Abstract
Background: MicroRNA-497(miR-497) has been studied for its irreplaceable role of predicting the prognosis of various cancers, but there has been no systematic study to summarize the data. Consequently, we performed this meta-analysis to reveal the association between the expression level of miR-497 and cancer prognosis systematically.
Materials and Methods: PubMed was searched for appropriate studies and a total of 12 eligible publications with 989 cancer patients were recruited into our analysis to assess the strength of the association. Hazard ratios (HRs) and odds ratios (ORs) were analyzed to finish this work.
Results: The cancer patients who have high expressing level of miR-497 are less possible to have lymph node metastasis (OR = 0.25, 95% CI: 0.16-0.40, P < 0.001) and more likely to have favourable tumor-node-metastasis stage (OR = 0.29, 95% CI: 0.17-0.49, P < 0.001). Also, high miR-497 expression level was notably connected to better overall survival (pooled HR = 0.41, 95% CI: 0.32-0.53, P < 0.001).
Conclusions: High expressing levels of miR-497 might be a potential biomarker which can be used to predict the better prognosis of different cancer types.
Keywords: prognosis, biomarker, microRNA-497, cancer, meta-analysis
Introduction
Although the amount of deaths owing to cancers continued to decline in recent years, in both developing and developed countries, the main cause of death is cancer especially in less developed regions which have the 82% of the world's population 1,2. In China, it is estimated that a total of 4292000 new cases of cancer and 2814000 deaths due to cancers had occurred in 20153. Although the previous diagnostic and treatment modalities have been greatly improved in recent years, the 5-year survival rate for patients with different cancer types still remains low 4. Patients will get better survival chances and higher quality of life, if cancers can be observed early and effective treatment can be given promptly. But when the patients are diagnosed with advanced cancer stage, the effectiveness of the treatment is limited. Therefore, there is urgent necessity for a new biomarker to predict tumor metastasis and prognosis.
A family of endogenous small non-coding RNAs has been named as MicroRNAs (miRNAs), whose target mRNAs expression have been suppressed by blinding to the 3'-UTR on the basis of both complementary sequence and potential target site. MiRNAs will inhibit target mRNAs translation or degradation to negatively regulate gene expression 5,6. There is a massive amount of evidence showed that many biological procedures for example cell proliferation, cell cycle, apoptosis, migration, invasion, and differentiation can be regulated by miRNAs 6,7. In addition, the emerging evidence suggested that miRNAs may play an important role in carcinogenesis or tumor suppression in humans 8,9.
It shows that the first intron of the MIR497HG (Gene ID: 10050675) gene on human chromosome 17p13.1 has encoded the MicroRNA-497 (miR-497) which is a highly conserved miRNA 10. MiR-497 was first reported in human breast cancer 11. Loss of miR-497 expression has been reported in many cancer types, whereas restoration of miR-497 expression has been shown to abrogate tumorigenesis 12-14. Also, several targets of miR-497 have been identified previously, for example, CCND1, CCNE1, Bcl-w and BDNF 15-18. Plenty of studies have discovered the down-regulation of miR-497 in various types of cancers, containing ovarian cancer 19,20, gastric cancer 21,22, clear cell renal cell carcinoma 23, diffuse large B-cell lymphoma 24, non-small cell lung cancer 25, hepatocellular carcinoma 26, osteosarcoma 27,28, gliomas29, thyroid cancer 30, renal cell carcinoma 31. Until now, numerous studies have demonstrated that clinicopathological features and the patients' overall survival time in various types of cancers are closely associated with the dysregulation of miR-497. However, most individual studies have their limits, for example, the sample size is small or obtaining controversial results, and so on. To further reveal the association between the miR-497 expression level and the clinical prognosis of cancer patients, we performed this meta-analysis.
Materials and Methods
Periodical search
We retrieved the PubMed database for reports published in English up to 31 January 2018. The following key words were used for the research: “microRNA-497” or “miR-497” and “cancer” or “carcinoma”. Primary publication in reference was also considered to acquire excess concerned articles.
Inclusion and exclusion criteria
If the study met the next criteria, would be included in this article: (1) the published articles were in English; (2) analyzing the prognostic role of miR-497 in cancer patients; (3) grouping the patients according to the expressing level of miR-497; (4) containing the association between clinicopathological parameters and miR-497 expression level; (5) providing adequate statistics to conjecture HRs with a corresponding 95% CI for OS. Research, reviews and case reports that did not provide enough data will not be contained in this meta-analysis. If the similar content is available more than once, only the most recent publications are contained.
Data extraction and quality valuation
Each study was revised by two authors, and they extracted enough valuable data based on the above criteria. The information below was gathered from the contained studies: the cognomen of first author; publication year; cancer types; nation; detecting miR-497 methods; over-all number of patients; quantity of LNM patients; the number of patients with different TNM stage in high and low miR-497 expression level groups; cut-off value; the follow-up time; HRs and the corresponding 95% CI for OS. Multivariate analysis effects are preferred, when the article provided both univariate and multivariate analysis. Any divergence was solved by reaching a consensus with other authors. To assess the quality of the pooled study, we performed the Newcastle-Ottawa Scale (NOS). High quality required an NOS score ≥6.
