Abstract Abstract
A large species diversity has recently been discovered in the genus Agaricus. Six subgenera and 23 sections are now recognised. In this study, three specimens collected from Thailand, formed a monophyletic clade in subgenus Pseudochitonia, based on analyses of ITS sequence data. Further analyses, based on multi-gene sequence data (ITS, LSU, tef1-α), using BEAST, revealed that this clade originated 26.7 Ma. According to their distinct morphological characteristics, phylogenetic position and relatively old divergence time, a new section Cymbiformes is proposed and this section is represented by a new species A.angusticystidiatus. This new section is characterised by the strong iodoform odour of basidiomes and cymbiform basidiospores. Descriptions, colour photographs and illustrations are presented.
Keywords: New taxa, Agaricaceae , Phylogeny, Taxonomy
Introduction
Agaricus L. 1753 (Agaricaceae, Agaricales) is a well-known genus. Many species in this genus are commercially cultivated and served as food. One of the popular edible mushrooms is A.bisporus (J.E. Lange) Imbach, which is the most extensively cultivated mushroom in the world, accounting for 38% of world production (ISMS Edible mushrooms 2017, http://www.isms.biz/edible-mushrooms/). Another popular edible mushroom, A.subrufescens Peck, is also a medicinal mushroom and contains abundant bioactive compounds, for example, some compounds extracted from the basidiomes can be used as antioxidant (De Silva et al. 2012, 2013a, b, Llarena-Hernández et al. 2017). In the field, Agaricus is easily recognised by its white or brown caps with fibrillose scales on the surface, free lamellae, brown spore print and annulate stipe. Under the microscope, it is characterised by brown basidiospores, single or multiseptate cheilocystidia and often lacks pleurocystidia. Habitats of Agaricus are various, the most common being forests and grasslands, such as A.campestris L. of section Agaricus, which can be found gregariously in small groups or in fairy rings in grasslands. Agaricus also exists in arid habitats, for example, A.colpeteorum T. Lebel and A.lamelliperditus T. Lebel & M.D. Barrett of section Minores, which were discovered in arid zones of Australia (Lebel 2013).
The taxonomic, systematic and species delimitation of Agaricus inferred by morphology are variable (Cappelli 1984, Singer 1986). In the 1990s, the application of molecular techniques brought new perspectives to fungal taxonomic research including the genus Agaricus (White et al. 1990). Using phylogenetic analyses, the taxonomy of Agaricus is becoming more and more stable. Zhao et al. (2011) used ITS sequence data from Agaricus specimens from temperate and tropical areas to build a phylogenetic topology for the genus, which revealed eleven new clades and indicated phylogenetic relationships between temperate and tropical species. Zhao et al. (2016) carried out multi-gene phylogenetic and evolutionary molecular clock analyses. In that study, Agaricus was segregated into five subgenera and 20 sections, according to the phylogenetic position and divergence time of each clade. With the recent discovery of an American subgenus and a new clade found in the Caribbean area, Agaricus now contains six subgenera and 23 sections (Zhao et al. 2016, Chen et al. 2017, Parra et al. 2018).
In this study three interesting specimens found near Chiang Mai, Thailand were analysed morphologically and molecularly. We provide a full description and analyses are presented to support the distinction of this material as a new species and section in subgenus Pseudochitonia.
Materials and methods
Morphological examination
Photographs were taken immediately in situ, in the field in Thailand. Basidiomes were wrapped in aluminium foil or kept in plastic boxes separately. Macro morphological characteristics were recorded when specimens were fresh. Every specimen was completely dried in an electrical food drier at 60 °C, then kept in a plastic ziplock bag and deposited in Herbarium Mycologicum Academiae Sinicae (HMAS), Mae Fah Luang University Herbarium (MFLU), Biotec Bandkok Herbarium (BBH) and the Thiers Herbarium at San Francisco State University (SFSU). Colour terms and notations in parentheses are those of Kornerup and Wanscher (1978). Anatomical and cytological characteristics including basidiospores, basidia, cystidia and pileipellis were observed using an Olympus CX31 microscope. Scanning electron microscope (SEM) photos for basidiospores were captured through a Hitachi SU8010 Field Emission SEM (Tokyo, Japan). Measurements were analysed and recorded as X = the mean of length by width ± SD, Q = the quotient of basidiospore length to width and Qm = the mean of Q values ± SD. All the protocols of morphological studies followed Largent’s methodology (Largent 1986).
DNA extraction and PCR
At the Institute of Microbiology Chinese Academy of Science, genomic DNA was extracted from dry specimens by using an E.Z.N.A. Forensic DNA Extraction Kit (D3591-01, Omega Bio-Tek) following the manufacturer’s protocol. PCR amplification was performed following He et al. (2017). Primers for the internal transcribed spacer (ITS), large ribosomal subunit (LSU) and translation elongation factor (tef1-α) were ITS4/ITS5, LR5/LROR and 983f/1567r, respectively (White et al. 1990, Moncalvo et al. 2000, 2002, Morehouse et al. 2003). PCR products were sent to a commercial company for sequencing and both directions were sequenced to ensure accuracy. At the Botanic Garden Meise (BR), genomic DNA was extracted from dry specimens using a CTAB isolation procedure adapted from Doyle (1990). Ca. 10 mg of tissue was ground with a Retsch 300 beadmill. ß-mercaptoethanol (0.2%) was added to the CTAB lysis buffer just prior to extraction; samples were lysed for 1 hour at 60 °C; proteins and polysaccharides were removed by two consecutive extractions with chloroform: isoamylalcohol (24:1), after which DNA was precipitated by the addition of 0.8 volume isopropanol to the aqueous phase. The pellet was washed once in 600 μl 70% ethanol, air-dried and suspended in 100 μl TE pH 8.0. RNA was then digested with RNase A. For PCR amplification of the ITS1-5.8S-ITS2 region of rDNA, ITS1-F (Gardes and Bruns 1993) and ITS4 (White et al. 1990) primers were used. Amplifications were performed in 20 μl reactions containing 2 µl 10× polymerase buffer, 0.2 μM of each dNTP, 200 μg μl-1 bovine serum albumin (BSA), 0.25 μM of forward and reverse primers and 0.5 U Taq polymerase (DreamTaq, Thermo Scientific, St. Leon-Rot, Germany). Cycling was carried out using the following programme: 3 min at 94 °C; 35 cycles of 30 s at 94 °C, 30 s at 52 °C, 60 s at 72 °C; 5 min at 72 °C. PCR products were purified by adding 1 U of Exonuclease I and 0.5 U FastAP Alkaline Phosphatase (Thermo Scientific, St. Leon-Rot, Germany) and incubated at 37 °C for 1 h, followed by inactivation at 80 °C for 15 min. Sequencing was performed by Macrogen Inc. (The Netherlands) with PCR primers.
