Abstract
The genetic architecture of asthma was relatively well explored. However, some work remains in the field to improve our understanding on asthma genetics, especially in non-Caucasian populations and with regards to commonly neglected genetic variants, such as Copy Number Variations (CNVs). In the present study, we investigated the contribution of CNVs on asthma risk among Latin Americans. CNVs were inferred from SNP genotyping data. Genome wide burden and association analyses were conducted to evaluate the impact of CNVs on asthma outcome. We found no significant difference in the numbers of CNVs between asthmatics and non-asthmatics. Nevertheless, we found that CNVs are larger in patients then in healthy controls and that CNVs from cases intersect significantly more genes and regulatory elements. We also found that a deletion at 6p22.1 is associated with asthma symptoms in children from Salvador (Brazil) and in young adults from Pelotas (Brazil). To support our results, we conducted an in silico functional analysis and found that this deletion spans several regulatory elements, including two promoter elements active in lung cells. In conclusion, we found robust evidence that CNVs could contribute for asthma susceptibility. These results uncover a new perspective on the influence of genetic factors modulating asthma risk.
Introduction
Asthma is a chronic inflammatory disorder of the airways characterized by reversible airflow obstruction. Asthma is clinically heterogeneous and patients may experience intermittent cough, dyspnea, wheezing, and chest tightness1. The pathophysiology of asthma is complex and typically involves airway eosinophilic inflammation, but many individuals can present a persistent noneosinophilic disease2,3. It is estimated that nearly 334 million people have asthma worldwide and its prevalence has been increasing in several regions of the planet4,5. In Latin America, the global prevalence of asthma symptoms in adolescents was estimated in approximately 16%6. Markedly, Brazil has one of the highest disease prevalence among Latin American countries, reaching 24,4% in 20027,8.
The asthma epidemic observed in the last decades has been essentially attributed to temporal changes in a set of different factors, among them diet, allergen exposure, microbiota diversity and occurrence of infections that occurred particularly in high income countries and in urban areas of low-to-middle income countries9–12. Nevertheless, it is important to note that such changes in social and environmental conditions operate on individuals or populations with variable degrees of genetic predisposition to asthma. The initial studies mapping candidate genes in the context of asthma identified more than 200 genetic variants associated with disease development and severity, many of these associations being replicated in different populations13,14. Later, several large-scale studies, applying mainly small nucleotide polymorphism (SNP) microarrays and whole genome sequencing, have identified multiple short variants (rare and common) associated with asthma in different loci, including: 1q31.3 (DENND1B), 2q12.1 (IL1RL1/IL18R1), 5q12.1 (PDE4D), 5q22.1 (TSLP/WDR36), 5q31.1 (IL13), 6p21.32 (HLA-DR/DQ), 9p24.1 (IL33), 14q11.2 (DAD1/OXAL1L), 15q22.2 (FOXB1) and 17q21.1 (ORMDL3/GSDMB)15–20.
Due to all these efforts, the genetic architecture of asthma is now relatively well known, with the genetic factors identified so far explaining a reasonable proportion of the heritability attributed to the disease (varying between 35% and 95%)21. However, some work remains in the field to improve our understanding on asthma genetics, especially in non-Caucasian populations and with regards to other variants found in the human genome, such as Copy Number Variations (CNVs). CNVs are large deletions or duplications that can encompass genes (and their regulatory elements) leading to dosage imbalances22. Estimates suggest that CNVs affect approximately 12% of the human genome23. These structural variations have been widely studied in several complex human traits, including immunological disorders such as type 1 Diabetes24,25 and rheumatoid arthritis24,26. However, few comprehensive studies explored the role of CNVs in asthma and only suggestive associations have been found27,28. Additionally, it was observed that genes involved in asthma pathogenesis are affected by CNVs29.
Here, we conducted a genome wide copy number variation study based in our previously published SNP genotyping data19 to investigate the contribution of CNVs on asthma risk in Latin American admixed populations.
Results
Global contribution of copy number variations on asthma outcome
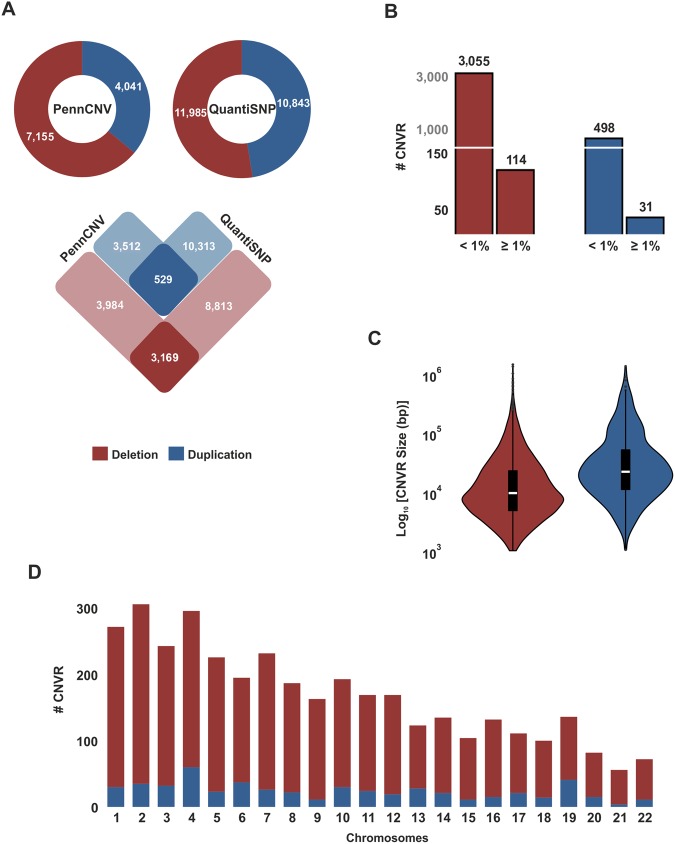
Copy number variations in the genome of admixed children from Salvador (Northeast Region of Brazil) (Table 1) were inferred from SNP genotyping data (Illumina HumanOmni 2.5–8v1 panel) using two distinct algorithms implemented in PennCNV and QuantiSNP. To combine CNVs corresponding to the same event, deletions or duplications showing sequence overlap were grouped into a single copy number variation region (CNVR). Only CNVRs detected by both programs were considered valid. After stringent quality control (detailed in Methods), a set of 3,698 CNVRs (3,169 deletions and 529 duplications) was identified in 872 individuals (Fig. 1A). Of these, only 114 deletions and 31 duplications presented frequencies ≥ 1% in this study population (Fig. 1B and Supplementary Table 1). Regarding the median size of the CNVRs, it was found that duplications (22.0 kb) are more than twice as large as deletions (9.6 kb) (Fig. 1C). Finally, we found that CNVRs were well dispersed across the genome and the distribution of these events reflects the size of the human chromosomes, with decreasing frequency of CNVRs from the first to the twenty-second autosomal chromosome (Fig. 1D).
