Abstract
The last years have witnessed a breakthrough in the development of cell-based tolerance-inducing cell therapies for the treatment of autoimmune diseases and solid-organ transplantation. Indeed, the use of tolerogenic dendritic cells (tolDC) and regulatory macrophages (Mreg) is currently being tested in Phase I and Phase II clinical trials worldwide, with the aim of finding an effective therapy able to abrogate the inflammatory processes causing these pathologies without compromising the protective immunity of the patients. However, there exists a wide variety of different protocols to generate human tolDC and Mreg and, consequently, the characteristics of each product are heterogeneous. For this reason, the identification of biomarkers able to define their functionality (tolerogenicity) is of great relevance, on the one hand, to guarantee the safety of tolDC and Mreg before administration and, on the other hand, to compare the results between different cell products and laboratories. In this article, we perform an exhaustive review of protocols generating human tolDC and Mreg in the literature, aiming to elucidate if there are any common transcriptomic signature or potential biomarkers of tolerogenicity among the different approaches. However, and although several effectors seem to be induced in common in some of the most reported protocols to generate both tolDC or Mreg, the transcriptomic profile of these cellular products strongly varies depending on the approach used to generate them.
Keywords: biomarkers, tolerogenic dendritic cells, regulatory macrophages, tolerance mechanisms, genetic markers, immune tolerance, regulatory dendritic cells
Introduction
The immune system develops complex and sophisticated reactions, which are able to differentiate between what is dangerous and what is innocuous for the host (1), thus specifically attacking pathogens and other potentially dangerous antigens while remaining unresponsive against whether non-dangerous or self-molecules. This balance between immunogenicity and tolerance is orchestrated in the periphery by professional antigen-presenting cells (APC), such as dendritic cells (DC) and, in a lesser extent, macrophages, which direct the immune response depending on the characteristics of the antigen and the cytokine milieu they encounter (2). Briefly, DC are in charge of both the initiation of the adaptive immune response and the control or abrogation of the inflammatory processes once the immunogenic antigen has been cleared. For this regulatory role, DC can deploy several mechanisms such as the induction of anergy or deletion of the activated immune cells, as well as, the activation of regulatory T cells (Treg) in an antigen-specific manner. Therefore, since DC have the potential to both stimulate or inhibit immune responses, the role of these cells in the immune system is complex and bidirectional (3–6). By their part, macrophages also play a minor role as APC, developing some of the regulatory processes mentioned above, although their main function consists in the clearance of cell debris, pathogens and other molecules after the immune response has concluded (7).
Eventually, the immune homeostasis can be disturbed due to a malfunction of the immune system, thus setting up immunogenic responses toward self-antigens from specific tissues and organs, which may lead to the development of autoimmune diseases. In the last years, there has been a significant progress in the knowledge of the mechanisms of immune regulation mediated by APC. Consequently, the development of novel autologous cell therapies capable of re-educating the immune system toward a tolerogenic profile has been postulated as a promising therapeutic alternative to conventional, unspecific immunomodulatory and immunosuppresive drugs, which often present severe side effects and a relatively poor efficacy (8).
So far, a wide variety of in vitro protocols has been established for the generation of immune tolerance-inducing DC—or tolerogenic DC (tolDC)—and regulatory macrophages (Mreg). Moreover, some of these cell products have been successfully translated from the bench to the bedside in the last few years, being tested in Phase I clinical trials in patients with autoimmune diseases—such as type 1 diabetes, rheumatoid arthritis or Crohn's disease—as well as, kidney transplantation, demonstrating in all cases that tolerogenic cell therapies are safe and well tolerated, without relevant side effects (9–13). In addition, many other studies are currently ongoing (14). These results, therefore, support the use of tolDC and Mreg as novel and safe approaches aiming to restore the immune tolerance. However, given the wide variety of protocols available for the generation of these cell products, finding objective and measurable biomarkers to characterize tolDC and Mreg and compare their characteristics between different approaches and laboratories remains one of the main obstacles to overcome.
In this context, the identification of differentially expressed (up- or down-modulated) genes in tolDC and/or Mreg constitutes one of the best tools for the definition of biomarkers of tolerogenicity, since they can provide more robust and reliable information compared to conventional methods such as phenotypical characterization by flow cytometry (with high variability) or functional studies (which require several days), as it will be further discussed below. In the case of tolDC and Mreg, these biomarkers would be able to guarantee the proper generation of the therapeutic cell product, ensuring that the cells are both safe and tolerogenic. Therefore, the ideal biomarker would be one that is selectively overexpressed or repressed in the tolerance-inducing cell product compared to its respective mature immunogenic steady-state control condition.
With that purpose, here we review the main human tolDC- and Mreg-inducing protocols reported on the literature. We specifically focus on the different agents and drugs used to generate these cell products, in order to define a catalog of genes and/or proteins induced by these stimuli and thus try to find potential and universal biomarkers of tolDC and Mreg.
Tolerogenic dendritic cells as key tolerance-inducing players and their transcriptomic signature
DC constitute an heterogeneous subset that includes classical, plasmacytoid, and monocyte-derived myeloid DC (15). In their immature state (iDC), DC are mainly antigen-capturing cells with tolerance-inducing functionality. However, once in the presence of a pro-inflammatory stimulus such as TNF-α, lipopolysaccharide (LPS) or IL-1β, they can differentiate into immunogenic mature DC (mDC). By their part, mDC are capable of priming and activating T cells to initiate an immune response after providing the three required activation signals of the immune synapsis once a specific and immunogenic antigen has been recognized. During this maturation process, an upregulation of the expression of human leukocyte antigen (HLA) molecules, as well as, of other costimulatory molecules such as CD40, CD80, CD83, or CD86 takes place, along with an increase in the production of IL-12 and other proinflammatory cytokines (2, 3, 8).
However, a third type of DC has been defined in the last years, combining immune tolerance-inducing properties with a stability against maturation stimuli, called tolerogenic DC (tolDC). It is not clear whether tolDC constitute a different DC subset by themselves or if they are mere maturation-impaired iDC, although there seems to be a consensus about which features they have to possess in order to develop their regulatory function. Thus, tolDC usually present one or more of these characteristics: a semi-mature phenotype, with low expression of co-stimulatory (CD80, CD86, CD83) and HLA molecules, a maintained CCR7-dependant migratory ability toward the secondary lymphoid organs, an increased IL-10 production accompanied by low or null IL-12 and IFN-γ secretion, a lowered T cell-proliferation priming capability, potential to induce Treg and stability against maturation in front of a proinflammatory milieu. Specifically the latter, which has been described in the majority of these studies, probably constitutes the most important feature among all of them (16–18).
Importantly, tolDC can be differentiated in vitro from peripheral blood monocytes in the presence of a determined tolerogenic-inducing agent. Indeed, a wide variety of protocols have emerged in the last 20 years describing the induction of tolDC with several stimuli, such as anti-inflammatory cytokines—IL-10 (19, 20), TGF-β (20, 21)—, pharmacological agents and immunosuppressant compounds—rapamycin (20, 22, 23), different corticosteroids (24), dexamethasone (20, 23, 25, 26), vitamin D3 (20, 23, 27) or a combination of both dexamethasone and vitamin D3 (28)—, several drugs and blocking molecules—aspirin (29), mitomycin C (30), the NF-κB inhibitor BAY11-7082 (11)—and other strategies, such as genetic engineering for the selective repression or induction of key molecules and pathways (10, 31), among many others further discussed below. Generally, most of these protocols share several features in common, such as the differentiation of monocytes in the presence of GM-CSF and IL-4, as well as, the addition of a maturation stimulus (which usually includes different combinations of LPS, monophosphoryl lipid A, TNF-α, IL-1β, prostaglandin E2, and/or IL-6), with few exceptions.
Either if we assume tolDC are a specific DC subset per se or just a modified state of iDC, there must be some footprint left by this condition. At the transcriptomic level, as already hinted, some obvious downregulated candidates would be the genes encoding co-stimulatory molecules or pro-inflammatory cytokines. However, those features would be shared with steady state iDC, thus making them useless in terms of differentially characterizing tolDC. In fact, ideally, a comparison against both immature and immunogenic control conditions should be taken into account in the search of specific genetic biomarkers, something that has not been considered in the majority of the reviewed studies. An ideal candidate should be, furthermore, clearly differentiated by a matter of full induction or repression, as a slight increase/decrease of its expression could be ambiguous and would always require the use of robust controls, which is not always possible.
Consequently, many research groups have been working on the identification of genetic markers for human tolDC, and deep transcriptomic studies are becoming more frequent each year. However, and although several studies have described a pool of markers for some specific tolerogenic cell products, common genetic biomarkers have not been found yet.
Glucocorticoids and immunomodulatory molecules in the generation of tolerogenic dendritic cells
Since mDC are immunogenic cells, or, in other words, promoters of inflammatory responses, the use of corticosteroids and other immunosuppressant drugs has been widely reported for the generation of tolDC. Rapamycin (20, 22, 23, 32, 33) and a combination of hydrocortisone and clobetasol-17-propionate (24), but especially dexamethasone (20, 23, 25, 26, 32–45), have all been used for the generation of tolDC. As a glucocorticoid-induced molecule, the expression of the gene encoding the anti-inflammatory mediator known as glucocorticoid-induced leucine zipper (GILZ) (46) has been reported strongly up-modulated in many of these studies, thus making it a good albeit predictable marker for tolDC generated with this kind of immunomodulatory agents. Furthermore, other molecules related with the complement and the immune system have been found commonly up- or down-modulated in several of these tolDC protocols, such as the anti-inflammatory cytokine IL-10 (up-regulated), the pro-inflammatory cytokine IL-12 or the fascin 1-encoding gene FSCN1 (both down-modulated), which are common features that define these cells (32). The full list of differentially expressed molecules reported for each of the abovementioned protocols and their respective references can be found on Table 1.
Table 1.