Sequence alignment, phylogenetic analyses and divergence time estimation
A total of 119 specimens representing 87 species were incorporated in phylogenetic analyses. Three new sequences representing A.angusticystidiatus were generated from this study. They are one ITS sequence from specimen BC088 and two LSU sequences from ZRL2085 and ZRL2043 separately. Details of all sequences are listed in Table 1. Sequences were checked in BioEdit V.7.0.4 first (Hall 2007). Alignments were made by Muscle (Edgar 2004) for each region separately, then adjusted by hand and ambiguous regions removed. Alignments were submitted to TreeBase (Submission ID: 22231). Two data matrices were made for different analyses. The first one is an ITS sequence dataset which contains 84 specimens, all belonging to subgenus Pseudochitonia and an outgroup A.campestris. This dataset was used for Bayesian and Maximum Likelihood analyses. Phylogenetic trees generated by Bayesian Inference (BI) analysis were performed in MrBayes 3.1.2. (Ronquist and Huelsenbeck 2003). Best model is GTR + I + G which was indicated by MrModeltest 2.2 (Nylander 2004). Ten million generations were run for six Markov chains and sampled every 100th generation resulting in 100,000 trees. Burn-in was determined in Tracer v1.6 with effective sample sizes (ESS) higher than 200 (http://tree.bio.ed.ac.uk/software/tracer). Remaining trees were used to calculate Bayesian posterior probabilities (PP). Maximum Likelihood (ML) analysis and bootstrap values calculation were performed in raxmlGUI 1.5b1 using GTRGAMMA model with 1000 replicates (Silvestro and Michalak 2012). The second dataset included 63 ITS, 61 LSU and 59 tef1-α gene sequences from specimens representing the six subgenera of Agaricus. The second multi-gene dataset was used for divergence time estimation. Model selections were performed in jModel Test v. 2 (Darriba et al. 2012) for each gene separately. An XML file was generated in BEAUTI v. 1.8. Priors were set according to the previous fossil-calibrated analysis of Zhao et al. (2016). An independent Monte Carlo Markov Chain of 50 million generations was run and log states every 5,000 generations by BEAST v1.8 (Drummond et al. 2012). The log file was checked in Tracer v. 1.6 (Rambaut et al. 2014) to ensure ESS (Effective Sample Sizes) value higher than 200. An ultrametric maximum-clade-credibility (MCC) tree was summarised using TreeAnnotator 1.8, discarding 10% of states as burn-in and annotating clades with ≥ 0.8 posterior probability.
Table 1.
Taxa information used in the phylogenetic analyses, new taxa are in bold, “T” refers to type.
| Species Name | Collection Number | LSU | ITS | tef1-α | Origin |
|---|---|---|---|---|---|
| Agaricus abruptibulbus | ZRL2012005 | KT951460 | KT951356 | KT951626 | Yunnan, China |
| A.albosquamosus T | LD2012192 | KT951520 | KT951394 | KT951636 | Thailand |
| A.amoenus T | ZRL2010072 | KT951524 | KT951348 | KT951638 | Yunnan, China |
| A. angusticystidiatus | BC088 | – | MG888054 | – | Thailand |
| A. angusticystidiatus | ZRL2085 | MG835413 | KT951434 | – | Thailand |
| A.angusticystidiatus T | ZRL2043 | MG835412 | JF691553 | – | Thailand |
| A. atrodiscus | LD2012185 | KT951473 | KT284912 | KT951653 | Thailand |
| A. benesii | LAPAG283 | – | JF797179 | – | Burgos, Spain |
| A. bernardiformis | CA433 | KT951467 | KT951321 | KT951577 | – |
| A. biannulatus | LAPAG611 | – | JF896229 | – | Sardinia, Italy |
| A. biberi | LAPAG687 | KR006614 | KM657919 | KR006642 | Hungary |
| A. bingensis | ADK1992 | – | KJ540954 | – | Atakora, Benin |
| A. bisporiticus | LD2012111 | KT951507 | KJ575611 | KT951650 | Thailand |
| A. bisporiticus | MCR25 | – | KJ575608 | – | Pakistan |
| A. bisporus | LAPAG446 | KR006611 | KM657920 | KR006640 | Burgos, Spain |
| A. bitorquis | CA427 | KT951491 | KT951320 | KT951646 | |
| A. bitorquis | WZR2012827 | KT951492 | KM657916 | KT951647 | Xingjiang, China |
