antibodies (Leica Biosystems, Buffalo Grove, IL), respectively, on paraffin embedded tissue sections on a Leica Bond system (Buffalo Grove, IL.) using the modified protocol F provided by the manufacturer. The section was pre-treated using heat mediated antigen retrieval with Tris-EDTA buffer (pH = 9, epitope retrieval solution 2) for 20 min and incubated with the antibodies for 15 min at room temperature. CD3, CD20 and CD68 were detected using an HRP conjugated compact polymer system and DAB as the chromogen. Each section was counterstained with haematoxylin and mounted with Leica Micromount. For each biopsy tissue sample, the total number of CD3, CD20 or CD68 positive cells in 5 high power fields (40X) in the nonsclerotic areas of the kidney allograft was counted and expressed as total number of cells per high power field. In the table, the median (IQR) number of CD3+ T cells, CD68+ monocytes/macrophages and CD20+ B cells for each diagnostic category is shown. Kuskal-Wallis test was used for comparisons across the three-biopsy diagnostic categories and the Dunn’s posttest was used for pair-wise comparisons. Statistically significant (P<0.05) results are shown in bold.
A graphical representation of the histological characteristics of the kidney allograft biopsies is illustrated in Figure 2. All 7 biopsies classified as active antibody mediated rejection and 5 of 10 (50%) biopsies classified as chronic active antibody mediated rejection were positive for peritubular capillary deposition of complement split product (C4d). Among the biopsies classified as chronic active antibody mediated rejection that were negative for C4d, all had at least moderate microvascular inflammation (g+ptc ≥2) as evidence of current/recent antibody interaction with vascular endothelium. Altogether, the data provided in Figure 2 demonstrate that the Banff criteria for biopsy categorization are fulfilled in each instance and that the biopsies included in this study are prototypical for the Banff biopsy classification (3). Importantly, the biopsies displayed varying degree of intragraft infiltration.
3.2. Nucleotide Variant Calling and Het/Hom Ratio Computation using the RNA Sequencing Data from the 1000 Genomes Project
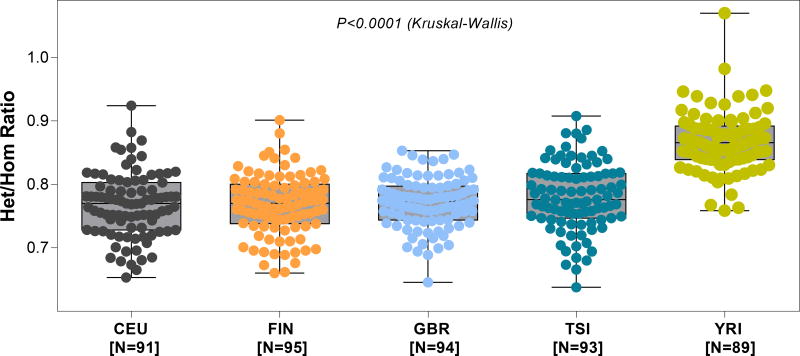
We leveraged the publically available gEUVADIS RNA sequence data to compute Het/Hom ratios and develop robust estimates of ratios of single genomes representing major races/ethnicities (Table 3). Figure 3 shows the distribution of Het/Hom ratios in these cells, stratified by continental ancestry. The median (IQR) ratio was 0.770 (0.730–0.800) for the CEU cohort, 0.770 (0.739–0.799) for the FIN cohort, 0.773 (0.744–0.797) for the GBR cohort, 0.776 (0.747–0.817) for the TSI cohort and 0.866 (0.839–0.890) for the YRI cohort. The median Het/Hom ratio was highest in the YRI cohort, in agreement with Het/Hom ratios obtained using DNA sequencing data (24). The differences in the Het/Hom ratios among the 5 major populations were statistically significant (P<0.0001 Kruskal-Wallis test). By Dunn’s multiple comparison test, differences in the Het/Hom ratios between YRI population and each of the other 4 population were statistically significant (all P<0.001) and that none of other pair-wise comparisons were significant (P>0.05).
Table 3.
Het/Hom ratio and number of variants called in the 462 lymphoblastoid cell lines
| Population* | Number of Individuals |
Number of Heterozygous Variants Median (IQR) |
Number of Homozygous Variants, Median (IQR) |
Het/Hom ratio, Median (IQR) |
|---|---|---|---|---|
|
| ||||
| CEU | 91 | 24,301 (21,185–28,283) | 32,199 (26,771–38,394) | 0.770 (0.730–0.800) |
| FIN | 95 | 26,473 (22,500–30,951) | 34,623 (28,876–41,536) | 0.770 (0.739–0.799) |
| GBR | 94 | 26,794 (23,991–33,319) | 35,563 (30,866–43,821) | 0.773 (0.744–0.797) |
| TSI | 93 | 25,750 (22,514–29,834) | 33,005 (27,534–38,355) | 0.776 (0.747–0.817) |
| YRI | 89 | 34,524 (29,252–41,074) | 38,646 (32,666–46,137) | 0.866 (0.839–0.890) |
Residents from Utah, USA with Northern and Western European ancestry (CEU), Finnish in Finland (FIN), British in England and Scotland (GBR), Tuscany in Italy (TSI), and Yoruba in Ibadan, Nigeria (YRI). The difference in the Het/Hom ratio among the groups was statistically significant (P<0.0001, Kruskal -Wallis test). By Dunn’s test, the difference in the Het/Hom ratio between YRI and each of the other four groups was statistically significant (P<0.05). None of the other pair-wise comparisons were statistically significant (all P>0.05).