Differentially up- and down-modulated genes and proteins in the most reported human tolDC-inducing protocols.
| Protocol | Type | Up-modulated molecules | Down-modulated molecules | References | |
|---|---|---|---|---|---|
| tolDC | Dexamethasone | Gene | ANXA1, C1QA, C1QC, C1QTNF1, C3AR1, CCL17, CD163, CD300LF, CD32, CFH, CLIC2, CSGALNACT1, CTSC, DCR3, EP2, EP3, F13A, FCGR2A, FCGR2B, FKBP5, FOXO3, FPR1, GILZ, GPX1, IDO1, IL10, IL12A, IL27B, IMDH2, JAG1, MERTK, MRC1, MT1, NCF1, OSF1, P2RY14, SLC39A8, SOD2, STAB1, TPP1, ZBTB16 | CCL22, CD1C, FCER1A, IDO1, IL12B, LAMP3, MMP12, ZNF366 | (26, 32, 34–36, 38, 39, 41) |
| Protein | CYP1B1, DAB2, DPYD, FCER1G, FCGR3A, FTL, GCLC, IVNS1ABP, LRRC25, MCTP1, MERTK, NUDT16, PDCD4, PECAM1, RNASE6, RNASET2, SIGLEC5, SLCO2B1 | FSCN1 | (12, 34, 37) | ||
| miRNA | miR-328-5P, miR-638, miR-663, miR-762, miR-1275, miR-1228, miR-1909 | miR-142-5p | (40) | ||
| Dexamethasone + rosiglitazone | Gene | FABP4, GILZ | (47) | ||
| Dexamethasone + vitamin D2 | Protein | ERK1/2, IDO, JNK/SAPK, mTOR, p38 MAPK, STAT3 | (48) | ||
| Dexamethasone + vitamin D3 | Gene | ACADM, ACADVL, ACO1, ACO2, ACOX2, ACSS1, ALDH2, ATP5G3, ATP5J, ATP5O, BLVRB, C1orf162, C1QA, CCR5, CD14, CD209, CD274, CD52, CLIC1, COX11, COX6A1, COX7A2, CTSB, CTSD, CTSH, CYC1, DHRS9, EIF3B, EIF3C, EIF3CL, EIF4A3, FBP1, FCGR2B, FCGR3A, FN1, FTH1, FTL, G6PD, GAPDH, IDH3A, IDH3B, ILT3, LDHB, LILRB4, MATK, MCEMP1, MDH2, ME1, ME3, NDUFB9, NDUFS1, NDUFS8, NOS3, PCK2, PDHA1, PDXK, PIK3R1, PKM2, PNP, PRDX3, PTPN6, RAC2, RGCC, RPS12, RPS19, RPS21, RPS6KA1, RPS6KA2, SDHA, SLC11A1, SLC27A5, SLC2A1, SLC2A5, SNCA, SUCLG1, SUCLG2, TCEB1, TGFB1, TP53, TPI1, UQCR10, UQCR11, UQCRB, UQCRC1 | ACTB, ADAM12, ADAM19, ANKRD33B, AOC1, CD25, CD40, CD80, CD83, CD86, DPYSL2, EHF, FSCN1, GPR157, ICOSLG, IKZF1, IKZF4, IL12B, IL2RA, ORMDL3, PIK3CG, PLEKHA5, PPP1R16B, PTPN2, SH2B3, TYK2, WDR1 | (49–51) | |
| Protein | ADK, AKR1A1, ALDH2, ALDOA, ATP5H, ECHS1, FBP1, FTL, G6PD, GPD2, GALK, MPDH2, PGAM, PGM1, PKM2, PNP, PRDX6, TALDO1, TKT, TPI1 | DPYSL2, ENO1, FSCN1, HSPD1, PDIA3 | (37) | ||
| Hepatocyte growth factor | Gene | IL10 | (52) | ||
| IFN-γ | Gene | IRF4, RELB, IL12p40 | (53, 54) | ||
| IL-10 | Gene | ANXA1, C1QC, CTSB, CTSC, CTSL, F13A, FTH1, GILZ, HLA-DOB, IL8, LILRB3, MRC1, STAB1, THBS1, TPP1 | CD74, LAMP3 | (32, 41, 55) | |
| IL-10 + IL-6 | Gene | CTSB, CTSL, FTH1, HLA-DOB, IL-8, THBS1 | CD74 | (55) | |
| Poly I:C | Gene | IDO1, PDL1 | (56, 57) | ||
| Rapamycin | Gene | ANXA1, C1QC, CTSC, GILZ, GPX1, IMDH2, OSF1, TPP1 | RALDH1 | (32) | |
| Retinoic acid | Gene | ALDH1A1, ALDH1A2, CD141, GARP | (58, 59) | ||
| TGF-β | Gene | ANXA1, CTSL, CXCL1, CXCR3, FTH1, HLA-DOB, IL8, LILRB3, THBS1 | CD74, STAB1 | (32, 55) | |
| TX527 (vitamin D3 analog) | Protein | ACADVL, ACO2, ACOX1, ATP5A1, CTSD, CTSS, COPG, FBP1, G6PD, HADHA, IDH3A, MnSOD, OGDH, PCK2, PKM2, PRX3, PTM, UQCRFS1 | ACAT1, ARCN1, DLD, PA28beta, PTM, RabGDI | (60) | |
| Vitamin D3 | Gene | ALOX5, ATP5A1, CAMP, CCL22, CD14, CD300LF, CMYC, CYP24, CYP24A1, CYP27B1, GILZ, GLUT3, HK3, ILT3, IRF8, LDHA, LGALS9, PDHA1, PFKFB4, PIK3CG, PRKAA1, THBD, VDR | CD1A, CD1C, CD1E, CD36, CD80, F13A, IER3, IRF4, LAMP3 | (32, 36, 41, 61–63) | |
| Protein | AKT, FTL, GSK-3b, mTOR | FSCN1, SOD2 | (37) | ||
| miRNA | miR-378 | (64) |
Genes validated by qPCR or proteins validated by western blot are shown in bold.
Dexamethasone-induced tolDC (dexa-tolDC) are one of the most widely implemented approaches worldwide for the generation of human tolDC, and are being or have been tested on clinical trials for the treatment of numerous autoimmune diseases, such as Crohn's disease (http://www.clinicaltrials.gov, NCT02622763) (12), rheumatoid arthritis (http://www.clinicaltrials.gov, NCT03337165; NCT03337165) and both multiple sclerosis or neuromyelitis optica (http://www.clinicaltrials.gov, NCT02283671). Several studies have reported the differential up-modulation of genes C1QA (encoding the C1q complement protein, chain A) (34, 35), CD163 (34, 35), GILZ (32, 35, 36), MERTK (encoding the MER Proto-Oncogene Tyrosine Kinase, also used as a marker in the abovementioned clinical trial for Crohn's disease) (12, 26, 35) and ZBTB16 (encoding zinc finger and BTB domain containing protein 16) (34, 35) in dexa-tolDC, thus making them the most relevant candidate biomarkers for this specific protocol. Additionally, the differential expression of IDO1, the gene encoding the indoleamine 2,3-dioxygenase —a molecule widely related to the induction of immune tolerance (65)—, has also been reported in dexa-tolDC. However, there is some controversy in this regard, as it has been found both up- (35) and down-modulated (32) in different studies. Besides, other induced genes described in studies using dexamethasone, relevant by their role in the modulation and mediation of different mechanisms of the immune system—with their respective encoded proteins in brackets—, are CD300LF (CD300 molecule-like, family member F), F13A (coagulation factor XIII A), FCGR2B (Fc fragment of IgG receptor IIb), FCGR3A (Fc fragment of IgG receptor IIIa), MRC1 (mannose receptor C-type 1), and STAB1 (stabilin 1), as well as, other non-immune related genes like FTL (ferritin light chain), IMDH2 (inosine monophosphate dehydrogenase 2), and SOD2 (superoxide dismutase 2). Furthermore, the combination of dexamethasone with rosiglitazone has also been reported for the generation of tolDC, highlighting the induction of FABP4 (fatty acid-binding protein 4) with this protocol, but specially also of GILZ gene (47).
The generation of human rapamycin-modulated tolDC (rapa-tolDC) is the second most reported protocol of this group of pharmacological and immunomodulatory agents. However, transcriptomic studies in tolDC generated with this strong immunosuppressant drug are not as predominant as those induced with dexamethasone. Yet, several genes have been postulated as candidate biomarkers for rapa-tolDC, both immune-related—ANXA1 (annexin 1), C1QC, CTSC (cathepsin C) and GILZ—and non-immune-related —GPX1 (Glutathione Peroxidase 1), IMDH2, OSF1 (pleiotrophin) and TPP1 (tripeptidyl peptidase 1)—. Interestingly, all these genes have also been described in common with dexa-tolDC (32).
Additionally, the immunostimulant TLR3 ligand polyinosinic:polycytidylic acid (poly I:C) has also been reported to induce human tolDC, although in an inconsistent and poorly efficient manner. Nevertheless, the differential up-modulation of both IDO1 and PD-L1, two genes involved in the induction and maintenance of immune tolerance, has been confirmed by quantitative PCR for these cells (56, 57). As for tolDC induced with hydrocortisone and clobetasol-17-propionate, no transcriptomic biomarkers have been reported.
Vitamins A and D modulate the transcriptomic footprint of tolerogenic dendritic cells
As reviewed by Mora et al. (66), vitamins A and D exert important immunomodulatory properties. While vitamin A and specifically its metabolite, retinoic acid, have been reported to have an influence in T cell differentiation and proliferation, as well as, Treg induction, vitamin D plays an important role as an immunoregulatory agent in the inhibition of T cell proliferation and the reduction of IL-2 and IFN-γ secretion. Furthermore, the absence or low levels of vitamin D in the organism has been widely linked to an increase in the incidence of autoimmune diseases.
The tolerogenic-inducing properties of 1,25-dihydroxycholecalciferol, the active form of vitamin D3, over DC (vitD3-tolDC) have been widely reported in vitro in many studies performed with murine (67–70) and even cattle cells (71), although we will only focus on biomarkers of human vitD3-tolDC (20, 23, 27, 32, 33, 36, 37, 41, 44, 61–64, 72–74). As a measurement of its relevance, such is the importance of vitD3-tolDC in the field of tolerogenic cell products that even two clinical trials are already ongoing for the treatment of multiple sclerosis using this cell product in Badalona, Spain (http://www.clinicaltrials.gov, NCT02903537) and in Antwerp, Belgium (http://www.clinicaltrials.gov, NCT02618902). Several transcriptomic and proteomic pre-clinical studies in human vitD3-tolDC have evidenced several genes and proteins strongly induced with this approach, including immune-related molecules—CCL22 (62, 63), ILT3 (immunoglobulin-like transcript 3) (36), CD300LF (62) or GILZ (32), these last two in common with dexa-tolDC—and oxidative metabolism enzymes and regulators—GLUT3 (glucose transporter 3), LDHA (lactate dehydrogenase A), mTOR (mammalian target of rapamycin), PDHA1 (pyruvate dehydrogenase E1, subunit alpha 1) or PFKFB4 (fructose-2,6-bisphosphatase) (63)—, as well as direct targets of the response to vitamin D3 through the interaction with its receptor, like CYP24A1 (cytochrome P450, family 24, subfamily A, member 1) (41, 61–63) and of course VDR (vitamin D receptor) (41). By their part, the repression of several co-stimulatory, pro-inflammatory, and antigen presenting genes and molecules like CD1A, CD1C, CD80, FSCN1 or the transcription factor IRF4 has been reported at the transcriptomic and proteomic levels (37, 62). Additionally, a synthetic structural analog of vitamin D3, TX527, has also been used for the induction of human tolDC (60). However, and although the up-modulation of the ATP synthase F1 subunit alpha-encoding gene (ATP5A1) was reported in common with vitD3-tolDC, the transcriptomic resemblance was more relevant with tolDC induced with a combination of dexamethasone and vitamin D3, a strategy that will be further discussed in the next section. Nevertheless, some of these induced molecules consist of mostly metabolic-related genes—ACADVL (Acyl-CoA dehydrogenase very long chain), ACO2 (aconitase 2), FBP1 (fructose bisphosphatase 1), IDH3A (isocitrate dehydrogenase 3, subunit alpha), PCK2 (phosphoenolpyruvate carboxykinase 2) and PKM2 (pyruvate kinase M2)—and CTSD, encoding the protease cathepsin D (37, 49, 50).