| A. bohusii | LAPAG562 | KR006613 | KM657928 | KR006641 | Madrid, Spain |
| A. boisseletii | CA123 | – | DQ182531 | – | – |
| A. brunneopictus | ADK2564 | – | JF514518 | – | Plateau Atlantique, Bénin |
| A.brunneopileatus T | ZRL2012115 | KT951489 | KT951404 | KT951587 | Yunnan, China |
| A. brunneosquamulosus | LD2012105 | – | KJ540968 | – | Thailand |
| A. brunneosquamulosus | ZRL4017 | – | JF691549 | – | Thailand |
| A. caballeroi | AH44503 | – | KJ575605 | – | Spain |
| A. campestris | LAPAG370 | KR006607 | KM657927 | KR006636 | Madrid, Spain |
| A. campestroides | LAPAF2 | – | JF727842 | – | Plateaux, Togo |
| A.candidolutescens T | LD2012129 | KT951525 | KT951335 | KT951616 | Thailand |
| A. cf. bernardi | CA383 | KT951469 | KT951319 | KT951576 | |
| A. cf. goossensiae | ADK2171 | – | JF514517 | – | Borgou, Benin |
| A. chiangmaiensis | NTS113 | – | JF514531 | – | Thailand |
| A. comtulus | LAPAG724 | KT951448 | KT951332 | KT951593 | Burgos, Spain |
| A.crassisquamosus T | ZRL2012607 | KT951510 | KT951376 | KT951645 | Tibet, China |
| A. cupressicola | LAPAG889 | KT951465 | KT951334 | KT951649 | Roma, Italy |
| A. desjardinii | WZR2012907 | KT951474 | KM657901 | KT951644 | Xinjiang, China |
| A.dilutibrunneus T | ZRL2012010 | KT951512 | KT951358 | KT951569 | Yunnan, China |
| A. dolichopus | ZRL2012715 | KT951502 | KT951382 | KT951573 | Tibet, China |
| A. dolichopus | ZRL2014120 | – | KT951433 | – | – |
| A. duplocingulatus | ZRL3064 | – | KJ540966 | – | Thailand |
| A.erectosquamosus T | LD2012165 | KT951509 | KT951338 | KT951565 | Thailand |
| A. erythrosarx | MURU6080 | – | JF495068 | – | – |
| A. freirei | CA186 | – | DQ185553 | – | – |
| A. fuscofibrillosus | WC913 | – | AY484684 | – | – |
| A. fuscopunctatus | LD2012115 | – | KJ575612 | – | Thailand |
| A. fuscovelatus | RWK2100 | – | KJ577973 | – | – |
| A. gennadii | CA339 | – | KT951318 | KT951575 | – |
| A.grandiomyces T | ZRL2012611 | KR006624 | KM657879 | KR006652 | Tibet, China |
| A. gratolens | ZRL3093 | KT951488 | JF691548 | – | Thailand |
| A. haematinus | ZRL2109 | – | KT951435 | – | Thailand |
| A. haematinus | ZRL2136 | – | JF691552 | – | Thailand |
| A. hondensis | RWK1938 | – | DQ182513 | – | USA |
| A. huijsmanii | LAPAG639 | KT951444 | KF447889 | KT951571 | Navarra, Spain |
| A. kunmingensis | ZRL2012015 | KT951506 | KT951361 | KT951642 | Yunnan, China |
| A. kunmingensis | ZRL2012007 | – | KT951427 | – | Yunnan, China |
| A.lamellidistans T | ZRL3099 | – | JF691556 | – | Thailand |
| A. laskibarii | LAPAG115 | – | AY943975 | – | Landes, France |
| A.leucocarpus T | LD2012159 | KX083981 | KU975101 | KX198048 | Thailand |
| A.leucolepidotus T | LD201214 | KT951519 | KT951336 | KT951635 | Thailand |
| A.linzhiensis T | ZRL2012618 | KT951503 | KT951378 | KT951582 | Tibet, China |
| A. litoralis | LAPAG420 | KT951483 | KT951327 | KT951572 | Burgos, Spain |
| A. litoraloides | ZRL2011249 | KT951523 | KT951353 | KT951580 | Yunnan, China |
| A. magnivelaris | F2389 | – | JF727851 | – | – |
| A. martinicensis | F2815 | KX084032 | JF727855 | KX198038 | MartiniqueFrance |
| A. megacystidiatus | LD2012179 | – | KF305946 | – | Thailand |
| A. microvolvatulus | LD201271 | KT951508 | KJ575614 | KT951651 | Thailand |
| A. murinocephalus | ZRL3044 | – | JF691555 | – | Thailand |
| A. nevoi | LAPAG257 | KR006606 | KM657922 | KR006635 | Burgos, Spain |
| A. nevoi | LAPAG535 | – | KT951330 | KT951574 | Teruel, Spain |
| A. nigrobrunnescens | DEH632 | – | JX308267 | – | Hawaii, USA |
| A.nigrogracilis T | ZRL2012014 | KR006621 | KM657882 | KR006647 | Yunnan, China |
| A. niveogranulatus | LD201124 | – | KJ540959 | – | Thailand |
| A. padanus | WZR2012903 | KR006616 | KM657903 | KR006644 | Xingjiang, China |
| A.pallidobrunneus T | ZRL2012358 | KT951471 | KT951370 | KT951566 | Yunnan, China |
| A. parvitigrinus | CA158 | – | AY899267 | – | – |
| A. pattersoniae | RWK1415 | – | AY943974 | – | – |
| A. phaeolepidotus | CA217 | – | DQ185552 | – | – |