Figure 3. Het/Hom ratio by population derived from mRNA sequencing of 462 lymphoblastoid cell lines.
In order to assess the variability of Het/Hom ratio at a population level for “normal” cells, we used the gEUVADIS (Genetic European Variation in Health and Disease), RNA sequencing project data. This is a European medical sequencing consortium with a publicly available database of mRNA and small RNA sequencing from 462 lymphoblastoid cell line samples from five populations of the 1000 Genomes Project. The five populations depicted in the above panels are: (i) CEU- Utah, USA, residents with Northern and Western European ancestry (N=92), (ii) FIN- Finnish in Finland (N=95), (iii) GBR- British in England and Scotland (N=94), (iv) TSI- Tuscany in Italy (N=93), and (v) YRI- Yoruba in Ibadan, Nigeria (N=89). The figure depicts the box plot of Het/Hom ratio derived from the RNA sequencing data of the five populations. The median Het/Hom ratio was 0.770 for CEU; 0.770 for FIN; 0.773 for GBR; 0.776 for TSI and 0.866 for YRI. The difference in the Het/Hom ratio among the groups was statistically significant (P<0.0001, Kruskal -Wallis test). By Dunn’s test, the difference in the Het/Hom ratio between YRI and each of the other four groups was statistically significant (P<0.05). None of the other pair-wise comparisons were statistically significant (all P>0.05).
3.3. Nucleotide Variants Calling and Het/Hom Ratios Computed from the RNA Sequencing Data of Human Kidney Allograft Biopsies
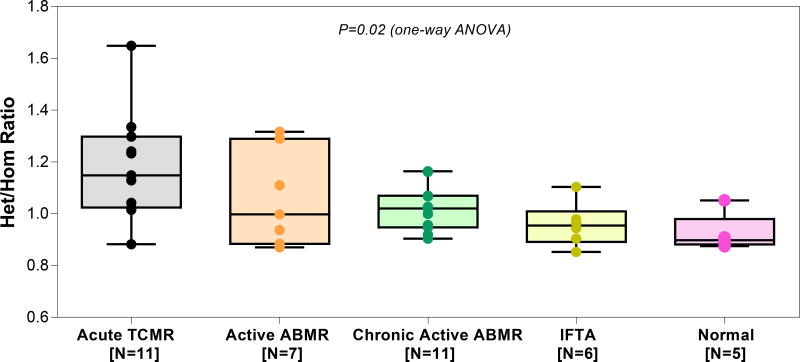
After establishing the range of Het/Hom ratios for single genomes from the 462 LCL included in the 1000 genome project, we next computed Het/Hom ratios for the kidney allograft biopsies representing major diagnostic categories (Table 4). Figure 4 shows the Het/Hom ratios, stratified by kidney allograft biopsy diagnosis and it is evident the number of heterozygous variants are strikingly higher in the biopsies compared to LCLs and among the biopsies, normal biopsies with minimal or no cellular invasion had the lowest number of heterozygous variants and acute TCMR biopsies with the highest degree of graft infiltration had the highest number of heterozygous variants (Table 3 vs. Table 4). Accordingly, the Het/Hom ratio was highest for biopsies that were categorized as acute TCMR and the median (IQR) ratio was 1.149 (1.033–1.269) and lowest for biopsies that were categorized as normal, 0.898 (0.888–0.908). The Het/Hom ratios were 0.997 (0.910–1.199) for active ABMR, 1.020 (0.952–1.068) for chronic active ABMR, and 0.954 (0.914–0.975) for IFTA. The differences in the Het/Hom ratios among the 5 diagnostic categories were statistically significant (P=0.02, Kruskal-Wallis test, Figure 4). By Dunn’s multiple comparison test, differences in the Het/Hom ratios between acute TCMR biopsies and Normal biopsies were statistically significant (P<0.05). The median Het/Hom ratio of normal biopsies (0.898 [0.888–0.908]) is closest to the values we calculated for the LCLs representing single genomes.
Table 4.