The use of vitamin A-derived molecules like retinoic acid, however, has not been so widely reported for the generation of human tolDC and only the selective up-regulation of ALDH1A1 and ALDH1A2 genes, encoding the aldehyde dehydrogenase 1 family members A1 and A2—involved the metabolism of retinoic acid—has been reported, as well as, the induction of CD141 and GARP genes (58, 59). Other differentially expressed genes induced by the protocols mentioned in this section are shown in Table 1.
The synergic effect of dexamethasone and vitamin D
Since dexamethasone and vitamin D treatments alone are able to generate tolDC, the combination of both of them is expected to induce synergic effects that would strengthen the tolerogenic functionality of these cells. Consequently, the simultaneous use of dexamethasone and vitamin D3, or vitamin D2 in a few cases (48, 75), has become one of the most widely reported human tolDC-generating protocols in vitro. Indeed, these cells have even reached the clinical phase for the treatment of rheumatoid arthritis, with successful results regarding the safety and tolerability of the product (http://www.clinicaltrials.gov, NCT01352858) (13).
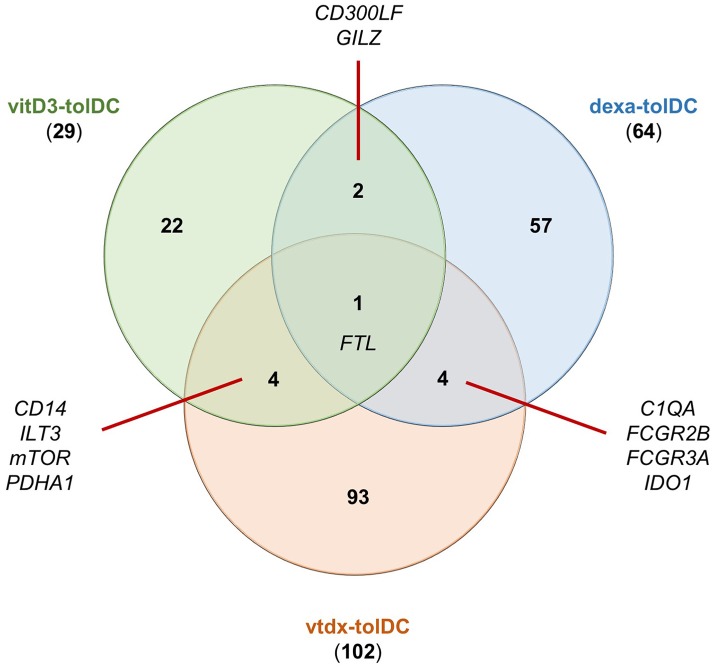
As expected, the genetic signature of dexamethasone + vitamin D-induced tolDC (vtdx-tolDC) reported in pre-clinical studies partially overlaps with that reported for each or both of these treatments alone to generate human dexa- and vitD3-tolDC. In fact, the analysis of the reported data for these protocols showed that C1QA, FCGR2B, FCGR3A and IDO1 genes were found induced in common with dexa-tolDC (34, 35, 38, 50, 48) and CD14, ILT3, mTOR and PDHA1 were shared with vitD3-tolDC (36, 48–50, 62, 63). Nevertheless, our analysis evidenced that the up-regulation of FTL and the suppression of FCSN1 genes were the only genetic modulations in common between these three protocols (34, 37, 63, 50). Interestingly, the function of the proteins encoded by all these genes is strongly related to the modulation of the immune system. Surprisingly, however, there was a pool of genes that were only described for vtdx-tolDC but not for either dexa-tolDC nor vitD3-tolDC, such as CTSB, DHRS9 (dehydrogenase/reductase 9), FTH1 (ferritin heavy chain 1), RGCC (regulator of cell cycle), SLC11A1 (solute carrier family 11 member 1), TBET or TGFB1 (49, 50, 51). Indeed, after our study, it is worth noting that out of 64 up-modulated genes and/or proteins reported for dexa-tolDC, 29 genes for vitD3-tolDC and 102 genes for vtdx-tolDC, only 4 genes could be found in common between vtdx-tolDC and each treatment separately, as shown in the Venn diagram in Figure 1. The chances are, however, that many of these genes could simply not be detected or were overlooked in the validation process of the separated protocols due to intrinsic limitations of the methodologies used, as it is known that biases frequently appear in high throughput transcriptomic and proteomic techniques. For this same reason, for instance, some already mentioned immune-related and metabolic genes were detected simultaneously induced in vtdx-tolDC and tolDC generated in the presence of the vitamin D3 analog TX527—ACADVL, ACO2, CTSD, FBP1, G6PD (glucose-6-phosphate dehydrogenase), IDH3A, PCK2, PKM2)—(37, 60, 49, 50). Although the down-modulation of genes is not as relevant toward the identification of transcriptomic biomarkers, it is nonetheless worth noting that the FSCN1 gene has been found repressed in vtdx-tolDC, dexa-tolDC, and vitD3-tolDC at the same time (37, 50). Table 1 shows a complete list of the differentially expressed genes and proteins reported in protocols using a combination of dexamethasone and vitamin D derivates.
Figure 1.
Common up-modulated genes in tolDC induced with either vitamin D3, dexamethasone or the combination of both. The numbers in the Venn diagram indicate the number of reportedly induced genes for each condition alone or in combination with one or both of the others. Dexa-tolDC: dexamethasone-induced tolDC; vitD3-tolDC: vitamin D3-induced tolDC; vtdx-tolDC: dexamethasone + vitamin D3-induced tolDC.
The effect of cytokines and growth factors in the induction of tolerogenic dendritic cells
Many different kinds of cytokines have been used for the induction of human tolDC, ranging from anti-inflammatory—IL-10 (19, 20, 32, 33, 41, 55, 76–78), TGF-β (21, 32, 33, 55, 79) or both (80)—to even immunostimulatory molecules—IFN-γ (53, 54) or a combination of IL-6 with IL-10 (55)—, but also several growth factors—hepatocyte growth factor (52) and low-doses of GM-CSF alone (81).
As previously mentioned, the secretion of IL-10 is one of the most sought features of tolDC due to its anti-inflammatory and regulatory properties. Consequently, the generation of tolDC in the presence of exogenous IL-10 (IL10-tolDC) constitutes one of the most implemented protocols for the generation of this type of regulatory cell products. In fact, many of the genes and molecules already cited for other protocols, with immune or metabolic involvement, have also been found induced in IL10-tolDC, such as, ANXA1, C1QC, CTSB, CTSC, CTSL (cathepsin L), F13A, FTH1, HLA-DOB, IL-8, LILRB3 (leukocyte immunoglobulin-like receptor B3), MRC1, STAB1, THBS1, TPP1 and, especially for its repeated prevalence, GILZ (32, 55). Also, and in line with the traditional concept of tolDC, the down-modulation of the antigen presenting molecule CD74 (also known as HLA-DR) (55) and LAMP3 (lysosomal-associated membrane protein 3), typically found on iDC (41), has been reported. Interestingly, the combined exposure in front of both of IL-10 and IL-6 for the generation of tolDC performed in one of the previously cited articles did not seem to change the transcriptomic profile of these cells, as many of the above mentioned genes were also found accordingly induced or repressed like they were in IL10-tolDC (55).
The use of TGF-β for the in vitro differentiation of tolDC is not as widely established as IL-10, but still some potential biomarkers have been described, both exclusively for this product (the immune related-genes CXCL1 and CXCR3) and in common with other regulatory cells (ANXA1, CTSL, FTH1, HLA-DOB, IL-8, LILRB3, THBS1) (32, 55). Just like in IL10-tolDC, CD74 appears differentially repressed in TGF-β-induced tolDC but, controversially, also does STAB1, reportedly up-modulated in the former protocol (55). As far as we are concerned, no potential transcriptomic or proteomic markers have been reported in cells induced with the combination of IL-10 and TGF-β for the generation of human tolDC.
Surprisingly, IFN-γ has also been described in a couple of publications for the generation of tolDC, even though it does not constitute the most obvious strategy due to its proinflammatory properties. Nevertheless, these studies have reported the selective reduction in the expression of the pro-inflammatory genes IRF4, RELB, and IL12p40 in this cell product (53, 54). Consequently, the down-modulation of these genes is in line with the expected anti-inflammatory profile for tolDC, and even IRF4 has also been reported as differentially repressed in vitD3-tolDC, as mentioned above (62). All the biomarkers described within the protocols mentioned in this section are shown in Table 1.
Finally, the differentiation of stable tolDC from monocytes in the presence of low doses of GM-CSF, and in the absence of IL-4 in the culture, has also been reported in humans (81), but also in animal models (82–84). In fact, their clinical use is being tested under the context of a multicentre trial named The ONE Study ATDC in living-donor renal transplantation (http://www.clinicaltrials.gov, NCT02252055) (85). However, any potential biomarker in human low-dose GM-CSF-induced tolDC has been reported yet.
Genetic changes induced in tolerogenic dendritic cells generated with other strategies
The pharmacological agents and factors mentioned so far comprise the most predominant strategies in the literature for the induction of tolDC, but there is still a wide variety of drugs, proteins and several treatments with the potential of generating this type of regulatory DC products. However, provided that the aim of this review is to look for universal biomarkers of immune tolerance, we have also considered these approaches. In fact, a significant amount of studies have reported the differential expression of several genes and molecules that could become potential biomarkers for their respective and specific protocols, generating tolDC in the presence of different organic compounds—such as the Aspergillus cell wall (32), curcumin (86), mitomycin C (87), paeoniflorin (88), phosphatidylserine liposomes imitating apoptotic bodies (89)—, other cell types—mast cells (90) and trophoblasts (91)—and a variety of agents, conditions and/or molecules—for instance a combination of the complement protein C5a and LPS (92), seminal plasma (93), the Wnt-5a protein (94) or even the deprivation of tryptophan in the culture (95)—. However, there are still many other different strategies without transcriptomic or proteomic studies reported in the literature that are, therefore, outside of the objective of this review. The full list of differentially expressed genes and molecules in the protocols mentioned in this section is presented in Table 2.
Table 2.