| A. pilosporus | LAPAG227 | – | KT951425 | – | Burgos, Spain |
| A. pseudolangei | ZRL3012 | – | JF691551 | – | Thailand |
| A. rufoaurantiacus | LAPAM15 | KX671708 | KT951313 | KT951641 | Dominican Republic |
| A. silvaticus | ALG07 213 | KT951307 | KT951567 | Algonquin, ON, Canada | |
| A. sinodeliciosus | WZR2012822 | KT951518 | KM657907 | KT951648 | Xingjiang, China |
| A. sordidocarpus | LD201237 | – | KJ540946 | – | Thailand |
| A. subrufescens | ZRL2012722 | KT951451 | KT951383 | KT951632 | Yunnan, China |
| A. subsaharianus | ADK4732 | – | JF440300 | – | Ouagadougou, Burkina Faso |
| A. sylvaticus | LAPAG382 | KR006608 | KM657929 | KR006637 | Burgos, Spain |
| A. sylvaticus | ZRL2012013 | KT951500 | KT951360 | KT951570 | Thailand |
| A. sylvaticus | ZRL2012568 | KT951501 | KT951371 | KT951568 | Tibet, China |
| A. tibetensis | ZRL2012585 | KR006633 | KM657895 | KR006658 | Tibet, China |
| A. tollocanensis | CA235 | – | AY703913 | – | – |
| A. toluenolens | CA911 | – | KJ540947 | – | – |
| A.trisulphuratus complex | LAPAF7 | KR006605 | KM657924 | KR006634 | Plateaux, Togo |
| A.trisulphuratus complex | Swk079 | KT951472 | KT951343 | KT951561 | Lanjak-Entimau, Malaysia |
| A.trisulphuratus complex | ZRL2014023 | – | KT951428 | – | China |
| A.trisulphuratus complex | ZRL2014024 | – | KT951429 | – | China |
| A.trisulphuratus complex | ZRL2014030 | – | KT951432 | – | China |
| A.trisulphuratus complex | ZRL2132 | – | JF691558 | – | Thailand |
| A. tytthocarpus | ZRLWXH3077 | KR006618 | KM657889 | KR006645 | Fujian, China |
| A. variabilicolor | ZRL4002 | – | KT951438 | – | Thailand |
| A. variabilicolor | ZRL4007 | – | KT951439 | – | Thailand |
| A. variabilicolor | ZRL4012 | – | KT951440 | – | Thailand |
| A. variicystis | LD201228 | – | KT951426 | – | Thailand |
| A.variicystis T | LD201234 | KT951517 | KT951339 | KT951562 | Thailand |
| A. xanthodermulus | CA160 | – | AY899273 | – | – |
| A. xanthodermus | CA15 | – | AY899271 | – | – |
| A. xanthodermus | LAPAG387 | KR006609 | KM657923 | KR006638 | Soria, Spain |
| A. xanthosarcus | Goossens5415 | – | JF514523 | – | – |
| A. sp. | CA486 | – | JF797189 | – | – |
| A. sp. | CA820 | – | JF727861 | – | – |
| A. sp. | LD2012162 | KT951493 | KT951337 | KT951563 | Thailand |
| A. sp. | NT020 | – | JF797197 | – | Thailand |
| A. sp. | Swk014 | KT951482 | KT951342 | KT951654 | Lanjak-Entimau, Malaysia |
| A. sp. | ZRL133 | KT951505 | KT951344 | KT951656 | Thailand |
| A. sp. | ZRL2010010 | KT951511 | KT951347 | KT951639 | Thailand |
| A. sp. | ZRL2010099 | KT951479 | KT951349 | KT951564 | Yunnan, China |
| A. sp. | ZRL2012267 | KT951504 | KT951368 | KT951655 | Yunnan, China |
| A. sp. | ZRL2012629 | KR006627 | KM657890 | KR006656 | Tibet, China |
| A. sp. | ZRLWXH3078 | KT951464 | KT951464 | KT951643 | Fujian, China |
| A. sp. | ZRLWXH3161 | KT951526 | KT951391 | KT951615 | Guangdong, China |
| A. sp. | ZRLWXH3140 | – | KT951441 | – | Guangdong, China |
| Heinemannomyces sp. | ZRL185 | KT951527 | KT951346 | KT951657 | Thailand |
Results
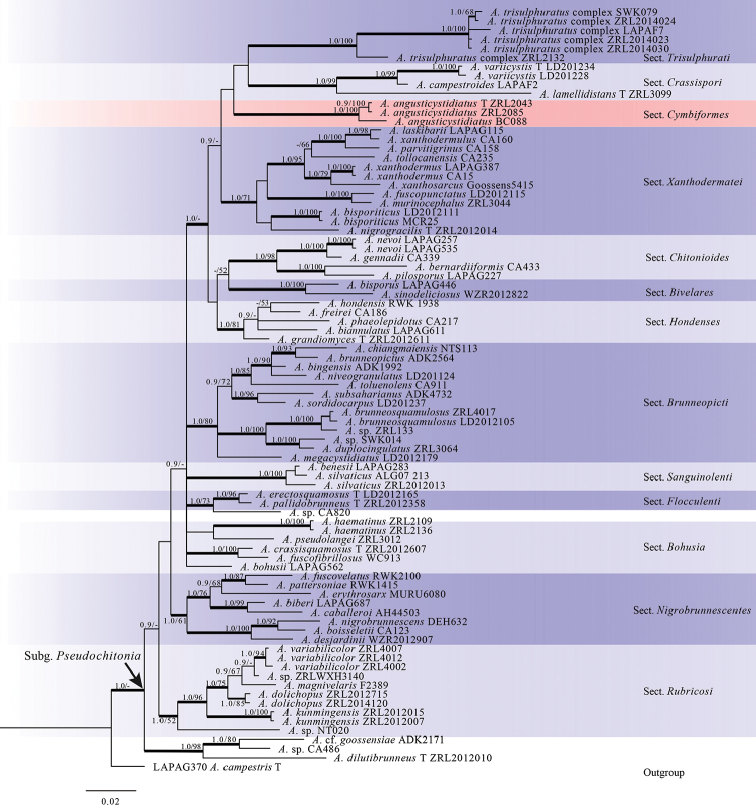
The Bayesian tree from ITS sequences is shown in Figure 1. A total of 84 sequences are represented from 12 sections of subg. Pseudochitonia and A.campestris was used as outgroup. All sections are well supported both by posterior probabilities (PP) and bootstrap (BS). Phylogenetic trees generated from Bayesian and ML analyses showed identical topologies and are also almost identical with those of Zhao et al. (2016) with the exception of A.dilutibrunneus R.L. Zhao, which clustered with two unknown specimens (A. sp./CA486 and A.cf.goossensiae/ADK2171) and formed a monophyletic clade in our analyses, isolated from all other species in the previous study (Zhao et al. 2016). Our three specimens (ZRL2043, ZRL2085 and BC088) formed a monophyletic clade in subg. Pseudochitonia which is fully supported both in PP and BS values and located at an isolated position (Fig. 1).