Het/Hom ratio and number of variants called in each of the 40 kidney allograft biopsies
| Sample # | Diagnostic Category |
Total Number of Variants |
Number of Heterozygous Variants |
Number of Homozygous Variants |
Het/Hom Ratior |
|---|---|---|---|---|---|
|
| |||||
| 1 | Acute TCMR | 92861 | 49246 | 43615 | 1.1291 |
| 2 | Acute TCMR | 85165 | 43090 | 42075 | 1.0241 |
| 3 | Acute TCMR | 108912 | 58221 | 50691 | 1.1485 |
| 4 | Acute TCMR | 98934 | 61577 | 37357 | 1.6483 |
| 5 | Acute TCMR | 78181 | 44701 | 33480 | 1.3351 |
| 6 | Acute TCMR | 71408 | 39541 | 31867 | 1.2408 |
| 7 | Acute TCMR | 67083 | 37043 | 30040 | 1.2331 |
| 8 | Acute TCMR | 88877 | 45349 | 43528 | 1.0418 |
| 9 | Acute TCMR | 91033 | 51414 | 39619 | 1.2977 |
| 10 | Acute TCMR | 103925 | 52362 | 51563 | 1.0154 |
| 11 | Acute TCMR | 105340 | 49366 | 55974 | 0.8819 |
| Median (IQR) | 91033 (81673–101430) | 49246 (43896–51888) | 42075 (35419–47153) | 1.1485 (1.0330–1.2693) | |
| 12 | Active ABMR | 82736 | 38505 | 44231 | 0.8705 |
| 13 | Active ABMR | 90847 | 51173 | 39674 | 1.2898 |
| 14 | Active ABMR | 106746 | 56156 | 50590 | 1.11 |
| 15 | Active ABMR | 89238 | 50710 | 38528 | 1.3161 |
| 16 | Active ABMR | 91478 | 45675 | 45803 | 0.9972 |
| 17 | Active ABMR | 80982 | 37988 | 42994 | 0.8835 |
| 18 | Active ABMR | 84407 | 40830 | 43577 | 0.9369 |
| Median (IQR) | 89238 (83572–91163) | 45675 (39668–50942) | 43577 (41334–45017) | 0.9972 (0.9102–1.1999) | |
| 19 | Chronic active ABMR | 71534 | 36128 | 35406 | 1.0203 |
| 20 | Chronic active ABMR | 71616 | 36277 | 35339 | 1.0265 |
| 21 | Chronic active ABMR | 64357 | 31463 | 32894 | 0.9564 |
| 22 | Chronic active ABMR | 53684 | 26824 | 26860 | 0.9986 |
| 23 | Chronic active ABMR | 84164 | 39953 | 44211 | 0.9036 |
| 24 | Chronic active ABMR | 75097 | 40358 | 34739 | 1.1617 |
| 25 | Chronic active ABMR | 93830 | 50468 | 43362 | 1.1638 |
| 26 | Chronic active ABMR | 78338 | 38119 | 40219 | 0.9477 |
| 27 | Chronic active ABMR | 82481 | 39485 | 42996 | 0.9183 |
| 28 | Chronic active ABMR | 89635 | 46282 | 43353 | 1.0675 |
| 29 | Chronic active ABMR | 84545 | 43688 | 40857 | 1.0692 |
| Median (IQR) | 78338 (71575–84355) | 39485 (36203–42023) | 40219 (35039–43175) | 1.0203 (0.9521–1.0684) | |
| 30 | IFTA | 104642 | 50840 | 53802 | 0.9449 |
| 31 | IFTA | 79814 | 41868 | 37946 | 1.1033 |
| 32 | IFTA | 100791 | 47843 | 52948 | 0.9035 |
| 33 | IFTA | 77884 | 38224 | 39660 | 0.9637 |
| 34 | IFTA | 79676 | 39401 | 40275 | 0.9782 |
| 35 | IFTA | 66019 | 30378 | 35641 | 0.8523 |
| Median (IQR) | 79745 (78332–95547) | 40635 (38518–46349) | 39968 (38375–49780) | 0.9543 (0.9139–0.9746) | |
| 36 | Normal | 73511 | 34302 | 39209 | 0.8748 |
| 37 | Normal | 50173 | 25715 | 24458 | 1.0513 |
| 38 | Normal | 90114 | 42633 | 47481 | 0.8978 |
| 39 | Normal | 89973 | 42318 | 47655 | 0.888 |
| 40 | Normal | 91219 | 43418 | 47801 | 0.9083 |
| Median (IQR) | 89973 (73511–90114) | 42318 (34302–42633) | 47481 (39209–47655) | 0.8978 (0.8880–0.9083) | |
Figure 4. Het/Hom ratio by diagnostic categories of the 40 allograft biopsies.
Boxplot and individual data points of the Het/Hom ratios computed from RNA sequencing data of 40 kidney allograft biopsy samples, stratified by kidney allograft biopsy diagnosis category. Acute TCMR, acute T-cell mediated rejection, Active ABMR, active antibody-mediated rejection, Chronic active ABMR, chronic and active antibody-mediated rejection, IFTA, interstitial fibrosis and tubular atrophy or normal allograft biopsy (Normal). Biopsies were categorized using the Banff 2017 update of the Banff ‘97 classification scheme. The median Het/Hom ratio was 1.149 in acute TCMR, 0.997 in active ABMR, 1.020 in chronic active ABMR, 0.954 in IFTA and 0.898 in Normal. The difference in Het/Hom ratio among the diagnostic categories was significant (P=0.02, Kruskal-Wallis test). By Dunn’s test, difference in Het/Hom ratio between acute TCMR and IFTA (P,0.05), and between acute TCMR and Normal (P<0.05) was statistically significant. None of the other pair-wise comparisons were statistically significant (all P>0.05).