Table 1.
Characteristics of the studied samples (after quality control).
| Sample Characteristics | Salvador, Brazil | Pelotas, Brazil | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cases | Controls | Cases | Controls | |||||||||
| Number of individuals | 188 | 684 | 367 | 1381 | ||||||||
| Sex (male/female) | 108/80 | 370/314 | 165/202 | 678/703 | ||||||||
| Median age, years (IQR) | 6 (4–11) | 6 (4–10) | 23 (22–23) | 23 (22–23) | ||||||||
| LRRSD, mediana | 0.155 | 0.152 | 0.155 | 0.151 | ||||||||
| Ancestry, median %b | EUR | AFR | NAT | EUR | AFR | NAT | EUR | AFR | NAT | EUR | AFR | NAT |
| 40.4 | 52.1 | 5.9 | 42.3 | 50.8 | 5.8 | 81.4 | 8.9 | 7.6 | 84.1 | 7.0 | 7.1 | |
aLog2 of R ratio standard deviation.
bAncestry proportions (ADMIXTURE method), EUR: European; AFR: African; NAT: Native American.
IQR, interquartile range.
Figure 1.
Copy number variation regions (CNVRs) detected in the genome of children from Salvador, Brazil. Copy number variations in the genome of 872 individuals from the SCAALA-Salvador cohort were inferred from SNP microarray data (Illumina HumanOmni 2.5–8v1 panel), using algorithms implemented in PennCNV v1.0.1 and QuantiSNP v2.0. Deletions or duplications showing sequence overlap were grouped into a single CNVR. (A) Number (#) of CNVRs detected by PennCNV and QuantiSNP (after quality control). Only CNVRs detected by both algorithms were considered for further analysis. (B) Frequency of deletions and duplications (<1% or ≥1%) in the study population. (C) Size of the deletions and duplications (bp, base pair), violin plots. (D) Number (#) of CNVRs by human autosomal chromosome.
After identifying CNVs in the genome of children from Salvador, analyses were conducted to evaluate the global impact of these structural variations on asthma outcome (Table 2). First, the number of CNVR per individual (CNVR count) was compared between patients and healthy controls and no significant difference was found. In average, it was observed 12.8 deletions and 3.6 duplications per asthmatic individual, while among non-asthmatic subjects we identified similar proportions, 14.3 deletions and 3.0 duplications per sample. Next, the size of CNVRs was compared between groups and it was found that structural variations (deletions + duplications) from asthmatic individuals are significantly larger than those presented by their controls (p = 5 × 10−3). The mean sizes of the deletions found in cases and controls were 35.6 kb and 26.4 kb, respectively (p = 0.03). Meanwhile, the average sizes of the duplications from cases and controls were 85.1 kb and 62.1 kb, respectively (p = 0.05). Based on this finding, we hypothesized that CNVRs from cases could mobilize more genes, regulatory and constrained elements than those from controls. To evaluate this assumption, CNVR positions were cross-referenced with DNA sequence annotations. As shown in Table 2, we found no significant differences regarding the number of constrained elements (sequence conservation across mammals) intersected by deletions and duplications from asthmatic and non-asthmatic individuals. On the other hand, CNVRs from cases mobilized significantly more genes (deletions, p = 0.01; duplications, p = 0.02; deletions + duplications, p = 2 × 10−4) and more regulatory elements (deletions, p = 0.03; duplications, p = 0.06; deletions + duplications, p = 7 × 10−4) than those from controls.
Table 2.
Global contribution of copy number variation regions (CNVRs) on asthma outcome.
| Deletions | Asthmatics | Non-asthmatics | Ratio | p | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | SD | Median | P25 | P75 | Mean | SD | Median | P25 | P75 | |||
| CNVR counta | 12.8 | 11.2 | 10.5 | 8.0 | 14.0 | 14.3 | 18.0 | 10.0 | 8.0 | 14.0 | 0.9 | 0.8 |
| Size (kb)b | 35.6 | 94.3 | 11.3 | 5.1 | 28.2 | 26.4 | 63.2 | 9.9 | 5.0 | 24.3 | 1.3 | 0.03 |
| Gene countc | 1.1 | 2.0 | 1.0 | 0.0 | 2.0 | 0.8 | 1.6 | 1.0 | 0.0 | 1.0 | 1.4 | 0.01 |
| Regulatory element countd | 5.1 | 11.6 | 2.0 | 0.0 | 5.0 | 3.9 | 8.7 | 2.0 | 0.0 | 4.0 | 1.3 | 0.03 |
| GERP element counte | 6.1 | 8.1 | 3.9 | 0.5 | 8.0 | 6.5 | 8.6 | 4.2 | 0.7 | 8.7 | 0.9 | 0.1 |
| Duplications | ||||||||||||
| CNVR counta | 3.6 | 4.5 | 2.0 | 1.0 | 4.0 | 3.0 | 3.7 | 2.0 | 1.0 | 4.0 | 1.2 | 0.2 |
| Size (kb)b | 85.1 | 161.2 | 28.1 | 13.6 | 75.8 | 62.1 | 118.3 | 22.9 | 11.2 | 53.1 | 1.4 | 0.05 |
| Gene countc | 2.8 | 4.2 | 1.0 | 0.0 | 3.0 | 2.1 | 3.5 | 1.0 | 0.0 | 2.0 | 1.3 | 0.02 |
| Regulatory element countd | 14.0 | 22.4 | 5.0 | 2.0 | 15.0 | 11.4 | 19.7 | 4.0 | 1.0 | 12.0 | 1.2 | 0.06 |
| GERP element counte | 7.5 | 8.9 | 4.7 | 2.1 | 9.7 | 7.6 | 8.2 | 4.9 | 2.4 | 10.2 | 1.0 | 0.5 |
| Deletions + Duplications | ||||||||||||
| CNVR counta | 16.4 | 12.4 | 13.0 | 10.0 | 18.0 | 17.3 | 18.4 | 13.0 | 10.0 | 18.0 | 1.0 | 0.5 |
| Size (kb)b | 43.0 | 108.4 | 13.0 | 5.8 | 32.9 | 31.6 | 74.8 | 11.4 | 5.5 | 28.9 | 1.4 | 5 × 10−3 |
| Gene countc | 1.4 | 2.7 | 1.0 | 0.0 | 2.0 | 1.0 | 2.1 | 1.0 | 0.0 | 1.0 | 1.4 | 2 × 10−4 |
| Regulatory element countd | 6.8 | 14.8 | 2.0 | 0.0 | 7.0 | 5.0 | 11.5 | 2.0 | 0.0 | 5.0 | 1.4 | 7 × 10−4 |
| GERP element counte | 6.3 | 8.3 | 4.0 | 0.7 | 8.3 | 6.6 | 8.5 | 4.3 | 1.0 | 8.9 | 1.0 | 0.1 |
Asthmatic and non-asthmatic children from Salvador-SCAALA were compared in terms of: acount of CNVRs per individual; bestimated size of CNVRs (kb, kilobase); cnumber of genes captured by CNVR; dnumber of regulatory elements captured by CNVR; enumber of GERP constrained elements captured by CNVR.
