Abstract
Background
No pharmacological therapy exists for calcific aortic valve disease (CAVD), which confers a dismal prognosis without invasive valve replacement. The search for therapeutics and early diagnostics is challenging since CAVD presents in multiple pathological stages. Moreover, it occurs in the context of a complex, multi-layered tissue architecture, a rich and abundant extracellular matrix phenotype, and a unique, highly plastic and multipotent resident cell population.
Methods
A total of 25 human stenotic aortic valves obtained from valve replacement surgeries were analyzed by multiple modalities, including transcriptomics and global unlabeled and label-based tandem-mass-tagged proteomics. Segmentation of valves into disease-stage-specific samples was guided by near infrared molecular imaging, and anatomical layer-specificity was facilitated by laser capture microdissection. Side-specific cell cultures were subjected to multiple calcifying stimuli, and their calcification potential and basal/stimulated proteomes were evaluated. Molecular (protein-protein) interaction networks were built and their central proteins and disease associations were identified.
Results
Global transcriptional and protein expression signatures differed between the non-diseased, fibrotic, and calcific stages of CAVD. Anatomical aortic valve microlayers exhibited unique proteome profiles that were maintained throughout disease progression and identified glial fibrillary acidic protein (GFAP) as a specific marker of valvular interstitial cells (VICs) from the spongiosa layer. CAVD disease progression was marked by an emergence of smooth muscle cell activation, inflammation, and calcification-related pathways. Proteins overrepresented in the disease-prone fibrosa are functionally annotated to fibrosis and calcification pathways, and we found that in vitro, fibrosa-derived VICs demonstrated greater calcification potential than those from the ventricularis. These studies confirmed that the microlayer-specific proteome was preserved in cultured VICs, and that VICs exposed to ALPL-dependent and ALPL-independent calcifying stimuli had distinct proteome profiles, both of which overlapped with that of the whole tissue. Analysis of protein-protein interaction networks found a significant closeness to multiple inflammatory and fibrotic diseases.
Conclusions
A spatially- and temporally-resolved multi-omics, and network and systems biology strategy identifies the first molecular regulatory networks in CAVD, a cardiac condition without a pharmacological cure, and describes a novel means of systematic disease ontology that is broadly applicable to comprehensive omics studies of cardiovascular diseases.
Keywords: aortic valve, stenosis, calcification, proteomics, transcriptomics, network medicine
Introduction
Calcific aortic valve disease (CAVD) resulting in aortic valve stenosis affects 25% of the population over the age of 651, and is the most common indication for invasive and costly valve replacement. Since no effective pharmacological therapy exists2, CAVD confers a high clinical and economic burden. The disease progression is rapid and while initially indolent, it results in heart failure and premature death if left untreated3. The underlying mechanisms of CAVD remain incompletely identified thereby limiting advances in biomedical research and clinical treatment in its earlier stages prior to cardiac damage. Owing to its high morbidity and mortality, a strong incentive exists to identify the key early molecular drivers contributing to the development of this disease.
Anatomically, the aortic valve (AV) possesses a three-layer architecture: the collagenous fibrosa layer facing the aortic side of the valve, the intermediate glycosaminoglycan-rich spongiosa layer, and the elastin-enriched ventricularis on the ventricular side4. CAVD is marked by inflammatory infiltration, fibrotic extracellular matrix synthesis by activated valvular interstitial cells (VICs), increased leaflet thickening and stiffness, and calcific mineral deposition. The resultant pathological remodeling ultimately impairs valve movement and obstructs blood flow across the narrowing valve orifice5. Mineralization predominantly originates at the base of the valve attachment to the aortic wall and in the fibrosa side of heart valve leaflets, and progresses toward the tip5. Distinct stages of disease progression are evident; a single AV leaflet may harbor non-diseased, fibrotic, and calcific areas. Together, this pathology leads to aortic valve stenosis, left ventricular remodeling, and the onset of symptoms.
In healthy valves, VICs are comprised of mostly quiescent fibroblasts that maintain valvular homeostasis and physiological leaflet mechanical properties. Under pathologic biochemical and biomechanical stimuli, VICs undergo differentiation towards myofibroblastic and osteoblastic phenotypes as a result of newly acquired expression of cytoskeletal or osteogenic genes. Myofibrogenesis drives ECM remodeling, fibrosis, and leaflet stiffening6, which, in turn, promote further myofibroblastic differentiation. Osteoblastic VICs actively deposit calcium-rich mineral, but the pathogenic mechanisms that trigger their maladaptive differentiation in vivo are unclear. To date, no complete ontology of VICs exists, and neither the relative contributions of VIC myofibrogenesis/osteogenesis to impaired valve function nor putative cross-talk between these two hallmarks of disease have been clearly elucidated.
Recent advances in protein-protein interactome-based systems biology and network medicine have shed novel light on inter-organ interplay, revolutionized target discovery, and are uniquely suited to capture a global view of highly complex pathobiology7. Despite this promise, systems medicine has not yet been applied to CAVD. Heart valve research in humans has traditionally been hindered by high intra- and inter-donor variability, a lack of accounting for the leaflet’s heterogeneous layered structure, and limitations of in vitro culture models. We hypothesized that using human AV samples in conjunction with a comprehensive holistic “macro-to-micro” approach encompassing i) disease stage-associated regions (CAVD Disease Stages), ii) anatomical tissue microlayers (CAVD Layers), and iii) VIC sub-types (CAVD VICs) would address prior limitations in temporal and spatial resolution8, 9, help to decipher the complexity of CAVD pathogenesis, and generate a highly specific dataset for subsequent novel systems biology investigations. Importantly, dissecting the molecular pathology from the tissue level, to tissue layers, and finally to layer-specific cells enabled the elucidation of the valvular proteome, encompassing highly abundant extracellular matrix proteins on the tissue level, down to the cellular fingerprints of the molecular CAVD pathology. Such an approach would address crucial questions with far-reaching implications for the field of CAVD research.
Furthermore, given the intricate anatomical structure and function of the AV, we employed not only a holistic approach encompassing disease stages, but also multiple complementary techniques to assess anatomical layers and heterogeneous cell populations to decode the complexities of the valvular pathobiology10. We used transcriptomics in conjunction with both label-free and label-based proteomics, with subsequent mapping by tissue immunofluorescence, and multi-dimensional network analysis (Fig. 1a). Network medicine thereafter aided in deriving biological insights from these omics datasets.
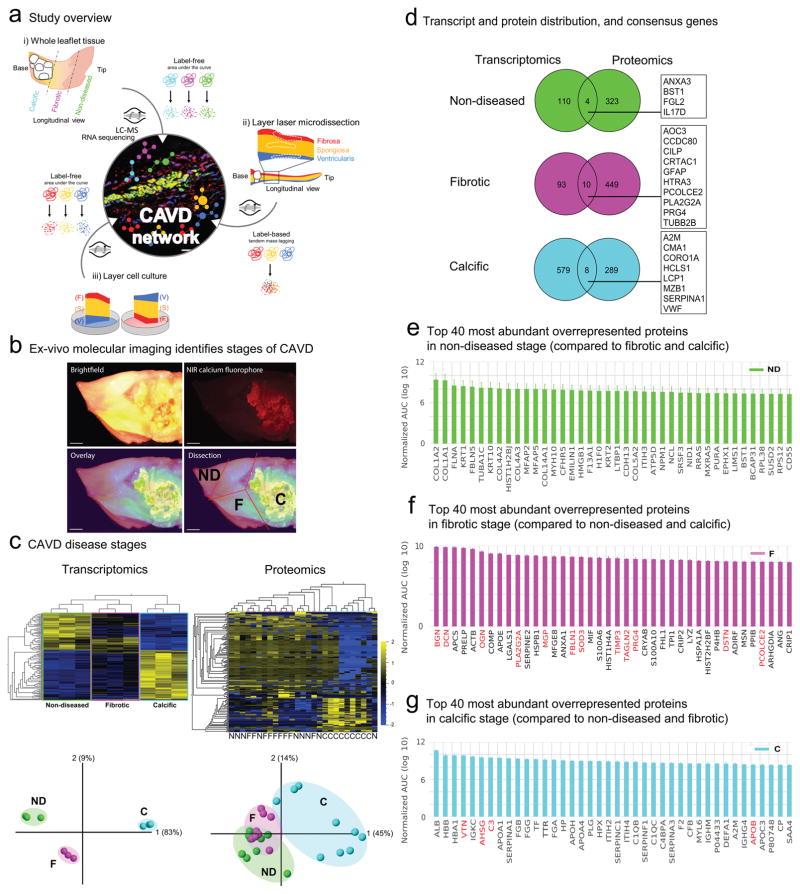
Figure 1. CAVD Omics Study Design and CAVD Disease-associated Stages.
a. CAVD mapping study overview: i) CAVD Disease Stages: human aortic valve (AV) whole leaflet tissue was separated into disease-associated stages guided by molecular imaging and transcriptomics and label-free proteomics were conducted (liquid-chromatography-mass spectrometry (LC-MS)); ii) CAVD Layers: human AV leaflet layer proteomics by label-based tandem mass tagging LC-MS; iii) CAVD VICs: layer-specific valvular interstitial cell (VIC) migration culture analysis by label-free LC-MS. Selected molecules form each sub-study were then mapped by immunofluorescence, and pathway and molecular network analyses. The area under the curve of the chromatographic peak intensity as a label-free measure of peptide abundance of up to the top three abundant peptides was used for quantification in the two label-free experiments.
b. Calcified human AV leaflets stained with near-infrared fluorescence calcium tracer was sectioned into 3 disease-associated stages (ND- non-diseased, F- fibrotic, C- calcific). Scale bar = 1mm.
c. CAVD Disease Stages: Transcriptomics and proteomics heat map and principal component analysis (PCA) for 3 stages on n=3 donors for transcriptomics and n=9 donors for proteomics, (transcripts and proteins filtered with q<0.2 for between-stage difference, N: non-diseased, F: fibrotic, C: calcific).
d. The number of overrepresented proteins and their percentages of each stage in the transcriptomics (left) and proteomics (right) CAVD Disease Stages data. Proteins/genes classified into the same stage in both omics datasets are shown in their respective boxes.
e–g. The top 40 most abundant ‘stage-specific’ proteins ranked according to the logarithm of their normalized mean AUCs for the three stages from the CAVD Disease Stages proteomics data. Proteins highlighted in red signify previously implicated proteins in AV fibrosis and calcification.