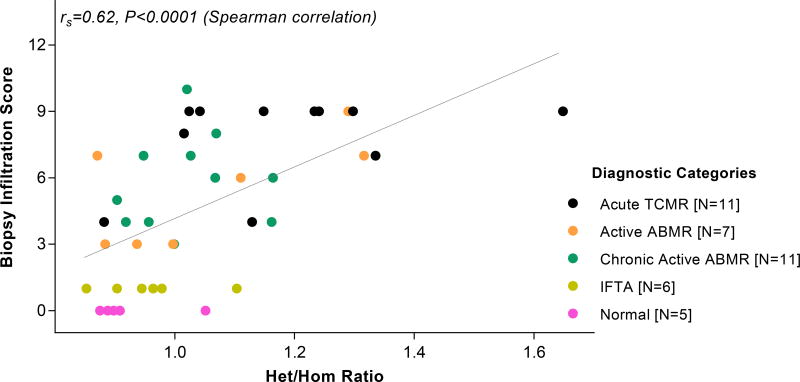
3.4. Het/Hom Ratios and Biopsy Infiltration Scores
For each kidney allograft biopsy sample, we summed the Banff lesion scores (0 to 3) for tubulitis, interstitial inflammation, glomerulities, peritubular capillary inflammation and vascular inflammation, and created a single numerical value, designated herein as the biopsy infiltration score. The score ranged from 0 to 15. We examined the relationship between the biopsy infiltration scores and the Het/Hom ratios in the 40 biopsies. The biopsy infiltration scores and Het/Hom ratios showed a significant association despite the semi quantitative nature of the Banff lesion scores (rs=0.62, P<0.0001, Spearman correlation, Figure 5). Het/Hom ratios vary not only across diagnoses but also within a diagnostic category and this later finding suggests that the ratio may help capture different degrees of graft infiltration.
Figure 5. Association between the Het/Hom ratio and the biopsy infiltration score of the 40 allograft biopsies.
Banff acute scores include the “t” [tubulitis] score, “i” [interstitial inflammation] score, “g” [glomerulitis] score, “ptc” [peritubular capillary inflammation] score and “v” [vascular inflammation] score. Each score ranges from 0 through 3. For each kidney allograft biopsy, we summed all the Banff acute scores and created a single numerical value, called the biopsy infiltration score, that ranges from 0 to 15. This figure depicts the infiltration score on the y-axis and Het/Hom ratio on the x-axis, for the 40 kidney allograft biopsy samples. The association between the Het/Hom ratio and the biopsy infiltration score was statistically significant (r=0.62, P<0.0001, Spearman rank-order correlation).
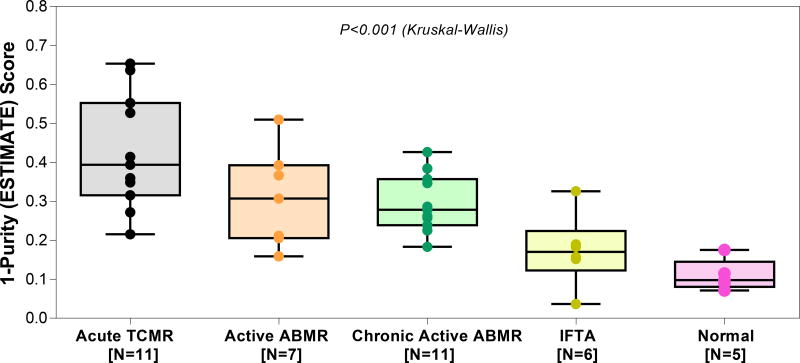
3.5. The ESTIMATE Scores
Normal allograft biopsies have the lowest ESTIMATE score of 0.098 (0.081–0.145) (Figure 6). The ESTIMATE score was much higher in rejection biopsies; 0.394 (0.315–0.552) for acute TCMR, 0.307 (0.206–0.393) for active ABMR, 0.278 (0.239–0.357) for chronic active ABMR, 0.170 (0.123–0.224) for IFTA. The difference in the ESTIMATE score among the biopsy diagnostic categories was significant (P<0.001, Kruskal-Wallis test). By Dunn’s multiple comparison test, differences in the ESTIMATE scores between acute TCMR biopsies and IFTA biopsies (P<0.01) and between acute TCMR biopsies and Normal biopsies (P<0.001) were statistically significant. We did not compute ESTIMATE score for RNA sequencing data from the LCLs as the algorithm includes a stromal score and cannot be interpreted for hematopoietic cell lines.
Figure 6. ESTIMATE score by diagnostic categories of the 40 allograft biopsies.
The ESTIMATE score was derived from the RNA sequencing data using the ESTIMATE algorithm. We used the raw sequencing read counts (as generated by STAR aligner in quant mode) as input for the ESTIMATE software (25), a tool for predicting tumor purity and the presence of infiltrating stromal/immune cells in tumor tissues using gene expression data. The ESTIMATE algorithm combines gene expression of 141 stromal and 141 immune genes and performs single-sample gene set-enrichment analysis for each set of selected genes, calculate a stromal score and an immune score to predict the level of infiltrating stromal and immune cells, respectively, and combines these individual scores to provide a final score called the ESTIMATE score which is converted to a tumor purity score, ranging from 0 to 1, with 1 denoting a highly pure sample. For simplicity, we represent ESTIAMTE score as 1-Purity score, so that a higher score represents increasing degrees of immune cell infiltration in the biopsy specimens. In the y-axis, the 1-Purity score ranges from 0 to 1, where 0 denotes a highly pure sample. Biopsies were categorized using the Banff 2017 update of the Banff ‘97 classification. The median 1-Purity score was 0.394 in acute TCMR, 0.307 in active ABMR, 0.278 in chronic active ABMR, 0.170 in IFTA and 0.098 in Normal. The difference in the 1-Purity score among the groups was statistically significant (P<0.001, Kruskal-Wallis test). By Dunn’s test, difference in ESTIMATE score between acute TCMR and IFTA (P<0.01), and between acute TCMR and Normal (P<0.001) was statistically significant. None of the other pair-wise comparisons were statistically significant (all P>0.05).
