Abstract Abstract
Sinohygrocybegen. nov., typified by S.tomentosipessp. nov., is described upon morphological and molecular evidence. The new genus is characterised by its sinuate to subdecurrent or short deccurent, usually furcate and interveined and relatively distant lamellae, dry and whitish tomentose stipe, thin-walled ellipsoid to oviod, non-constricted basidiospores and particularly elongated basidia and a ratio of basidiospore to basidium length of >5 to 8; it is close to genera Chromosera and Gloioxanthomyces of the tribe Chromosereae, but morphologically differs from Chromosera in less umbilicate basidiomata, tomentose stipe and usually longer basidia and differs from Gloioxanthomyces in more robust basidioma and less glutinous pileus and/or stipe surface. Phylogenetic analyses, with ITS-LSU-RPB2 data, also indicate that Sinohygrocybe forms a very distinct and independent clade at the generic level. In addition, a Chinese new record G.nitidus is described here.
Keywords: East Asia, new record species, new taxa, phylogeny overview
Introduction
Hygrophoraceae Lotsy (Hymenomycetes, Basidiomycota) is a large family in Agaricales, including 26 genera and over 600 species (Lodge et al. 2014). In a six-gene phylogenetic tree of Agaricales, Hygrophoraceae, Pterulaceae Corner, Typhulaceae Jülich and some small groups formed a Hygrophoroid clade, which is one of the six largest clades in Agaricales (Matheny et al. 2006); and in a genome based mushroom tree of life, Hygrophoraceae and Clavariaceae Chevall. are representative families of the suborder Hygrophorineae Aime, Dentinger & Gaya, which is one of the seven suborders of the Agaricales (Dentinger et al. 2016). Traditionally, the family Hygrophoraceae referred to a group of agaricoid, waxy-gilled and white-spored mushrooms; and a majority of the members are classified in the type genus Hygrophorus Fr. and genus Hygrocybe (Fr.) P. Kumm. Morphological characters of the Hygrophoraceae taxa are relatively simple (usually without annulus or volva and a cystidiate) amongst the agaric fungi and their basidioma colours are often very susceptible to the environmental conditions and developmental stages, making their classification and identification difficult, so it is often challenging to make correct identification and taxonomy of them just according to morphological recognition (Young 2005). Modern molecular techniques have been revolutionising the taxonomy and phylogeny of Hygrophoraceae.
Lodge et al. (2014) had conducted the most comprehensive molecular phylogenetic study on the family until now, therefore their systematic viewpoint on Hygrophoraceae is adopted in this paper. According to their study, the family could be divided into four groups at subfamily level, i.e. subfamily Hygrophoroideae E. Larss., Lodge, Vizzini, Norvell & S.A. Redhead, Hygrocyboideae Padamsee & Lodge, Lichenomphalioideae Lücking & Redhead and Cuphophylloid grade. The subfamily Hygrocyboideae could be divided into three tribes, i.e. tribe Chromosereae, Humidicuteae and Hygrocybeae; and the tribe Chromosereae included two sister genera, Chromosera Redhead, Ammirati & Norvell and Gloioxanthomyces Lodge, Vizzini, Ercole & Boertm.
Chromosera, the type genus of the tribe Chromosereae, was erected to accommodate Omphalinacyanophylla (Fr.) Quél. which was originally described from Sweden and combined as C.cyanophylla (Fr.) Redhead, Ammirati & Norvell (Redhead et al. 1995, 2012). Now, another four species, formerly placed in Hygrocybe or Hygrophorus, are also classified into Chromosera, i.e. C.citrinopallida (A.H. Sm. & Hesler) Vizzini & Ercole originally described from USA, C.lilacina (P. Karst.) Vizzini & Ercole originally described from the northern Fennoscandia, C.viola (J. Geesink & Bas) Vizzini & Ercole originally described from Belgium and C.xanthochroa (P.D. Orton) Vizzini & Ercole originally described from Scotland (Lodge et al. 2014).
