Abstract
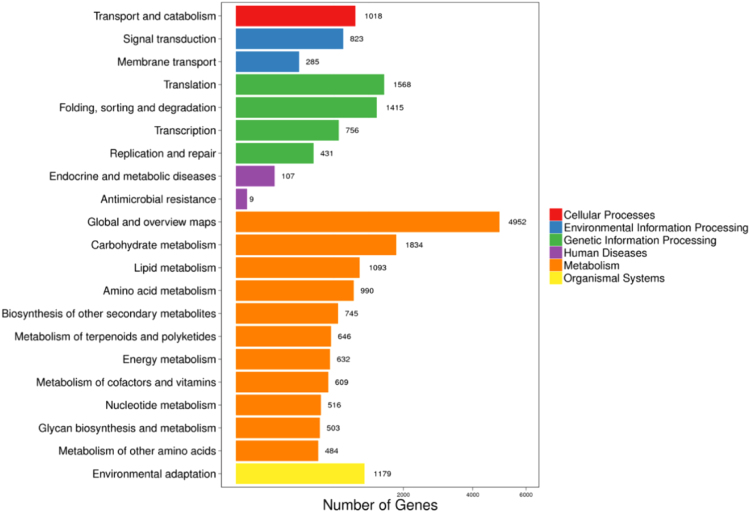
In this paper, we present the transcriptome profiles of the A. venetum L. by RNA-Seq approach. A total of 6.57 Gb raw data were obtained, and 52,983 unigenes with an average length of 1009 bp and N50 of 1632 bp were annotated with the 7 databases. The unigenes annotated to KEGG database were divided into 21 categories from 6 main groups. Among these, 4952 (22.21%) unigenes were clustered to “Global and overview maps”, and 1834 (8.23%) unigenes were clustered to “Carbohydrate metabolism”. In addition, 6340 unigenes containing 7579 SSRs were identified and the mononucleotide, dinucleotide, trinucleotide motifs were the most common motif type (95.59%), accounting for 39.62%, 36.02%, and 19.95%, respectively.
Specifications table
| Subject area | Biology |
| More specific subject area | Plant biology; Bioinformatics |
| Type of data | Table, text file, graph, figure |
| How data was acquired | RNA sequencing, Illumina HiSeq. 2000 and BGISEQ. 500 platform |
| Data format | Raw |
| Experimental factors | The leaves of A. venetum were collected for RNA sequencing |
| Experimental features | The sterilized seeds of Apocynum venetum L. were allowed to germinate and grow for 30 days in half-strength MS agar medium inside a growth chamber with a 14 h light/10 h dark cycle, air temperature of 25 °C, photon flux density (PFD) of 280 mol m−2 s−1. The leaves of A. venetum were collected, immediately frozen in liquid nitrogen, and stored at -80 °C until use. In order to increase the transcriptome coverage, a mixture of samples from these chambers were pooled for RNA sequencing. |
| Data source location | The seeds of A. venetum were collected in Xinjiang Province, China |
| Data accessibility | Data are with this article and available at https://www.ncbi.nlm.nih.gov/sra/SRP151546. |
| Related research article [1] | Xie W, Zhang X, Wang T, Hu J. Botany, traditional uses, phytochemistry and pharmacology of Apocynum venetum L. (Luobuma): a review. J. Ethnopharmacol., 2012;141(1): 1–8. |
Value of the data
-
•
Apocynum venetum (luobuma) is a common fiber and medicinal plant widely distributed in the salt marish, desert margins, alluvia flats and riversides [2], [3], which makes it an invaluable model for bast fiber development and plant stress resistance research.
-
•
The genetic information and gene sequences about the A. venetum in public databases are scanty.
-
•
The large dataset of transcripts and unigenes can be useful as it provides abundant genetic information for identifying of A. venetum genes.
-
•
The unigenes obtained provide a good resource for SSRs application in evolutionary genetic from A. venetum.
1. Data
Here we report a de novo transcriptome assembly of A. venetum. Our aim was to obtain a high quality reference transcriptome of A. venetum leaves, elucidate the molecular pathway of fiber and flavonoids synthetize, stress resistance, and find candidate genes of these process (see Table 1, Table 2, Table 3 and Fig. 1, Fig. 2, Fig. 3).
Table 1.