This study expedites the translation of basic research findings into the clinic by integrating post-operative molecular imaging and pathology with proteomics, transcriptomics, and network analysis, thereby creating the first integrated map of human CAVD. More broadly, we present a methodological blueprint that can be applied to multi-omics studies on a plethora of tissue types with complex tissue microarchitecture and temporal variation in disease evolution.
Methods
Detailed methods are provided in the Supplemental Material. The data are available through https://cics.bwh.harvard.edu/multiomics_databases11.
In total, 25 AVs were used in this study. AV leaflets were obtained from AV replacement surgeries for severe AV stenosis (Brigham and Women’s Hospital (BWH) approved IRB protocol number: 2011P001703). Written informed consent was provided. In brief, human stenotic AVs were segmented into stages of disease progression: (1) non-diseased, (2) fibrotic, and (3) calcific under the guidance of near-infrared molecular imaging. Transition zones were excluded from all analyses. In total, 27 sub-samples were prepared for label-free proteomics and 9 for transcriptomics.
AVs obtained from three additional patients with severe aortic valve stenosis were used for tissue layer tandem mass tagging (TMT) proteomics and AVs from autopsy donors served as controls. Anatomical layer-specificity was facilitated by laser capture microdissection.
Side-specific in vitro layer calcification potential was evaluated through a migration assay on AV leaflets from eight additional patients with severe AV stenosis after inspection by a pathologist to distinguish the fibrosa from the ventricularis side, and calcification was assessed by Alizarin Red staining at day 21. All cells which underwent proteomics were cultured and passaged in vitro prior to protein collection.
AV whole tissue label-free peptide samples were examined with the Q Exactive mass spectrometer. AV tissue layer TMT and in vitro migration label-free peptide samples were analyzed with the LTQ-Orbitrap Elite mass spectrometer.
For pathway analysis, the protein sets corresponding to each layer and stage were tested for enrichment by a hypergeometric test and adjusted for multiple comparisons using the Benjamini-Hochberg method for controlling the false discovery rate (FDR). Pathway networks were constructed based on their gene overlap. Layer- and stage-specific subnetworks were generated from literature-curated physical protein interactions. The closeness of the calcific stage subnetwork to human diseases was evaluated using average shortest network distance.
Results
OMICs Studies of Disease Stages in CAVD
Global transcriptional and protein expression signatures differ in each CAVD disease stage
AV leaflets were obtained from six patients undergoing surgical AV replacement due to aortic stenosis. To map the molecular expression signatures that associate with disease progression, with the aid of ex-vivo molecular imaging we dissected each leaflet into the three segments: 1) grossly non-diseased (ND), which is typified by a thin, transparent and pliable appearance; 2) fibrotic (F), which is thicker and stiffer than the non-diseased segment; and 3) calcific (C), which is visualized with a near-infrared fluorescence calcium tracer (Fig. 1b).
Transcriptomics (n=3 donors) and (label-free) whole tissue proteomics (n=9 donors) provided additional support for our CAVD stage classification (Fig. 1c). For example, hierarchical clustering of the q-adjusted relative gene expression profiles (see Supplemental Methods) demonstrated that the calcific stage is distinct from the non-diseased, with the fibrotic stage representing an intermediate profile. This notion is underscored by a principal component analysis (PCA) plot (Fig. 1c, left panel). Similarly, the proteomics data also captured these distinct stages and clearly demarcated non-diseased from calcific tissue (Fig. 1c, right panel).
Consistent trends and dissimilarities in gene and protein expression across disease stages
To resolve the valve transcriptome and proteome into the three stages of disease, we determined the unique sets of transcripts and proteins overrepresented in each stage, where overrepresentation is defined as a ≥1.2-fold increase of one stage relative to both of the other two stages (see Supplemental Methods). RNA sequencing revealed a total of 114 transcripts to be overrepresented in the non-diseased stage, 103 in the fibrotic stage, and 587 in the calcific stage relative to the other two stages (Fig. 1d, Supplemental Figure 1 for full lists). Correspondingly, 327 proteins were overrepresented in the non-diseased stage, 459 proteins in the fibrotic stage, and another 297 proteins in the calcific stage relative to the other two stages (Fig. 1d, the pie charts reflect the percentages of overrepresented molecules above; Supplemental Figure 2 for full lists). Transcriptomics and proteomics correlated weakly in their individual expression profiles (Spearman R2=0.25, Supplemental Figure 3a, b; Supplemental Figure 3c for overlap). To our knowledge, this is the first report of such a correlation analysis in AV data. The low correlation observed between transcriptomics and proteomics data has been previously reported for other tissues and systems12, 13. Nonetheless, despite the limited overlap, we identified common novel molecules between the transcriptomics and proteomics datasets: non-diseased stage (ANXA3, BST1, FGL2, IL17D), fibrotic stage (AOC3, CCDC80, CILP, CRTAC1, GFAP, HTRA3, PCOLCE2, PLA2G2A, PRG4, TUBB2B), and calcific stage (A2M, CMA1, CORO1A, HCLS1, LCP1, MZB1, SERPINA1, VWF) (Fig. 1d).
Overall, transcriptomics identified a higher proportion of overrepresented transcripts in the calcific stage, whereas proteomics identified similar proportions of overrepresented proteins in all stages (Fig. 1d). The high proportion of calcific stage-overrepresented transcripts may indicate a more diverse cellular environment potentially due to infiltration of inflammatory cells that would be more readily detected by the high sensitivity of mRNA-sequencing, in contrast to the relatively lower limit in the dynamic range of a typical proteomics analysis14. Gene expression profiling is frequently used to investigate disease states, however proteins are the major functional entity driving diseases15, for which reason we elected to pursue the remainder of the study with a specific focus on the valvular proteome.
Unveiling the valvular proteome in three disease stages
For a closer look at the CAVD stage proteomics, we provide the top 40 overrepresented proteins per CAVD stage, ranked according to their abundance (Fig. 1e–g; Supplemental Figure 2 shows all proteins). In the non-diseased stage, the proteome was dominated by homeostatic collagens (COL1A2, COL1A1, COL4A2, COL4A3, COL4A1, COL5A2, COL6A6, COL6A3; Fig. 1e, Supplemental Figure 2; 8 out of the 12 detected collagens, p=0.027, Fisher exact test), rendering it a suitable control group. By contrast and consistent with a pathological fibrotic phenotype, myofibrogenic VIC differentiation (MYH11 and TAGLN2), oxidative stress (SOD3), a member of the phospholipase family (PLA2G2A), an inhibitor of calcification (MGP), and members of the small leucine-rich proteoglycan family (BGN, DCN and OGN; known to bind oxidized lipids and pathological growth factors16) were abundant in fibrotic tissue that we refer to as the fibrotic disease stage (highlighted in red in Fig. 1f, Supplemental Figure 2). Incidentally, the fibrotic transcriptome was marked by high expression levels of CLU, PCOLCE2, and GFAP as well as CILP, PRG4, and CRTAC (Supplemental Figure 1), the latter suggesting the involvement of a chondrocytic cell type in this intermediate stage of disease, consistent with overrepresentation of proteoglycans in this stage described above.
In the calcific stage of the AV tissue (as characterized by near-infrared imaging, Fig. 1b), the glycoproteins vitronectin and fibronectin-1 were enriched, as were calcification-related proteins (TGM2, fetuin A [AHSG] and tissue-nonspecific alkaline phosphatase [ALPL]), a marker of inflammation (CRP), and pro-inflammatory apolipoproteins (APOB, APO(a) [LPA]; Fig. 1g, Supplemental Figure 2 for full list). Two other large families were also present: complement proteins (C1QB, C1QC, C1R, C1S, C3, C4A, C4B, C5, C6, C7, C8A, C8G, CFB, CFD, CFHR2, CFHR4, CFI, 17 out of the 21 detected complement proteins, p<0.001, Fisher exact test) and the SERPIN proteases (SERPINA1, A3, A4, A6, A7, A10, C1, D1, E1, F1, F2, and G1; 12 out of the 15 detected SERPIN proteases, p<0.001, Fisher exact test) (Fig. 1g, with known calcification modulators in red). To define further the specific resident proteome of the calcific stage, we excluded previously identified human plasma proteins17 (Supplemental Figure 2, panel ‘Calcific stage, serum proteins excluded’). In the transcriptomics data, CD74, APOE, A2M, SPP1, LAPTM5, VWF, IBSP, and MMP9 were among the highly overrepresented genes in the calcific stage (Supplemental Figure 1). These results indicate that, from the distinct proteins that have emerged thus far, we are able both to identify novel and to confirm known molecules, thus supporting our notion that distinct pathogenic stages exist in a single leaflet.