Gloioxanthomyces is a small genus with only two known species, the type species G.vitellinus (Fr.) Lodge, Vizzini, Ercole & Boertm. originally described from Europe and G.nitidus (Berk. & M.A. Curtis) Lodge, Vizzini, Ercole & Boertm. from North America (Crous et al. 2004, Lodge et al. 2014). Before the recognition of Gloioxanthomyces, those two species were usually placed in the genus Hygrocybe as H.vitellina (Fr.) P. Karst and H.nitida (Berk. & M.A. Curtis) Murrill, respectively. Morphologically, the main differences between the two species were in their basidiospore sizes: G.nitidus had ellipsoid to oblong basidiospores, measuring 7–10 × 5–6 μm with Q = 1.3–1.8; while G.vitellinus had subglobose basidiospores, measuring 6.5–8.5 × 5–7 μm with Q=1.1–1.6 (Boertmann 1990). Since their differences were limited, the two taxa seemed to be conspecific (Boertmann 2011). However, according to the phylogenetic analyses with ITS data by Boertmann (2012), the European collections clearly clustered together as the G.vitellinus species clade, while the North American materials independently formed another group as the G.nitidus species clade, thus they could actually be sharply defined as two separated sister species.
During the studies on the Chinese Hygrophoraceae in recent years, some collections morphologically corresponding to tribe Chromosereae were collected. Comprehensive observation and analyses revealed some interesting findings, which can contribute to the taxonomic knowledge of the tribe. In this paper, we aim to: 1) formally describe a new genus of tribe Chromosereae from East Asia based upon morphological and molecular analyses and present a Chinese new record of Gloioxanthomycesnitidus; 2) reconstruct the phylogeny of the family Hygrophoraceae using 3 gene regions, i.e. the internal transcribed spacer region (ITS), the large subunit nuclear ribosomal RNA region (nrLSU) and the nuclear RPB2 6F to 7.1R region (RPB2). Detailed studies were therefore conducted and the results are presented as follows.
Materials and methods
Morphological studies
Specimens were photographed and annotated in the field and then dried in an electric drier. Macroscopic descriptions were gained from the original field notes and photographs. Colour descriptions followed Kornerup and Wanscher (1978). Tissue sections were immersed in 5% potassium hydroxide (KOH) and/or 1% Congo Red solution for microscopical examinations, but in distilled water for colour descriptions of basidia, pileipellis and stipitipellis. From a mature specimen, over 40 basidiospores and 20 basidia were randomly selected and measured under a light microscope in KOH. The notation (a)b–c(d) was used to describe dimensions where the range b–c representing 90% or more of the measured values and a, d were the extreme values. The length/width ratio of spores was presented as Q and the mean ratio was presented as Qm. The studied specimens were deposited in the Fungal Herbarium of Guangdong Institute of Microbiology (GDGM), Guangzhou, China.
Molecular studies
Genomic DNA was extracted from the herbarium specimens using the Sangon Fungus Genomic DNA Extraction kit (Sangon Biotech Co., Ltd., Shanghai, China) according to the manufacturer’s instructions. The ITS, LSU and RPB2 gene regions were amplified by Polymerase Chain Reaction, using universal primers ITS1F/ITS5 and ITS4 (White et al. 1990; Gardes and Bruns 1993), LR0R and LR5 (http://biology.duke.edu/fungi/mycolab/primers.htm) and RPB2-6F and RPB2-7.1R (Matheny 2005), respectively. Amplified products were sequenced by Beijing Genomic Institute (BGI) using the same primers. The abi format sequences were assembled by SeqMan version 7.1.0 (DNAStar, Inc.) and then the assembled sequences were submitted to GenBank.