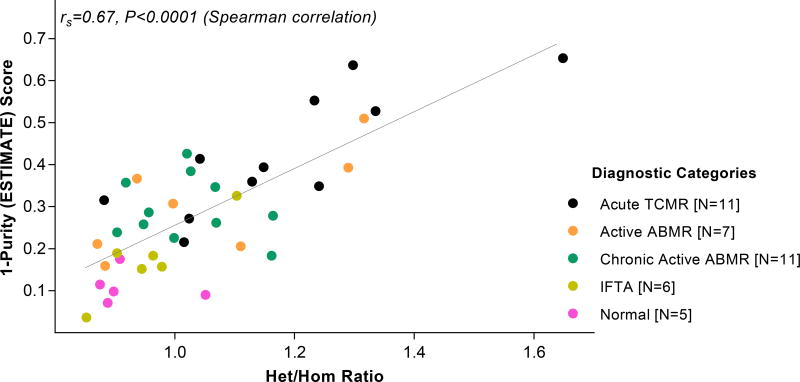
The biopsy infiltration scores and ESTIMATE scores showed a significant association (rs=0.77, P<0.0001). The ESTIMATE score was positively associated (rs=0.54, P<0.01) with the sum of graft infiltrating CD3+ cells, CD68+ cells and CD20+cells, and among the two components contributing to the ESTIMATE score, the immune score was more strongly associated (rs=0.55, P<0.01) than the stromal score (rs=0.37, P=0.47) with the sum of graft infiltrating cells. Graft infiltration determined using the ESTIMATE score correlated with Het/Hom ratio for the 40-kidney allograft biopsies (r=0.67, P<0.0001, Spearman correlation, Figure 7).
Figure 7. Association between Het/Hom ratio and ESTIMATE score of the 40 allograft biopsies.
The Het/Hom ratio and the ESTIMATE score was derived from the RNA sequencing data. We show the ESTIAMTE score as a 1-Purity score. The association between Het/Hom ratio and 1-Purity score was statistically significant (r=0.67, P<0.0001, Spearman rank-order correlation).
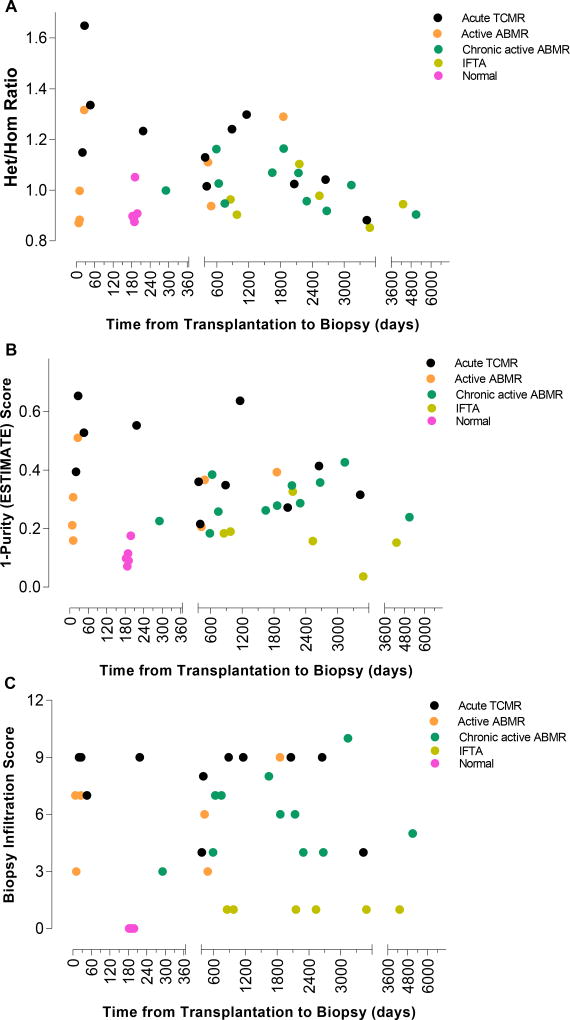
3.6. Time from Transplantation to Biopsy and Het/Hom Ratios or the ESTIMATE Scores
We assessed whether time from transplantation to biopsy affects the Het/Hom ratio. Among the 29 biopsies with rejection (acute TCMR, active ABMR and chronic active ABMR), 9 (31%) biopsies were done within the first 12 months of transplantation. Within each diagnostic category, the correlation between Het/Hom ratio and time from transplantation to biopsy was not statistically significant (P>0.05, Spearman correlation, Figure 8). Similarly, the relation between ESTIMATE score, or the biopsy infiltration score, and time from transplantation to biopsy was not statistically significant (P>0.05, Spearman correlation, Figure 8).