Figure 1.
Phylogenetic tree of AgaricussubgenusPseudochitonia generated from Bayesian analysis of ITS sequences, rooted with A.campestris. Bayesian posterior probability (PP) values ≥ 0.9 or Bootstrap support (BS) values ≥ 50% are indicated at the internodes (PP/BS). The branches in bold mean the related PP > 0.95, “T” refers to sequences from type specimen.
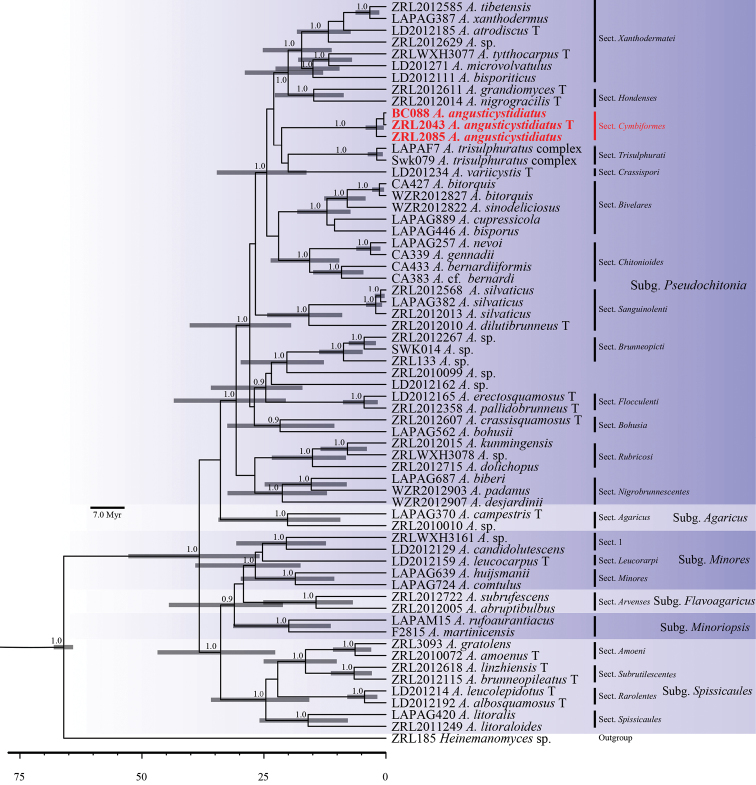
The multi-gene MCC tree is shown in Figure 2. It was conducted based on the dataset of multi-gene sequences. A total of 63 specimens were included, comprising 43 specimens used in ITS analysis, 19 specimens from five subgenera and an outgroup Heinemannomyces sp. All subgenera and sections are well-supported statistically. Agaricus diverged at the stem age 66 Ma (million years ago), all subgenera diverged between 29.2–33.9 Ma and sections diverged between 20–26.9 Ma. Our three specimens formed a new monophyletic clade in subg. Pseudochitonia with strong PP support and this clade diverged at 26.7 Ma.
Figure 2.
Maximium Clade Credibility tree of genus Agaricus based on ITS, LSU and tef1-α gene sequences with the outgroup Heinemannomyces sp. Posterior probability values equal or above 0.9 are annotated at the internodes. The 95% highest posterior density of divergence time estimation are marked by horizontal bars.
Taxonomy
Agaricus (Pseudochitonia) section Cymbiformes
M.Q. He & R.L. Zhao sect. nov.
MB824147
Type species.
Agaricusangusticystidiatus M.Q. He, Desjardin., K.D. Hyde & R.L. Zhao
Etymology.
In reference to the cymbiform basidiospores.
Original description.
KOH reaction negative, Schäffer’s reaction negative on dry specimens. No discolouration on touching, but discolouration reddish-brown on cutting. Annulus membranous. Smell strong iodoform. Basidiospores cymbiform and cheilocystidia narrow with variable shapes.
Agaricus angusticystidiatus
M.Q. He, Desjardin, K.D. Hyde & R.L. Zhao sp. nov.