Statistical methods
The association between the expressing level of miR-497 and clinical prognosis was assessed by using ORs or HRs with 95% CIs. The heterogeneity contained in the pooled studies was evaluated by I2 based Q test and I2 index. We prioritized the use of fixed-effects models, if the Q test showed no significant heterogeneity in qualitative studies, defined as P heterogeneity > 0.05 and / or I2 < 50% 32. Else, we used a random-effects model 33. We conducted sensitivity analysis to assess the contribution of the individual study to the pooled HR, so that, we could estimate the stability of our results. The Kaplan-Meier curve provided the HRs and 95% CIs for OS with the help of the Engauge Digitizer 4.1 software 34. We evaluate the publication bias through the Begg's funnel plot, and P < 0.05 means obvious publication bias 35. Stata software was used to complete the statistical analyses (version 13.0, Stata Corp, College Station, USA).
Consequences
Features of studying
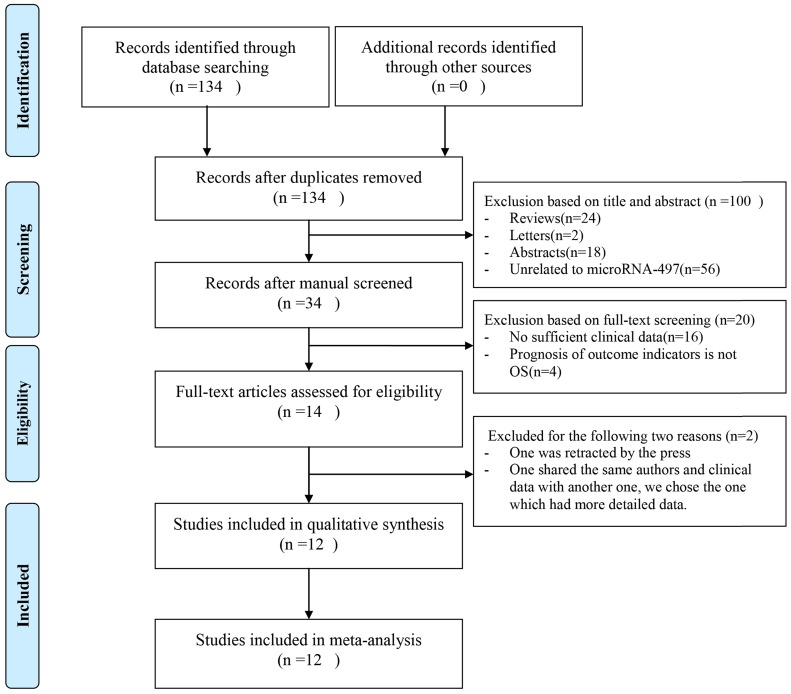
As shown in Figure 1, 134 publications were acquired from PubMed databases. 120 publications were excluded because of the lack of necessary data that we need and 14 publications were selected for further full-text review after we reviewed the abstracts and the full texts. One of the left 14 publications was retracted by the press, so it was excluded. In the left 13 publications, two publications were written by the same author and shared the same clinical data, so we chose the one which was the most recent and had more detailed data that we needed. At last, 12 studies were enrolled. Among these studies, the cancer types were ovarian cancer 19, gastric cancer 21,22, clear cell renal cell carcinoma 23, diffuse large B-cell lymphoma 24, non-small cell lung cancer 25, hepatocellular carcinoma 26, osteosarcoma 27,28, gliomas 29, thyroid cancer 30, renal cell carcinoma 31. In table 1, the features of the involved studies were indicated.
Figure 1.
Flowchart of included publications.
Table 1.
Characteristics of studies included in this meta-analysis
| Study | Country | Year | Cancer type | MicroRNA-497 expression | MicroRNA-497 assay | HR statistic | HR (95%CI) high/low | Survival analysis | NOS | Cut-off value | Follow-up (months) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | ||||||||||||||||
| Total | LNM | Ⅲ/Ⅳ | Total | LNM | Ⅲ/Ⅳ | ||||||||||||
| Wang 19 | China | 2014 | OC | 38 | 17 | 27 | 58 | 40 | 38 | qRT-PCR | Survival curve | 0.92 (0.42-2.01) | Univariate | 8 | - | 60 | |
| Li 21 | China | 2014 | GC | 48 | 22 | 24 | 49 | 36 | 30 | qRT-PCR | Survival curve | 0.37 (0.20-0.68) | Univariate | 8 | Median value | 60 | |
| Zhao 23 | China | 2015 | ccRCC | 38 | 2 | 8 | 48 | 11 | 26 | qRT-PCR | Date in paper | 0.39 (0.16-0.59) | Multivariate | 8 | Mean level | 60 | |
| Katharina 24 | Austria | 2015 | DLBCL | 29 | - | 16 | 29 | - | 17 | qRT-PCR | Date in paper | 0.36 (0.12-1.01) | Univariate | 7 | Median value | 120 | |
| Huang 25 | China | 2015 | NSCLC | 25 | 7 | - | 26 | 13 | - | qRT-PCR | Survival curve | 0.44 (0.22-0.91) | Univariate | 7 | Median value | 60 | |
| Zhang 26 | China | 2016 | HCC | 43 | - | 4 | 43 | - | 15 | qRT-PCR | Date in paper | 0.47 (0.28-0.81) | Multivariate | 7 | Median value | 60 | |
| Pang 27 | China | 2016 | OS | 94 | - | 20 | 91 | - | 54 | qRT-PCR | Date in paper | 0.26 (0.12-0.50) | Multivariate | 7 | Median value | 70 | |
| Feng 29 | China | 2016 | Gliomas | 52 | - | 16 | 58 | - | 48 | qRT-PCR | Date in paper | 0.30 (0.14-0.91) | Multivariate | 7 | Median value | 60 | |
| Song 28 | China | 2017 | OS | 19 | - | 3 | 27 | - | 16 | qRT-PCR | - | - | - | 6 | Mean level | 72 | |
| Liu 22 | China | 2017 | GC | 38 | - | 17 | 48 | - | 34 | qRT-PCR | - | - | - | 6 | Mean level | - | |
| Wang 30 | China | 2017 | TC | 21 | 1 | 2 | 27 | 14 | 15 | qRT-PCR | - | - | - | 7 | Mean level | - | |
| Sun 31 | China | 2017 | RCC | 14 | 1 | 2 | 26 | 12 | 10 | qRT-PCR | - | - | - | 7 | - | - | |
Note: The dashes mean no data.