Differentially up- and down-modulated genes and proteins in other human tolDC- and Mreg-inducing protocols.
| Protocol | Type | Up-modulated molecules | Down-modulated molecules | References | |
|---|---|---|---|---|---|
| tolDC | Aspergillus cell wall | Gene | ANXA1, STAB1, GILZ, IDO, RALDH1, RALDH2 | F13A, MRC1 | (32) |
| C5a and LPS | Gene | RGCC, FERMT2, SLC39A14, TNFSF14, TGFB1 | IL12B, FOXO1 | (92) | |
| Curcumin | Gene | RELB | (86) | ||
| Mast cells | Gene | IDO1, NFKB1, NFKB2, RELB, SOCS5 | SOCS3 | (90) | |
| Mitomycin C | Gene | ADM, CSF2RA, DDIT3, FDXR, GAB2, LILRB4, LRDD, MAFB, MAP4K4, PERP, TNFRSF10B, TRAF4, TSC22D3 | CFLAR (FLAME-1, I-FLICE, Usurpin), NRG2 | (87) | |
| Paeoniflorin | Gene | IDO1 | (88) | ||
| Phosphatidyl-serine lyposomes | Gene | CLCN6, CYTH4, IFNLR1, LAIR1, LDLR, MFSD2A, NFKBIA, PLAUR, PPME1, SHB, SLC43A3, TNFAIP3, TNFSF14, VEGFA | ALKBH1, ATP10D, AURKA, BCL2L1, BLCAP, BST1, BTBD3, BTK, BUB1, C9orf64, CASP3, CBX4, CD1D, CDC23, CDC42SE1, CDK13, CDYL2, CKAP2, CLCN3, CSRP2BP, CUL3, DAPP1, DCAF12, DCAF7, DCLRE1A, DCTD, DDO, DYRK2, EHBP1, ERLIN1, FBXO25, FBXO36, FRAT2, FZD5, GIMAP4, GLRX, GOLPH3L, GTF2B, HHEX, HPGD, ICK, KBTBD6, KIF11, KIF20B, LMNB1, LNX2, MAPRE2, MCM4, MCPH1, MDM1, MEF2C, MEGF9, MIER3, MLH1, MNDA, MSH2, MYB, N4BP1, NCAPG2, NET1, NFIA, NSMCE4A, NUP160, PAQR8, PARG, PAXIP1, PCNA, PMP22, PROS1, RAB32, RAD51C, RCSD1, RMDN1, RMND5A, SCYL3, SEC22C, SKI, SLAMF6, SLC10A7, SLC40A1, SMC2, SNN, SNX18, SOCS2, STIM2, STX3, TIMMDC1, TNFRSF11A, TPK1, TRIM5, UBE2E3, UBFD1, UNC50, VWA5A, WRNIP1, ZBED3, ZBTB39, ZBTB5, ZFP36L2, ZNF436 | (89) | |
| Seminal plasma | Gene | COX2, TGFB1 | CD1A | (93) | |
| Trophoblasts | Gene | IDO1 | (91) | ||
| Tryptophan-deprived | Gene | CHOP, ILT3 | (95) | ||
| Wnt5a | Gene | ID3, IRF1, IRF2, SOCS3, TLR1 | ID2, IRF8, TLR3, TLR4, TLR5 | (94) | |
| Mreg | M-CSF + IFN-γ | Gene | ALDH1A1, ALDH1A2, CD1D, DHRS9 | (96) | |
| M-CSF + LPS | Protein | IL-12p35, IL-12p40, IL23-p19 | (97) |
Genes validated by qPCR or proteins validated by western blot are shown in bold.
A totally different approach to generate tolDC consists in using targeted genetic engineering in order to achieve cells with specific functional features either silenced or induced. There are several strategies reported in this regard, ranging from the impairment of immunogenic properties—such as silencing the expression of CD40, CD80, and CD86, already tested in type 1 diabetes patients, which was the first clinical trial using a tolerogenic cell therapy (10, 98) (http://www.clinicaltrials.gov, NCT00445913)—to selectively inducing the production of several anti-inflammatory cytokines like IL-10 and TGF-β (31, 99), overexpressing the IL-12 and IL-23-suppressor factor SOCS-3 (100) or transfecting the cells with a modified CTLA4 construct that inhibits the expression of the co-stimulatory molecules CD80 and CD86 (101). Surprisingly, some approaches using genetic manipulation achieved to generate human IL-10-producing DC through the induction of, a priori, immunogenic functions such as the CD40-CD40L signaling pathway (102). However, the definition of transcriptomic biomarkers for tolDC induced by genetic engineering would not be of much utility, provided that the differentially expressed genes or proteins to check would be precisely those that have been specifically induced or repressed by the procedure itself.
Generation of regulatory macrophages: differences and similarities with tolerogenic dendritic cells
Mreg constitute one of the three main macrophage subtypes, being the other two the classical macrophages and the so-called alternatively activated macrophages, or M2 macrophages. However, and as reviewed by Fleming and Mosser (103), Mreg present unique features: they are characterized by their ability to modulate the immune system toward a regulatory TH2 response through the production of IL-10 and a limited or absent secretion of IL-12 mediated by the activation of the ERK cascade. In addition, these cells present an increased antigen-presenting functionality with an elevated expression of HLA class II and B7 co-stimulatory molecules. And this is, probably, the crucial feature in which Mreg and M2 macrophages differ the most, because although both subsets exhibit immunoregulatory properties, the ability to induce antigen-specific responses is limited in M2 macrophages due to their low HLA expression. However, Mreg are considered to deploy their potent T cell suppressor functions mainly through three non-antigen-specific mechanisms: via IFN-γ-induced IDO activity, by a contact-dependent deletion of activated T lymphocytes or mediating the induction of TIGIT+ FoxP3+ Treg (9, 96, 104, 105).
Just like tolDC, human macrophages can be generated in vitro by differentiating them from monocytes. Briefly, classical macrophages are obtained in the presence of GM-CSF, and M2 and Mreg macrophages are generated in the presence of M-CSF, but with different supplementary treatments. While M2 macrophages are normally achieved using M-CSF + IL-4 and/or other TH2 cytokines, Mreg are treated with M-CSF + LPS or IFN-γ for a brief period of time (106, 107). This combination of M-CSF and a short and complementary pro-inflammatory treatment is precisely the responsible for the strong induction of IL-10 production, something that both stimuli alone fail to achieve in macrophages (108, 109).
The generation of human Mreg is not as widely extended as tolDC, and consequently the number of protocols describing the differentiation of these cells is much more reduced. However, several molecules have already been postulated as potential biomarkers for these cell products. So far, transcriptomic studies have only been performed over LPS (LPS-Mreg) and IFN-γ-activated Mreg (IFN-Mreg). The former, LPS-Mreg, were initially described as IL-10-producing M2 macrophages, and their impaired IL-12 and IL-23 production was confirmed by qPCR (97, 110). However, IFN-Mreg are more widely reported and studied, especially considering their translation into the clinic, where they have already been used for the treatment of living-donor renal transplant-recipient patients (http://www.clinicaltrials.gov, NCT00223067 and NCT02085629) (9, 105). This product is obtained by the stimulation of M-CSF-differentiated macrophages with IFN-γ, after 7 days of culture (9, 105, 111), and a strong up-modulation of ALDH1A1, ALDH1A2 and CD1D genes has been reported, as well as the induction of DHRS9 as an specific IFN-Mreg biomarker (96). The detailed list of markers reported in pre-clinical human Mreg protocols is shown in Table 2.
Nevertheless, although the list of genetic biomarkers described in Mreg is short, the identification of DHRS9 in IFN-Mreg achieves a high relevance in the context of immune tolerance biomarkers, provided that the enzyme encoded by these gene seems to be involved in the biosynthesis of retinoic acid (112). As commented above, this compound is a vitamin A-derived molecule that can be used to differentiate human monocytes into tolDC. Interestingly, both ALDH1A1 and ALDH1A2 genes have been identified as differentially induced in retinoic acid-generated tolDC (58) as well as IFN-Mreg, making them two interesting candidates for the characterization of at least this couple of different tolerance-inducing cell products. Furthermore, the differential up-modulation of DHRS9 has also been reported in vtdx-tolDC, also discussed above (49). Consequently, since these cells are generated with both dexamethasone and vitamin D3, a clear relation between the transcriptomic profile of both IFN-Mreg and tolDC induced with either vitamin A or D is likely to exist. For this reason, further studies and validations in this direction could be of great interest, as potential common biomarkers of two different immune-regulatory myeloid cell-derived products could be identified.
Summary and concluding remarks
The identification of robust biomarkers for the characterization of tolerogenic and immunoregulatory cell products constitutes one of the last steps needed to take the final leap toward the broad application of these novel autologous antigen-specific therapies in the clinic. Specifically, their key importance resides in their capability to provide a fast and reliable quality control of the proper generation, functionality and safety of tolDC and Mreg.
In this article we have performed an exhaustive review of the currently published human tolDC- and Mreg-generating protocols that have reported potential biomarkers for these cells, with the aim of elucidating if a common transcriptomic or proteomic pattern relating all of them could be drawn. However, as it has been discussed, albeit many genes and molecules have been found separately induced using different strategies to generate these immunoregulatory cell products, so far, there is not a biomarker or a pool of biomarkers that can functionally characterize or at least identify the entirety of the studied protocols. Nonetheless, this is not necessarily bad news, as the chances of identifying a common biomarker were slim given the overwhelming variety of approaches and cell types reported in this review. As already mentioned above, the immune system can deploy several strategies for the induction of tolerance that modulate many different immune and non-immune related pathways and transcriptomic cascades, thus making this goal even more unlikely. However, it is also worth noting that finding biomarkers provided only by the tolerance-inducing mechanisms could also be misleading; for instance, DC subtypes like iDC are capable of developing some tolerogenic functions, but still they could not be applied as a therapeutic approach in autoimmune diseases provided their lack of stability against pro-inflammatory stimuli, as discussed above.
Still, despite the consideration of such a wide variety and heterogenicity of protocols for the induction of regulatory cells, a significant amount of differentially expressed genes encoding several anti-inflammatory and immunomodulatory molecules has been reported in very different protocols, for instance IDO1 (in 7 approaches) GILZ (in 6 approaches) or ANXA1 (in 5 approaches). Similarly, the down-modulation of the pro-inflammatory cytokine IL-12 has been reported in 5 different tolDC-inducing strategies. In other words, in this review we have gathered all the genes and proteins that have been described separately with each of the approaches for the generation of tolDC and Mreg in the literature, and we have subsequently compared and put them all together in order to evidence potential common biomarkers between them. The complete list of the genes that have been reported in studies with at least two different approaches for the generation of human tolDC and/or Mreg are shown in Table 3. Therefore, the general idea that lies behind these reported molecules is that all the considered tolerogenic-inducing agents are modulating the cells toward a regulatory profile that might be partially shared between some approaches, but that is often achieved through different mechanisms and biological pathways that are strongly dependent on the stimuli used to generate them.
Table 3.