Figure 8. Lack of association between the time from transplantation to biopsy and Het/Hom ratio, ESTIMATE score, or the biopsy infiltration score.
Scatterplot depicts the relationship between the time from transplantation to biopsy and Het/Hom ratio (Panel A), ESTIMATE score (1-Purity) (Panel B), and biopsy infiltration score (Panel C) for all 40 biopsies. Within each diagnostic category, there was no significant correlation between the time from transplantation to biopsy and Het/Hom ratio, ESTIMATE score, or the biopsy infiltration score (P>0.5, Spearman rank-order correlation).
3. Discussion
With the use of RNA sequencing data from 40 unique biopsy samples representing clinically relevant diagnostic categories of human kidney allograft pathology, we tested the hypothesis that the ratio of heterozygosity to non-reference genome homozygosity (Het/Hom ratio) can serve as a yardstick of allograft infiltration by recipient’s cells. In accord with our postulate, we found that kidney biopsies classified as acute T-cell mediated rejection biopsy had the highest Het/Hom ratio and normal surveillance biopsies with no or minimal infiltration had the lowest Het/Hom ratio and a positive correlation was demonstrable between the Het/Hom ratio and the biopsy infiltration score.
Single nucleotide polymorphisms have traditionally been identified using DNA sequencing information. Recent advances in computational biology have led to the identification of genomic variants from RNA-Seq data (20, 21). Apriori, there are no reasons that variant calling using RNA-seq data should be more accurate compared to variant calling using DNA sequence data. However, RNA-Seq data has the additional advantage of charecterizing gene expression patterns.
The ESTIMATE score has hitherto been used to reflect tumor purity and is based on the expression of 141 selected stromal signature genes and of 141 selected immune signature genes. In accord with the notion that graft “contamination” by varying degrees of infiltrating cells would impact the ESTIMATE score, it was highest for acute rejection biopsies, lowest for normal allograft biopsies, and differed significantly among biopsy diagnoses characterized by different degrees of graft infiltration. Deconvolution of bulk RNA sequencing data can yield information regarding cell composition of complex tissues based on their gene expression patterns. The ESTIMATE score computed in this study is a composite score of immune and stromal signature scores. We demonstrate that the ESTIMATE score is positively associated with the sum of graft infiltrating CD3+ cells, CD68+ cells and CD20+cells, and that between the two components contributing to the ESTIMATE score, the immune score is more strongly associated than the stromal score with the sum of graft infiltrating cells. Further deconvolution, i.e., identification of cell subtypes such as T cell subsets, is not feasible using the ESTIMATE score and requires apriori availability of reference gene expression profiles (34) and statistical workflows such as DeconRNASeq (35) to deconvolve the bulk RNA sequencing data generated in our study.
As anticipated, the biopsy Het/Hom ratios and the ESTIMATE scores were highly correlated. Taken together, these observations confirm our hypothesis that the total burden of cellular invasion of the allograft during allograft rejection can be interrogated using the ratio between genetic material derived from the donor cells and the genetic material derived from the recipient cells, and that the Het/Hom ratio may represent a precisely quantifiable parameter of cellular invasion into the allograft.
Many features of our investigation are worthy of emphasis. The use of RNA sequencing data of human kidney allograft biopsies for variant calling and applying the Het/Hom ratios, hitherto used as quality control parameter of DNA sequencing data (23), to infer cellular traffic into the allograft is unprecedented. Another innovation is the first successful application of the ESTIMATE algorithm, until now utilized to estimate tumor purity, to gauge cellular infiltration into the kidney allograft. It is readily acknowledged that RNA sequencing characterizes whole transcriptome at an unprecedented level of precision. It is therefore reasonable to hypothesize that computations based on RNA sequencing data of kidney allograft biopsies would yield robust estimates of cellular infiltration in to an allograft.
Our study is significant from both biologic and clinical perspectives. Not only the execution phase but also the priming phase of allograft rejection is likely to be dependent upon the physical contact between the recipient’s immune cells and graft parenchymal cells and in all probabilities assessment using mRNA is likely to be more sensitive than using proteins or cells as parameters of graft infiltration. From a clinical perspective, the concerns related to variations in the interpretation of biopsy findings, less than optimal reproducibility for grading severity of histological lesions, and poor sensitivity for detecting tubulitis using conventional microscopy are all mitigated using RNA sequencing data.
Our study has several limitations. Although the kidney allograft biopsies included in this study were selected to represent most of the major types of graft pathology, our biopsy sample size of 40 biopsies is not large enough to capture the full spectrum of graft infiltration and associated variability in Het/Hom ratios and ESTIMATE scores. Our study cohort was also insufficient to investigate the contribution of ethnicity of the organ donor and the recipient to the observed variability in Het/Hom ratios; we note however that that only 3 of the 40 organ donors were categorized as black and 8 of 40 recipients were classified as black. Another limitation is that we did not include allograft biopsies manifesting viral infections such as BK virus or CMV. Changes in RNA expression may influence the ESTIMATE scores. Such biases are not correctable using ESTIMATE score. However, the bulk RNA-seq data used in this study to generate the ESTIMATE score can be deconvolved to elucidate cell types infiltrating the allograft. This would require single cell RNA sequencing of human kidney allograft biopsies to develop reference gene expression patterns and appropriate statistical work flow to deconvolve bulk RNA-seq data. We used conventional histopathology as the gold standard for classifying the phenotypes. It should be noted that inter-observer variability remains an important issue in the Banff classification (5).