SD: standard deviation; P25: 25% percentile; P75: 75% percentile; Ratio: case/ctrl ratio (mean); p values ≤ 0.05 were considered statistically significant [Mann-Whitney U test (two-sided)].
Association of copy number variations with asthma in salvador
In the discovery association phase, analyses were conducted to evaluate the effect of specific structural variations on asthma risk in children from Salvador. The association of CNVRs with asthma was investigated by comparing frequencies of low-to-common variations (minor allele frequency ≥1%) between asthmatic and non-asthmatic individuals, under an additive model. Sex and age, which are considered classic risk factors for asthma, were included as covariates in the logistic regression analysis. Additionally, Log2 of R ratio standard deviation (LRRSD), to account for potential differences in sample and/or call quality between cases and controls, and the first three principal components, to correct for population stratification, were included in the regression model. This initial screening stage revealed several deletions and duplications that were nominally associated with asthma (p ≤ 0.05). Supplementary Table 2 shows the results for all CNVRs evaluated in the discovery study. Remarkably, only one deletion with approximately 41.6 kb of size, located at 6p22.1 (6:29,889,788–29,931,412) (Supplementary Fig. 2A), was significantly associated with the disease (OR = 3.0, p = 2 × 10−4) (Table 3), overcoming the significance level established for this discovery phase (p ≤ 3.4 × 10−4).
Table 3.
A deletion region located in the locus 6p22.1 is associated with asthma in two independent Brazilian populations.
| CNVR | CNVR class | CNVR size (bp) | Freq. Case | Freq. Ctrl | OR | 95% CI | p | Power |
|---|---|---|---|---|---|---|---|---|
| Salvador, Brazil (Discovery) | ||||||||
| 6:29,889,788–29,931,412 | Deletion | 41,624 | 6.6 | 2.4 | 3.0 | 1.7–5.2 | 2 × 10 −4 | 0.902 |
| Pelotas, Brazil (Replication) | ||||||||
| 6:29,881,842–29,931,412 | Deletion | 49,570 | 4.0 | 1.6 | 1.9 | 1.2–2.8 | 4 × 10 −3 | 0.837 |
| Random-effects meta-analysis | ||||||||
| OR | p | |||||||
| 2.3 | 3 × 10 −6 | |||||||
CNVR: Copy number variation region, chromosome:start-end; bp: base pair; Frequency of CNVR (%); Case: asthmatic; Ctrl: non-asthmatic; OR, odds ratio; SE, SE of odds ratio (OR); 95% CI, 95% confidence interval; p, p value (additive model). The significance threshold established for the discovery phase was p ≤ 3.4 × 10−4; The significance level applied in the replication study was p = 0.05; Power: A posteriori statistical power.
Covariates in multivariate analysis: sex, age, Log2 of R ratio standard deviation (LRRSD) and principal components (PC1, PC2 and PC3).
Human genome assembly: GRCh38.
Replication study and association in different ancestry compositions
We then attempted to replicate the association signal at 6p22.1 in another admixed Brazilian sample, composed of 1,748 young adults from the city of Pelotas, located in the Southern Region of Brazil (Table 1). CNVRs located in the locus 6p22.1 were also inferred from SNP genotyping data (Illumina HumanOmni 2.5–8v1 panel) using PennCNV and QuantiSNP. Interestingly, both algorithms detected a 49.6 kb deletion (6:29,881,842–29,931,412) (Supplementary Fig. 2B) whose limits overlap those of the deletion associated with asthma in Salvador, representing, therefore, a single CNVR. As show in Table 3, the association of this structural variation with asthma was replicated in this second Brazilian cohort (OR = 1.9, p = 4 × 10−3), with p value below the significance threshold assumed for the replication phase (p = 0.05).
Next, we conducted a meta-analysis on Salvador and Pelotas samples, by applying a random-effects model that assumes significant inter-study variability (Table 3). This analysis confirmed association of this deletion with the disease (OR = 2.3; p = 3 × 10−6), providing support for the notion that structural variations could represent risk factors for asthma.
Additional experiments were conducted to evaluate the effect of the deletion at 6p22.1 in subjects with different ancestry. First, our data sets were dichotomized in groups of individuals with proportion of European ancestry above or below the median. Next, we carried out association tests in these subgroups and, despite the reduced sample sizes, the deletion was nominally associated (p ≤ 0.05) with asthma in both situations (proportion of European ancestry above or below the median) (Supplementary Table 3).