The Proteomes of Distinct Aortic Valve Microlayers
Anatomical aortic valve microlayers exhibit unique proteome profiles
The aortic valve can also be interrogated in yet another dimension. In addition to the pathological breakdown above, more information can be gleaned from the distinct anatomical microlayers of the valve helping to answer the lingering issue of the predominant disease susceptibility of the fibrosa layer. To assess the proteomes of the three AV layers (fibrosa, spongiosa, ventricularis), we examined leaflets from non-diseased valves from autopsy (n = 2 donors) and CAVD valves from surgical explants for aortic stenosis (n = 3 donors). A typical non-diseased valve is <1mm in thickness; in contrast, fibrotic and calcified valves are significantly thicker5 (Supplemental Figure 4a; Supplemental Figure 4b indicates the color scheme used throughout Fig. 2). The layer architecture is signified by the collagen-rich fibrosa (yellow by Movat’s staining), the proteoglycan-rich spongiosa (blue-green), and the elastin-rich ventricularis (black) (Supplemental Figure 4c). CAVD valve thickening is primarily triggered by ECM remodeling and calcification expansion originating from the fibrosa layer. In order to obtain layer-specific protein profiles, we used laser-capture microdissection to excise each tissue layer (Supplemental Figure 4d) for proteomics.
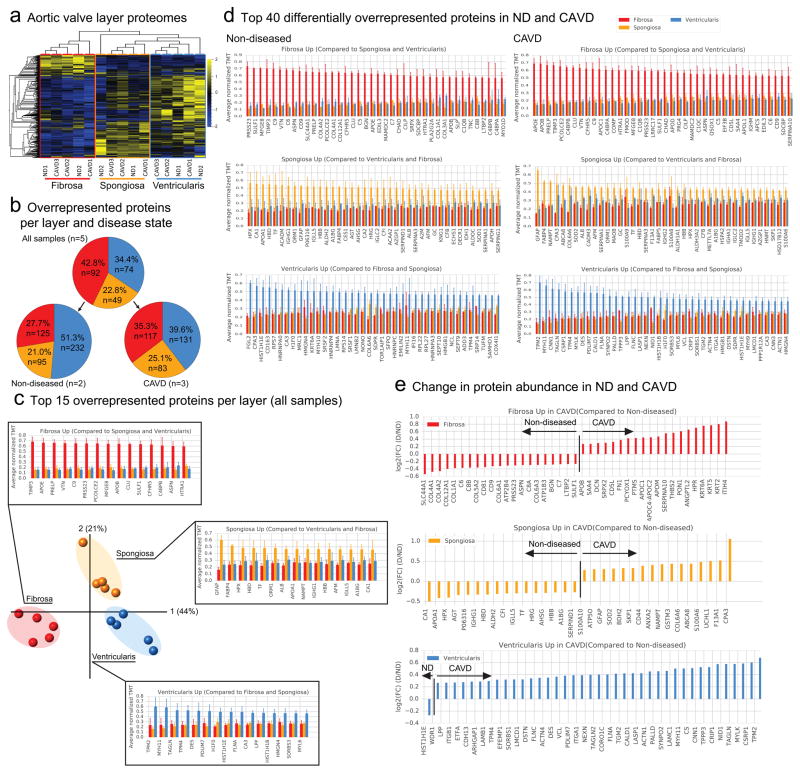
Figure 2. CAVD OMICs in the Three Leaflet Microlayers (CAVD Layers).
a. Proteomics heatmap of 5 donor samples (2 ND and 3 CAVD) per layer.
b. The number and relative percentage of proteins highly expressed in the fibrosa (red), spongiosa (yellow), and ventricularis (blue) layer for non-diseased (ND) tissue only (left), diseased (CAVD) tissue only (right).
c. PCA of 5 donor samples (2 ND and 3 CAVD) per layer and the top 15 “layer-specific” proteins per layer, ranked according to their average normalized TMT in all three layers as a consistent signal regardless of the disease state. Error bars indicate standard deviation (SD).
d. The top 40 “layer-specific” proteins ranked according to their average normalized TMT in all three layers for ND (left) and CAVD (right) tissues. Error bars indicate SD.
e. The “layer-specific” proteins ranked according to their fold change abundance from ND to CAVD [denoted FC(D/ND]. The proteins on the left of the vertical line are more expressed in ND tissue whereas the proteins to the right of the vertical line are more expressed in CAVD tissue. Proteins with FC(D/ND) > 1.2 and FC(D/ND) < 1/1.2 are shown for brevity.
From the combined proteomics data (see Supplemental Methods, TMT proteomics), we quantified 1,872 proteins, of which 856 were common across the TMT sub-experiments and analyzed further. Even across non-diseased and CAVD samples, the proteomes of the individual layers were conserved and distinct from one other, with the fibrosa layer being the most distinctive among the three layers, as demonstrated by a hierarchical heat map (Fig. 2a).
To determine which proteins define the individual layers, we filtered for proteins with a fold increase of at least 1.2 in one layer relative to both of the other two layers (as done for the CAVD stage data) using both non-diseased and CAVD samples (Fig. 2b). The distribution of these layer-specific proteins was similar for the combined and individual sample types (Fig. 2b). Specifically, in the non-diseased AVs, 125 proteins were overrepresented in the fibrosa, 95 in the spongiosa, and 232 in the ventricularis (Fig. 2b). In the diseased AV samples 117, 83, and 131 proteins were overrepresented in the fibrosa, spongiosa, and ventricularis, respectively (Fig. 2b).
We present the top 15 most enriched proteins per layer for the common distribution pie chart (Fig. 2b) (Fig. 2c and Supplemental Figure 5 for full list), and the top 40 proteins for the corresponding non-diseased and CAVD samples (Fig. 2d, Supplemental Figure 6 and S7 for full lists). In the combined dataset, TIMP3, APOE, PRELP, VTN, C9, PRSS23, PCOLCE2, MFGE8, APOB, CLU, SULF1, CFHR5, C4BP8, ASPN, and HTRA1 dominate in the fibrosa layer. In the spongiosa, GFAP and the adipocyte marker FABP4 were among the highest overrepresented proteins. The ventricularis layer proteome was signified by myofibroblast activation markers (TPM2, MYH11, TAGLN, TPM4, and MYL6). The proteins are mapped to their respective quadrants in the PCA plot (Fig. 2c), which was generated from the 856 common proteins (Fig. 2a).
In order to find proteins that are uniquely enriched in either non-diseased or CAVD samples, we performed a sub-analysis on each sample type separately (Fig. 2d). Although the heatmap suggests a high degree of conservation in the proteome across the sample types, a closer inspection of the top 40 proteins reveals distinct differences. For example, a general trend towards a higher proportion of apolipoproteins in the diseased fibrosa and proteins signifying myofibroblast activation in the diseased ventricularis appears evident (Fig. 2d). For a more systematic view of the differences between non-diseased and CAVD samples, we quantified the differential expression of all proteins within each layer (Fig. 2f). In the fibrosa, DCN, APOB, APOM, APOC1, FN1, ANGPTL2, and PON1 were enriched in the diseased compared with the non-diseased samples, while the relative contribution of collagens was reduced (Fig. 2e). In the diseased spongiosa, GFAP was higher in relative abundance compared to non-diseased samples (Fig. 2e), which is consistent with its enrichment in both the fibrotic stage transcriptome and proteome (Fig. 1d). An enrichment of proteins signifying myofibroblast activation was observed in the diseased ventricularis (MYH11, TAGLN2, CNN1, TAGLN, MYLK, TPM2, and TPM4) (Fig. 2e).
CAVD Pathway Enrichment Analysis
The wealth of information contained within the valvular proteomics data is best exploited by implementation of pathway and network analyses, thereby permitting us to investigate potential global disease pathways that promote CAVD. In order to highlight the shared proteins of the biological processes represented by these enriched pathways, we constructed pathway networks. These pathway networks can provide a global view of the multiple processes involved in CAVD where clusters of similar pathways emerge naturally due to strong interconnections between them, allowing us to gather the predominant biological themes in each respective stage or layer. We elected to focus the pathway network analysis on the calcific stage and the diseased fibrosa layer in order to study the most advanced disease phenotypes.
CAVD disease progression is marked by an emergence of smooth muscle cell activation, inflammation, and calcification-related pathways
We first determined the enriched pathways for the overrepresented proteins in the CAVD disease stages identified above (Supplemental Figure 2). In total, 125 significantly enriched (p-value < 0.05) pathways were recovered from the analysis in the non-diseased stage proteome, 226 in the fibrotic stage proteome, and 249 in the calcific stage proteome (Fig. 3a) (see Supplemental Table 1 and Supplemental Figure 8 for the entire pathway lists). The identified pathways of the fibrotic stage included pathways associated with myofibroblastic activation (e.g. ‘smooth muscle contraction’), vesicular transport, integrin signaling, and oxidative stress (Fig. 3a, Supplemental Figure 8, and Supplemental Table 1). Among the 249 pathways in the calcific stage were immune system-, lipid-, integrin-, MAPK-signaling-, retinoid-, and vesicle-related pathways, all of which are known contributors to cardiovascular calcification (Fig. 3a, Supplemental Table 1).
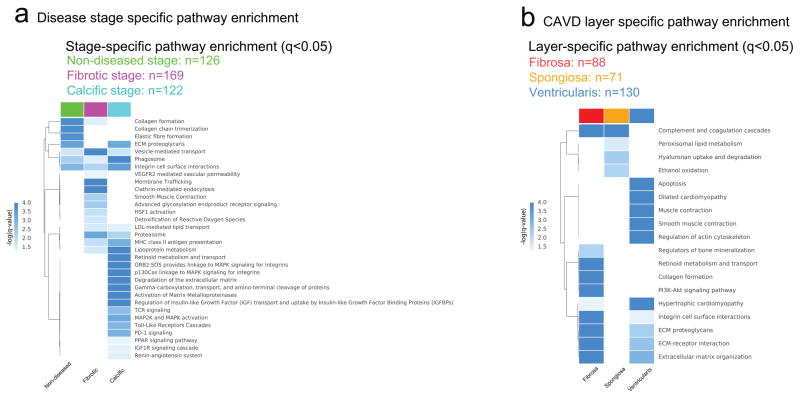
Figure 3. CAVD Disease Stages and CAVD Layers Functional Analysis.
a.. Pathways that are significantly enriched (q < 0.05) in the non-diseased (ND, green), fibrotic (F, magenta), and calcific (C, cyan) stages, obtained from proteomics data (n=9 donors). Selected pathways are listed for the non-diseased, fibrotic and calcific stages (Full pathway lists are available in Supplemental Figure 9, Supplemental Table 3).
b. Pathways that are significantly enriched (q < 0.05) in the fibrosa (red), spongiosa (orange), and ventricularis (blue) layers, obtained from proteomics data (n=3 CAVD AV samples). Selected pathways are listed for the three layers (Full pathway lists are available in Supplemental Figure 11, Supplemental Table 4).