III/IV: TNM stage III/IV; LNM: lymph node metastasis; OC: ovarian cancer; GC: gastric cancer; ccRCC: clear cell renal cell carcinoma; DLBL: diffuse large B-Cell lymphoma; NSCLC: non-small cell lung cancer; HCC: hepatocellular carcinoma; OS: osteosarcoma; TC: thyroid cancer; RCC: renal cell carcinoma
Association between expression level of miR-497 and TNM stage (III/IV vs. I/II)
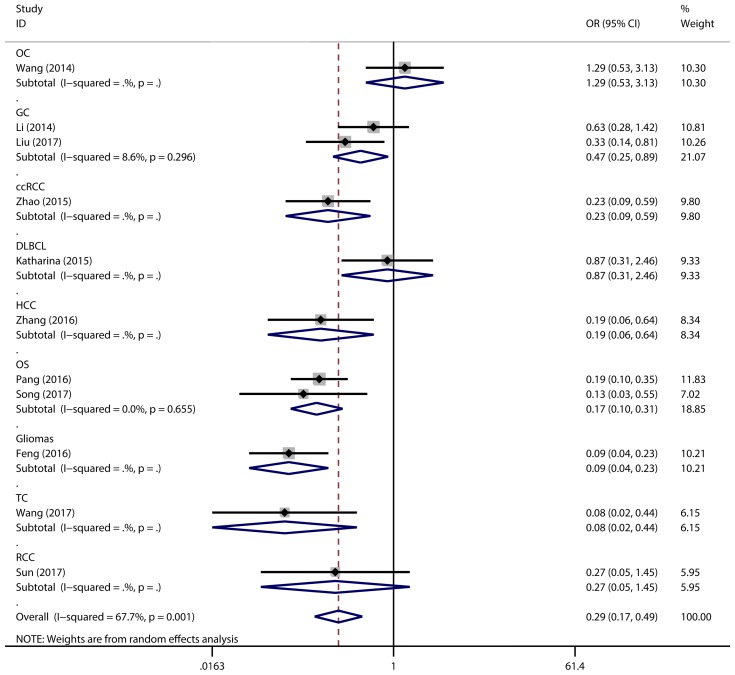
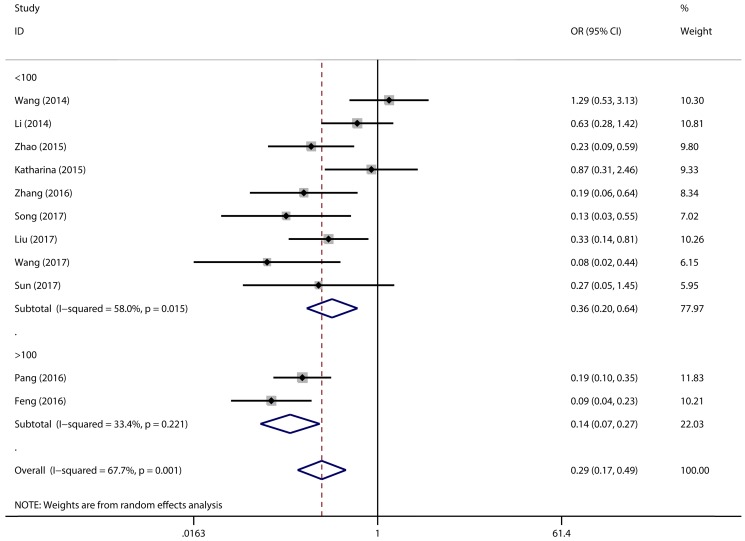
Figure 2 has showed that a random-effects model was used to analyze the pooled odds ratio (OR) and its 95% CI, as the result of the obvious heterogeneity among these eleven reports (I2 = 67.7%, P = 0.001). Consequently, we detected that the high expressing level of microRNA-497 (miR-497) was predictive of favorable tumour-node-metastasis (TNM) stage (OR = 0.29, 95% CI: 0.17-0.49, P < 0.001). Because of the discovery of the heterogeneity, we performed the subgroup analysis of the data by cancer types and sample size. The results showed that the high miR-497 expressing level was related with lower TNM stage in gastric cancer (OR = 0.47, 95% CI: 0.25-0.89, P = 0.02), osteosarcoma (OR = 0.17, 95% CI: 0.10-0.31, P < 0.001), but not existed in ovarian cancer (OR = 1.29, 95% CI: 0.53-3.13, P = 0.57). At the same time, we performed another subgroup analysis by sample sizes. The sample size of the contained studies ranged from 40 to 185. We find a significant association between TNM stage and the miR-497 expression level in a subgroup with simple sizes > 100 (OR = 0.14, 95% CI: 0.07-0.27, P < 0.001) (Figure 3). During our work, if the 50 and 90 were chosen as the classification criteria for the sample size, we found an obvious heterogeneity in the smaller sample size group (data not shown).