Fine-mapping of the 6p22.1 region
Considering that 6p22.1 is a very complex region, making association signals difficult to interpret, we performed a fine-mapping of the entire locus (6:27,100,000–30,500,000; RefSeq: GRCh38). We focused in the identification of SNPs that could explain the association signal found in this region (Supplementary Fig. 3). Notably, no robust linkage disequilibrium (r2 > 0.6) was found between our deletion and any evaluated SNP in the region. In addition, none of the SNPs investigated in this region was significantly associated with asthma risk in Salvador [locus p-value threshold = 8 × 10−6 (0.05/6057 SNPs)] and Pelotas [locus p-value threshold = 9 × 10−6 (0.05/5782 SNPs)]. We also carried out conditional tests to evaluate the possibility that our deletion and any other SNP tested could be capturing the same association signal. Remarkably, we found that association signals for the SNPs at 6p22.1 are not influenced by the signal of the reported deletion, i.e., the −log10 (p values) after adjustment for the deletion genotypes were strongly correlated with −log10 (p values) without adjustment [Pearson correlation: Salvador (r2 = 0.97; p-value < 10−4); Pelotas (r2 = 0.98; p-value < 10−4)].
In silico functional analyses
To investigate the regulatory potential of the deletion at 6p22.1, the region was cross-referenced with genomic and epigenomic annotations, obtained from the Ensembl database. This region was evaluated in terms of transcripts location, binding sites for transcription factors, sequence constraint, chromatin segmentation state (evidences of promoter and enhancer marks) and enrichment for marks of open chromatin (DNase I hypersensitive sites). In Fig. 2, it is possible to observe the limits found in Salvador and Pelotas for the asthma-associated deletion. This deletion region may have relevant functional consequences, since it covers a region with seven transcripts, numerous constrained sequences and several regulatory elements (including promoter and promoter flanking regions, transcription factor binding sites and an open chromatin element). In addition, it is close to HLA genes (HLA-G and HLA-A) and intersects a SNP (rs2523809) previously associated with dysregulation of plasma IgE concentrations in Europeans30. Collectively, these data support the biological plausibility of our findings.
Figure 2.
In silico functional study on the regulatory potential of the deletion at 6p22.1. Schematic representation of the locus containing the deletion region associated with asthma in Brazilian populations (expanded view: 6:29,801,381–30,018,756; RefSeq: GRCh38). This region was cross-referenced with DNA sequence annotations, including: location of protein coding or non-protein coding genes (GENCODE 26); presence of large structural variations identified by the 1000 genomes project, phase 3 (SV – 1KG 3); position of putative regulatory elements (regulatory build); location of constrained elements for 40 eutherian mammals (GERP, Genomic Evolutionary Rate Profiling); presence of SNPs and short indels associated with any human phenotype in previous studies. Limits of the deletions found in samples from Salvador or Pelotas are symbolized by a yellow or a green bar, respectively. Image created using the Ensembl genome browser (http://www.ensembl.org).
Discussion
Initially, we conducted an exploratory analysis, based in our previously published high-density SNP genotyping data19, to detect copy number variations throughout the genome of children from Salvador, Brazil. After stringent quality control, the algorithm implemented in PennCNV identified 7,155 deletions and 4,041 duplications, while QuantiSNP detected 11,985 deletions and 10,843 duplications, in 872 individuals. To avoid false discoveries, we focused only on the variations simultaneously detected by the two programs, remaining 3,169 deletions and 529 duplications. These results highlight an imbalanced ratio between the numbers of deletions and duplications. This can be explained primarily by limitations related to the PennCNV algorithm for the detection of duplication events, which are normally inferred by increased number of peaks in the BAF distribution, as well as increased LRR values. Wang and colleagues (2007)31 obtained similar results when testing the PennCNV package. In their data set, deletions were approximately twofold more frequent than duplications. Furthermore, they also found that deletions presented smaller sizes than duplications.
Then, we tested the hypothesis that the cumulative effect of multiple structural variations through an individual’s genome could increase asthma risk. Initially, we found no significant difference in the numbers of CNVRs between asthmatic and non-asthmatic individuals. Nevertheless, we found that CNVRs were larger in cases when compared to controls and that CNVRs from cases intersected significantly more genes and regulatory elements. Despite the modest differences found, this may be increasing the risk of presenting asthma symptoms. To date, the only genome-wide burden analysis associating asthma and CNVs found no evidences on the global contribution of these variations in disease risk27. However, it is important to note that this cited study was carried out among Australian children (European descent), using a less dense SNP chip (Illumina 610 K array). Besides that, their analyses were restricted to large (100 to 1.000 kb) and common CNVs [minor allele frequency (MAF) >5%]. In the present study, more robust conditions were created to detect the joint effect of structural variations on asthma risk by applying a much higher density SNP platform (with 2,237,482 SNPs) and by using broader spectra of CNV size (ranging from 1 to 1,430 kb) and frequency (rare to common). Similar results have already been described for other human traits, such as schizophrenia32 and obesity33.
Individual effects of CNVRs were also evaluated and we found that a deletion located at 6p22.1 was significantly associated with asthma symptoms in Salvador. The SCAALA-Salvador cohort has the largest proportion of African ancestry (50.8%) among the EPIGEN-Brazil populations34, with 42.9% and 6.4% of European and Native American ancestries, respectively. This association was replicated in another Brazilian admixed population from the EPIGEN-Brazil program, composed of young adults from the city of Pelotas. Global ancestry in Pelotas is 76.1% European, 15.9% African, and 8% Native American. Even though genetic ancestry at locus-segment level needs to be investigated, this consistent effect found in populations with different genetic backgrounds suggests functional relevance for this deletion or strong linkage to a causal variant yet to be identified.
Despite our sample size in the discovery study (188 asthma cases and 684 controls), our analysis was well powered (>80%) to detect the effect found for the deletion at 6p22.1 (OR = 3.0). This was possible because we evaluated only 145 low-to-common (MAF ≥ 1%) CNVRs that placed the significance level at 3.4 × 10−4. Even though not achieving statistical significance following adjustment for multiple tests, several other CNVRs were nominally associated (p ≤ 0.05) with asthma symptoms in the discovery phase (Supplementary Table 2). Although further studies using larger samples are necessary to confirm these results, we investigated if these nominal associations occurred in loci associated with asthma symptoms in our previous study19. Notably, no CNVRs were identified in the regions 14q11.2 (DAD1/OXAL1L genes) and 15q22.2 (FOXB1 gene) that could explain SNP associations. Furthermore, we found no deletions or duplications nominally associated with asthma symptoms in loci consistently associated with the disease in previous studies, including: DENND1B, IL1RL1, PDE4D, TSLP, IL13, HLA-DR/DQ and IL33 regions.