We did not include patients with subclinical rejection in this study. Whether the Het/Hom ratio or the ESTIMATE score is diagnostic of subclinical rejection is not known. Because subclinical rejection is diagnosed by the presence of graft infiltrating cells and because RNA-Seq is a highly sensitive and precise technique, there is no apriori reason that subclinical rejection will not be associated with the Het/Hom ratio or the ESTIMATE score.
An important unresolved question is whether the Het/Hom ratio or the ESTIMATE score computed from RNA-sequencing of kidney allografts is useful for the differential diagnosis of biopsies manifesting different types of rejection, e.g., discrimination of acute TCMR from active ABMR. We believe that this is feasible but would require single cell RNA sequencing of human kidney allograft biopsies with different biopsy diagnosis to develop reference gene expression patterns for the deconvolution of bulk RNA-seq data with appropriate statistical work flow.
To our knowledge, this is the first study to assess the Het/Hom ratio and ESTIMATE score as measures of immune cell invasion of the kidney allograft. If validated in larger cohorts, and with increasing use of RNA sequencing and accordingly falling costs, these novel measures have a wide-ranging potential for assessing rejection events in organ allograft recipients. For instance, it should also be possible to derive Het/Hom ratios also from transcriptome and genome sequencing data of urinary cells, which would provide a noninvasive assessment of immune cells invasion of the kidney allograft. In the future, it may then be possible to derive strong objective classifiers by combining both approaches.
Acknowledgments
Supported in part, by awards from the National Institutes of Health (NIH MERIT Award, R37-AI051652 to M. Suthanthiran, K08-DK087824 to T. Muthukumar, and UL1TR000457 Clinical and Translational Science Center Award to Weill Cornell Medical College), by the Biomedical Research Program at Weill Cornell Medical College in Qatar, a program funded by the Qatar Foundation, by an award from the American Society of Transplantation (AST-Faculty Development Grant to T. Muthukumar), and by an award from the Mendez National Institute of Transplantation Foundation (to M. Suthanthiran).
Abbreviations
- Active ABMR
Active antibody mediated rejection
- Acute TCMR
Acute T-cell mediated rejection
- BAM
Binary Alignment/Map
- CD
Cluster of differentiation
- cDNA
Complementary deoxyribonucleic acid
- Chronic active ABMR
Chronic active antibody mediated rejection
- dbSNP 138
Single Nucleotide Polymorphism Database build 138
- DNA
Deoxyribonucleic acid
- ESTIMATE
Estimation of Stromal and Immune cells in Malignant Tumors using Expression Data
- FS
Fisher Strand value
- gEUVADIS
Genetic European Variation in Health and Disease
- GATK
Genome Analysis Tool Kit
- GRCh37
Genome Reference Consortium human genome build 37
- Het/Hom ratio
Heterozygosity to non-reference genome homozygosity ratio
- IFTA
Interstitial fibrosis and tubular atrophy
- LCL
Lymphoblastoid cell line
- mRNA
Messenger ribonucleic acid
- PCR
Polymerase chain reaction
- RNA
Ribonucleic acid
- SNP
Single nucleotide polymorphism
- STAR aligner
Spliced Transcripts Alignment to a Reference aligner
Footnotes
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
References
- 1.Racusen LC, Solez K, Colvin RB, Bonsib SM, Castro MC, Cavallo T, et al. The Banff 97 working classification of renal allograft pathology. Kidney international. 1999;55(2):713–723. doi: 10.1046/j.1523-1755.1999.00299.x. [DOI] [PubMed] [Google Scholar]
- 2.Loupy A, Haas M, Solez K, Racusen L, Glotz D, Seron D, et al. The Banff 2015 Kidney Meeting Report: Current Challenges in Rejection Classification and Prospects for Adopting Molecular Pathology. Am J Transplant. 2017;17(1):28–41. doi: 10.1111/ajt.14107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haas M, Loupy A, Lefaucheur C, Roufosse C, Glotz D, Seron D, et al. The Banff 2017 Kidney Meeting Report: Revised diagnostic criteria for chronic active T cell-mediated rejection, antibody-mediated rejection, and prospects for integrative endpoints for next-generation clinical trials. Am J Transplant. 2018;18(2):293–307. doi: 10.1111/ajt.14625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legendre C, Canaud G, Martinez F. Factors influencing long-term outcome after kidney transplantation. Transpl Int. 2014;27(1):19–27. doi: 10.1111/tri.12217. [DOI] [PubMed] [Google Scholar]
- 5.Furness PN, Taub N Convergence of European Renal Transplant Pathology Assessment Procedures P. International variation in the interpretation of renal transplant biopsies: report of the CERTPAP Project. Kidney international. 2001;60(5):1998–2012. doi: 10.1046/j.1523-1755.2001.00030.x. [DOI] [PubMed] [Google Scholar]
- 6.Marcussen N, Olsen TS, Benediktsson H, Racusen L, Solez K. Reproducibility of the Banff classification of renal allograft pathology. Inter- and intraobserver variation. Inter- and intraobserver variation. Transplantation. 1995;60(10):1083–1089. doi: 10.1097/00007890-199511270-00004. [DOI] [PubMed] [Google Scholar]