In this study, two datasets were constructed. The first one is an ITS-LSU-RPB2 matrix of the family Hygrophorceaeae for making a comprehensive phylogenetic tree and analysing the positions of the new taxa; most known species of Hygrophoraceae with available sequences from reliable sources were included in the dataset, each of them having at least an LSU sequence and Typhulaphacorrhiza (Reichard) Fr. was selected as the outgroup referred from Yang et al. (2013) and Lodge et al. (2014). The second dataset is an ITS matrix of the tribe Chromosereae and Hygrocybeconica (Schaeff.) P. Kumm. and H.conicavar.conicoides (P.D. Orton) Boertm. were chosen as outgroups. Each gene was independently aligned on the online MAFFT service (Katoh et al. 2017), then combined by the Geneious software (Biomatters Ltd.) for the first dataset. Maximum likelihood phylogenetic trees were generated by the RAxML software (Stamatakis 2014) on the CIPRES service (Miller et al. 2010) with 1000 bootstrap replications using the default options.
Results
Molecular phylogenetic results
The combined 3-gene dataset composed of 120 samples (Table 1), including 5 newly sequenced samples and 115 published ones. In the final matrix, the ITS, LSU and RPB2 regions comprised positions 1 to 1751, 1752 to 2873, 2874 to 3759, respectively. In the 3-gene Maximum Likelihood tree (Fig. 1), the four Chinese collections (GDGM43351 and GDGM43347 from Sichuan province, GDGM50075 and GDGM50149 from Hunan province) formed a strong monophyletic clade with 100% bootstrap support, which was near the Chromosera-Gloioxanthomyces clade composed of members of Chromosera and Gloioxanthomyces with 76% bootstrap support.
Table 1.
Sequences information of samples used for the ITS-LSU-RPB2 combined tree. Newly generated sequences were bold.
| Species name | Isolate/voucher ID | ITS | LSU | RPB2 |
|---|---|---|---|---|
| Acantholichen albomarginatus | MDF543 | KT429797 | KT429809 | – |
| Acantholichen campestris | DIC595b | KT429798 | KT429810 | KT429818 |
| Acantholichen galapagoensis | MDF057 | KT429784 | KT429799 | KT429811 |
| Acantholichen galapagoensis | MDF058 | KT429785 | KT429800 | KT429812 |
| Acantholichen galapagoensis | MDF089 | KT429786 | KT429801 | – |
| Acantholichen galapagoensis | MDF090 | KT429787 | KT429802 | KT429813 |
| Acantholichen galapagoensis | MDF093 | KT429790 | KT429803 | KT429814 |
| Acantholichen galapagoensis | MDF094 | KT429791 | KT429804 | KT429815 |
| Acantholichen galapagoensis | MDF100 | KT429792 | KT429805 | KT429816 |
| Acantholichen pannarioides | MDF352 | KT429795 | KT429807 | KT429817 |
| Acantholichen pannarioides | Bungartz 5593 | EU825953 | EU825953 | – |
| Acantholichen sorediatus | DIC335 | KT429794 | KT429806 | – |
| Acantholichen variabilis | MDF679 | KT429796 | KT429808 | – |
| Ampulloclitocybe clavipes | DJL06TN40 | – | KF381542 | KF407938 |
| Ampulloclitocybe clavipes | AFTOL-ID 542 | AY789080 | AY639881 | AY780937 |
| Arrhenia auriscalpium | Lutzoni Lamoure 910824-3 | U66428 | U66428 | – |
| Arrhenia lobata | Lutzoni Lamoure 910824-1 | U66429 | U66429 | – |
| Cantharellula umbonata | RDY-1366 (SFSU) | KF381519 | AF261443 | – |
| Cantharocybe brunneovelutina | DJL-BZ-1883 (holotype) | KX452404 | HM588721 | – |