Figure 2.
Forest plot for the association between expressing levels of miR-497 and TNM stage: subgroup analysis by cancer type.
Figure 3.
Forest plot for the association between expressing levels of miR-497 and TNM stage: subgroup analysis by sample size.
Association between miR-497 expression level and lymph node metastasis
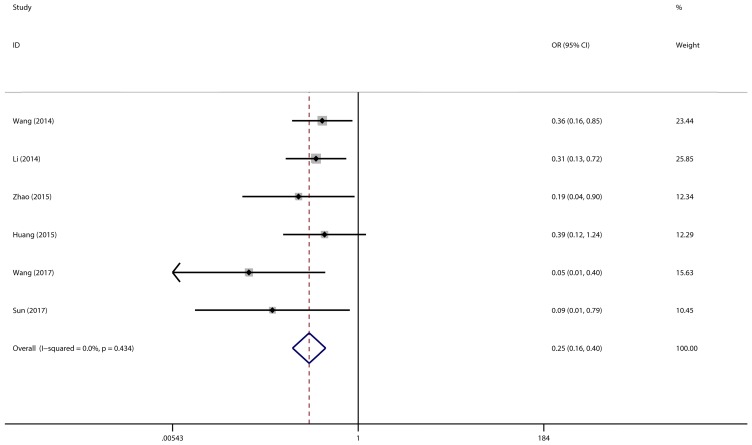
A total of 6 studies with 418 cancer patients were involved in this lymph node metastasis (LNM) analysis. We chose a fixed-effect model to complete the comparison between high miR-497 expression level and low miR-497 expression level (I2 = 0.0%, P = 0.434). The results showed that patients with low miR-497 expression level were more likely to have lymph node metastases (OR = 0.25, 95% CI: 0.16-0.40, P < 0.001) (Figure 4).
Figure 4.
Forest plot for the association between miR-497 and LNM.
Association between expression level of miR-497 and overall survival
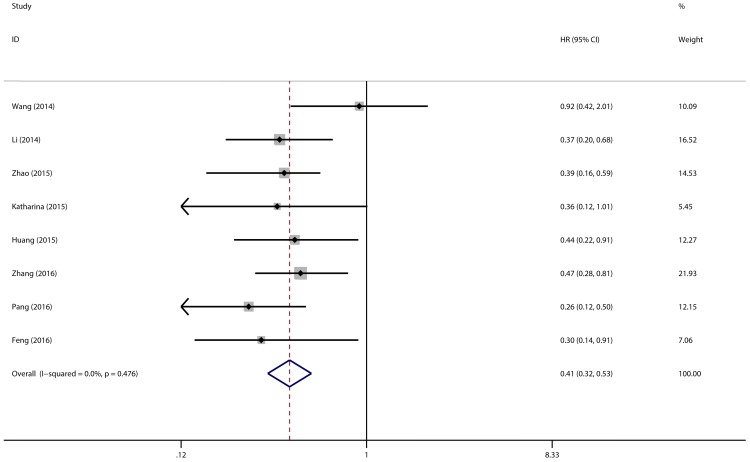
Eight studies with 769 patients were included to gain a pooled HR and 95% CI for overall survival (OS) in various types of cancer. In this analysis, a fixed-effects model had been adopted because no heterogeneity was discovered in the OS group (I2 = 0.0%, P = 0.476). Through this analysis, we discovered a significant association between high expression level of miR-497 and better overall survival (pooled HR = 0.41, 95% CI: 0.32-0.53, P < 0.001) (Figure 5).
Figure 5.
Forest plot of pooled HRs of high expression level of miR-497 for OS.