Next, focusing on the most advanced CAVD stage, the calcific stage, we investigated the relationship of its pathways (Supplemental Figure 8) by incorporating them in a network where pathways are connected to each other based on the number of shared proteins as reflected in the proteomics measurements (see Supplemental Methods). For the pathways enriched in the calcific stage, this network revealed 6 distinct pathway clusters (Supplemental Figure 9). By inspecting these densely interconnected clusters, the underlying biological processes can be inferred. For example, the ‘MAPK signaling’ cluster or subnetwork is based on the presence of pathways involved in ‘MAPK family signaling cascades’ (Supplemental Figure 10), and the ‘complement’ cluster comprised the ‘complement cascade.’ Therefore, the overall network can be described as containing the following sub-networks: 1. MAPK signaling, 2. lipid metabolism, 3. coagulation, 4. complement system, 5. inflammation, and 6. extracellular matrix. The network representation of enriched pathways, thus, provides us with a concise overview of potential mechanisms underlying CAVD pathogenesis.
Proteins overrepresented in the diseased fibrosa are functionally annotated to fibrosis and calcification pathways
Consistent with the approach from ‘macro-to-micro’, we next hypothesized that pathways implicated in the calcific stage would similarly appear in conjunction with the diseased fibrosa. Since CAVD occurs preferentially in the fibrosa layer5, we focused on the diseased layers in the subsequent analyses and identified a total of 98 significantly enriched (p-value < 0.05) pathways (Fig. 3b; Supplemental Figure 10, Supplemental Table 2 for the entire pathway list). A significant pathway enrichment was observed for ‘integrin cell surface interactions,’ ‘PI3K-Akt signaling pathway;’ ‘retinoid metabolism and transport,’ and ‘regulators of bone mineralization,’ signifying a putative role in CAVD disease progression. The high enrichment of blood coagulation and oxidation-related pathways in the spongiosa suggests a novel and active involvement of this layer in disease progression (Fig. 3b). Pathways with the highest enrichment scores in the diseased ventricularis were associated with smooth muscle contraction and (cardio) myopathy (Fig. 3b).
Consistent with the calcific stage pathway network (Supplemental Figure 9), coagulation- and complement pathways as well as lipoprotein-, and extracellular matrix-related pathways were enriched in the CAVD fibrosa pathway network. In addition, PI3K-Akt signaling, endocrine system, platelet pathways, glycosaminoglycans, integrin- and collagen-related pathways were the most prominent subnetworks (Supplemental Figure 11).
Microlayer-specific VIC Migration Proteome
The microlayer-specific proteome is preserved in VICs
Further progressing though our ‘macro-to-micro’ view of the AV, we next focused on the VICs, the predominant valvular cell type. Current models posit that VIC osteogenic transition contributes to valvular calcification18. Because valvular calcification is initiated in the fibrosa layer, we hypothesized that VICs from the fibrosa and ventricularis layers possess different phenotypes and distinct calcific potential. To test this hypothesis, we isolated fibrosa- and ventricularis-specific VIC populations by the outgrowth method (Fig. 4a) from eight CAVD donor leaflets and conducted an in vitro calcification assay (Fig. 4b). To simulate valvular calcification in vitro, we employed two commonly used calcifying media conditions19. In both alkaline phosphatase-dependent osteogenic medium (OM) and alkaline phosphatase-independent pro-calcifying medium (PM) conditions, mineralization, as assessed by Alizarin Red staining, was enhanced in the fibrosa-derived VICs vs. those from the ventricularis (Fig. 4b).
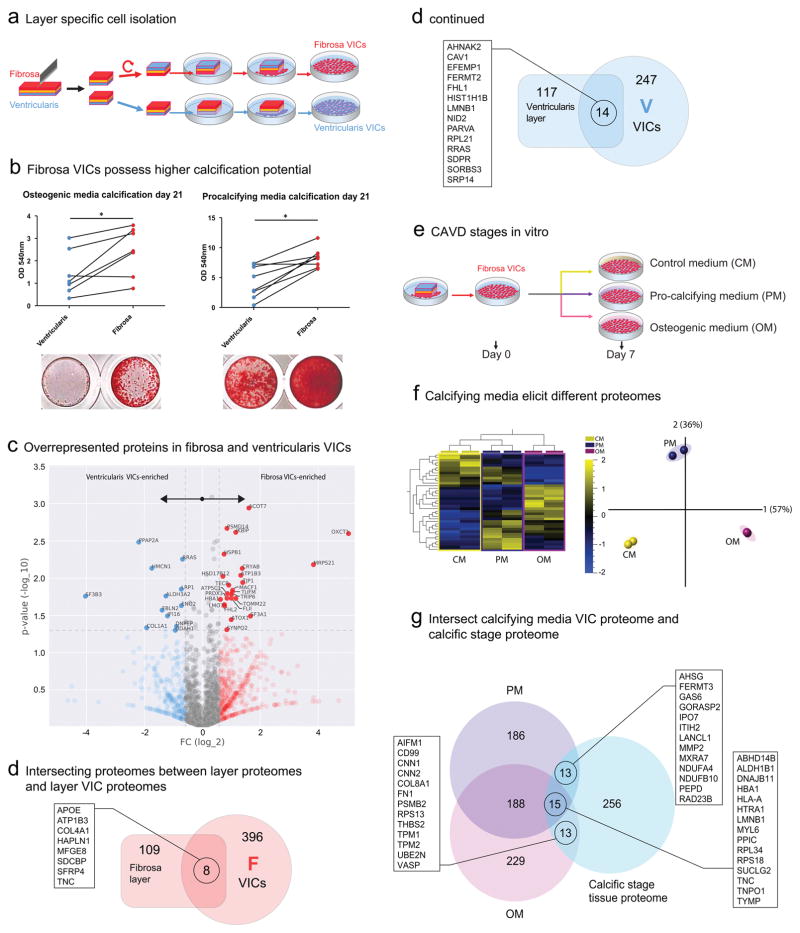
Figure 4. CAVD using VICs Isolated from Fibrosa and Ventricularis Microlayers.
a. Layer specific cell isolation: AV leaflet samples were dissected in halves, and the fibrosa or ventricularis side placed downwards. The VICs migrate out and expand the layer-specific cultures.
b. Fibrosa VICs display a higher propensity to calcification in osteogenic (OM, n=7 individual donors) and procalcifying media (PM, n=8 donors); calcification was evaluated by Alizarin Red staining and quantitatively evaluated; * indicates p<0.05.
c. Volcano plot for the Day 0 CAVD VICs proteomics data. Blue and red markers indicate “layer-specific” proteins highly expressed in ventricularis and fibrosa, respectively, with a fold change (FC) cutoff of 1.5. The significantly enriched (p<0.05) layer-specific proteins are denoted with their names.
d. Venn diagrams showing the intersection of proteins from tissue proteomics (CAVD Layers, rectangle on the left) and VIC proteomics (CAVD VICs, circle on the right) for the fibrosa (red) and ventricularis (blue) layers. Proteins at the intersection of each pair of sets are shown.
Comparison of Calcification-associated Disease Stage with Fibrosa-derived VICs Cultured in Osteogenic and Procalcifying Environment
e. Fibrosa-derived VICs were cultured in control (CM), osteogenic (OM), and procalcifying (PM) media, and protein samples were collected after 7 days in the respective media.
f. Heatmap and PCA of the proteomes of fibrosa VICs (from 2 independent donors) cultured in CM, OM, and PM for 7 days (q<0.05-filtered data for difference between media conditions, data derived from the CAVD VICs dataset).
g. The intersection of the VIC-PM and VIC-OM proteome after seven days of media treatment with the calcific stage CAVD Disease Stages proteome (n=9) from whole tissue proteomics. Proteins at the intersection regions are marked.
We hypothesized that the difference in calcific potential is a consequence of the difference in the proteomes of VIC populations derived from either the fibrosa or the ventricularis. We first analyzed the baseline proteomes of the outgrown fibrosa and ventricularis VIC populations at their calcifying state (~2 months in culture, passages 2–3). Inspecting their differentially expressed proteins (Fig. 4c), we found twelve proteins that were significantly elevated in the ventricularis-VICs. In the fibrosa-VICs, 24 proteins were overrepresented (Fig. 4c). In the ventricularis-VICs, the identified proteins were functionally annotated to ‘glycolysis and gluconeogenesis’ and ‘ECM organization,’ whereas the fibrosa-VICs showed an association of their enriched proteome to mitochondrial, fatty acid, and ketone body-associated pathways potentially pointing towards differences in metabolic state between fibrosa and ventricularis VICs (Supplemental Figure 12). We next mapped all of the overrepresented proteins per VIC layer origin (F/V FC<1/1.5) onto the protein-protein interaction (PPI) network and ranked them by their betweenness centrality (see Supplemental Methods, Supplemental Figure 13/14). This analysis revealed a stress fiber-associated protein (ARF6) and the proto-oncogenes FYN and GSK3B to be of high centrality in fibrosa-VICs. EGFR, PRKCA, FN1, RPL21, and AKT1 projected the highest betweenness centrality in the ventricularis-VIC PPI network.