| Cantharocybe gruberi | AFTOL-ID 1017 | DQ200927 | DQ234540 | DQ385879 |
| Cantharocybe gruberi | AH24539 | JN006422 | JN006420 | – |
| Cantharocybe virosa | TENN 63483(holotype) | KX452405 | JX101471 | – |
| Chromosera citrinopallida | DUKE8895 | U66435 | U66435 | – |
| Chromosera citrinopallida | D. Boertmann 2006/2 | KF291072 | KF291073 | – |
| Chrysomphalina chrysophylla | AFTOL-ID 1523 | – | DQ457656 | DQ192180 |
| Chrysomphalina chrysophylla | S.A. Redhead 7700 | – | U66430 | U66430 |
| Chrysomphalina grossula | OSC 113667 | – | EU652372 | EU644703 |
| Chrysomphalina grossula | OSC 113683 | – | EU652373 | EU644704 |
| Cora minor | Luecking 15243 | EU825968 | EU825968 | – |
| Cuphophyllus acutoides var. pallidus | CFMR TN-257 | – | KF291097 | – |
| Cuphophyllus adonis | MES-152 | – | KF291036 | KF291037 |
| Cuphophyllus aff. pratensis | PBM-752 | – | DQ457650 | KF442252 |
| Cuphophyllus aurantius | CFMR PR-6601 | – | KF291100 | KF291102 |
| Cuphophyllus bicolor | DJL-PR-2 | – | KF291056 | – |
| Cuphophyllus flavipes | Hattori-JP-6 | – | KF291045 | KF291047 |
| Cuphophyllus fornicatus | D. Boertmann 2009/94 | – | KF291124 | – |
| Cuphophyllus pratensis | DJL-Scot-8 | – | KF291058 | – |
| Cuphophyllus sp. | AM01 | – | HM026542 | – |
| Dictyonema glabratum | AFTOL-ID 1995 | DQ917656 | DQ917661 | – |
| Dictyonema glabratum | Luecking 15581 | EU825958 | EU825958 | – |
| Dictyonema glabratum | Luecking 16563 | EU825956 | EU825956 | – |
| Dictyonema glabratum | R06 | EU825959 | EU825959 | – |
| Dictyonema glabratum | R11 | EU825960 | EU825960 | – |
| Dictyonema glabratum | R18 | EU825961 | EU825961 | – |
| Dictyonema glabratum | R20 | EU825963 | EU825963 | – |
| Gliophorus aff. psittacinus | CFMR JP-4 | KF291079 | KF291080 | – |
| Gliophorus graminicolor | TJB-10048 | KF381520 | KF381545 | KF407936 |
| Gliophorus psittacinus | D. Boertmann 2002/10 | KF291075 | KF291076 | KF291078 |
| Gloioxanthomyces nitidus | GDGM41710 | MG712283-4 | MG712282 | MG711911 |
| Haasiella splendidissima | Herbarium Roux n. 3666 | JN944398 | JN944399 | – |
| Haasiella splendidissima | Herbarium Roux n. 4044 | JN944400 | JN944401 | – |
| Haasiella splendidissima | JVG1071013-1 | JN944395 | JN944396 | – |
| Haasiella venustissima | A. Gminder 971488 | KF291092 | KF291093 | – |
| Haasiella venustissima | E.C. 08191 | JN944393 | JN944394 | – |
| Humidicutis sp. 2 | CFMR PR4047 | – | KF291151 | KF291149 |
| Humidicutis sp. 2 | DJL-2103 CFMR PR-6524 | KF291150 | KF291151 | |
| Humidicutis sp. 3 | D.J. Lodge DJL-BZ-3 | KF291110 | KF291111 | – |
| Hygroaster albellus | AFTOL ID 1997 | KF381521 | EF551314 | KF381510 |
| Hygroaster nodulisporus | AFTOL-ID 2020 | – | EF561625 | KF381511 |
| Hygrocybe acutoconica f. japonica | CFMR JP-2 | KF291161 | KF291162 | |
| Hygrocybe aff. citrinovirens | DJL05TN10 | KF291090 | KF291091 | – |
| Hygrocybe aff. conica | PBM 918 | AY854074 | DQ071739 | AY803747 |
| Hygrocybe aff. prieta | DJL-BZ-65 | KF291168 | KF291169 | |
| Hygrocybe caespitosa | DMWV-03-737 | KF291104 | KF291105 | KF291107 |
| Hygrocybe cantharellus | AFTOL-ID 1714 | DQ490628 | DQ457675 | |
| Hygrocybe ceracea | D. Boertmann 2002/7 | KF291108 | KF291109 | – |
| Hygrocybe cf. acutoconica | DJL04NC2 | KF291117 | KF291118 | KF291120 |