Summary of the transcriptome sequencing and assembly.
| Assembly statistic | Apocynum venetum |
|---|---|
| Total Clean Bases(Gb) | 6.57 |
| Clean Read Q20 (%) | 95.97 |
| Number of assembled reads | 63,906 |
| Total Length of assembled reads | 58,605,303 |
| Number of unigenes | 50,957 |
| Total Length of unigene (bp) | 51,426,191 |
| Average unigene length (bp) | 1009 |
| GC (%) | 40.42 |
| Unigene N50 (bp) | 1632 |
Table 2.
Unigenes functional annotation by various databases.
| Index | Apocynum venetum |
|---|---|
| Unigenes annotated in Nr | 31,250 |
| Unigenes annotated in Nt | 21,507 |
| Unigenes annotated in Swissport | 20,148 |
| Unigenes annotated in KEEG | 22,294 |
| Unigenes annotated in KOG | 23,492 |
| Unigenes annotated in Interpro | 24,553 |
| Unigenes annotated in GO | 10,483 |
Table 3.
General statistics of SSR identified transcriptome.
| Item | Marker |
|---|---|
| Total number of identified SSR | 7579 |
| Number of SSR containing sequences | 6340 |
| Number of sequences containing> 1 SSR | 1040 |
| Mononucleotide | 3003 |
| Dinucleotide | 2730 |
| Trinucleotide | 1512 |
| Tetranucleotide | 43 |
| Pentanucleotide | 112 |
| Hexanucleotide | 179 |
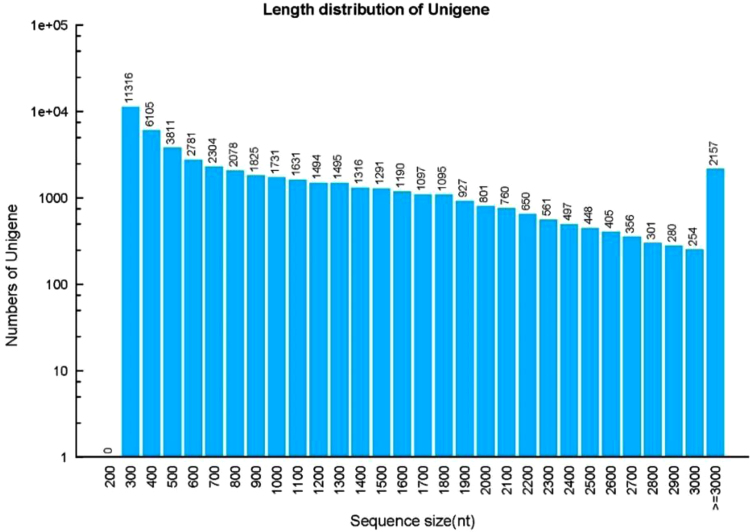
Fig. 1.
Length distributions of the de novo assembly for unigenes. The length distribution of unigenes were counted with an interval of every 100 bp from 300 bp to 3000 bp. X-axis indicates sequence size (nt), Y-axis indicates number of assembled contigs and unigenes.
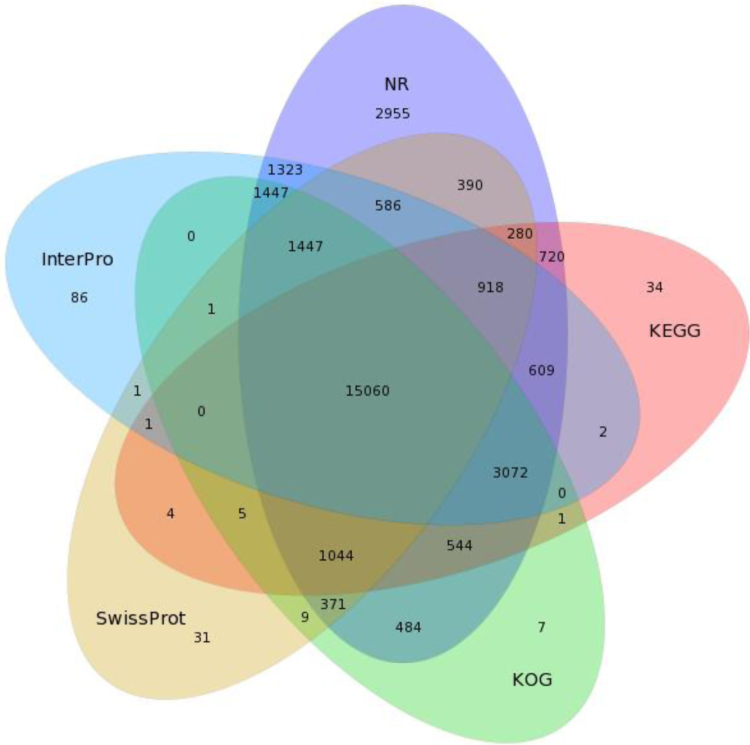
Fig. 2.