MB825177
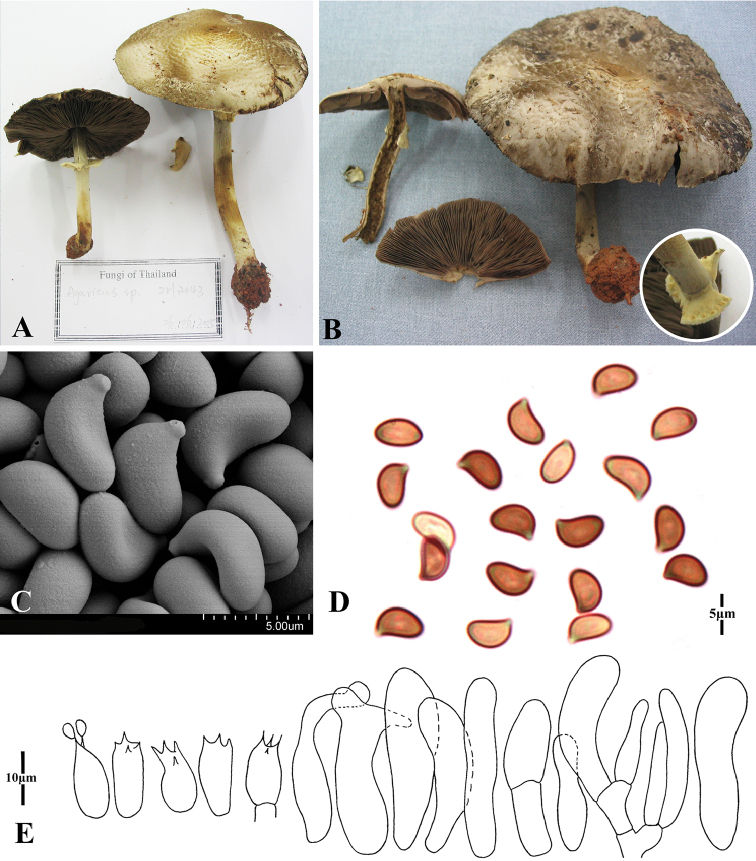
Figure 3.
Morphology of AgaricusangusticystidiatusA, B basidiomes C, D basidiospores E basidia and cheilocystidia.
Etymology.
refers to the narrow clavate cheilocystidia.
Type.
Thailand, Chiang Mai Province, Mae Taeng, Baan Mae Sae village, on Hwy 1095 near 50 km marker, 19°14.599'N, 98°39.456'E, alt. 960 m. In rain forest dominated by Castanopsisarmata, Castanopsis sp., Pinus sp., Lithocarpus sp., 26 June 2005, collected by Jennifer Kerekes. Holotype: ZRL2043 (HMAS279593); Isotype: BBH19428 and SFSUZRL2043,
Original description.
Pileus 40–80 mm diam., plano-convex, applanate, broadly umbonate; surface concentric squamulose with small skull-cup at disc, appressed, slightly fissured, light brown (6D8), brown (7E3), greyish-brown (5D5), dark brown (6D6) against the grey (8E3) background. Context 4–5 mm thick at disc, fragile, white to grey (8E3) in age. Lamellae free, crowded, lamellulae with 3–4 lengths, 3–4 mm broad, normal to slightly ventricose, brown (7E5) to dark brown (7F7-8), edge colour similar to the gill itself. Stipe 55–100 × 5–8 (base 8–15) mm, cylindrical bulbous, with rhizomorphs in most cases, hollow, surface glabrous to silky, white to dark brown (6D6). Annulus pendent or percurrent; single; upper side membranous, white; lower side surface powdery, light yellow (4B2) grain-like dots in circulate; superior, persistent, edge entire, up to 5 mm broad. Smell of iodoform. No colour change on touching; light dull red, greyish brown (7D4) on cutting.
KOH reaction: negative. Schäffer’s reaction: negative on dry specimens.
Basidiospores 5–6.5 × 3–4 (–4.5) µm [X = 5.6 ± 0.5 × 3.8 ± 0.4, Q = 1.1–2.2, Qm = 1.52 ± 0.7, n = 20], cymbiform, some endosporium, no germ pore, brown. Basidia 10–15 × 5.5–7 µm, clavate, hyaline, smooth, 4-spored. Pleurocystidia absent. Cheilocystidia 20–30 (–45) × 5–8 µm, occasionally one septum, narrowly clavate to clavate, some with elongated top, rarely subcapitate, hyaline, smooth. Pileipellis cutis consisting of 3–5 µm diam. hyphae, hyaline, smooth, non-constricted at septa. Annulus hyphae same as pileipellis.
Habit.
Gregarious on soil in rain forest which is mainly dominated by Castanopsisarmata, Castanopsis sp., Pinus sp., Lithocarpus sp.
Distribution.
Thailand, Chiang Mai Province (type distribution).
Other materials examined.
Thailand, Chiang Mai Province, Mae Taeng, Ban Mae Sae Village, on Hwy 1095 near 50 km marker, 19°14.599'N, 98°39.456'E, elev. ca. 960 m, 3 July 2004, collected by Thitiya Boonpratuang, ZRL2085 (HMAS279594, BBH19468 and SFSUZRL2085); Thailand, Chiang Mai Province, Mae Taeng, Mushrooms research center, 30 July 2014, collected by Boontiya Chuankid, BC088 (MFLU 14-0903).
Notes.
This new species is morphologically distinguished from other Agaricus species by its strong iodoform smell, context reddish-brown discolouration on cutting, cymbiform basidiospores and narrow cheilocystidia with variable shapes. Phylogenetic analyses confirmed it is a member of the subgenus Pseudochitonia with an isolated phylogenetic position in Agaricus. This new species is similar to A.iodolens Heinem. & Gooss.-Font. of section Xanthodermatei, because both have relatively slender basidiomes and odour of iodine (Naritsada et al. 2014). However, this new species has cymbiform basidiospores and a bulbous stipe, while those of A.iodolens are ellipsoid and an equal stipe (Zoberi 1972). Agaricuslamellidistans R.L. Zhao and A.variicystis L.J. Chen, K. D. Hyde & R. L. Zhao of section Crassispori resemble this new species, because all have greyish-brown pilei and cymbiform basidiospores. These species lack discolouration on cutting, while those of A.angusticystidiatus have dull red discolouration on cutting (Zhao et al. 2016).