| Hygrocybe chloochlora | DJL-BZ-32 | EU435147 | EU435147 | – |
| Hygrocybe chlorophana | Boertmann 2002/9 | EU435148 | EU435148 | KF381513 |
| Hygrocybe coccinea | AFTOL-ID 1715 | DQ490629 | DQ457676 | DQ472723 |
| Hygrocybe coccinea | Boertmann02/8 | EU435146 | EU435146 | KF291114 |
| Hygrocybe constrictospora | D. Boertmann 2007/38 | KF291115 | KF291116 | |
| Hygrocybe glutinipes var. rubra | DJL05NC9 | EU435149 | EU435149 | – |
| Hygrocybe helobia | AK-124 | KF291182 | KF291183 | – |
| Hygrocybe hypohaemacta | DJL-BZ-105 | EU435150 | EU435150 | KF291165 |
| Hygrocybe konradii var. konradii | Boertmann 2004/6 | KF306329 | KF306330 | – |
| Hygrocybe lepida | Boertmann 2002/2 | KF306333 | KF306334 | – |
| Hygrocybe melleofusca | DJL-PR-EV | KF291154 | KF291155 | – |
| Hygrocybe miniata | AK-110 | KF291179 | KF291180 | |
| Hygrocybe miniata f. longipes | AFTOL-ID 1891 | DQ490630 | DQ457677 | DQ472724 |
| Hygrocybe noninquinans | DJL-PR-1 | KF291127 | KF291129 | KF291128 |
| Hygrocybe occidentalis var. occidentalis | Cancerel PR 02 | EU435151 | EU435151 | – |
| Hygrocybe punicea | DJL-SCOT-B2 | KF291133 | KF291134 | – |
| Hygrocybe purpureofolia | DJL04NC1 | KF291192 | KF291193 | |
| Hygrocybe reidii | DJL-ENG-15-2006 | KF291158 | KF291159 | |
| Hygrocybe rosea | DJL-PR-4 | KF291197 | KF291198 | – |
| Hygrophorus agathosmus | EL2-00 | – | AY586660 | – |
| Hygrophorus cossus | SJ94064 | AY548963 | AY548963 | |
| Hygrophorus hyacinthinus | SJ950830 | – | HM143012 | – |
| Hygrophorus olivaceoalbus | SJ91060 | – | AY586662 | – |
| Hygrophorus russula | JP-3 | KF291216 | KF291217 | KF291219 |
| Hygrophorus sordidus | AFTOL-1338 | DQ490632 | AF042562 | – |
| Lichenomphalia umbellifera | J. Geml-2 | U66445 | U66445 | KF381515 |
| Neohygrocybe ingrata | GWG H. ingrata 23-10-06 (ABS) | KF291225 | KF291226 | – |
| Neohygrocybe ingrata | TN-62 voucher DJL05TN62 | KF381525 | KF381558 | KF381516 |
| Neohygrocybe ingrata | CFMR NY-43 | – | KF291223 | KF291224 |
| Neohygrocybe ovina | K(M) 187568 | KF291228 | KF291229 | – |
| Neohygrocybe ovina | GWG H. ovina Rhosisaf (ABS) | KF291233 | KF291234 | KF291236 |
| Neohygrocybe subovina | WRWV04-752 (DEWV 5366) | – | KF291142 | KF291138 |
| Neohygrocybe subovina | CFMR NC-61 | KF291136 | KF291137 | – |
| Neohygrocybe subovina | DJL04TN16 (GRSM 77065) | KF291140 | KF291141 | – |
| Omphalina epichysium | Redhead3140 | U66442 | U66442 | – |
| Omphalina grossula | Gulden 417/75 | – | U66444 | U66444 |
| Omphalina hudsoniana | LUTZ-920728.4a | U66446 | U66446 | – |
| Omphalina obscurata | Lam L73-101 | U66448 | U66448 | – |
| Omphalina philonotis | LUTZ930804-5 | U66449 | U66449 | – |
| Omphalina sphagnicola | LUTZ930810 | U66453 | U66453 | – |
| Omphalina velutina | LUTZ-930812.1 | U66454 | U66454 | – |
| Omphalina velutipes Lamoure | L77 | U66455 | U66455 | – |
| Omphalinawynniae A. H. Smith | 82899 | – | U66457 | U66457 |
| Porpolomopsis aff. calyptriformis | DJL05TN80 | KF291246 | KF291247 | KF291249 |
| Porpolomopsis calyptriformis | EB-ENG-3 | KF291242 | KF291243 | KF291245 |
| Porpolomopsis lewelliniae | TJB-10034 | KF291238 | KF291239 | KF291241 |
| Pseudoarmillariella bacillaris | HKAS76377 | KC222315 | KC222316 | – |