Venn diagram shows commonality and difference of annotation based on NR, KEGG, Swiss-Prot, InterPro and KOG.
Fig. 3.
KEGG annotation of unigenes.
The de novo transcriptome assembly of A. venetum L., and the SRA records is accessible with the following link: https://www.ncbi.nlm.nih.gov/sra/SRP151546.
2. Experimental design, materials and methods
2.1. Plant materials
The seeds of A. venetum were collected from Xinjiang Province, China, in November 2016. Seeds were surface-sterilized by rinsing in 70% (v/v) ethanol for 60 s, then in 5% (v/v) sodium hypochlorite (NaClO) for 30 min while rocking on a platform, and washed in distilled water for 8 min. The seeds were allowed to germinate and grow for 30 days in half-strength MS agar medium inside a growth chamber with a 14 h light/10 h dark cycle, air temperature of 25 °C, photon flux density (PFD) of 280 mol m−2 s−1. The leaves of A. venetum were collected, immediately frozen in liquid nitrogen, and stored at -80 °C until use. Total RNA was extracted using TRIzol Reagent (Invitrogen, LifeTechnologies, USA) following the manufacturer׳s instructions, then rtreated with DNase I (Invitrogen, Life Technologies, USA). The RNA integrity was verified using an Agilent 2100 BioAnalyzer (Agilent, USA).
2.2. RNA sequencing
RNA-Seq libraries were constructed using the RNA Library Prep Kit for Illumina using to the manufacturer׳s instructions (NEB, USA). Library quality was assessed on the Agilent Bioanalyzer 2100 system. The libraries were sequenced on the BGIEQ-500 platform (BGI, CHN) based on sequencing by synthesis with 100 bp paired-end reads (BGI Technologies, Shenzhen). All RNA-Seq data were deposited in National Center for Biotechnology Information (NCBI) with the accession number SRP151546.
2.3. Leaf transcriptome assembly and gene functional annotation
The raw reads were firstly filtered and combined to form longer fragments, then de novo assembled into unigenes using the short read assembly program Trinity with default settings [4], [5]. Functional annotation of the unigenes was performed by searching the following databases: Nr; Pfam; KOG/COG; Swiss Prot; KEGG; and GO. The information on the annotation was summarized and the distribution of unigenes was illustrated by Venn diagram (Fig. 2).
2.4. Identification of SSR markers
Using the MISA software [6], 6,340 unigenes containing 7,579 SSRs were identified, of which 1040 sequences contained more than one SSR.
Acknowledgements
This research work was supported by the “Agricultural Science and Technology Innovation Project of Chinese (CAAS-ASTIP-2018)”.
Footnotes
Transparency data associated with this article can be found in the online version at https://doi.org/10.1016/j.dib.2018.08.207.
Transparency document. Supplementary material
Supplementary material
References
- 1.Xie W., Zhang X., Wang T., Hu J. Botany, traditional uses, phytochemistry and pharmacology of Apocynum venetum L. (Luobuma): a review. J. Ethnopharmacol. 2012;141(1):1–8. doi: 10.1016/j.jep.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 2.Xiong Q.B., Fan W.Z., Tezuka Y., Adnyana I.K., Stampoulis P. Hepatoprotective effect of Apocynum venetum and its active constituents. Planta Med. 2000;66:127–133. doi: 10.1055/s-2000-11135. [DOI] [PubMed] [Google Scholar]
- 3.Mohanty A.K., Misra M., Drzal L.T. Natural Fibers, Biopolymers, and Biocomposites. CRC Press; Boca Raton: 2005. Cellulose-based nanocomposites; pp. 807–832. [Google Scholar]
- 4.Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011;29(7):644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pertea G., Huang X., Liang F., Antonescu V., Sultana R., Karamycheva S. TIGR gene indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19(5):651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 6.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 2004;5(6):435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material