Discussion
Based on phylogenetic and morphological studies, we propose A.angusticystidiatus as a new species in subgenus Pseudochitonia. Furthermore, the dating analysis, based on multi-gene sequences, indicated that A.angusticystidiatus diverged at 26.7 Ma which is slightly older than other sections in Agaricus (18–26 Ma, in Zhao et al. 2016). Therefore, a new section Cymbiformes is proposed, which presently only contains species A.angusticystidiatus. Thus up to now, there are six subgenera and 24 sections in the genus Agaricus (Zhao et al. 2016; Chen et al. 2017; Parra et al. 2018).
Zhao et al. 2016 had conducted a reconstruction of the taxonomic system of Agaricus. In that study, they used the following criteria to recognise subgenera and sections: “(i) they must be monophyletic and statistically well-supported in the multi-gene analyses; (ii) their respective stem ages should be roughly equivalent and subgenera stem ages must be older than section stem ages; and (iii) they should be identifiable phenotypically, whenever possible” (Zhao et al. 2016). That means divergence time has been used as an additional criterion to rank taxa of above species level in Agaricus. Later, the criterion of divergence time, along with phylogenetic, monophyletic and morphological support, has been accepted in other new subgenus and section recognitions in Agaricus, such as a new subgenus Minoriopsis (Chen et al. 2017); and a new section Kerrigania (Parra et al. 2018).
As mentioned before, this proposed new section Cymbiformes has a closely phylogenetic relationship with sections Trisulphurati and Crassispori. In morphology, all of them differed with other sections of Agaricus by the combination of negative Schäffer’s reaction, chemical odours such as phenol, ink or carbolic acid and basidiospores endosporium and often cymbiform. However, section Trisulphurati has woolly squamules on the surfaces of the pileus and stipe and the other two sections only have appressed squamules at the centre of the pileus. Furthermore, this new section Cymbiformes could be separated from section Crassispori by its negative KOH reaction and developed annulus (the latter is positive KOH reaction and with fragile annulus) (Zhao et al. 2016).
So far, section Cymbiformes is only known from a tropical area. The cymbiform basidiospores are rare in Agaricus species. Presently there are three Agaricus species from tropical areas which have this kind of basidiospores. They are A.angusticystidiatus of section Cymbiformes and A.lamellidistans and A.variicystis of section Crassispori (Zhao et al. 2016). In phylogenetic analyses, these two sections also show a close phylogenetic position, which is similar to previous studies (specimens ZRL2043 and ZRL2085 were treated as A. sp. in Zhao et al. 2011; Zhao et al. 2016). The presence of cymbiform basidiospores is a common character in another genus Micropsalliota of Agaricaceae. In phylogenetic analyses, Agaricus is sister to Hymenagaricus, then sister to Chlorophyllum, Heinemannomyces and Micropsalliota (Zhao et al. 2017) and all of them have tropical distribution habitats. Thus we hypothesised that cymbiform basidiospores have formed at least twice in evolutionary events and are associated with tropical environments.
Supplementary Material
Acknowledgements
This project was conducted under the financial support of the National Key R&D Program of China (Project No. 2018YFD0400200), the National Natural Science Foundation of China (Project ID:31470152 and 31360014) and Beijing Innovative Consortium of Agriculture Research System (Project ID: BAIC05-2018).
Citation
He M-Q, Chuankid B, Hyde KD, Cheewangkoon R, Zhao R-L (2018) A new section and species of Agaricus subgenus Pseudochitonia from Thailand. MycoKeys 40: 53–67. https://doi.org/10.3897/mycokeys.40.26918
Contributor Information
Ratchadawan Cheewangkoon, Email: ratchadawan.c@cmu.ac.th.com.
Rui-Lin Zhao, Email: zhaorl@im.ac.cn.
References
- Capelli A. (1984) Agaricus, Fungi Europaei. Libreria editrice Biella Giovanna, I – 21047, Saronno, Italy 1, 1–560.
- Chen J, Callac P, Parra LA, Karunarathna SC, He MQ, Moinard M, De Kesel A, Raspé O, Wisitrassameewong K, Hyde KD, Zhao RL. (2017) Study in AgaricussubgenusMinores and allied clades reveals a new American subgenus and contrasting phylogenetic patterns in Europe and Greater Mekong Subregion. Persoonia 38: 170–196. 10.3767/003158517X695521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Zhao RL, Parra LA, Guelly AK, De Kesel A, Rapior S, Hyde KD, Chukeatirote E, Callac P. (2015) AgaricussectionBrunneopicti: a phylogenetic reconstruction with descriptions of four new taxa. Phytotaxa 192(3): 145–168. 10.11646/phytotaxa.192.3.2 [DOI] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. (2012) jModelTest 2: more models, new heuristics and parallel computing. Nature methods 9: 772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed]
- De Silva DD, Rapior S, Fons F, Bahkali AH, Hyde KD. (2012) Medicinal mushrooms in supportive cancer therapies: an approach to anti-cancer effects and putative mechanisms of action. Fungal Diversity 55(1): 1–35. 10.1007/s13225-012-0151-3 [DOI] [Google Scholar]
- De Silva DD, Rapior S, Hyde KD, Bahkali AH. (2013a) Medicinal mushrooms in prevention and control of diabetes mellitus. Fingal Diversity 56(1): 1–29. 10.1007/s13225-012-0187-4 [DOI] [Google Scholar]
- De Silva DD, Rapior S, Sudarman E, Stadler M, Xu JC, Alias SA, Hyde KD. (2013b) Bioactive metabolites from macrofungi: ethnopharmacology, biological activities and chemistry. Fungal Diversity 62(1): 1–40. 10.1007/s13225-013-0265-2 [DOI] [Google Scholar]
- Doyle JJ. (1990) Isolation of plant DNA from fresh tissue. Focus 12: 13–15. [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. (2012) Bayesian phylogenetics with BEAUti and BEAST 1.7. Molecular Biology and Evolution 29(8): 1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32(5): 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for basidiomycetes—application to the identification of mycorrhizae and rusts. Molecular ecology 2(2): 113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Hall T. (2007) BioEdit v7. Ibis Biosciences, Carlsbad Available from: http://www.mbio.ncsu.edu/BioEdit/BioEdit.html
- He MQ, Chen J, Zhou JL, Ratchadawan C, Hyde KD, Zhao RL. (2017) Tropic origins, a dispersal model for saprotrophic mushrooms in AgaricussectionMinores with descriptions of sixteen new species. Scientific Reports 7(1): 5122. 10.1038/s41598-017-05203-5 [DOI] [PMC free article] [PubMed]
- Kornerup A, Wanscher JH. (1978) Methuen handbook of colour (3rd edn). Eyre Methuen, London.