| Pseudoarmillariella ectypoides | AFTOL-ID 1557 | DQ192175 | DQ154111 | DQ474127 |
| Sinohygrocybe tomentosipes | GDGM43351 | MG685872 | MG696901 | MG696905 |
| Sinohygrocybe tomentosipes | GDGM43347 | – | MG696900 | MG696904 |
| Sinohygrocybe tomentosipes | GDGM50075 | MG685873 | MG696902 | MG696906 |
| Sinohygrocybe tomentosipes | GDGM50149 | MG685874 | MG696903 | MG675232 |
| Typhula phacorrhiza | TP21 | AF134710 | AF393079 | AY218525 |
Figure 1.

Phylogenetic overview of the family Hygrophoraceae inferred from ITS-LSU-RPB2 data using Maximum Likelihood (ML) method. Typhulaphacorrhiza was selected as outgroup. Bootstrap values (≥50%) were presented around the branches. The newly generated sequences are shown in bold.
The ITS dataset included 30 samples of all known taxa of tribe Chromosereae and 2 Hygrocybe sequences chosen as the outgroups, the matrix length is 679 bp. In the ITS Maximum Likelihood tree (Fig. 2), collections of the species G.nitidus and G.vitellinus were clustered together with 93% and 100% support values, respectively and the North American and the East Asian G.nitidus were clustered as sister groups with 93% support value; all the members of Chromosera (except C.viola), Gloioxanthomyces and Sinohygrocybe were clustered together with 95%, 93% and 100% support values, respectively; and the Chromosera-Gloioxanthomyces clade was presented as the sister clade of the Sinohygrocybe clade with strong support value (100%).
Figure 2.
Phylogenetic overview of the tribe Chromosereae inferred from ITS data using ML method. Two Hygrocybeconica sequences were rooted as outgroups. Bootstrap values (≥50%) are shown around the branches. GenBank accession numbers of downloaded sequences were added after the species name and the collection locations were added at the ends. NA, EA and EU referred to North America, East Asia and Europe, respectively. The newly generated sequences are shown in bold.
Taxonomy
Sinohygrocybe
C.Q. Wang, Ming Zhang & T.H. Li gen. nov.
MB824821
Diagnosis.
Differs from Chromosera and Gloioxanthomyces by its less omphalioid, more robust basidiomata, dry to subviscid pileus, dry and white tomentose stipe, more elongated basidia, higher length ratio (up to 8 times) of basidia to basidospores.
Etymology.
Sino- refers China, the holotype’s location of the genus; -hygrocybe indicates that it is a Hygrocybe-like genus.
Type species.
Sinohygrocybetomentosipes C.Q. Wang, Ming Zhang & T.H. Li
Description.
Basidiomata medium-sized, subcaespiotose. Pileus convex to applanate, slightly depressed in the centre, yellow, orangish-yellow to orange, dry to subviscid, slightly when wet, never strongly gelatinised or glutinous. Lamellae adnate to decurrent, concolorous with pileus, with usually furcate and interveined lamellulae. Stipe yellow to whitish or almost concolorous with pileus, yellow or covered by white to yellowish-white tomentum. Basidiospores ellipsoid to oblong, ovoid, Qm = 1.6-1.7, not constricted, thin-walled, inamyloid, hyaline, smooth; basidia usually 4-sterigmate, 41–80 μm long, ratio of basidia to basidiospore length over 5 (up to 8), with basal clamp connection. Pileipellis and stipitipellis a cutis. Lamellar trama subregular. Clamp connections present throughout.