Differentially expressed genes reported in at least two different protocols for the generation of human tolDC and/or Mreg.
| Gene | Name | Modulation | Repeats | Protocols | References |
|---|---|---|---|---|---|
| ACADVL | Acyl-CoA Dehydrogenase Very Long Chain | Up | 2 | Dexa+vitD3, TX527 | (60, 49) |
| ACO2 | Aconitase 2 | Up | 2 | Dexa+vitD3, TX527 | (60, 49) |
| ALDH1A1 | Retinaldehyde Dehydrogenase 1 | Up | 3 | Asp, IFNg Mreg, RA | (58, 96) |
| ALDH1A2 | Retinaldehyde Dehydrogenase 2 | Up | 2 | IFNg Mreg, RA | (58, 96) |
| ANXA1 | Annexin A1 | Up | 5 | Asp, dexa, IL10, rapa, TGFb | (32) |
| ATP5A1 | ATP Synthase 5 Alpha Subunit 1 | Up | 2 | TX527, vitD3 | (63, 60) |
| C1QA | Complement C1q A Chain | Up | 2 | Dexa, dexa+vitD3 | (34, 35, 50) |
| C1QC | Complement C1q C Chain | Up | 3 | Dexa, IL10, rapa | (32) |
| CD14 | Cluster of Differentiation 14 | Up | 2 | dexa+vitD3, vitD3 | (62, 50) |
| CD1A | CD1a Receptor | Down | 2 | Sem, vitD3 | (50, 93) |
| CD1C | CD1c Receptor | Down | 2 | Dexa, vitD3 | (35, 62) |
| CD300LF | CD300 Molecule Like Family Member F | Up | 2 | Dexa, vitD3 | (34, 62) |
| CD80 | Cluster of Differentiation 80 | Down | 2 | Dexa+vitD3, vitD3 | (62, 50) |
| CTSB | Cathepsin B | Up | 3 | Dexa+vitD3, IL10, IL10+6 | (50, 55) |
| CTSC | Cathepsin C | Up | 3 | Dexa, IL10, rapa | (32) |
| CTSD | Cathepsin D | Up | 2 | Dexa+vitD3, TX527 | (60, 49) |
| CTSL | Cathepsin L | Up | 3 | IL10, IL10+6, TGFb | (55) |
| DHRS9 | Dehydrogenase/Reductase 9 | Up | 2 | Dexa+vitD3, IFNg Mreg | (49, 96) |
| F13A | Coagulation Factor XIII A | Up | 2 | Dexa, IL10 | (32) |
| Down | 2 | Asp, vitD3 | (32) | ||
| FBP1 | Fructose-Bisphosphatase 1 | Up | 2 | Dexa+vitD3, TX527 | (37, 60, 49) |
| FCGR2B | Fc Fragment Of IgG Receptor IIb | Up | 2 | Dexa, dexa+vitD3 | (34, 49) |
| FCGR3A | Fc Fragment Of IgG Receptor IIIa | Up | 2 | Dexa, dexa+vitD3 | (34, 49) |
| FSCN1 | Fascin Actin-Bundling Protein 1 | Down | 3 | Dexa, dexa+vitD3, vitD3 | (37, 50) |
| FTH1 | Ferritin Heavy Chain | Up | 4 | Dexa+vitD3, IL10, IL10+6, TGFb | (50, 55) |
| FTL | Ferritin Light Chain | Up | 3 | Dexa, dexa+vitD3, vitD3 | (34, 37, 50) |
| G6PD | Glucose-6-Phosphate Dehydrogenase | Up | 2 | Dexa+vitD3, TX527 | (37, 60, 50) |
| GILZ | Glucocorticoid-Induced Leucine Zipper | Up | 6 | Asp, dexa, RGZ, IL10, rapa, vitD3 | (32, 35, 36, 47) |
| GPX1 | Glutathione Peroxidase 1 | Up | 2 | Dexa, rapa | (32) |
| HLA-DOB | Human Leukocyte Antigen Class II, DO Beta Chain | Up | 3 | IL10, IL10+6, TGFb | (55) |
| IDH3A | Isocitrate Dehydrogenase 3 Alpha | Up | 2 | Dexa+vitD3, TX527 | (60, 49) |
| IDO1 | Indoleamine 2,3-Dioxygenase | Up | 7 | Asp, dexa, dexa+vitD2, mast, pae, pIC, tropho | (32, 35, 57, 48, 88, 90, 91) |
| IL-10 | Interleukin 10 | Up | 2 | Dexa, hepa | (35, 52) |
| IL-12 | Interleukin 12 | Down | 5 | C5a, dexa, dexa+vitD3, IFNg, LPS Mreg | (35, 38, 50, 53, 54, 92, 97) |
| IL-8 | Interleukin 8 | Up | 2 | IL10, IL10+6 | (55) |
| ILT3 | Immunoglobulin-Like Transcript 3 | Up | 4 | Dexa+vitD3, mitC, tryp, vitD3 | (36, 50, 87, 95) |
| IMDH2 | Inosine Monophosphate Dehydrogenase 2 | Up | 2 | Dexa, rapa | (32) |
| IRF4 | Interferon Regulatory Factor 4 | Down | 2 | IFNg, vitD3 | (62, 54) |
| LAMP3 | Lysosome-Associated Membrane Protein 3 | Down | 3 | Dexa, IL10, vitD3 | (41) |
| LILRB3 | Leukocyte Immunoglobulin Like Receptor B3 | Up | 2 | IL10, TGFb | (55) |
| MRC1 | Mannose Receptor C-Type 1 | Up | 2 | Dexa, IL10 | (32) |
| mTOR | Mammalian Target Of Rapamycin | Up | 2 | Dexa+vitD2, vitD3 | (63, 48) |
| OSF1 | Pleiotrophin | Up | 2 | Dexa, rapa | (32) |
| PCK2 | Phosphoenolpyruvate Carboxykinase 2 | Up | 2 | Dexa+vitD3, TX527 | (60, 49) |
| PDHA1 | Pyruvate Dehydrogenase E1 Alpha 1 Subunit | Up | 2 | Dexa+vitD3, vitD3 | (60, 49) |
| PIK3CG | Phosphatidylinositol-3-Kinase Subunit Gamma | Up | 2 | Dexa+vitD3, vitD3 | (63, 49, 50) |
| PKM2 | Pyruvate Kinase Muscle Isozyme M2 | Up | 2 | Dexa+vitD3, TX527 | (37, 60, 49) |
| RELB | RelB Transcription Factor, NF-κB Subunit | Down | 2 | Cur, IFNg | (53, 54, 86) |
| RGCC | Regulator Of Cell Cycle | Up | 2 | C5a, dexa+vitD3 | (50, 92) |
| STAB1 | Stabilin 1 | Up | 3 | Asp, dexa, IL10 | (32) |
| TGFB | Transforming Growth Factor Beta | Up | 3 | C5a, dexa+vitD3, sem | (51, 92, 93) |
| THBS1 | Thrombospondin 1 | Up | 3 | IL10, IL10+6, TGFb | (55) |
| TNFSF14 | TNF Superfamily Member 14 | Up | 2 | C5a, lipo | (88, 92) |
| TPP1 | Tripeptidyl Peptidase 1 | Up | 3 | Dexa, IL10, rapa | (32) |
The column “Modulation” indicates if a determined gene has been found up- or down-modulated, and the field “Repeats” indicates the amount of different protocols in which each gene or protein has been described. The abbreviations stand for either tolDC induced with asp, Aspergillus cell wall; C5a, C5a, and LPS; cur, curcumin; dexa, dexamethasone; dexa+vitD2, dexamethasone + vitamin D2; dexa+vitD3, dexamethasone + vitamin D3; hepa, hepatocyte growth factor; IFNg, IFN-γ; IL10, IL-10; IL10-6, IL-6 + IL-10; mast, mast cells; mitC, mitomycin C; pae, paeoniflorin; pIC, Polyinosinic:polycytidylic acid; RA, retinoic acid; rapa, rapamycin; RGZ, rosiglitazone; sem, seminal plasma; TGFb, TGF-β; tropho, trophoblasts; tryp, tryptophan deprivation; vitD3, vitamin D3; or regulatory macrophages induced with IFNg Mreg, IFN-γ; LPS Mreg, lipopolysaccharide.
Consequently, this review evidences that the definition of strong biomarkers for tolDC and Mreg is still needed, but also that, although a universal transcriptomic profile of immune tolerance induction might not be achievable, the elaboration of useful panels of biomarkers can still be feasible for determined pools of tolerogenic products. Bearing that in mind, our work could therefore serve as a starting point for developing and guiding further research in this field. For instance, one of the next steps that could be taken in this regard could be to specifically try to validate some of the above discussed genes in different protocols in which they have not been explicitly reported, either because they have been already identified in several approaches—like IDO1 or GILZ—or because the stimuli used to induce the tolerogenic status share some functional or structural resemblance that might translate into the induction of common pathways and metabolic processes. In other words, with this review we intend to provide a useful reference of currently described biomarkers from which direct the investigation of new genes and proteins, most likely protocol-specific.
Thus, the combination of both stimulus-specific and some other partially-common differentially expressed genes could potentially lead to the development of transcriptomic panels of tolerogenic functionality. After all, provided that the relevance of tolerance-inducing cell therapies in the treatment of autoimmune diseases and solid organ transplantation rejection is becoming hugely relevant in the last years, the need for adequate and objective biomarkers is increasing accordingly. And in this context, the definition of panels of tolerogenic functionality for at least a limited pool of protocols would consequently provide a robust tool for the establishment of reliable quality and safety controls for trials using tolDC- and/or Mreg-based therapies in the near future, which would also allow to properly compare them and therefore to dramatically accelerate their translation into the clinic.
Author contributions
EM-C, JN-B, and MM conceived the manuscript. JN-B wrote the manuscript. EM-C and MM reviewed the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by projects PI14/01175 and PI17/01521, integrated in the Plan Nacional de I+D+I and co-supported by the ISCIII-Subdirección General de Evaluación and the Fondo Europeo de Desarrollo Regional (FEDER), and by Project IWT-TMB Grant 140191 (Belgium). JN-B was beneficiary of a FI-DGR PhD contract from AGAUR (Agència de Gestió d'Ajuts Universitaris i de Recerca), supported by the Government of Catalonia (02/2015-01/2018). This work has been supported by positive discussion through A FACTT network (Cost Action BM1305; www.afactt.eu). COST is supported by the EU Framework Programme Horizon 2020.