Sensitivity analysis and publication bias
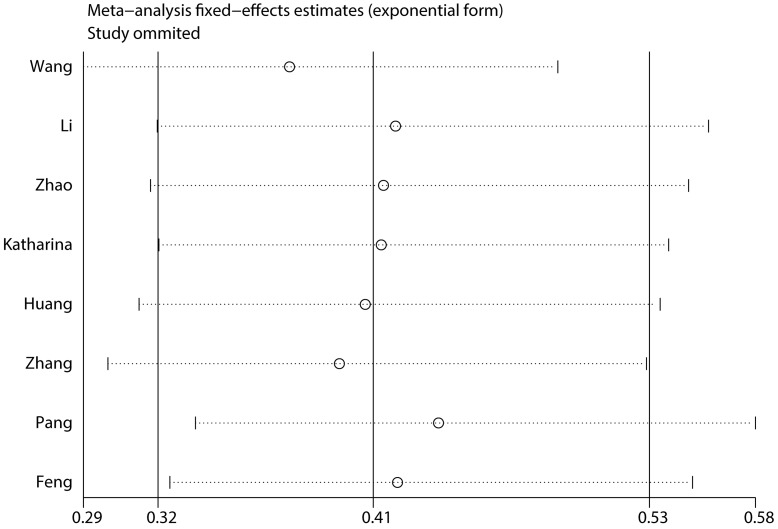
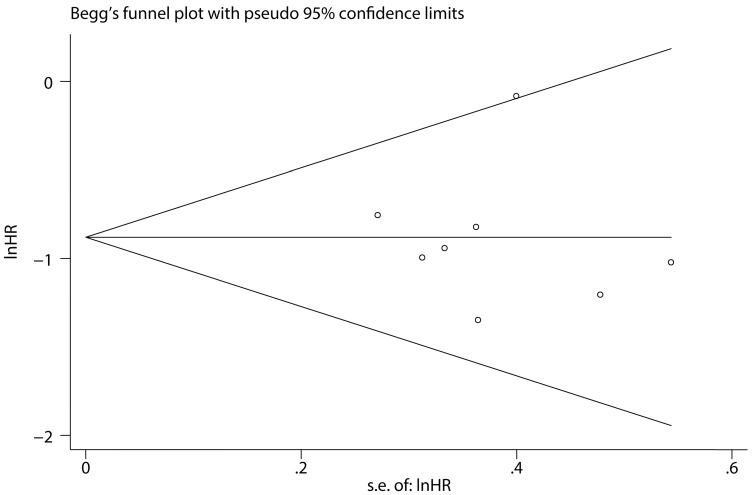
To assess the steadiness and reliability of our meta-analysis, we analyzed the sensitivity of the OS group through the way that we omitted each study from our analysis one at a time. The results showed no individual study affected the pooled HR (Figure 6). Furthermore, analysis of sensitivity indicated that there were no significant effects on the association between the pooled OR for elevated miR-497 and TNM stage or LNM when we excluded any of the studies (data not shown). Publication bias for the meta-analysis was assessed by Begg's funnel plot analysis (P = 0.711) and Egger's linear regression test was used to verify the accuracy of the Begg's funnel plot (P = 0.804). No significant publication bias was found through the two tests (Figure 7).
Figure 6.
Consequence of sensitivity analysis
Figure 7.
Funnel plot analysis of potential publication bias (Begg's test).
Discussion
As one of small regulatory RNA molecules, miRNA can moderate the expression of their target genes. MiRNAs can blind to the 3'UTR and 5'UTR of their target mRNAs so that they can mediate degradation or translational repression of the mRNAs 36. Studies have demonstrated that miRNAs have broad effects on numerous biological processes, including differentiation, metastasis, apoptosis, metabolism and maturation 37,38. The importance of miRNAs expression levels in various cancers and their potential utility as prognostic factors have become apparent. Plenty of evidence has suggested that miRNAs can regulate tumor proliferation and invasion, and they are widely analyzed in diagnosis, therapies and prognosis of various cancers 39,40. A quantity of miRNAs have been discovered to be associated with the clinical characteristics of various cancers, due to the obvious impact of miRNAs on gene expression. These miRNAs include miR-21841, miR-21 42, and miR-539 43. This means that miRNAs have the potential value to become the tumor biomarker, forecasting prognosis, therapeutic efficacy.
MiR-497 has been studied for many years, but it is still a popular studying topic currently. Previous studies have suggested that miR-497 was a tumor-associated miRNA in various cancers such as oral colorectal cancer, non-small cell lung cancer and squamous cell carcinoma et al 44-46. The mechanism by which miR-497 promotes tumor progression has been complicated, also, more studies are needed to reveal this. Liu et al. 47 found that miR-497 might promote tumor progression in breast cancer, which could target SMAD family number 7 (SMAD7) mRNA to inhibit the breast cancer cell growth and invasion. Xu et al.48 found that miR-497 served a significant role in suppressing colorectal cancer (CRC) cell growth via inhibition of insulin receptor substrate 1 (IRS1). Yu et al.49 discovered that miR-497 was down-regulated in samples of multiple myeloma (MM) and cell lines, also, it could directly target pre-B-cellleukemia transcription factor 3 (PBX3) to promote the malignant progression in MM. Zhang et al.50 found that miR-497 suppresses the proliferation and invasion of the bladder transitional cell carcinoma (BTCC) cells, possibly through modulating the target gene E2F transcription factor 3 (E2F3).