- Largent DL. (1986) How to identify mushrooms to genus vol. I–V. Mad River Press Eureka.
- Lebel T. (2013) Two new species of sequestrate Agaricus (sectionMinores) from Australia. Mycological Progress 12(4): 699–707. 10.1007/s11557-012-0879-x [DOI] [Google Scholar]
- Llarena-Hernández RC, Renouf E, Vitrac X, Mérillon JM, Savoie JM. (2017) Antioxidant Activities and Metabolites in Edible Fungi, a Focus on the Almond Mushroom Agaricussubrufescens. In: Mérillon J-M, Ramawat KG. (Eds) Fungal Metabolites.Springer International Publishing AG, Cham, 739–760.
- Moncalvo JM, Lutzoni FM, Rehner SA, Johnson J, Vilgalys R. (2000) Phylogenetic relationships of Agaric fungi based on nuclear large subunit ribosomal DNA sequences. Systematic biology 49(2): 278–305. 10.1093/sysbio/49.2.278 [DOI] [PubMed] [Google Scholar]
- Moncalvo JM, Vilgalys R, Redhead SA, Johnson JE, James TY, Aime MC, Hofstetter V, Verduin SJW, Larsson E, Baroni TJ, Thorn RG, Jacobsson S, Clémençon H, Miller Jr OK. (2002) One Hundred and Seventeen Clades of Euagarics. Molecular phylogenetics and evolution 23(3): 357–400. 10.1016/S1055-7903(02)00027-1 [DOI] [PubMed] [Google Scholar]
- Morehouse EA, James TY, Ganley ARD, Vilgalys R, Berger L, Murphy PJ, Longcore JE. (2003) Multilocus sequence typing suggests that the chytrid pathogen of amphibians is a recently emerged clone. Molecular Ecology 12(2): 395–403. 10.1046/j.1365-294X.2003.01732.x [DOI] [PubMed] [Google Scholar]
- Naritsada T, Nawaz R, Khalid AN, Chen J, Hyde KD, Zhao R, Parra LA, Hanif M, Moinard M, Callac P. (2014) Morphological and molecular characterization of three Agaricus species from tropical Asia (Pakistan, Thailand) reveals a new group in section Xanthodermatei. Mycologia 106(6): 1220–1232. 10.3852/14-076 [DOI] [PubMed] [Google Scholar]
- Nylander J. (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Parra LA, Angelini C, Ortiz-Santana B, Mata G, Billette C, Rojo C, Chen J, Callac P. (2018) The genus Agaricus in the Caribbean. Nine new taxa mostly based on collections from the Dominican Republic. Phytotaxa 345(3): 219–271. 10.11646/phytotaxa.345.3.2 [DOI] [Google Scholar]
- Rambaut A, Suchard MA, Xie D, Drummond AJ. (2014) Tracer v1.6. Available from http://beast.bio.ed.ac.uk/Tracer [accessed on 24 July 2015]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12): 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Silvestro D, Michalak I. (2012) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12(4): 335–337. 10.1007/s13127-011-0056-0 [DOI] [Google Scholar]
- Singer R. (1986) The Agaricales in modern taxonomy. ed. 4. J. Cramer, Vaduz.
- White TJ, Bruns T, Lee SJ, Taylor JL. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky J, White TJ. (Eds) PCR Protocols: a Guide to Methods and Applications.Academic Press, San Diego, 315–322.
- Zhao RL, Karunarathna SC, Raspé O, Parra LA, Guinberteau J, Moinard M, De Kesel A, Barroso G, Courtecuisse R, Hyde KD, Guelly AK. (2011) Major clades in tropical Agaricus. Fungal Diversity 51(1): 279–296. 10.1007/s13225-011-0136-7 [DOI] [Google Scholar]
- Zhao RL, Zhou JL, Chen J, Margaritescu S, Sánchez-Ramírez S, Hyde KD, Callac P, Parra LA, Li GJ, Moncalvo JM. (2016) Towards standardizing taxonomic ranks using divergence times–a case study for reconstruction of the Agaricus taxonomic system. Fungal Diversity 78(1): 239–292. 10.1007/s13225-016-0357-x [DOI] [Google Scholar]
- Zhao RL, Li GJ, Sánchez-Ramírez S, Stata M, Yang ZL, Wu G, Dai YC, He SH, Cui BK, Zhou JL, Wu F. (2017) A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal Diversity 84(1): 43–74. 10.1007/s13225-017-0381-5 [DOI] [Google Scholar]
- Zoberi MH. (1972) Tropical Macrofungi: some common species. Palgrave Macmillan, London, 99–108. 10.1007/978-1-349-01618-1_20 [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.