References
- 1.Matzinger P. The danger model: a renewed sense of self. Science (2002) 296:301–5. 10.1126/science.1071059 [DOI] [PubMed] [Google Scholar]
- 2.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature (1998) 392:245–52. 10.1038/32588 [DOI] [PubMed] [Google Scholar]
- 3.Banchereau J, Briere F, Caux C, Davoust J, Lebecque S, Liu Y-J, et al. Immunobiology of Dendritic Cells. Annu Rev Immunol. (2000) 18:767–811. 10.1146/annurev.immunol.18.1.767 [DOI] [PubMed] [Google Scholar]
- 4.Mueller DL. Mechanisms maintaining peripheral tolerance. Nat Immunol. (2010) 11:21–7. 10.1038/ni.1817 [DOI] [PubMed] [Google Scholar]
- 5.Ganguly D, Haak S, Sisirak V, Reizis B. The role of dendritic cells in autoimmunity. Nat Rev Immunol. (2013) 13:566–77. 10.1038/nri3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopp A-K, Rupp A, Lukacs-Kornek V. Self-antigen presentation by dendritic cells in autoimmunity. Front Immunol. (2014) 5:55. 10.3389/fimmu.2014.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ovchinnikov Dmitry A. Macrophages in the embryo and beyond: much more than just giant phagocytes. Genesis (2008) 46:447–62. 10.1002/dvg.20417 [DOI] [PubMed] [Google Scholar]
- 8.Hilkens CMU, Isaacs JD, Thomson AW. Development of dendritic cell-based immunotherapy for autoimmunity. Int Rev Immunol. (2010) 29:156–183. 10.3109/08830180903281193 [DOI] [PubMed] [Google Scholar]
- 9.Hutchinson JA, Riquelme P, Sawitzki B, Tomiuk S, Miqueu P, Zuhayra M, et al. Cutting edge: immunological consequences and trafficking of human regulatory macrophages administered to renal transplant recipients. J Immunol. (2011) 187:2072–8. 10.4049/jimmunol.1100762 [DOI] [PubMed] [Google Scholar]
- 10.Giannoukakis N, Phillips B, Finegold D, Harnaha J, Trucco M. Phase I (safety) study of autologous tolerogenic dendritic cells in type 1 diabetic patients. Diabetes Care (2011) 34:2026–32. 10.2337/dc11-0472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benham H, Nel HJ, Law SC, Mehdi AM, Street S, Ramnoruth N, et al. Citrullinated peptide dendritic cell immunotherapy in HLA risk genotype-positive rheumatoid arthritis patients. Sci Transl Med. (2015) 7:290ra87. 10.1126/scitranslmed.aaa9301 [DOI] [PubMed] [Google Scholar]
- 12.Jauregui-Amezaga A, Cabezón R, Ramírez-Morros A, España C, Rimola J, Bru C, et al. Intraperitoneal administration of autologous tolerogenic dendritic cells for refractory crohn's disease: a phase I study. J Crohns Colitis (2015) 9:1071–8. 10.1093/ecco-jcc/jjv144 [DOI] [PubMed] [Google Scholar]
- 13.Bell GM, Anderson AE, Diboll J, Reece R, Eltherington O, Harry RA, et al. Autologous tolerogenic dendritic cells for rheumatoid and inflammatory arthritis. Ann Rheum Dis. (2017) 76:227–34. 10.1136/annrheumdis-2015-208456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ten Brinke A, Hilkens CMU, Cools N, Geissler EK, Hutchinson JA, Lombardi G, et al. Clinical use of tolerogenic dendritic cells-harmonization approach in european collaborative effort. Med Inflamm. (2015) 2015:471719 10.1155/2015/471719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satpathy AT, Wu X, Albring JC, Murphy KM. Re(de)fining the dendritic cell lineage. Nat Immunol (2012) 13:1145–54. 10.1038/ni.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adler HS, Steinbrink K. Tolerogenic dendritic cells in health and disease: friend and foe! Eur J Dermatol EJD (2007) 17:476–91. 10.1684/ejd.2007.0262 [DOI] [PubMed] [Google Scholar]
- 17.Morelli AE, Thomson AW. Tolerogenic dendritic cells and the quest for transplant tolerance. Nat Rev Immunol. (2007) 7:610–21. 10.1038/nri2132 [DOI] [PubMed] [Google Scholar]
- 18.Suwandi JS, Nikolic T, Roep BO. Translating mechanism of regulatory action of tolerogenic dendritic cells to monitoring endpoints in clinical trials. Front Immunol. (2017) 8:1598. 10.3389/fimmu.2017.01598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steinbrink K, Wölfl M, Jonuleit H, Knop J, Enk AH. Induction of tolerance by IL-10-treated dendritic cells. J Immunol. (1997) 159:4772–80. [PubMed] [Google Scholar]
- 20.Boks MA, Kager-Groenland JR, Haasjes MSP, Zwaginga JJ, van Ham SM, Ten Brinke A. IL-10-generated tolerogenic dendritic cells are optimal for functional regulatory T cell induction–a comparative study of human clinical-applicable DC. Clin Immunol. (2012) 142:332–42. 10.1016/j.clim.2011.11.011 [DOI] [PubMed] [Google Scholar]
- 21.Fogel-Petrovic M, Long JA, Misso NL, Foster PS, Bhoola KD, Thompson PJ. Physiological concentrations of transforming growth factor β1 selectively inhibit human dendritic cell function. Int Immunopharmacol. (2007) 7:1924–33. 10.1016/j.intimp.2007.07.003 [DOI] [PubMed] [Google Scholar]
- 22.Fedoric B, Krishnan R. Rapamycin downregulates the inhibitory receptors ILT2, ILT3, ILT4 on human dendritic cells and yet induces T cell hyporesponsiveness independent of FoxP3 induction. Immunol Lett. (2008) 120:49–56. 10.1016/j.imlet.2008.06.009 [DOI] [PubMed] [Google Scholar]
- 23.Naranjo-Gómez M, Raïch-Regué D, Oñate C, Grau-López L, Ramo-Tello C, Pujol-Borrell R, et al. Comparative study of clinical grade human tolerogenic dendritic cells. J Transl Med. (2011) 9:89. 10.1186/1479-5876-9-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Jong EC, Vieira PL, Kalinski P, Kapsenberg ML. Corticosteroids inhibit the production of inflammatory mediators in immature monocyte-derived DC and induce the development of tolerogenic DC3. J Leukoc Biol. (1999) 66:201–4. [DOI] [PubMed] [Google Scholar]
- 25.Xia C-Q, Peng R, Beato F, Clare-Salzler MJ. Dexamethasone induces IL-10-producing monocyte-derived dendritic cells with durable immaturity. Scand J Immunol. (2005) 62:45–54. 10.1111/j.1365-3083.2005.01640.x [DOI] [PubMed] [Google Scholar]
- 26.Cabezón R, Carrera-Silva EA, Flórez-Grau G, Errasti AE, Calderón-Gómez E, Lozano JJ, España C, et al. MERTK as negative regulator of human T cell activation. J Leukoc Biol. (2015) 97:751–60. 10.1189/jlb.3A0714-334R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penna G, Adorini L. 1 Alpha,25-dihydroxyvitamin D3 inhibits differentiation, maturation, activation, and survival of dendritic cells leading to impaired alloreactive T cell activation. J Immunol. (2000) 164:2405–11. 10.4049/jimmunol.164.5.2405 [DOI] [PubMed] [Google Scholar]
- 28.Anderson AE, Sayers BL, Haniffa MA, Swan DJ, Diboll J, Wang X-N, et al. Differential regulation of naïve and memory CD4+ T cells by alternatively activated dendritic cells. J Leukoc Biol. (2008) 84:124–33. 10.1189/jlb.1107744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buckland M, Jago C, Fazekesova H, George A, Lechler R, Lombardi G. Aspirin modified dendritic cells are potent inducers of allo-specific regulatory T-cells. Int Immunopharmacol. (2006) 6:1895–1901. 10.1016/j.intimp.2006.07.008 [DOI] [PubMed] [Google Scholar]
- 30.Jiga LP, Bauer TM, Chuang J-J, Opelz G, Terness P. Generation of tolerogenic dendritic cells by treatment with mitomycin C: inhibition of allogeneic T-cell response is mediated by downregulation of ICAM-1, CD80, and CD86. Transplantation (2004) 77:1761–4. 10.1097/01.TP.0000131165.37177.6E [DOI] [PubMed] [Google Scholar]
- 31.Lu L, Lee WC, Takayama T, Qian S, Gambotto A, Robbins PD, et al. Genetic engineering of dendritic cells to express immunosuppressive molecules (viral IL-10, TGF-beta, and CTLA4Ig). J Leukoc Biol. (1999) 66:293–6. [DOI] [PubMed] [Google Scholar]
- 32.Zimmer A, Bouley J, Le Mignon M, Pliquet E, Horiot S, Turfkruyer M, et al. A regulatory dendritic cell signature correlates with the clinical efficacy of allergen-specific sublingual immunotherapy. J Allergy Clin Immunol. (2012) 129:1020–30. 10.1016/j.jaci.2012.02.014 [DOI] [PubMed] [Google Scholar]
- 33.Adnan E, Matsumoto T, Ishizaki J, Onishi S, Suemori K, Yasukawa M, et al. Human tolerogenic dendritic cells generated with protein kinase C inhibitor are optimal for functional regulatory T cell induction — a comparative study. Clin Immunol. (2016) 173:96–108. 10.1016/j.clim.2016.09.007 [DOI] [PubMed] [Google Scholar]
- 34.Gueguen C, Bouley J, Moussu H, Luce S, Duchateau M, Chamot-Rooke J, et al. Changes in markers associated with dendritic cells driving the differentiation of either TH2 cells or regulatory T cells correlate with clinical benefit during allergen immunotherapy. J Allergy Clin Immunol. (2016) 137:545–58. 10.1016/j.jaci.2015.09.015 [DOI] [PubMed] [Google Scholar]
- 35.García-González PA, Schinnerling K, Sepúlveda-Gutiérrez A, Maggi J, Mehdi AM, Nel HJ, et al. Dexamethasone and monophosphoryl lipid a induce a distinctive profile on monocyte-derived dendritic cells through transcriptional modulation of genes associated with essential processes of the immune response. Front Immunol. (2017) 8:1350. 10.3389/fimmu.2017.01350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chamorro S, García-Vallejo JJ, Unger WWJ, Fernandes RJ, Bruijns SCM, Laban S, et al. TLR triggering on tolerogenic dendritic cells results in TLR2 up-regulation and a reduced proinflammatory immune program. J Immunol. (2009) 183:2984–94. 10.4049/jimmunol.0801155 [DOI] [PubMed] [Google Scholar]
- 37.Ferreira GB, Kleijwegt FS, Waelkens E, Lage K, Nikolic T, Hansen DA, et al. Differential protein pathways in 1,25-dihydroxyvitamin d(3) and dexamethasone modulated tolerogenic human dendritic cells. J Proteome Res. (2012) 11:941–71. 10.1021/pr200724e [DOI] [PubMed] [Google Scholar]
- 38.Dixon KO, van der Kooij SW, Vignali DAA, van Kooten C. Human tolerogenic dendritic cells produce IL-35 in the absence of other IL-12 family members. Eur J Immunol. (2015) 45:1736–47. 10.1002/eji.201445217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Flórez-Grau G, Cabezón R, Borgman KJE, España C, Lozano JJ, Garcia-Parajo MF, et al. Up-regulation of EP2 and EP3 receptors in human tolerogenic dendritic cells boost the immunosuppressive activity of PGE2. J Leukoc Biol. (2017) 102:881–95. 10.1189/jlb.2A1216-526R [DOI] [PubMed] [Google Scholar]
- 40.Lombardi V, Luce S, Moussu H, Morizur L, Gueguen C, Neukirch C, et al. Effector and regulatory dendritic cells display distinct patterns of miRNA expression. Immun Inflamm Dis. (2017) 5:310–7. 10.1002/iid3.165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malaguarnera L, Marsullo A, Zorena K, Musumeci G, Di Rosa M. Vitamin D3 regulates LAMP3 expression in monocyte derived dendritic cells. Cell Immunol. (2017) 311:13–21. 10.1016/j.cellimm.2016.09.013 [DOI] [PubMed] [Google Scholar]