To support our results, we carried out an in silico functional analysis of the deletion at 6p22.1. Remarkably, this structural variation region was previously identified through DNA sequencing in populations from several continents by the 1000 genomes project, phase 3 (Del 6:29,882,895–29,937,238, RefSeq: GRCh38; DGVa ID: esv3608493). We evaluated several genomic annotations in this region and found that the sequence covered by the asthma-associated deletion spans essentially pseudogenes. Nevertheless, it deletes several regulatory elements in this region, including two promoters active in lung cells (empirical data from the ENCODE project)35 that could be involved in local gene expression regulation. Indeed, this deletion is located near the HLA-A and HLA-G genes and could impact on their transcriptional regulations. The HLA-A product, as a classical MHC I antigen, is responsible for initiating cell-mediated immunity36. On the other hand, HLA-G protein, a non-classical MHC I antigen, has immunoinhibitory functions and the loss of HLA-G immune-mediated control seems to be involved in the onset of inflammatory diseases37. Interestingly, Granada and colleagues (2012)30 found several SNPs near the HLA-A and HLA-G genes as potential determinants of atopy and IgE production among Europeans. In the aforementioned study, the SNP rs2523809, which is located at approximately 59 kb 5′ of the HLA-A gene and is intersected by our asthma-associated deletion, was strongly associated (4 × 10−8) with dysregulation of plasma IgE concentrations. Linkage disequilibrium between the SNP rs2523809 and the deletion at 6p22.1 was investigated in our cohorts and very low values were found (r2 < 0.1). Additionally, a recent meta-analysis identified another SNP (rs1233578) in the region 6p22.1 that was strongly associated with asthma risk in individuals from ethnically diverse populations38. This SNP is located more than 1 Mb away from the 5′ end of the CNV reported in our study and they are not in linkage disequilibrium in our cohorts (r2 < 0.2). In addition, the association of this SNP with asthma was not replicated in Salvador (p-value = 0.38) and in Pelotas (p-value = 0.42).
Another important aspect is that we identified a genetic variant that confers susceptibility to asthma in populations with very different ages: children from Salvador (4–11 years of age) and young adults from Pelotas (22–23 years of age). Asthma has various clinical phenotypes that are age-related39 and several evidences indicate that although some genetic variations can influence risk of both childhood and adult-onset asthma, other loci are exclusively associated to each group18. Although we cannot establish that the appearance of asthma symptoms in patients from Pelotas occurred in adult life, it is possible to affirm that the deletion at 6p22.1 is a genetic risk factor for current asthma in both age groups. Furthermore, phenotyping was conducted in the present study by using the phase II ISAAC questionnaire on asthma symptoms, a tool that has already been applied in hundreds of studies and has proved to be useful to determine asthma prevalence worldwide40. However, we did not distinguish atopic from non-atopic asthma. Considering that atopic asthma represents a minor proportion of the cases reported in Latin America41 and that the 6p22.1 locus is potentially involved in IgE response30, the associations found in our data sets may be underestimated by phenotypic heterogeneity.
In conclusion, we found robust evidence that CNVs could contribute for asthma susceptibility. More specifically and to the best of our knowledge, for the first time we identified a deletion that confers susceptibility to asthma in Latin American children and young adults. These results uncover a new perspective on the influence of genetic factors modulating asthma risk.
Methods
Study design and populations
Discovery cohort (Salvador)
As previously described34, the SCAALA-Salvador (Social Changes, Asthma and Allergy in Latin America) is one of the three population-based cohorts from the EPIGEN-Brazil initiative on population genomics and genetic epidemiology. Originally, the SCAALA-Salvador is a longitudinal study that comprises children living in Salvador (Bahia State), a city of approximately 3 million inhabitants in Northeastern Brazil. Further details on the original cohort and the procedures for collecting data are described by Barreto and colleagues42.
Replication cohort (Pelotas)
The replication of the association findings was conducted in a cohort of Brazilians from the city of Pelotas, Rio Grande do Sul State. Pelotas is located in the Southern region of Brazil with approximately 340,000 inhabitants. Throughout 1982, the three maternity hospitals in the city were visited daily and births were recorded, corresponding to 99.2% of all births in the city. The live-born infants whose families lived in the urban area constituted the original cohort. Further details on the Pelotas (1982) birth cohort can be seen in Victora and Barros43.
Ethics statement and accordance with guidelines and regulations
The SCAALA-Salvador study was approved by the ethics committee of the Institute of Collective Health (ISC) of the Federal University of Bahia (UFBA). For the Pelotas project, the Ethical Review Board of the Federal University of Pelotas (UFPel) approved all phases of the study. Genotyping of individuals from both cohorts was approved by Brazil’s National Research Ethics Committee (CONEP), as part of the EPIGEN-Brazil project (resolution number: 15895). Informed consent was obtained from all participants at baseline and at all follow-up interviews. Participants signed an informed consent form and authorized their genotyping. All methods and protocols were performed in accordance with the principles of the Declaration of Helsinki.
Definition of asthma symptoms
Definition of asthma symptoms and phenotyping were performed in the same way for both discovery (Salvador) and replication (Pelotas) studies. Parents or caregivers of children from Salvador (resurveyed in 2005, 4–11 years of age) and young adults from Pelotas (resurveyed in 2004, 22–23 years of age) answered Portuguese-adapted questionnaires from The International Study of Asthma and Allergies in Childhood (ISAAC) project40. The interviews were carried out by appropriately trained researchers and individuals were classified as asthmatic when wheezing was reported in the 12 months prior to the questionnaire application and by reporting any one of the following situations: (1) diagnosis of asthma ever; (2) wheezing during exercise in the last 12 months; (3) four or more episodes of wheezing in the last 12 months; or (4) waking up at night because of wheezing in the last 12 months. All other individuals were classified as current non-asthmatics.