- 7.Elshafie M, Furness PN. Identification of lesions indicating rejection in kidney transplant biopsies: tubulitis is severely under-detected by conventional microscopy. Nephrol Dial Transplant. 2012;27(3):1252–1255. doi: 10.1093/ndt/gfr473. [DOI] [PubMed] [Google Scholar]
- 8.Consortium GT, Coordinating Center -Analysis Working G, Statistical Methods groups-Analysis Working G, Enhancing Gg, Fund NIHC et al. Genetic effects on gene expression across human tissues. Nature. 2017;550(7675):204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saha A, Kim Y, Gewirtz ADH, Jo B, Gao C, McDowell IC, et al. Co-expression networks reveal the tissue-specific regulation of transcription and splicing. Genome Res. 2017;27(11):1843–1858. doi: 10.1101/gr.216721.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.e GP. Enhancing GTEx by bridging the gaps between genotype, gene expression, and disease. Nat Genet. 2017 doi: 10.1038/ng.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mele M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348(6235):660–665. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Dov IZ, Muthukumar T, Morozov P, Mueller FB, Tuschl T, Suthanthiran M. MicroRNA sequence profiles of human kidney allografts with or without tubulointerstitial fibrosis. Transplantation. 2012;94(11):1086–1094. doi: 10.1097/TP.0b013e3182751efd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dorr C, Wu B, Guan W, Muthusamy A, Sanghavi K, Schladt DP, et al. Differentially expressed gene transcripts using RNA sequencing from the blood of immunosuppressed kidney allograft recipients. PLoS One. 2015;10(5):e0125045. doi: 10.1371/journal.pone.0125045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurian SM, Velazquez E, Thompson R, Whisenant T, Rose S, Riley N, et al. Orthogonal Comparison of Molecular Signatures of Kidney Transplants With Subclinical and Clinical Acute Rejection: Equivalent Performance Is Agnostic to Both Technology and Platform. Am J Transplant. 2017;17(8):2103–2116. doi: 10.1111/ajt.14224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47(9):1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pirinen M, Lappalainen T, Zaitlen NA, Consortium GT, Dermitzakis ET, Donnelly P, et al. Assessing allele-specific expression across multiple tissues from RNA-seq read data. Bioinformatics. 2015;31(15):2497–2504. doi: 10.1093/bioinformatics/btv074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wheeler HE, Shah KP, Brenner J, Garcia T, Aquino-Michaels K, Consortium GT, et al. Survey of the Heritability and Sparse Architecture of Gene Expression Traits across Human Tissues. PLoS Genet. 2016;12(11):e1006423. doi: 10.1371/journal.pgen.1006423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cummings BB, Marshall JL, Tukiainen T, Lek M, Donkervoort S, Foley AR, et al. Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci Transl Med. 2017;9(386) doi: 10.1126/scitranslmed.aal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan MH, Li Q, Shanmugam R, Piskol R, Kohler J, Young AN, et al. Dynamic landscape and regulation of RNA editing in mammals. Nature. 2017;550(7675):249–254. doi: 10.1038/nature24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Piskol R, Ramaswami G, Li JB. Reliable identification of genomic variants from RNA-seq data. Am J Hum Genet. 2013;93(4):641–651. doi: 10.1016/j.ajhg.2013.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez-Maestre H, Brinza L, Marchet C, Kielbassa J, Bastien S, Boutigny M, et al. SNP calling from RNA-seq data without a reference genome: identification, quantification, differential analysis and impact on the protein sequence. Nucleic Acids Res. 2016;44(19):e148. doi: 10.1093/nar/gkw655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paul MR, Levitt NP, Moore DE, Watson PM, Wilson RC, Denlinger CE, et al. Multivariate models from RNA-Seq SNVs yield candidate molecular targets for biomarker discovery: SNV-DA. BMC Genomics. 2016;17:263. doi: 10.1186/s12864-016-2542-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo Y, Ye F, Sheng Q, Clark T, Samuels DC. Three-stage quality control strategies for DNA re-sequencing data. Brief Bioinform. 2014;15(6):879–889. doi: 10.1093/bib/bbt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang J, Raskin L, Samuels DC, Shyr Y, Guo Y. Genome measures used for quality control are dependent on gene function and ancestry. Bioinformatics. 2015;31(3):318–323. doi: 10.1093/bioinformatics/btu668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501(7468):506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aken BL, Ayling S, Barrell D, Clarke L, Curwen V, Fairley S, et al. The Ensembl gene annotation system. Database (Oxford) 2016:2016. doi: 10.1093/database/baw093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11–33. doi: 10.1002/0471250953.bi1110s43. 11 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics (Oxford, England) 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schelker M, Feau S, Du J, Ranu N, Klipp E, MacBeath G, et al. Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat Commun. 2017;8(1):2032. doi: 10.1038/s41467-017-02289-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gong T, Szustakowski JD. DeconRNASeq: a statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics. 2013;29(8):1083–1085. doi: 10.1093/bioinformatics/btt090. [DOI] [PubMed] [Google Scholar]