To further validate the cellular protein signatures, we compared the VIC proteomes from the fibrosa- and ventricularis-derived VICs with the tissue proteomes from the fibrosa and ventricularis layers (Fig. 4d). Seven proteins were common to both the diseased fibrosa and fibrosa-derived VICs (APOE, ATP1B3, COL4A1, HAPLN1, SDCBP, SFRP4, TNC). In contrast, 14 proteins were shared by the ventricularis-VICs and their native tissue layer (AHNAK2, CAV1, CIRBP, EFEMP1, FERMT2, FHL1, HIST1H1B, LMNA, LMNB1, NID2, PARVA, SDPR, SORBS3, SRP14).
In vitro Proteome of CAVD Disease-associated Stages
Calcifying VICs have distinct proteome profiles
After determining that fibrosa-derived VICs have higher calcification potential, we further evaluated their protein profiles under calcifying media conditions (Fig. 4e). Using human primary VIC donors (n=2), we quantified the stimulated proteomes after seven days of calcifying stimulation (‘Day 7’, Fig. 4f). The q-adjusted relative protein abundance heat map demonstrated that the OM and PM proteomes were distinct both from each other and from that of control medium (CM) (Fig. 4f).
We next sought to evaluate similarities between the calcification-prone fibrosa-VIC proteomes (OM and PM; CAVD VICs dataset) and the calcific disease stage proteome (Supplemental Figure 2; CAVD Disease Stages). We filtered for proteins with a 1.2-fold increase of one medium condition relative to the other two (Fig. 4g). OM and calcific disease stage overlapped with 13 proteins (including FN1), PM and calcific with 13 proteins (i.e. AHSG and MMP2), and all three datasets shared 15 proteins (including TNC) (Fig. 4g). This finding suggests that both media conditions simulate components of valvular calcification in vivo.
A closer look at the intersection between the CAVD disease stage transcriptomics (FPKM ≥1 in at least one stage), CAVD disease stage proteomics (≥2 unique peptides), and the VIC in vitro proteome (≥2 unique peptides) revealed a set of 133 proteins detected in whole tissue proteomics that were neither detected in the VIC proteomics, nor in the CAVD disease stage transcriptomics (Supplemental Figure 15a, b). This set was enriched in plasma proteins (APO(a) [LPA], APOC3, FETUB, APOA2, PLG, CPN1, FGA, LRG1, CRP; full list in Supplemental Figure 15c) when queried against a human plasma proteome17, suggesting the potential contribution of plasma proteins to CAVD. As one such example, Lp(a) (APO(a) [LPA]) has previously been shown to promote CAVD20.
A Map of Human CAVD
This study identified several novel proteins that had not previously been implicated in the structure of normal and diseased AVs and the pathogenesis of CAVD. Based on the protein lists generated form the perspectives of the CAVD proteome, disease stages, and leaflet layers, we mapped the presence and expression of a subset of newly identified proteins via immunofluorescence staining and confocal microscopy. We first examined protein expression as a function of the CAVD stage (Fig. 5a–c).
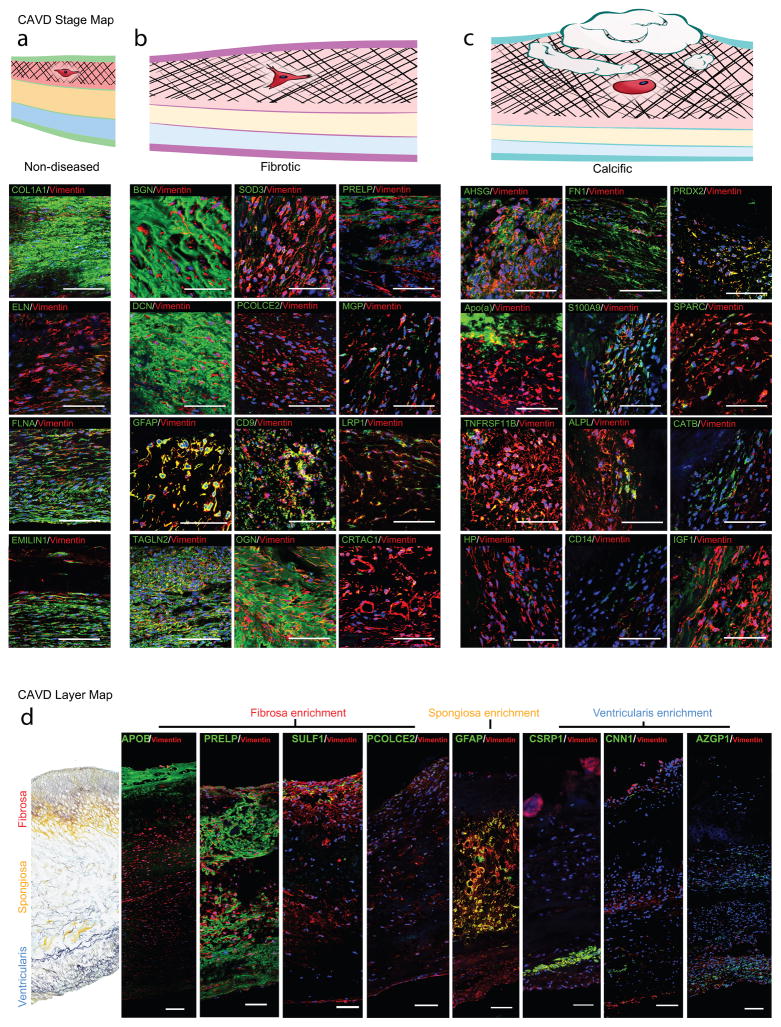
Figure 5. CAVD Protein Maps.
a–c. CAVD disease stage map: Immunofluorescence staining (IF) of proteins enriched in the (a) non-diseased, (b) fibrotic, and (c) calcific stage. Vimentin indicates VICs. DAPI stains nuclei. Scale bar indicates 100μm.
d. CAVD microlayer map: IF of representative proteins for high layer specificity in the layer proteomics; ApoB, PRELP, SULF1, and PCOLCE2 were detected primarily in the fibrosa; GFAP specifically in the spongiosa, and CSRP1, CNN1, and AZGP1 in the ventricularis VICs; scale bar indicates 100μm.
In the non-diseased stage proteomics, COL1A1 was detected, and its presence also confirmed in non-diseased CAVD tissue portions (Fig. 5a). Proteins that were overexpressed in the fibrotic stage include BGN, SOD3, PRELP, DCN, PCOLCE2, MGP, GFAP, CD9, LRP1, TAGLN2, OGN, and CRTAC1 (Fig. 5b); and in the calcific stage, AHSG, FN1, PRDX2, APO(a) [LPA], S100A9, SPARC, TNFRSF11B, ALPL, CATB, HP, CD14, and IGF1 (Fig. 5c).
We then mapped proteins across the layers (Fig. 5d). We confirmed expression of APOB, PRELP, SULF-1, and PCOLCE2 in the fibrosa layer, while CNN1, CSRP1, and AZGP1 were mapped onto VICs in the ventricularis layer (Fig. 5d). Proteomics identified the astrocyte marker glial fibrillary acidic protein (GFAP) to be enriched in the spongiosa, in both the non-diseased and diseased AV leaflets (Fig. 2d). Cardiac valves have previously been shown to harbor areas of GFAP-positivity21, 22. We determined by immunostaining that GFAP was cell-specific, and GFAP+/vimentin+ cells were exclusively localized to the spongiosa (Fig. 5d). This finding demonstrates the heterogeneous origin and phenotype of the mesenchymal VICs, is the first identification of a layer-specific VIC marker, and points toward a neural crest-derived origin of the spongiosa VIC population.
Human CAVD Networks and Disease Classification
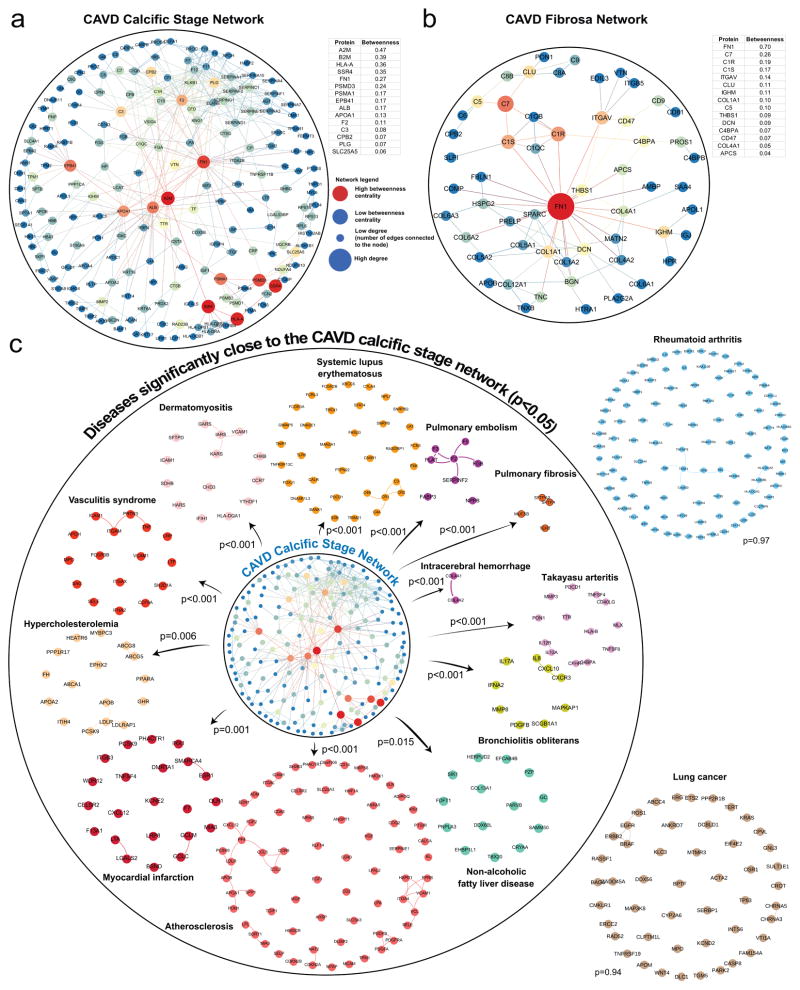
To determine the key pathological contributors to CAVD by their physical interactions at the molecular level, we focused on the calcific stage (Fig. 1g) and the CAVD fibrosa layer (Fig. 2d) proteomes. We mapped their respective overrepresented proteins onto the human PPI network23 comprised of interactions with experimental evidence curated from multiple sources (see Supplemental Methods). This analysis yielded the CAVD calcific stage (Fig. 6a) and CAVD fibrosa layer (Fig. 6b) protein subnetworks. We then interrogated the biological importance of the proteins within these subnetworks by means of their betweenness centrality, which signifies the “bottleneck” nodes that are crucial to the communication within the network24. High betweenness centrality proteins are, hence, prominent intermediary proteins that have been shown to play essential functional and dynamical roles7 as well as be potential drug targets25. We found that fibronectin-1 (FN1) played a pivotal role in both networks (Fig. 6a, b; betweenness centrality ranked 1st in CAVD fibrosa network: 0.70; ranked 5th in CAVD calcific stage network: 0.27). Beside the prominent role of FN1 in both networks, PSMD3 and PSMA1 from the calcific stage network belong to the canonical TGFβ-, canonical wnt-, MAPK and NOTCH-signaling pathways, all of which have been shown to contribute to CAVD. THBS1 and DCN from the CAVD fibrosa network are part of the TGF β-signaling pathway, as well, and COL1A1 and ITGAV are annotated to osteoblast signaling, further highlighting that this network strategy yields relevant key molecules.