Recently, a variety of reports have suggested the association between the expressing level of miR-497 and the various cancers prognosis. To verify the potential predictive value of miR-497 expression level, we conducted this meta-analysis of 12 studies including 989 cancer patients. We noticed that the lower expressing level of miR-497 significantly means unfavorable LNM and TNM stage for cancer patients. Through merging HRs from included studies, we noticed that the higher expressing level of miR-497 was associated with the longer overall survival time. Also, patients with higher level expressing of miR-497 had longer OS time compared with those with lower expressing levels. Yet, our meta-analysis is not impeccable because of the limitation of the data in the included publications. Because of the lack of some necessary data, we were not allowed to perform the meta-analysis to assess the association between the miR-497 expression level and survival without disease, survival without progression, survival without event. Obvious heterogeneity was discovered when we conducted the TNM stage group analysis. In order to find out the origin of heterogeneity, we performed an analysis in subgroups by cancer types and size of sample. No obvious heterogeneity was found in the subgroup with sample size > 100, and when we left the Wang 19 study out of the subgroup with size of sample < 100. There is also no observation of significant heterogeneity. So we reflected that the main source of heterogeneity was the variance in size of sample and the Wang 19 study. In his study, the sample size of patients with low expression is much larger than the ones with high expression. Also, we considered that Wang 19 study has more distinct individual variation than the other included studies. However, we could not be very sure the origin of the heterogeneity because the entire sample size of the involved studies is small. In a word, through our meta-analysis, we found that miR-497 might be vital in tumor progression and could be very valuable to work as a new biomarker to predict clinical outcomes in patients with cancer.
As far as we know, our work is the first study to use meta-analysis to assess the clinical value of the expressing level of miR-497 in cancer patients. Nonetheless, our study also had the limitation that cannot be ignored. Firstly, only 12 studies with 989 patients were pooled in our analysis, so it is less reliable to some extent. Secondly, because of the limited publications, we ignored the subgroup analysis according to age, sex, and so on, but cancer progression is associated with many factors. Thirdly, direct access to some HRs in the study cannot be achieved, so we had to estimate and extract HRs through the survival curves or calculate HRs through the reported data. This reduced the credibility of our results. Last but not least, the cut-off value distinguishing high or low levels of miR-497 differed from these studies. To solve this, more studies and larger sample sizes were needed.
In summary, as shown in the results of our meta-analysis that high miR-497 expression level in different cancer types is significantly associated with favourable TNM stage, no LNM, and longer OS time. Therefore, miR-497 may become a new and valuable biomarker for predicting well prognosis in cancer patients. We hope that this result can be paid enough attention to, so that, more cancer patients can benefit from our scientific research.
References
- 1.Jemal A, Ward EM, Johnson CJ, Cronin KA, Ma J, Ryerson AB, Mariotto A, Lake AJ, Wilson R, Sherman RL, Anderson RN, Henley SJ, Kohler BA, Penberthy L, Feuer EJ, Weir HK. Annual Report to the Nation on the Status of Cancer, 1975-2014, Featuring Survival. JNCI: Journal of the National Cancer Institute; 2017. p. 109. (9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA: A Cancer Journal for Clinicians. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA: A Cancer Journal for Clinicians. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 4.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. INT J CANCER. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 5.de Planell-Saguer M, Rodicio MC. Analytical aspects of microRNA in diagnostics: A review. ANAL CHIM ACTA. 2011;699(2):134–152. doi: 10.1016/j.aca.2011.05.025. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. CELL. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. DEV CELL. 2006;11:441–450. doi: 10.1016/j.devcel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Calin GA, Croce CM. MicroRNA signatures in human cancers. NAT REV CANCER. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. DEV BIOL. 2007;302(1):1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 10.Itesako T, Seki N, Yoshino H, Chiyomaru T, Yamasaki T, Hidaka H, Yonezawa T, Nohata N, Kinoshita T, Nakagawa M, Enokida H. The microRNA expression signature of bladder cancer by deep sequencing: the functional significance of the miR-195/497 cluster. PLOS ONE. 2014;9(2):e84311. doi: 10.1371/journal.pone.0084311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, Zeng YX, Shao JY. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14(11):2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo ST, Jiang CC, Wang GP, Li YP, Wang CY, Guo XY, Yang RH, Feng Y, Wang FH, Tseng HY, Thorne RF, Jin L, Zhang XD. MicroRNA-497 targets insulin-like growth factor 1 receptor and has a tumour suppressive role in human colorectal cancer. ONCOGENE. 2013;32(15):1910–1920. doi: 10.1038/onc.2012.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu J, Wang T, Cao Z, Huang H, Li J, Liu W, Liu S, You L, Zhou L, Zhang T, Zhao Y. MiR-497 downregulation contributes to the malignancy of pancreatic cancer and associates with a poor prognosis. ONCOTARGET. 2014;5(16):6983–6993. doi: 10.18632/oncotarget.2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li D, Zhao Y, Liu C, Chen X, Qi Y, Jiang Y, Zou C, Zhang X, Liu S, Wang X, Zhao D, Sun Q, Zeng Z, Dress A, Lin MC, Kung HF. et al. Analysis of MiR-195 and MiR-497 expression, regulation and role in breast cancer. CLIN CANCER RES. 2011;17(7):1722–1730. doi: 10.1158/1078-0432.CCR-10-1800. [DOI] [PubMed] [Google Scholar]
- 15.Lehmann U, Streichert T, Otto B, Albat C, Hasemeier B, Christgen H, Schipper E, Hille U, Kreipe HH, Langer F. Identification of differentially expressed microRNAs in human male breast cancer. BMC CANCER. 2010;10:109. doi: 10.1186/1471-2407-10-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen L, Li J, Xu L, Ma J, Li H, Xiao X, Zhao J, Fang L. miR-497 induces apoptosis of breast cancer cells by targeting Bcl-w. EXP THER MED. 2012;3(3):475–480. doi: 10.3892/etm.2011.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo Q, Li X, Gao Y, Long Y, Chen L, Huang Y, Fang L. MiRNA-497 regulates cell growth and invasion by targeting cyclin E1 in breast cancer. CANCER CELL INT. 2013;13(1):95. doi: 10.1186/1475-2867-13-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang P, Meng X, Huang Y, Lv Z, Liu J, Wang G, Meng W, Xue S, Zhang Q, Zhang P, Chen G. MicroRNA-497 inhibits thyroid cancer tumor growth and invasion by suppressing BDNF. ONCOTARGET. 2017;8(2):2825–2834. doi: 10.18632/oncotarget.13747. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Wang W, Ren F, Wu Q, Jiang D, Li H, Peng Z, Wang J, Shi H. MicroRNA-497 inhibition of ovarian cancer cell migration and invasion through targeting of SMAD specific E3 ubiquitin protein ligase 1. BIOCHEM BIOPH RES CO. 2014;449(4):432–437. doi: 10.1016/j.bbrc.2014.05.053. [DOI] [PubMed] [Google Scholar]
- 20.Wang W, Ren F, Wu Q, Jiang D, Li H, Shi H. MicroRNA-497 suppresses angiogenesis by targeting vascular endothelial growth factor A through the PI3K/AKT and MAPK/ERK pathways in ovarian cancer. ONCOL REP. 2014;32(5):2127–2133. doi: 10.3892/or.2014.3439. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Jin X, Deng X, Zhang G, Zhang B, Ma L. The putative tumor suppressor microRNA-497 modulates gastric cancer cell proliferation and invasion by repressing eIF4E. BIOCHEM BIOPH RES CO. 2014;449(2):235–240. doi: 10.1016/j.bbrc.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Li Y, Zou Y, Zhang J, An J, Guo J, Ma M, Dai D. MicroRNA-497 acts as a tumor suppressor in gastric cancer and is downregulated by DNA methylation. ONCOL REP. 2017;38(1):497–505. doi: 10.3892/or.2017.5698. [DOI] [PubMed] [Google Scholar]
- 23.Zhao X, Zhao Z, Xu W, Hou J, Du X. Down-regulation of miR-497 is associated with poor prognosis in renal cancer. Int J Clin Exp Pathol. 2015;8(1):758–764. [PMC free article] [PubMed] [Google Scholar]
- 24.Troppan K, Wenzl K, Pichler M, Pursche B, Schwarzenbacher D, Feichtinger J, Thallinger G, Beham-Schmid C, Neumeister P, Deutsch A. miR-199a and miR-497 Are Associated with Better Overall Survival due to Increased Chemosensitivity in Diffuse Large B-Cell Lymphoma Patients. INT J MOL SCI. 2015;16(8):18077–18095. doi: 10.3390/ijms160818077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang C, Ma R, Yue J, Li N, Li Z, Qi D. MiR-497 Suppresses YAP1 and Inhibits Tumor Growth in Non-Small Cell Lung Cancer. CELL PHYSIOL BIOCHEM. 2015;37(1):342–352. doi: 10.1159/000430358. [DOI] [PubMed] [Google Scholar]
- 26.Zhang L, Yu Z, Xian Y, Lin X. microRNA-497 inhibits cell proliferation and induces apoptosis by targeting YAP1 in human hepatocellular carcinoma. FEBS OPEN BIO. 2016;6(2):155–164. doi: 10.1002/2211-5463.12032. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Pang PC, Shi XY, Huang WL, Sun K. miR-497 as a potential serum biomarker for the diagnosis and prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2016;20(18):3765–3769. [PubMed] [Google Scholar]
- 28.Song J, Wu X, Liu F, Li M, Sun Y, Wang Y, Wang C, Zhu K, Jia X, Wang B, Ma X. Long non-coding RNA PVT1 promotes glycolysis and tumor progression by regulating miR-497/HK2 axis in osteosarcoma. BIOCHEM BIOPH RES CO. 2017;490(2):217–224. doi: 10.1016/j.bbrc.2017.06.024. [DOI] [PubMed] [Google Scholar]