SNP genotyping and quality control
Procedures for SNP genotyping and quality control (QC) were extensively described in Kehdy et al.44. Briefly, 1,307 children from Salvador and 1,841 young adults from Pelotas, who fully answered the asthma survey, were successfully genotyped as part of the EPIGEN-Brazil project using the Illumina HumanOmni 2.5–8v1 BeadChip panel (comprising 2,237,482 autosomal SNPs; Illumina, San Diego, CA). Stringent post-genotyping QC procedures and filtering were performed for both populations separately and 1 individual from Salvador and 20 from Pelotas were excluded due to inconsistency between the sex registered and the genetic sex, based on X-chromosome markers (using PLINK v1.945; –check-sex). Fifty seven samples from Salvador and 71 from Pelotas were eliminated from further analysis because of close relationship estimated by kinship coefficients for each pair of individuals, using a method implemented in the REAP software (Relatedness Estimation in Admixed Populations)46. Pairs of individuals were considered closely related if the estimated kinship coefficient between them was ≥0.1. Finally, we eliminated 1 individual from Salvador and 2 from Pelotas presenting more than 1% of undetermined genotypes, using PLINK v1.9 (−mind 0.01). QC was also performed to eliminate autosomal SNPs showing significant deviation from the Hardy-Weinberg equilibrium [p < 10−3 (−hwe 0.001), based on controls only; 56,496 in Salvador and 82,307 in Pelotas] and SNPs with more than 1% of undetermined genotypes (−geno 0.01) in Salvador (112,230) and in Pelotas (99,419). These last two QC stages were also carried out using PLINK v1.9.
Copy number variation calling and quality control
Intensity values from autosomal SNP probes that passed SNP QC were used to detect genomic structural variations based on algorithms implemented in two of the most used programs in the literature for the detection of copy number variations from SNP arrays: PennCNV v1.0.131 and QuantiSNP v2.047. Both PennCNV and QuantiSNP evaluate deviations in signal intensity patterns to identify changes in number of copies of DNA segments.
Two intensity values were obtained for each probe (using Genome Studio software v2011.1): LRR (Log2 of R ratio, where R is the value of the total intensity for the two SNP alleles) and BAF (B allele frequency, a measure of allelic intensity ratio for each SNP). Intensity values were quantile-normalized in order to avoid batch effects. SNP arrays may show variations in hybridization intensity. An algorithm described by Diskin and colleagues48 and implemented in PennCNV (genomic_wave.pl option; -adjust argument) was applied to adjust signal intensity values from samples showing a waveness factor (WF value) less than -0.04 or higher than 0.04.
To limit the occurrence of false discoveries in the initial phase, only CNVs ≥ 1 kb and overlapping at least 5 SNP probes were taken into account49. Considering that telomeric and centromeric regions show excessive spurious CNV calls31, CNVs with at least 1 bp (base pair) overlap with centromeric or telomeric regions (500 kb+/−) were not included in our analyses. Additionally, in MHC region (6:28,510,120–33,480,577, RefSeq: GRCh38), a highly repetitive locus, CNV calls with greater than 70% repeat coverage were excluded. RepeatMasker software (v4.0.6; default options) was used to screen interspersed repeats and low complexity DNA sequences. Following the QC procedures, 235 samples from Salvador were excluded on the basis of large variation in LRR intensities at genome-wide level [standard deviation (SD) >0.20]. Also, 141 samples from Salvador were eliminated from further analysis due to large number of CNVs called (2 SD from the mean) or large CNV sizes (2 SD from the mean). This CNV-based genomic QC was not applied to the Pelotas cohort, since analysis in the replication stage was restricted to the 6p22.1 region.
Definition of copy number variation regions (CNVRs)
In order to combine structural variations corresponding to the same event, the duplications or deletions detected in the genome of the individuals were grouped into copy number variation regions (CNVRs). CNVs overlapping at least 1 base-pair were merged into a single CNVR50, using CNVRuler software51. To avoid overestimation of CNVR size and frequency, regional density (recurrence) of participating CNVs were checked and sparse areas not satisfying the density threshold (10%) were trimmed. Only CNVRs called by both PennCNV and QuantiSNP were considered valid.
Sequence annotations
The regulatory potential of CNVs associated with asthma was evaluated in silico. Comparative genomic data and regulatory features for the region 6:29,881,842–29,931,412 (RefSeq: GRCh38) were obtained from the Ensembl database (http://www.ensembl.org). The position of the deletion at 6p22.1 was cross-referenced with DNA sequence annotations, including: (1) transcripts location (introns, exons, 3′ and 5′ untranslated regions); (2) presence of consensus sequences for transcription factors; (3) genomic evolutionary rate profiling–constrained elements for 40 eutherian mammals (GERP)52; (4) chromatin segmentation state35; and (5) indicative of chromatin accessibility (DNase I hypersensitive sites)35.
Population structure analyses
To explore the admixed nature of our samples, we conducted principal components analysis (PCA) of ancestry, using PLINK v1.9. In Salvador (Supplementary Fig. 1A,B) and Pelotas (Supplementary Fig. 1C,D), only the first three principal components (PCs) account each one for more than 2% of data variance. So, these three more informative PCs were used to adjust for population stratification in the association tests. Additionally, the ADMIXTURE method53 was applied to dissect the ancestry composition of asthma cases and controls (Table 1). Based on the results of ADMIXTURE with number of ancestral clusters (K) = 3, we were able to differentiate the main continental parental groups that contributed to the formation of the Brazilian population: Europeans, Africans and Native Americans. These analyses were previously detailed in Kehdy et al.44.
Statistical analysis
Burden analysis
Burden analyses were conducted to evaluate the global impact of CNVs on asthma outcome. Cases and controls from the discovery cohort were compared in terms of: (1) number of CNVRs per individual (CNVR count); (2) estimated size of CNVRs; (3) number of genes overlapped by a CNVR (at least 1 bp overlapped with any genic region); (4) number of regulatory regions overlapped by a CNVR (at least 1 bp overlapped with regulatory elements: promoter and promoter flanking region, enhancer, open chromatin and transcription factor binding site); and (5) number of constrained elements captured by a CNVR (at least 1 bp overlapped with GERP elements). Size of CNVRs and number of genes, regulatory and constrained regions covered by CNVRs are related to the total for all CNVRs per individual. Gene, regulatory and constrained element annotations were obtained from the Ensembl Biomart tool (http://www.ensembl.org/biomart; Ensembl Genes 88, RefSeq: GRCh38). All comparisons were performed with the non-parametric Mann-Whitney U test (two-sided), using SPSS statistics software v20.0 (IBM). Significance level used in this analysis was α = 0.05.