- 42.García-González P, Morales R, Hoyos L, Maggi J, Campos J, Pesce B, et al. A short protocol using dexamethasone and monophosphoryl lipid A generates tolerogenic dendritic cells that display a potent migratory capacity to lymphoid chemokines. J Transl Med. (2013) 11:128. 10.1186/1479-5876-11-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Escobar A, Aguirre A, Guzmán MA, González R, Catalán D, Acuña-Castillo C, et al. Tolerogenic dendritic cells derived from donors with natural rubber latex allergy modulate allergen-specific T-cell responses and IgE production. PLoS ONE (2014) 9:e85930. 10.1371/journal.pone.0085930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spallanzani RG, Torres NI, Avila DE, Ziblat A, Iraolagoitia XLR, Rossi LE, et al. Regulatory dendritic cells restrain NK cell IFN-γ production through mechanisms involving NKp46, IL-10, and MHC class I–specific inhibitory receptors. J Immunol. (2015) 195:2141–8. 10.4049/jimmunol.1403161 [DOI] [PubMed] [Google Scholar]
- 45.Maggi J, Schinnerling K, Pesce B, Hilkens CM, Catalán D, Aguillón JC. Dexamethasone and monophosphoryl lipid A-modulated dendritic cells promote antigen-specific tolerogenic properties on naive and memory CD4+ T Cells. Front Immunol. (2016) 7:359. 10.3389/fimmu.2016.00359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ronchetti S, Migliorati G, Riccardi C. GILZ as a mediator of the anti-inflammatory effects of glucocorticoids. Front Endocrinol. (2015) 6:170. 10.3389/fendo.2015.00170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Obreque J, Vega F, Torres A, Cuitino L, Mackern-Oberti JP, Viviani P, et al. Autologous tolerogenic dendritic cells derived from monocytes of systemic lupus erythematosus patients and healthy donors show a stable and immunosuppressive phenotype. Immunology (2017) 152:648–59. 10.1111/imm.12806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dánová K, Klapetková A, Kayserová J, Šedivá A, Špíšek R, Jelínková LP. NF-κB, p38 MAPK, ERK1/2, mTOR, STAT3 and increased glycolysis regulate stability of paricalcitol/dexamethasone-generated tolerogenic dendritic cells in the inflammatory environment. Oncotarget (2015) 6:14123–38. 10.18632/oncotarget.4234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malinarich F, Duan K, Hamid RA, Bijin A, Lin WX, Poidinger M, et al. High mitochondrial respiration and glycolytic capacity represent a metabolic phenotype of human tolerogenic dendritic cells. J Immunol. (2015) 194:5174–86. 10.4049/jimmunol.1303316 [DOI] [PubMed] [Google Scholar]
- 50.Nikolic T, Woittiez NJC, van der Slik A, Laban S, Joosten A, Gysemans C, et al. Differential transcriptome of tolerogenic versus inflammatory dendritic cells points to modulated T1D genetic risk and enriched immune regulation. Genes Immun. (2017) 18:176–83. 10.1038/gene.2017.18 [DOI] [PubMed] [Google Scholar]
- 51.Anderson AE, Swan DJ, Wong OY, Buck M, Eltherington O, Harry RA, et al. Tolerogenic dendritic cells generated with dexamethasone and vitamin D3 regulate rheumatoid arthritis CD4+T cells partly via transforming growth factor-β1. Clin Exp Immunol. (2017) 187:113–23. 10.1111/cei.12870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rutella S, Bonanno G, Procoli A, Mariotti A, de Ritis DG, Curti A, et al. Hepatocyte growth factor favors monocyte differentiation into regulatory interleukin (IL)-10++IL-12low/neg accessory cells with dendritic-cell features. Blood (2006) 108:218–27. 10.1182/blood-2005-08-3141 [DOI] [PubMed] [Google Scholar]
- 53.Rojas D, Krishnan R. IFN-γ generates maturation-arrested dendritic cells that induce T cell hyporesponsiveness independent of Foxp3+ T-regulatory cell generation. Immunol Lett. (2010) 132:31–7. 10.1016/j.imlet.2010.05.003 [DOI] [PubMed] [Google Scholar]
- 54.Rojas-Canales D, Krishnan R, Jessup CF, Coates PT. Early exposure of interferon-γ inhibits signal transducer and activator of transcription-6 signalling and nuclear factor κB activation in a short-term monocyte-derived dendritic cell culture promoting ‘FAST' regulatory dendritic cells. Clin Exp Immunol. (2012) 167:447–58. 10.1111/j.1365-2249.2011.04537.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torres-Aguilar H, Aguilar-Ruiz SR, González-Pérez G, Munguía R, Bajaña S, Meraz-Ríos MA, et al. Tolerogenic dendritic cells generated with different immunosuppressive cytokines induce antigen-specific anergy and regulatory properties in memory CD4+ T cells. J Immunol. (2010) 184:1765–75. 10.4049/jimmunol.0902133 [DOI] [PubMed] [Google Scholar]
- 56.Gröschel S, Piggott KD, Vaglio A, Ma-Krupa W, Singh K, Goronzy J, et al. TLR-mediated induction of negative regulatory ligands on dendritic cells. J Mol Med. (2008) 86:443–55. 10.1007/s00109-008-0310-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pavlović B, Tomić S, Dokić J, Vasilijić S, Vučević D, Lukić J, et al. Fast dendritic cells matured with Poly (I:C) may acquire tolerogenic properties. Cytotherapy (2015) 17:1763–76. 10.1016/j.jcyt.2015.08.001 [DOI] [PubMed] [Google Scholar]
- 58.Bakdash G, Vogelpoel LT, van Capel TM, Kapsenberg ML, de Jong EC. Retinoic acid primes human dendritic cells to induce gut-homing, IL-10-producing regulatory T cells. Mucosal Immunol. (2015) 8:265–78. 10.1038/mi.2014.64 [DOI] [PubMed] [Google Scholar]
- 59.Agrawal S, Ganguly S, Tran A, Sundaram P, Agrawal A. Retinoic acid treated human dendritic cells induce T regulatory cells via the expression of CD141 and GARP which is impaired with age. Aging (2016) 8:1223–35. 10.18632/aging.100973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferreira GB, van Etten E, Lage K, Hansen DA, Moreau Y, Workman CT, et al. Proteome analysis demonstrates profound alterations in human dendritic cell nature by TX527, an analogue of vitamin D. Proteomics (2009) 9:3752–64. 10.1002/pmic.200800848 [DOI] [PubMed] [Google Scholar]
- 61.Penna G, Amuchastegui S, Giarratana N, Daniel KC, Vulcano M, Sozzani S, et al. 1,25-Dihydroxyvitamin D3 selectively modulates tolerogenic properties in myeloid but not plasmacytoid dendritic cells. J Immunol. (2007) 178:145–53. 10.4049/jimmunol.178.1.145 [DOI] [PubMed] [Google Scholar]
- 62.Széles L, Keresztes G, Töröcsik D, Balajthy Z, Krenács L, Póliska S, et al. 1,25-dihydroxyvitamin D3 is an autonomous regulator of the transcriptional changes leading to a tolerogenic dendritic cell phenotype. J Immunol. (2009) 182:2074–83. 10.4049/jimmunol.0803345 [DOI] [PubMed] [Google Scholar]
- 63.Ferreira GB, Vanherwegen A-S, Eelen G, Gutiérrez ACF, Van Lommel L, Marchal K, et al. Vitamin D3 induces tolerance in human dendritic cells by activation of intracellular metabolic pathways. Cell Rep. (2015) 10:711–25. 10.1016/j.celrep.2015.01.013 [DOI] [PubMed] [Google Scholar]
- 64.Pedersen AW, Holmstrøm K, Jensen SS, Fuchs D, Rasmussen S, Kvistborg P, et al. Phenotypic and functional markers for 1α,25-dihydroxyvitamin D3-modified regulatory dendritic cells. Clin Exp Immunol. (2009) 157:48–59. 10.1111/j.1365-2249.2009.03961.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mbongue JC, Nicholas DA, Torrez TW, Kim N-S, Firek AF, Langridge WHR. The role of indoleamine 2, 3-dioxygenase in immune suppression and autoimmunity. Vaccines (2015) 3:703–29. 10.3390/vaccines3030703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mora JR, Iwata M, von Andrian UH. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat Rev Immunol. (2008) 8:685–98. 10.1038/nri2378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ferreira GB, Gysemans CA, Demengeot J, da Cunha JP, Vanherwegen AS, Overbergh L, et al. 1,25-dihydroxyvitamin D3 promotes tolerogenic dendritic cells with functional migratory properties in NOD mice. J Immunol. (2014) 192:4210–20. 10.4049/jimmunol.1302350 [DOI] [PubMed] [Google Scholar]
- 68.Huang Y, Zhao Y, Ran X, Wang C. Increased expression of herpesvirus entry mediator in 1,25-dihydroxyvitamin D3-treated mouse bone marrow-derived dendritic cells promotes the generation of CD4+CD25+Foxp3+ regulatory T cells. Mol Med Rep. (2014) 9:813–8. 10.3892/mmr.2013.1874 [DOI] [PubMed] [Google Scholar]
- 69.Mansilla MJ, Sellès-Moreno C, Fàbregas-Puig S, Amoedo J, Navarro-Barriuso J, Teniente-Serra A, et al. Beneficial effect of tolerogenic dendritic cells pulsed with MOG autoantigen in experimental autoimmune encephalomyelitis. CNS Neurosci Ther. (2015) 21:222–30. 10.1111/cns.12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mansilla MJ, Contreras-Cardone R, Navarro-Barriuso J, Cools N, Berneman Z, Ramo-Tello C, et al. Cryopreserved vitamin D3-tolerogenic dendritic cells pulsed with autoantigens as a potential therapy for multiple sclerosis patients. J Neuroinflamm. (2016) 13:113. 10.1186/s12974-016-0584-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Corripio-Miyar Y, Mellanby RJ, Morrison K, McNeilly TN. 1,25-Dihydroxyvitamin D3 modulates the phenotype and function of Monocyte derived dendritic cells in cattle. BMC Vet Res. (2017) 13:390. 10.1186/s12917-017-1309-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raïch-Regué D, Naranjo-Gómez M, Grau-López L, Ramo C, Pujol-Borrell R, Martínez-Cáceres E, et al. Differential effects of monophosphoryl lipid A and cytokine cocktail as maturation stimuli of immunogenic and tolerogenic dendritic cells for immunotherapy. Vaccine (2012) 30:378–87. 10.1016/j.vaccine.2011.10.081 [DOI] [PubMed] [Google Scholar]
- 73.Raïch-Regué D, Grau-López L, Naranjo-Gómez M, Ramo-Tello C, Pujol-Borrell R, Martínez-Cáceres E, et al. Stable antigen-specific T-cell hyporesponsiveness induced by tolerogenic dendritic cells from multiple sclerosis patients. Eur J Immunol. (2012) 42:771–82. 10.1002/eji.201141835 [DOI] [PubMed] [Google Scholar]
- 74.Lee W-P, Willekens B, Cras P, Goossens H, Martínez-Cáceres E, Berneman ZN, et al. Immunomodulatory effects of 1,25-dihydroxyvitamin D3 on dendritic cells promote induction of T cell hyporesponsiveness to myelin-derived antigens. J Immunol Res. (2016) 2016:5392623. 10.1155/2016/5392623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dánová K, Grohová A, Strnadová P, Funda DP, Šumník Z, Lebl J, et al. Tolerogenic dendritic cells from poorly compensated type 1 diabetes patients have decreased ability to induce stable antigen-specific T cell hyporesponsiveness and generation of suppressive regulatory T cells. J Immunol. (2017) 198:729–40. 10.4049/jimmunol.1600676 [DOI] [PubMed] [Google Scholar]