- 29.Feng F, Kuai D, Wang H, Li T, Miao W, Liu Y, Fan Y. Reduced expression of microRNA-497 is associated with greater angiogenesis and poor prognosis in human gliomas. HUM PATHOL. 2016;58:47–53. doi: 10.1016/j.humpath.2016.04.022. [DOI] [PubMed] [Google Scholar]
- 30.Wang P, Meng X, Huang Y, Lv Z, Liu J, Wang G, Meng W, Xue S, Zhang Q, Zhang P, Chen G. MicroRNA-497 inhibits thyroid cancer tumor growth and invasion by suppressing BDNF. ONCOTARGET. 2017;8(2):2825–2834. doi: 10.18632/oncotarget.13747. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Pengcheng S, Ziqi W, Luyao Y, Xiangwei Z, Liang L, Yuwei L, Lechen L, Wanhai X. MicroRNA-497 suppresses renal cell carcinoma by targeting VEGFR-2 in ACHN cells. BIOSCIENCE REP. 2017;37(3):R20170270. doi: 10.1042/BSR20170270. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.MANTEL N, HANSZEL W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 33.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7(3):177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 34.Lan X. Impact of human leukocyte antigen mismatching on outcomes of liver transplantation: A meta-analysis. WORLD J GASTROENTERO. 2010;16(27):3457. doi: 10.3748/wjg.v16.i27.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Egger M, Davey SG, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. LANCET ONCOL. 2012;13:e249–e258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 37.Mendell JT. MicroRNAs: critical regulators of development, cellular physiology and malignancy. CELL CYCLE. 2005;4(9):1179–1184. doi: 10.4161/cc.4.9.2032. [DOI] [PubMed] [Google Scholar]
- 38.Schickel R, Boyerinas B, Park SM, Peter ME. MicroRNAs: key players in the immune system, differentiation, tumorigenesis and cell death. ONCOGENE. 2008;27(45):5959–5974. doi: 10.1038/onc.2008.274. [DOI] [PubMed] [Google Scholar]
- 39.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. CANCER SCI. 2010;101(10):2087–2092. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie F, Yuan Y, Xie L, Ran P, Xiang X, Huang Q, Qi G, Guo X, Xiao C, Zheng S. miRNA-320a inhibits tumor proliferation and invasion by targeting c-Myc in human hepatocellular carcinoma. Onco Targets Ther. 2017;10:885–894. doi: 10.2147/OTT.S122992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li N, Wang L, Tan G, Guo Z, Liu L. MicroRNA-218 inhibits proliferation and invasion in ovarian cancer by targeting Runx2. ONCOTARGET. 2017;8(53):91530–91541. doi: 10.18632/oncotarget.21069. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.LeMay-Nedjelski L, Mason-Ennis J, Taibi A, Comelli E, Thompson L. Omega-3 Polyunsaturated Fatty Acids Time-Dependently Reduce Cell Viability and Oncogenic MicroRNA-21 Expression in Estrogen Receptor-Positive Breast Cancer Cells (MCF-7) INT J MOL SCI. 2018;19(1):244. doi: 10.3390/ijms19010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo J, Gong G, Zhang B. miR-539 acts as a tumor suppressor by targeting epidermal growth factor receptor in breast cancer. SCI REP-UK. 2018;8(1):2073. doi: 10.1038/s41598-018-20431-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang Y, Meng Q, Qi J, Shen H, Sun S. MiR-497 promotes metastasis of colorectal cancer cells through Nrdp1 inhibition. Tumour Biol. 2015;36(10):7641–7647. doi: 10.1007/s13277-015-3489-9. [DOI] [PubMed] [Google Scholar]
- 45.Gu A, Lu J, Wang W, Shi C, Han B, Yao M. Role of miR-497 in VEGF-A-mediated cancer cell growth and invasion in non-small cell lung cancer. Int J Biochem Cell Biol. 2016;70:118–125. doi: 10.1016/j.biocel.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 46.Hu J, Xu JF, Ge WL. MiR-497 enhances metastasis of oral squamous cell carcinoma through SMAD7 suppression. AM J TRANSL RES. 2016;8(7):3023–3031. [PMC free article] [PubMed] [Google Scholar]
- 47.Liu J, Zhou Y, Shi Z, Hu Y, Meng T, Zhang X, Zhang S, Zhang J. microRNA-497 Modulates Breast Cancer Cell Proliferation, Invasion, and Survival by Targeting SMAD7. DNA CELL BIOL. 2016;35(9):521–529. doi: 10.1089/dna.2016.3282. [DOI] [PubMed] [Google Scholar]
- 48.Xu Y, Chen J, Gao C, Zhu D, Xu X, Wu C, Jiang J. MicroRNA-497 inhibits tumor growth through targeting insulin receptor substrate 1 in colorectal cancer. ONCOL LETT. 2017;14(6):6379–6386. doi: 10.3892/ol.2017.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu T, Zhang X, Zhang L, Wang Y, Pan H, Xu Z, Pang X. MicroRNA-497 suppresses cell proliferation and induces apoptosis through targeting PBX3 in human multiple myeloma. AM J CANCER RES. 2016;6(12):2880–2889. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Zhang Y, Zhang Z, Li Z, Gong D, Zhan B, Man X, Kong C. MicroRNA-497 inhibits the proliferation, migration and invasion of human bladder transitional cell carcinoma cells by targeting E2F3. ONCOL REP. 2016;36(3):1293–1300. doi: 10.3892/or.2016.4923. [DOI] [PubMed] [Google Scholar]