Association analysis
CNVRs were defined as low-to-common if their frequencies were ≥1% in our cohorts (cases and controls) and only low-to-common variants were evaluated at this point. For the discovery and replication phases, association of CNVRs with asthma risk was evaluated using PLINK v1.9. Distribution of genomic copy number segments was compared between cases and controls under an additive genetic model (0, 1 or 2 allele copies for deletions; 2, 3 or 4 allele copies for duplications). No CNVR with 5 or more allele copies has passed CNV-based QC. Classical risk factors for asthma, such as sex and age, were included as covariates from the logistic regression model. In addition, Log2 of R ratio standard deviation (LRRSD), to account for potential differences in sample and/or call quality between cases and controls, and the first three principal components from PCA (Supplementary Fig, 1A,C), to correct for eventual population stratification, were included in the regression model. Results are described as estimates of odds ratio (OR) and confidence interval (CI). In the discovery phase, a multiple test threshold (Bonferroni) was applied to the p values to control the probability of observing false-positive results. After that, p values ≤ 3.4 × 10−4 (0.05/145) were taken as significant. In the replication study, since only one CNVR was tested, the significance level was α = 0.05. To combine the association results found in both cohorts, a random-effects meta-analysis (assuming inter-study variability) was carried out using PLINK v1.9. A posteriori statistical power was estimated using the GAS Power Calculator tool. Linkage disequilibrium calculations (r2) were conducted using PLINK v1.9. Pearson correlations were carried out using SPSS statistics software v20.0.
Electronic supplementary material
Acknowledgements
We thank Pedro M. Meirelles for his help with the plots of the Figure 1. This work was funded by the Department of Science and Technology (DECIT, Ministry of Health, Brazil), National Fund for Scientific and Technological Development (FNDCT, Ministry of Science and Technology, Brazil), Funding of Studies and Projects (FINEP, Ministry of Science and Technology, Brazil), the Brazilian National Research Council (CNPq). Pablo Oliveira received a post-doctoral fellowship from the CNPq Foundation, the Ministry of Science, Technology, Innovation and Communication, Brazil.
Author Contributions
P.O., R.L.F. and M.L.B. conceived the project. P.O., G.N.O.C., A.K.A.D. and G.C.G.B. performed the burden and association analysis. P.O., F.P.H., C.A.F., R.C.R.-S., A.P., M.F.L.-C., F.S.K., E.T.-S., B.L.H., L.C.R., R.L.F. and M.L.B. participated in the data collection and interpretation of results. All authors contributed to the writing and editing of the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-32837-w.
References
- 1.Fergeson JE, Patel SS, Lockey RF. Acute asthma, prognosis, and treatment. J. Allergy Clin. Immunol. 2017;139:438–447. doi: 10.1016/j.jaci.2016.06.054. [DOI] [PubMed] [Google Scholar]
- 2.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat. Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 3.Martinez FD, Vercelli D. Asthma. Lancet. 2013;382:1360–1372. doi: 10.1016/S0140-6736(13)61536-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.To T, et al. Global asthma prevalence in adults: findings from the cross-sectional world health survey. BMC Public Health. 2012;12:204. doi: 10.1186/1471-2458-12-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asher I, Pearce N. Global burden of asthma among children. Int. J. Tuberc. Lung Dis. 2014;18:1269–1278. doi: 10.5588/ijtld.14.0170. [DOI] [PubMed] [Google Scholar]
- 6.Lai CK, et al. Global variation in the prevalence and severity of asthma symptoms: phase three of the International Study of Asthma and Allergies in Childhood (ISAAC) Thorax. 2009;64:476–483. doi: 10.1136/thx.2008.106609. [DOI] [PubMed] [Google Scholar]
- 7.Pearce N, et al. Worldwide trends in the prevalence of asthma symptoms: phase III of the International Study of Asthma and Allergies in Childhood (ISAAC) Thorax. 2007;62:758–766. doi: 10.1136/thx.2006.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solé D, et al. Changes in the prevalence of asthma and allergic diseases among Brazilian schoolchildren (13-14 years old): comparison between ISAAC Phases One and Three. J. Trop. Pediatr. 2007;53:13–21. doi: 10.1093/tropej/fml044. [DOI] [PubMed] [Google Scholar]
- 9.Devereux G, Seaton A. Diet as a risk factor for atopy and asthma. J. Allergy Clin. Immunol. 2005;115:1109–1117. doi: 10.1016/j.jaci.2004.12.1139. [DOI] [PubMed] [Google Scholar]
- 10.Huang YJ, Boushey HA. The microbiome in asthma. J. Allergy Clin. Immunol. 2015;135:25–30. doi: 10.1016/j.jaci.2014.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper PJ, et al. Risk factors for asthma and allergy associated with urban migration: background and methodology of a cross-sectional study in Afro-Ecuadorian school children in Northeastern Ecuador (Esmeraldas-SCAALA Study) BMC Pulm. Med. 2006;6:24. doi: 10.1186/1471-2466-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rook GA. The hygiene hypothesis and the increasing prevalence of chronic inflammatory disorders. Trans. R. Soc. Trop. Med. Hyg. 2007;101:1072–1074. doi: 10.1016/j.trstmh.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 13.Ober C, Hoffjan S. Asthma genetics 2006: the long and winding road to gene discovery. Genes Immun. 2006;7:95–100. doi: 10.1038/sj.gene.6364284. [DOI] [PubMed] [Google Scholar]
- 14.Meyers DA. Genetics of asthma and allergy: what have we learned? J. Allergy Clin. Immunol. 2010;126:439–446. doi: 10.1016/j.jaci.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moffatt MF, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 16.Himes BE, et al. Genome-wide association analysis identifies PDE4D as an asthma-susceptibility gene. Am. J. Hum. Genet. 2009;84:581–593. doi: 10.1016/j.ajhg.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sleiman PM, et al. Variants of DENND1B associated with asthma in children. N. Engl. J. Med. 2010;362:36–44. doi: 10.1056/NEJMoa0901867. [DOI] [PubMed] [Google Scholar]