- 76.Bellinghausen I, König B, Böttcher I, Knop J, Saloga J. Inhibition of human allergic T-helper type 2 immune responses by induced regulatory T cells requires the combination of interleukin-10-treated dendritic cells and transforming growth factor-beta for their induction. Clin Exp Allergy J Br Soc Allergy Clin Immunol. (2006) 36:1546–55. 10.1111/j.1365-2222.2006.02601.x [DOI] [PubMed] [Google Scholar]
- 77.Bellinghausen I, Reuter S, Martin H, Maxeiner J, Luxemburger U, Türeci Ö, et al. Enhanced production of CCL18 by tolerogenic dendritic cells is associated with inhibition of allergic airway reactivity. J Allergy Clin Immunol. (2012) 130:1384–93. 10.1016/j.jaci.2012.08.039 [DOI] [PubMed] [Google Scholar]
- 78.Kryczanowsky F, Raker V, Graulich E, Domogalla MP, Steinbrink K. IL-10–modulated human dendritic cells for clinical use: identification of a stable and migratory subset with improved tolerogenic activity. J Immunol. (2016) 197:3607–17. 10.4049/jimmunol.1501769 [DOI] [PubMed] [Google Scholar]
- 79.Abediankenari S, Ghasemi M. Generation of immune inhibitory dendritic cells and CD4+T regulatory cells inducing by TGF-beta. Iran J Allergy Asthma Immunol. (2009) 8:25–30. 10.08.01/ijaai.2530 [DOI] [PubMed] [Google Scholar]
- 80.Torres-Aguilar H, Blank M, Kivity S, Misgav M, Luboshitz J, Pierangeli SS, et al. Tolerogenic dendritic cells inhibit antiphospholipid syndrome derived effector/memory CD4+ T cell response to β2GPI. Ann Rheum Dis. (2012) 71:120–8. 10.1136/annrheumdis-2011-200063 [DOI] [PubMed] [Google Scholar]
- 81.Chitta S, Santambrogio L, Stern LJ. GMCSF in the absence of other cytokines sustains human dendritic cell precursors with T cell regulatory activity and capacity to differentiate into functional dendritic cells. Immunol Lett. (2008) 116:41–54. 10.1016/j.imlet.2007.11.013 [DOI] [PubMed] [Google Scholar]
- 82.Segovia M, Cuturi MC, Hill M. Preparation of mouse bone marrow-derived dendritic cells with immunoregulatory properties. Methods Mol Biol. (2011) 677:161–8. 10.1007/978-1-60761-869-0_11 [DOI] [PubMed] [Google Scholar]
- 83.Baas MC, Kuhn C, Valette F, Mangez C, Duarte MS, Hill M, et al. Combining autologous dendritic cell therapy with CD3 antibodies promotes regulatory T cells and permanent islet allograft acceptance. J Immunol. (2014) 193:4696–703. 10.4049/jimmunol.1401423 [DOI] [PubMed] [Google Scholar]
- 84.Pletinckx K, Vaeth M, Schneider T, Beyersdorf N, Hünig T, Berberich-Siebelt F, et al. Immature dendritic cells convert anergic nonregulatory T cells into Foxp3- IL-10+ regulatory T cells by engaging CD28 and CTLA-4. Eur J Immunol. (2015) 45:480–91. 10.1002/eji.201444991 [DOI] [PubMed] [Google Scholar]
- 85.Geissler EK. The ONE study compares cell therapy products in organ transplantation: introduction to a review series on suppressive monocyte-derived cells. Transplant Res. (2012) 1:11. 10.1186/2047-1440-1-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rogers NM, Kireta S, Coates PTH. Curcumin induces maturation-arrested dendritic cells that expand regulatory T cells in vitro and in vivo. Clin Exp Immunol. (2010) 162:460–73. 10.1111/j.1365-2249.2010.04232.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Terness P, Oelert T, Ehser S, Chuang JJ, Lahdou I, Kleist C, et al. Mitomycin C-treated dendritic cells inactivate autoreactive T cells: toward the development of a tolerogenic vaccine in autoimmune diseases. Proc Natl Acad Sci USA. (2008) 105:18442–7. 10.1073/pnas.0807185105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen D, Li Y, Wang X, Li K, Jing Y, He J, et al. Generation of regulatory dendritic cells after treatment with paeoniflorin. Immunol Res. (2016) 64:988–1000. 10.1007/s12026-015-8773-7 [DOI] [PubMed] [Google Scholar]
- 89.Rodriguez-Fernandez S, Pujol-Autonell I, Brianso F, Perna-Barrull D, Cano-Sarabia M, Garcia-Jimeno S, et al. Phosphatidylserine-liposomes promote tolerogenic features on dendritic cells in human type 1 diabetes by apoptotic mimicry. Front Immunol. (2018) 9:253. 10.3389/fimmu.2018.00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rodrigues CP, Ferreira ACF, Pinho MP, de Moraes CJ, Bergami-Santos PC, Barbuto JAM. Tolerogenic IDO+ dendritic cells are induced by PD-1-expressing mast cells. Front Immunol. (2016) 7:9. 10.3389/fimmu.2016.00009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Salamone G, Fraccaroli L, Gori S, Grasso E, Paparini D, Geffner J, et al. Trophoblast cells induce a tolerogenic profile in dendritic cells. Hum Reprod. (2012) 27:2598–606. 10.1093/humrep/des208 [DOI] [PubMed] [Google Scholar]
- 92.Zaal A, Nota B, Moore KS, Dieker M, van Ham SM, Ten Brinke A. TLR4 and C5aR crosstalk in dendritic cells induces a core regulatory network of RSK2, PI3Kβ, SGK1, and FOXO transcription factors. J Leukoc Biol. (2017) 102:1035–54. 10.1189/jlb.2MA0217-058R [DOI] [PubMed] [Google Scholar]
- 93.Lenicov FR, Rodrigues CR, Sabatté J, Cabrini M, Jancic C, Ostrowski M, et al. Semen promotes the differentiation of tolerogenic dendritic cells. J Immunol. (2012) 189:4777–86. 10.4049/jimmunol.1202089 [DOI] [PubMed] [Google Scholar]
- 94.Valencia J, Hernández-López C, Martínez VG, Hidalgo L, Zapata AG, Vicente Á, et al. Wnt5a skews dendritic cell differentiation to an unconventional phenotype with tolerogenic features. J Immunol. (2011) 187:4129–39. 10.4049/jimmunol.1101243 [DOI] [PubMed] [Google Scholar]
- 95.Brenk M, Scheler M, Koch S, Neumann J, Takikawa O, Häcker G, et al. Tryptophan deprivation induces inhibitory receptors ILT3 and ILT4 on dendritic cells favoring the induction of human CD4+CD25+ Foxp3+ T regulatory cells. J Immunol. (2009) 183:145–54. 10.4049/jimmunol.0803277 [DOI] [PubMed] [Google Scholar]
- 96.Riquelme P, Amodio G, Macedo C, Moreau A, Obermajer N, Brochhausen C, et al. DHRS9 is a stable marker of human regulatory macrophages. Transplantation (2017) 101:2731–8. 10.1097/TP.0000000000001814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Verreck FA, de Boer T, Langenberg DM, Hoeve MA, Kramer M, Vaisberg E, et al. Human IL-23-producing type 1 macrophages promote but IL-10-producing type 2 macrophages subvert immunity to (myco)bacteria. Proc Natl Acad Sci USA. (2004) 101:4560–5. 10.1073/pnas.0400983101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Di Caro V, Phillips B, Engman C, Harnaha J, Trucco M, Giannoukakis N. Retinoic acid-producing, ex-vivo-generated human tolerogenic dendritic cells induce the proliferation of immunosuppressive B lymphocytes. Clin Exp Immunol. (2013) 174:302–17. 10.1111/cei.12177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gorczynski RM, Bransom J, Cattral M, Huang X, Lei J, Xiaorong L, et al. Synergy in induction of increased renal allograft survival after portal vein infusion of dendritic cells transduced to express TGFβ and IL-10, along with administration of CHO Cells expressing the regulatory molecule OX-2. Clin Immunol. (2000) 95:182–9. 10.1006/clim.2000.4860 [DOI] [PubMed] [Google Scholar]
- 100.Li Y, Chu N, Rostami A, Zhang G-X. Dendritic cells transduced with SOCS-3 exhibit a tolerogenic/DC2 phenotype that directs type 2 Th cell differentiation in vitro and in vivo. J Immunol. (2006) 177:1679–88. 10.4049/jimmunol.177.3.1679 [DOI] [PubMed] [Google Scholar]
- 101.Tan PH, Yates JB, Xue S-A, Chan C, Jordan WJ, Harper JE, et al. Creation of tolerogenic human dendritic cells via intracellular CTLA4: a novel strategy with potential in clinical immunosuppression. Blood (2005) 106:2936–43. 10.1182/blood-2005-05-1826 [DOI] [PubMed] [Google Scholar]
- 102.Tuettenberg A, Fondel S, Steinbrink K, Enk AH, Jonuleit H. CD40 signalling induces IL-10-producing, tolerogenic dendritic cells. Exp Dermatol. (2010) 19:44–53. 10.1111/j.1600-0625.2009.00975.x [DOI] [PubMed] [Google Scholar]
- 103.Fleming BD, Mosser DM. Regulatory macrophages: setting the threshold for therapy. Eur J Immunol. (2011) 41:2498–502. 10.1002/eji.201141717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hutchinson JA, Riquelme P, Geissler EK, Fändrich F. Human regulatory macrophages. Methods Mol Biol. 2011) 677:181–92. 10.1007/978-1-60761-869-0_13 [DOI] [PubMed] [Google Scholar]
- 105.Riquelme P, Haarer J, Kammler A, Walter L, Tomiuk S, Ahrens N, et al. TIGIT + iTregs elicited by human regulatory macrophages control T cell immunity. Nat Commun. (2018) 9:2858. 10.1038/s41467-018-05167-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Akagawa KS. Functional heterogeneity of colony-stimulating factor-induced human monocyte-derived macrophages. Int J Hematol. (2002) 76:27–34. 10.1111/j.1440-1843.2006.00805.x [DOI] [PubMed] [Google Scholar]
- 107.Li G, Kim Y-J, Broxmeyer HE. Macrophage colony-stimulating factor drives cord blood monocyte differentiation into IL-10(high)IL-12absent dendritic cells with tolerogenic potential. J Immunol. (2005) 174:4706–17. 10.4049/jimmunol.174.8.4706 [DOI] [PubMed] [Google Scholar]
- 108.Lucas M, Zhang X, Prasanna V, Mosser DM. ERK activation following macrophage FcgammaR ligation leads to chromatin modifications at the IL-10 locus. J Immunol. (2005) 175:469–77. 10.4049/jimmunol.175.1.469 [DOI] [PubMed] [Google Scholar]
- 109.Zhang X, Edwards JP, Mosser DM. Dynamic and transient remodeling of the macrophage IL-10 promoter during transcription. J Immunol. (2006) 177:1282–8. 10.4049/jimmunol.177.2.1282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Verreck FA, de Boer T, Langenberg DM, van der Zanden L, Ottenhoff TH. Phenotypic and functional profiling of human proinflammatory type-1 and anti-inflammatory type-2 macrophages in response to microbial antigens and IFN-gamma- and CD40L-mediated costimulation. J Leukoc Biol. (2006) 79:285–93. 10.1189/jlb.0105015 [DOI] [PubMed] [Google Scholar]
- 111.Hutchinson JA, Ahrens N, Geissler EK. MITAP-compliant characterization of human regulatory macrophages. Transpl Int Off J Eur Soc Organ Transplant. (2017) 30:765–75. 10.1111/tri.12988 [DOI] [PubMed] [Google Scholar]
- 112.Napoli JL. Physiological insights into all-trans-retinoic acid biosynthesis. Biochim Biophys Acta (2012) 1821:152–67. 10.1016/j.bbalip.2011.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]