Figure 6. CAVD Disease-associated Stage and CAVD Fibrosa Leaflet Microlayer Protein Networks in Relation to CAVD Calcific Stage Disease Module.
a. The largest connected component of the calcific stage subnetwork (calcific stage of the CAVD Disease Stage proteome). The subnetwork is generated by mapping the calcific stage proteome onto the protein-protein interaction (PPI) network. The node size indicates the number of connections (degree) and the node color indicates the betweenness centrality (blue: low betweenness centrality, red: high betweenness centrality). The key proteins with the highest betweenness centrality values shown in the table are marked by the red nodes in the subnetwork.
b. The largest connected component of the CAVD fibrosa subnetwork (CAVD fibrosa layer of the CAVD Layers proteome).
c. Disease associations of the CAVD calcific stage network to cardiovascular, malignant, metabolic, autoimmune diseases were measured in terms of the average network distance (see Methods for details). Disease modules are built by mapping the respective disease-related genes onto the PPI network. Diseases inside the circle represent the significant disease associations, with empirical p <0.05, whereas the diseases outside the circle are not significantly associated. Empirical p-values are calculated based on 1,000 randomizations.
In light of the increasing evidence that disease proteins tend to form closely interacting subgraphs (disease modules) within the cellular interactome, and that disease-disease relationships can be uncovered from the network proximity of their disease modules23, we next sought to establish the association of CAVD with a diverse array of human diseases, including metabolic, cardiovascular, inflammatory, and malignant diseases (see Supplemental Table 6 for a full list). Based on the average shortest (non-Euclidean) distance between disease modules (see Supplemental Methods, Supplemental Figure 16), we identified that vasculitis, dermatomyositis, systemic lupus erythematosus, pulmonary embolism, pulmonary fibrosis, intracerebral hemorrhage, Takayasu arteritis, hypercholesteremia, bronchiolitis obliterans, non-alcoholic fatty liver disease, atherosclerosis, myocardial infarction, and hypercholesteremia were closest23 to the calcific stage subnetwork (empirical p-value < 0.05; Fig. 6c; Supplemental Figure 17 for the direct overlap between the calcific disease stage network and these diseases). These diseases fall under the categories of inflammatory and fibrotic diseases26. In contrast, oncological diseases did not possess significant closeness to the calcific stage module. In general, despite the current limitations regarding the incompleteness of the interactome, the proximal location of a disease to diseases in certain parts of the interactome and the lack of connectivity to diseases in other parts provides important clues about the pathobiological relationships between these diseases, since topologically overlapping or proximal diseases tend to show significant gene co-expression, symptom similarity, and comorbidity, whereas topologically separated diseases do not. These findings, hence, point towards shared pathogenic contributors between a subset of diseases and calcification in CAVD, and may ultimately enable a protein-based disease ontology23.
Discussion
Advances in individual omics technologies and network medicine are revolutionizing preclinical and clinical medicine. As these high-throughput technologies mature, integration of multiple types of omics and subsequent downstream analyses into coherent and informative datasets that allow for conclusive answers to questions of causative changes in disease initiation, progression, and treatment is required27. CAVD is a prototypical disease that exists at the confluence of both highly complex pathophysiology and high clinical need. We present the first integrated atlas of the proteome and transcriptome of human CAVD, using a comprehensive undertaking that addresses the complexity of AV structural architecture and evolution of disease. We do so through incorporating macroscopic signatures of CAVD progression, anatomical layers, the in vitro VIC proteome, and cellular heterogeneity, followed by bioinformatics and multi-dimensional network analyses, and state-of-the-art systems biology approach of disease categorization. Our macro (whole-tissue and microlayers)- to micro (VICs)- strategy allowed for a high degree of temporal and spatial resolution, while the global proteomics and transcriptomics analyses have enabled us to build unbiased expression profiles of the AV proteome across a wide-ranging set of experimental conditions. Furthermore, this macro to micro strategy allowed us to overcome the limitation of proteomics that highly abundant peptides are identified more readily than those with low abundance. High abundant plasma and matrix proteins that dominated the whole valve proteome became less prominent at the layer proteome and even lesser so at the VIC culture proteome, allowing us to uncover low abundant cellular-derived protein signatures of CAVD. This work provides an important new basis for hypothesis generation and mechanistic insight in heart valve pathobiology and allows for identification of novel pharmacological and imaging targets for early diagnosis and treatment of CAVD.
Together, our multi-pronged omics assessment of CAVD has led us to a number of new insights into the pathobiology of this poorly understood disease. The enrichment in myofibrotic and calcification-related proteins and pathways validates our proposed regional approach and confirms that disease progression mapping is a valid technique on the level of a single heart valve leaflet. Identification of these annotated pathways may further guide the exploration of mechanisms governing CAVD and consideration of these pathways for pharmacological interventions to target CAVD. Our assessment of the stage-specific valvular proteome and transcriptome revealed classical fibrotic and calcific signatures. The fibrotic stage-specific proteome annotated pathways implicated players of early/fibrotic CAVD, including VIC myofibrogenesis28 and oxidative stress29. In contrast, the calcific stage-specific network included a number of established modulators of valvular calcification, including ALPL19, AHSG30, APO(a) [LPA]20 and APOB31, and was annotated to pathways known to be associated with VIC pathogenesis, including MMP activation32, and MAPK-signaling33. The presence of these previously described proteins confirmed the relevancy of our multi-omics approach. We also observed a prominent inflammatory signal in both the diseased fibrosa proteome (e.g. C8A, C8B, SLPI) and the calcific stage of CAVD (ELANE, HLA-DRA, MNDA, CD14, MPO), lending further credence to the debated notion that unresolved inflammation is a major contributor to human CAVD progression34. Importantly, our study identified several novel proteins that could serve as potential targets to treat CAVD during the stages of both fibrosis and calcification. Prior studies of valve pathobiology in animal models and cultured VICs have identified roles for individual components of MAPK signaling33, PI3K-Akt signaling35, lipoproteins/lipids20, and vesicular processes36. Here, we demonstrate for the first time both a degree of novel clustering and novel interconnectedness between these pathways in the human AV, thus establishing a basis for future investigations.
Data from histological studies of human CAVD using quantitative analysis of microcalcifications (precursors of macrocalcification37) suggested that calcification initiates from the fibrosa layer5. Despite this widely held view, until now there has been no layer-specific study of the valvular proteome. Our experimental approach allowed us to identify significant differences in the proteome of the calcification-prone fibrosa layer as compared to the spongiosa and ventricularis, and to establish the presence of layer-specific alterations in advanced CAVD. This discovery supports our hypothesis that both structural matrix (e.g., FN1, VTN, PRELP, PCOLCE2, COMP) and secreted (e.g. TIMP3, CLU, HTRA1, TNC, SPARC) VIC proteins in the fibrosa contribute to its calcification propensity and suggest that proteins enriched in the fibrosa layer may be therapeutic targets to halt CAVD.
In order to identify VIC intracellular molecular drivers of AV calcification that could otherwise be masked by the highly abundant extracellular matrix in proteomics, we isolated VICs specifically from the fibrosa region and compared their calcific potential to VICs derived from the calcification-protected ventricularis in vitro. Consistent with our leaflet layer proteomes, we discovered a heightened predilection for calcification in VICs derived from the fibrosa layer, further implicating fibrosa-derived VICs as the main drivers for calcification in CAVD. As a first indication of dysfunction in the fibrosa, we identified differential metabolic programming in glycolytic and fatty-acid metabolism between layer-derived VIC populations. In addition, we showed that the proteome of the diseased fibrosa was rich in pathogenic pathways, including collagen biosynthesis and modification5, extracellular matrix degradation38, glycosaminoglycan metabolism39, and lipid metabolism40.