- 18.Meyers DA, Bleecker ER, Holloway JW, Holgate ST. Asthma genetics and personalised medicine. Lancet Respir. Med. 2014;2:405–415. doi: 10.1016/S2213-2600(14)70012-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa GN, et al. A genome-wide association study of asthma symptoms in Latin American children. BMC Genet. 2015;16:141. doi: 10.1186/s12863-015-0296-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith D, et al. A rare IL33 loss-of-function mutation reduces blood eosinophil counts and protects from asthma. PLoS Genet. 2017;13:e1006659. doi: 10.1371/journal.pgen.1006659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ober C, Yao TC. The genetics of asthma and allergic disease: a 21st century perspective. Immunol. Rev. 2011;242:10–30. doi: 10.1111/j.1600-065X.2011.01029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat. Rev. Genet. 2015;16:172–183. doi: 10.1038/nrg3871. [DOI] [PubMed] [Google Scholar]
- 23.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grayson BL, et al. Genome-wide analysis of copy number variation in type 1 diabetes. PLoS One. 2010;5:e15393. doi: 10.1371/journal.pone.0015393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wellcome Trust Case Control Consortium et al. Genome-wide association study of CNVs in 16,000 cases of eight common diseases and 3,000 shared controls. Nature464, 713–720 (2010). [DOI] [PMC free article] [PubMed]
- 26.Uddin M, Sturge M, Rahman P, Woods MO. Autosome-wide copy number variation association analysis for rheumatoid arthritis using the WTCCC high-density SNP genotype data. J. Rheumatol. 2011;38:797–801. doi: 10.3899/jrheum.100758. [DOI] [PubMed] [Google Scholar]
- 27.Ferreira MA, et al. Association between ORMDL3, IL1RL1 and a deletion on chromosome 17q21 with asthma risk in Australia. Eur. J. Hum. Genet. 2011;19:458–464. doi: 10.1038/ejhg.2010.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rogers AJ, et al. Copy number variation prevalence in known asthma genes and their impact on asthma susceptibility. Clin. Exp. Allergy. 2013;43:455–462. doi: 10.1111/cea.12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vishweswaraiah, S. et al. Copy number variation burden on asthma subgenome in normal cohorts identifies susceptibility markers. Allergy Asthma Immunol Res. 7, 265–275 (2015). [DOI] [PMC free article] [PubMed]
- 30.Granada M, et al. A genome-wide association study of plasma total IgE concentrations in the Framingham Heart Study. J. Allergy Clin. Immunol. 2012;129:840–845. doi: 10.1016/j.jaci.2011.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang K, et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 2007;17:1665–1674. doi: 10.1101/gr.6861907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature455, 237–241 (2008). [DOI] [PMC free article] [PubMed]
- 33.Wheeler E, et al. Genome-wide SNP and CNV analysis identifies common and low-frequency variants associated with severe early-onset obesity. Nat. Genet. 2013;45:513–517. doi: 10.1038/ng.2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lima-Costa MF, et al. Genomic ancestry and ethnoracial self-classification based on 5,871 community-dwelling Brazilians (The Epigen Initiative) Sci. Rep. 2015;5:9812. doi: 10.1038/srep09812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature489, 57–74 (2012). [DOI] [PMC free article] [PubMed]
- 36.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 37.Morandi F, Rizzo R, Fainardi E, Rouas-Freiss N, Pistoia V. Recent Advances in Our Understanding of HLA-G Biology: Lessons from a Wide Spectrum of Human Diseases. J. Immunol. Res. 2016;2016:4326495. doi: 10.1155/2016/4326495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Demenais F, et al. Multiancestry association study identifies new asthma risk loci that colocalize with immune-cell enhancer marks. Nat. Genet. 2018;50:42–53. doi: 10.1038/s41588-017-0014-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirota T, et al. Genome-wide association study identifies three new susceptibility loci for adult asthma in the Japanese population. Nat. Genet. 2011;43:893–896. doi: 10.1038/ng.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Asher MI, et al. International Study of Asthma and Allergies in Childhood (ISAAC): rationale and methods. Eur. Respir. J. 1995;8:483–491. doi: 10.1183/09031936.95.08030483. [DOI] [PubMed] [Google Scholar]
- 41.Weinmayr G, et al. Atopic Sensitization and the International Variation of Asthma Symptom Prevalence in Children. Am. J. Respir. Crit. Care Med. 2007;176:565–574. doi: 10.1164/rccm.200607-994OC. [DOI] [PubMed] [Google Scholar]
- 42.Barreto ML, et al. Risk factors and immunological pathways for asthma and other allergic diseases in children: background and methodology of a longitudinal study in a large urban center in Northeastern Brazil (SCAALA-Salvador study) BMC Pulm. Med. 2006;6:15. doi: 10.1186/1471-2466-6-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Victora CG, Barros FC. Cohort profile: the 1982 Pelotas (Brazil) birth cohort study. Int. J. Epidemiol. 2006;35:237–242. doi: 10.1093/ije/dyi290. [DOI] [PubMed] [Google Scholar]
- 44.Kehdy FS, et al. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc. Natl. Acad. Sci. USA. 2015;112:8696–8701. doi: 10.1073/pnas.1504447112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang CC, et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience. 2015;4:7. doi: 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thornton T, et al. Estimating Kinship in Admixed Populations. Am. J. Hum. Genet. 2012;91:122–138. doi: 10.1016/j.ajhg.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Colella S, et al. QuantiSNP: an Objective Bayes Hidden-Markov Model to detect and accurately map copy number variation using SNP genotyping data. Nucleic Acids Res. 2007;35:2013–2025. doi: 10.1093/nar/gkm076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diskin SJ, et al. Adjustment of genomic waves in signal intensities from whole-genome SNP genotyping platforms. Nucleic Acids Res. 2008;36:e126. doi: 10.1093/nar/gkn556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carter NP. Methods and strategies for analyzing copy number variation using DNA microarrays. Nat. Genet. 2007;39:S16–21. doi: 10.1038/ng2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim JH, et al. CNVRuler: a copy number variation-based case–control association analysis tool. Bioinformatics. 2012;28:1790–1792. doi: 10.1093/bioinformatics/bts239. [DOI] [PubMed] [Google Scholar]
- 52.Cooper GM, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–1664. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.