Although the ventricularis is generally understood to be protected from disease development, perhaps due to high shear stresses on the ventricular side of the leaflet41, we identified a dysregulated myo-contractile response in the diseased ventricularis, consistent with a prior study that found greater upregulation of myofibrogenesis in ventricularis-derived VICs than those from the fibrosa upon exposure to mechanical strain or TGF-β1 stimuli42. While the ventricularis layer itself may not calcify, there remains the intriguing possibility that pro-myofibrogenic changes in this layer alter how cyclic mechanical forces are transmitted throughout the leaflet, contributing to pathological VIC differentiation in calcification-prone regions of the fibrosa.
Multiple calcifying media formulations exist in the CAVD literature. We provide evidence that culture models of VIC calcification which recapitulate either ALPL-mediated osteogenic calcification or ALPL-independent hyperphosphatemia associated with mineral imbalance (e.g., chronic kidney disease19) stimulates calcific proteomes in vitro and share a high degree of similarity with that of the calcific stage proteome in vivo. This finding suggests that investigation of both ALPL-mediated and ALPL-independent culture conditions will be important to future studies of cellular behaviors in CAVD.
Furthermore, our holistic omics approach followed by comprehensive multi-dimensional network analysis discovered several common proteins enriched in diseased stages and disease-prone layers. Among these is fibronectin-1, which was identified in both the CAVD fibrosa and calcific stage proteomes, as well as the corresponding betweenness-centrality-weighted PPI networks. Fibronectin-1 has previously been implicated in VIC myofibrogenesis in vitro due to its potential binding to TGF-β143, thus suggesting an additional novel role for fibronectin-1 in modulating valvular calcification, and demonstrating the potential of these networks for hypothesis generation. We further utilized this network approach to construct the first protein-based disease ontology of CAVD and compared it to 38 curated human disease modules. As a positive control, we showed significant overlap with calcification-related diseases. Of note, we found significant overlap between the CAVD disease module and those of a variety of fibrotic, inflammatory, and hypercholesterolemia disease modules. Our network analysis also identified a significant overlap between our CAVD calcific stage network and that of atherosclerosis. Although this overlap may appear contradictory given potentially different mechanisms of calcification between CAVD and atherosclerosis44 and the unfavorable findings of a number of randomized clinical trials of statin treatment in CAVD45, statins have been shown to inhibit calcification in cultured VICs46 and there are some indications that the application of statins to CAVD may require different temporal implementation than for atherosclerosis45, 47. This overlap may, therefore, motivate future studies of anti-fibrotic and anti-inflammatory drugs towards the treatment of CAVD.
The current study describes the first integrated molecular atlas of human CAVD. We provide evidence of multiple discrete, yet co-existing, pathological processes that may act in parallel to promote valvular fibrosis and calcification. Our assessment of disease stage- and layer-specific proteomes is the first to our knowledge to examine CAVD with a tissue resolution beyond that of the whole organ, thereby elucidating key drivers of valvular mineralization within the region most predisposed to disease – the fibrosa. Furthermore, we show that VICs derived from the fibrosa and ventricularis are phenotypically and functionally different, with differential proteomes reflecting their pro-fibrotic and pro-calcific potential. This work also identifies disease stage- and layer-specific VIC markers, including the GFAP-positive VIC phenotype as a specific marker for spongiosa VICs. Together, these fundamental results comprise the disease-specific protein network of the AV and provide a rich resource for future studies of human CAVD, thereby paving the way for clinically meaningful investigations and development of novel therapeutic and diagnostic imaging approaches that better address the intricate mixture of homeostatic and pathological processes at play in the AV. This framework for integration of spatiotemporal multi-omics and systems biology holds promise as a universal template for analysis of complex pathobiologies in a wide variety of disease states.
Supplementary Material
Clinical Perspective.
What is new?
First application of multi-omics and network medicine to CAVD
A high-resolution spatiotemporal atlas of the combined valvular tissue, layer and cell proteome, and a comprehensive repository of molecular drivers of CAVD
First demonstration of side-specific valve cellular calcification potential and identification of a novel layer-specific cell marker
Comparative-omics based identification of optimal cell culture models to recapitulate in vivo valvular calcification
Network-based disease comparison of CAVD with other human diseases reveals a novel protein-based disease ontology
What are the clinical implications?
Identification of novel targets for diagnostic molecular clinical imaging and pharmacological intervention in early or advanced CAVD
Network medicine-based rationale for putative utility of anti-fibrotic and anti-inflammatory therapeutics for the treatment of CAVD
Roadmap for the multi-omics study of complex cardiovascular diseases
Acknowledgments
The authors acknowledge Jennifer R. Wen, Jung Choi and Michael Creager for their histological assistance.
Sources of Funding: National Institutes of Health grants (R01 HL114805, R01 HL136431 and HL119798 to E.A.; R01 HL114823 to S.C.B.; and R01 HL61795 and GM107618 to J.L.)
Footnotes
Disclosures: None declared.
References
- 1.Stewart BF, Siscovick D, Lind BK, Gardin JM, Gottdiener JS, Smith VE, Kitzman DW, Otto CM. Clinical factors associated with calcific aortic valve disease. J Am Coll Cardiol. 1997;29:630–634. doi: 10.1016/s0735-1097(96)00563-3. [DOI] [PubMed] [Google Scholar]
- 2.Nishimura RA, Otto CM, Bonow RO, Carabello BA, Erwin JP, 3rd, Guyton RA, O’Gara PT, Ruiz CE, Skubas NJ, Sorajja P, Sundt TM, 3rd, Thomas JD. 2014 aha/acc guideline for the management of patients with valvular heart disease: Executive summary: A report of the american college of cardiology/american heart association task force on practice guidelines. Circulation. 2014;129:2440–2492. doi: 10.1161/CIR.0000000000000029. [DOI] [PubMed] [Google Scholar]
- 3.Ross J, Braunwald E. Aortic stenosis. Circulation. 1968;38:V-61–V-67. doi: 10.1161/01.cir.38.1s5.v-61. [DOI] [PubMed] [Google Scholar]
- 4.Schoen FJ. Evolving concepts of cardiac valve dynamics. Circulation. 2008;118:1864. doi: 10.1161/CIRCULATIONAHA.108.805911. [DOI] [PubMed] [Google Scholar]
- 5.Otto CM, Kuusisto J, Reichenbach DD, Gown AM, O’Brien KD. Characterization of the early lesion of ‘degenerative’ valvular aortic stenosis. Histological and immunohistochemical studies. Circulation. 1994;90:844–853. doi: 10.1161/01.cir.90.2.844. [DOI] [PubMed] [Google Scholar]
- 6.Rabkin E, Aikawa M, Stone JR, Fukumoto Y, Libby P, Schoen FJ. Activated interstitial myofibroblasts express catabolic enzymes and mediate matrix remodeling in myxomatous heart valves. Circulation. 2001;104:2525–2532. doi: 10.1161/hc4601.099489. [DOI] [PubMed] [Google Scholar]
- 7.Barabasi AL, Gulbahce N, Loscalzo J. Network medicine: A network-based approach to human disease. Nat Rev Genet. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gil-Dones F, Martin-Rojas T, Lopez-Almodovar LF, Cuesta Fdl, Darde VM, Alvarez-Llamas G, Juarez-Tosina R, Barroso G, Vivanco F, Padial LR, Barderas MG. Valvular aortic stenosis: A proteomic insight. Clinical Medicine Insights: Cardiology. 2010;4:1–7. doi: 10.4137/cmc.s3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin-Rojas T, Mourino-Alvarez L, Alonso-Orgaz S, Rosello-Lleti E, Calvo E, Lopez-Almodovar LF, Rivera M, Padial LR, Lopez JA, Cuesta Fdl, Barderas MG. Itraq proteomic analysis of extracellular matrix remodeling in aortic valve disease. 2015;5:17290. doi: 10.1038/srep17290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yutzey KE, Demer LL, Body SC, Huggins GS, Towler DA, Giachelli CM, Hofmann-Bowman MA, Mortlock DP, Rogers MB, Sadeghi MM, Aikawa E. Calcific aortic valve disease: A consensus summary from the alliance of investigators on calcific aortic valve disease. Arterioscler Thromb Vasc Biol. 2014;34:2387–2393. doi: 10.1161/ATVBAHA.114.302523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brigham and Women’s Hospital. Harvard Medical School. Center for Interdisciplinary Cardiovascular Sciences; 2018. https://cics.bwh.harvard.edu/multiomics_databases. [Google Scholar]
- 12.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kustatscher G, Grabowski P, Rappsilber J. Pervasive coexpression of spatially proximal genes is buffered at the protein level. Molecular Systems Biology. 2017;13:937. doi: 10.15252/msb.20177548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh SA, Aikawa E, Aikawa M. Current trends and future perspectives of state-of-the-art proteomics technologies applied to cardiovascular disease research. Circ J. 2016;80:1674–1683. doi: 10.1253/circj.CJ-16-0499. [DOI] [PubMed] [Google Scholar]
- 15.Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 16.Neufeld EB, Zadrozny LM, Phillips D, Aponte A, Yu ZX, Balaban RS. Decorin and biglycan retain ldl in disease-prone valvular and aortic subendothelial intimal matrix. Atherosclerosis. 2014;233:113–121. doi: 10.1016/j.atherosclerosis.2013.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geyer PE, Kulak NA, Pichler G, Holdt LM, Teupser D, Mann M. Plasma proteome profiling to assess human health and disease. Cell Syst. 2016;2:185–195. doi: 10.1016/j.cels.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 18.Rajamannan NM, Subramaniam M, Rickard D, Stock SR, Donovan J, Springett M, Orszulak T, Fullerton DA, Tajik AJ, Bonow RO, Spelsberg T. Human aortic valve calcification is associated with an osteoblast phenotype. Circulation. 2003;107:2181–2184. doi: 10.1161/01.CIR.0000070591.21548.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hutcheson JD, Blaser MC, Aikawa E. Giving calcification its due: Recognition of a diverse disease: A first attempt to standardize the field. Circ Res. 2017;120:270–273. doi: 10.1161/CIRCRESAHA.116.310060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bouchareb R, Mahmut A, Nsaibia MJ, Boulanger MC, Dahou A, Lepine JL, Laflamme MH, Hadji F, Couture C, Trahan S, Page S, Bosse Y, Pibarot P, Scipione CA, Romagnuolo R, Koschinsky ML, Arsenault BJ, Marette A, Mathieu P. Autotaxin derived from lipoprotein(a) and valve interstitial cells promotes inflammation and mineralization of the aortic valve. Circulation. 2015;132:677–690. doi: 10.1161/CIRCULATIONAHA.115.016757. [DOI] [PubMed] [Google Scholar]
- 21.Hainfellner JA, Voigtlander T, Strobel T, Mazal PR, Maddalena AS, Aguzzi A, Budka H. Fibroblasts can express glial fibrillary acidic protein (gfap) in vivo. J Neuropathol Exp Neurol. 2001;60:449–461. doi: 10.1093/jnen/60.5.449. [DOI] [PubMed] [Google Scholar]
- 22.Oki T, Fukuda N, Kawano T, Iuchi A, Tabata T, Manabe K, Kageji Y, Sasaki M, Yamada H, Ito S. Histopathologic studies of innervation of normal and prolapsed human mitral valves. J Heart Valve Dis. 1995;4:496–502. [PubMed] [Google Scholar]
- 23.Menche J, Sharma A, Kitsak M, Ghiassian SD, Vidal M, Loscalzo J, Barabasi AL. Disease networks. Uncovering disease-disease relationships through the incomplete interactome. Science. 2015;347:1257601. doi: 10.1126/science.1257601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pavlopoulos GA, Secrier M, Moschopoulos CN, Soldatos TG, Kossida S, Aerts J, Schneider R, Bagos PG. Using graph theory to analyze biological networks. BioData Min. 2011;4:10. doi: 10.1186/1756-0381-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Csermely P, Korcsmaros T, Kiss HJ, London G, Nussinov R. Structure and dynamics of molecular networks: A novel paradigm of drug discovery: A comprehensive review. Pharmacol Ther. 2013;138:333–408. doi: 10.1016/j.pharmthera.2013.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ghiassian SD, Menche J, Chasman DI, Giulianini F, Wang R, Ricchiuto P, Aikawa M, Iwata H, Müller C, Zeller T, Sharma A, Wild P, Lackner K, Singh S, Ridker PM, Blankenberg S, Barabási A-L, Loscalzo J. Endophenotype network models: Common core of complex diseases. Scientific Reports. 2016;6:27414. doi: 10.1038/srep27414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biology. 2017;18:83. doi: 10.1186/s13059-017-1215-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rabkin-Aikawa E, Farber M, Aikawa M, Schoen FJ. Dynamic and reversible changes of interstitial cell phenotype during remodeling of cardiac valves. J Heart Valve Dis. 2004;13:841–847. [PubMed] [Google Scholar]
- 29.Miller JD, Chu Y, Brooks RM, Richenbacher WE, Pena-Silva R, Heistad DD. Dysregulation of antioxidant mechanisms contributes to increased oxidative stress in calcific aortic valvular stenosis in humans. J Am Coll Cardiol. 2008;52:843–850. doi: 10.1016/j.jacc.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koos R, Brandenburg V, Mahnken AH, Muhlenbruch G, Stanzel S, Gunther RW, Floege J, Jahnen-Dechent W, Kelm M, Kuhl HP. Association of fetuin-a levels with the progression of aortic valve calcification in non-dialyzed patients. Eur Heart J. 2009;30:2054–2061. doi: 10.1093/eurheartj/ehp158. [DOI] [PubMed] [Google Scholar]
- 31.Weiss RM, Ohashi M, Miller JD, Young SG, Heistad DD. Calcific aortic valve stenosis in old hypercholesterolemic mice. Circulation. 2006;114:2065–2069. doi: 10.1161/CIRCULATIONAHA.106.634139. [DOI] [PubMed] [Google Scholar]
- 32.Fondard O, Detaint D, Iung B, Choqueux C, Adle-Biassette H, Jarraya M, Hvass U, Couetil JP, Henin D, Michel JB, Vahanian A, Jacob MP. Extracellular matrix remodelling in human aortic valve disease: The role of matrix metalloproteinases and their tissue inhibitors. Eur Heart J. 2005;26:1333–1341. doi: 10.1093/eurheartj/ehi248. [DOI] [PubMed] [Google Scholar]
- 33.Munjal C, Jegga AG, Opoka AM, Stoilov I, Norris RA, Thomas CJ, Smith JM, Mecham RP, Bressan GM, Hinton RB. Inhibition of mapk-erk pathway in vivo attenuates aortic valve disease processes in emilin1-deficient mouse model. Physiol Rep. 2017;5:e13152. doi: 10.14814/phy2.13152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hjortnaes J, Butcher J, Figueiredo JL, Riccio M, Kohler RH, Kozloff KM, Weissleder R, Aikawa E. Arterial and aortic valve calcification inversely correlates with osteoporotic bone remodelling: A role for inflammation. Eur Heart J. 2010;31:1975–1984. doi: 10.1093/eurheartj/ehq237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poggio P, Branchetti E, Grau JB, Lai EK, Gorman RC, Gorman JH, Sacks MS, Bavaria JE, Ferrari G. Osteopontin–cd44v6 interaction mediates calcium deposition via phospho-akt in valve interstitial cells from patients with noncalcified aortic valve sclerosis. Arterioscler Thromb Vasc Biol. 2014;34:2086–2094. doi: 10.1161/ATVBAHA.113.303017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cui L, Rashdan NA, Zhu D, Milne EM, Ajuh P, Milne G, Helfrich MH, Lim K, Prasad S, Lerman DA, Vesey AT, Dweck MR, Jenkins WS, Newby DE, Farquharson C, Macrae VE. End stage renal disease-induced hypercalcemia may promote aortic valve calcification via annexin vi enrichment of valve interstitial cell derived-matrix vesicles. J Cell Physiol. 2017;232:2985–2995. doi: 10.1002/jcp.25935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hutcheson JD, Goettsch C, Bertazzo S, Maldonado N, Ruiz JL, Goh W, Yabusaki K, Faits T, Bouten C, Franck G, Quillard T, Libby P, Aikawa M, Weinbaum S, Aikawa E. Genesis and growth of extracellular vesicle-derived microcalcification in atherosclerotic plaques. Nature Materials. 2016;15:335–343. doi: 10.1038/nmat4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Satta J, Oiva J, Salo T, Eriksen H, Ohtonen P, Biancari F, Juvonen TS, Soini Y. Evidence for an altered balance between matrix metalloproteinase-9 and its inhibitors in calcific aortic stenosis. Ann Thorac Surg. 2003;76:681–688. doi: 10.1016/s0003-4975(03)00529-0. discussion 688. [DOI] [PubMed] [Google Scholar]
- 39.Stephens EH, Saltarrelli JG, Baggett LS, Nandi I, Kuo JJ, Davis AR, Olmsted-Davis EA, Reardon MJ, Morrisett JD, Grande-Allen KJ. Differential proteoglycan and hyaluronan distribution in calcified aortic valves. Cardiovasc Pathol. 2011;20:334–342. doi: 10.1016/j.carpath.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mahmut A, Boulanger MC, Fournier D, Couture C, Trahan S, Page S, Arsenault B, Despres JP, Pibarot P, Mathieu P. Lipoprotein lipase in aortic valve stenosis is associated with lipid retention and remodelling. Eur J Clin Invest. 2013;43:570–578. doi: 10.1111/eci.12081. [DOI] [PubMed] [Google Scholar]
- 41.Yap CH, Saikrishnan N, Yoganathan AP. Experimental measurement of dynamic fluid shear stress on the ventricular surface of the aortic valve leaflet. Biomech Model Mechanobiol. 2012;11:231–244. doi: 10.1007/s10237-011-0306-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moraes C, Likhitpanichkul M, Lam CJ, Beca BM, Sun Y, Simmons CA. Microdevice array-based identification of distinct mechanobiological response profiles in layer-specific valve interstitial cells. Integr Biol (Camb) 2013;5:673–680. doi: 10.1039/c3ib20254b. [DOI] [PubMed] [Google Scholar]
- 43.Cushing MC, Liao JT, Anseth KS. Activation of valvular interstitial cells is mediated by transforming growth factor–beta1 interactions with matrix molecules. Matrix Biol. 2005;24:428–437. doi: 10.1016/j.matbio.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 44.Hutcheson JD, Aikawa E, Merryman WD. Potential drug targets for calcific aortic valve disease. Nature Reviews Cardiology. 2014;11:218–231. doi: 10.1038/nrcardio.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Y, Nicoll R, He Yh, Henein MY. The effect of statins on valve function and calcification in aortic stenosis: A meta-analysis. Atherosclerosis. 2016;246:318–324. doi: 10.1016/j.atherosclerosis.2016.01.023. [DOI] [PubMed] [Google Scholar]
- 46.Osman L, Yacoub MH, Latif N, Amrani M, Chester AH. Role of human valve interstitial cells in valve calcification and their response to atorvastatin. Circulation. 2006;114:I547–552. doi: 10.1161/CIRCULATIONAHA.105.001115. [DOI] [PubMed] [Google Scholar]
- 47.Marquis-Gravel G, Redfors B, Leon MB, Généreux P. Medical treatment of aortic stenosis. Circulation. 2016;134:1766–1784. doi: 10.1161/CIRCULATIONAHA.116.023997. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.