Sinohygrocybe tomentosipes
C.Q. Wang, Ming Zhang & T.H. Li sp. nov.
MB824824
Figure 3.
Basidiomata of Sinohygrocybetomentosipes (a–b GDGM43351 c–d GDGM43352 e GDGM43347 f GDGM50075 g–h GDGM50149). Scale bars: 2 cm.
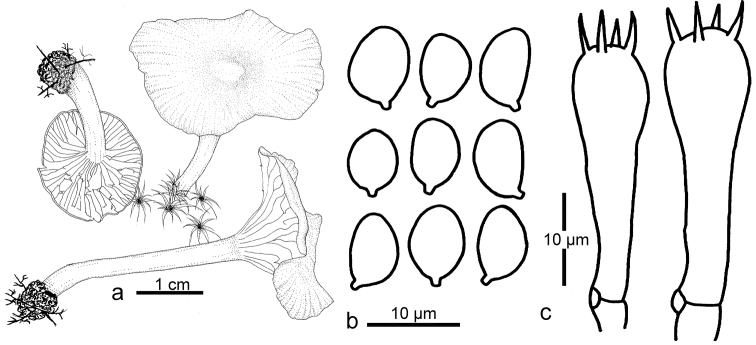
Figure 4.
Line drawings of Sinohygrocybetomentosipes. a Basidiomata b Basidiospores c Basidia d Elements of pileipellis cells e Elements of gill trama.
Diagnosis.
Differs from the other members of the tribe Chromosereae by its larger and more robust basidiomata, concolorous yellow pileus, lamellae and the subsurface of stipe, usually furcate and interveined lamellae and lamellulae, white fibrillose stipe surface, long basidia (up to 80 μm), ratio of basidia to basidiospore length over 5 and even up to 8.
Etymology.
The species epithet tomentosipes refers to the tomentose stipe.
Type.
China. Sichuan Province, Panzhihua City, Yanbian County, Gesala Eco-tourism Area, at 27°16'N, 101°26'E, alt. 3100 m, 24 Aug 2013, Ming Zhang (GDGM43351, holotype).
Description.
Basidiomata small to medium-sized. Pileus 2.5–6 cm diam., convex to applanate, usually slightly depressed in the centre, smooth, dry but subviscid when wet, light yellow to vivid yellow (3A5–8) or to deep yellow (4A5–8), or light orange to dark orange (5A5–8), becoming paler when dry; margin even, straight or upturned and occasionally split when mature. Lamellae up to 7 mm wide, adnate to sinuate or decurrent, distant, 17–22 lamellae per pileus, with 1–3 lamellulae between two complete lamellae, usually furcate, often interveined or anastomosing at lamella base, thick, concolorous with the pileus; lamellar base and lamellulae irregular and occasionally the whole hymenophore irregular; lamellar edge even and concolorous. Context concolorous with lamellae and pileus, unchanged when cut. Stipe 4–6.5 × 0.6–1.2 cm, central or occasionally eccentric, subcylindrical, moderately to densely covered with white tiny adpressed fibres. Odour indistinct.
Basidiospores 8–10(–10.5) × (4.5–)5–7(–7.5) μm, Q = (1.3–)1.5–1.8, Qm = 1.6–1.7, ellipsoid to ellipsoid-oblong, ovoid, not constricted, thin-walled, hyaline, smooth. Basidia 41–80 × 4–10 μm, strongly elongated, narrow clavate, 4-spored, thin-walled; sterigmata up to 10 μm long; ratio of basidia to basidiospore length over 5 and up to 8. Hymenophoral trama subregular, yellow, made up of thin-walled hyphae 3–15 µm wide and usually less than 100 μm long and some conducting elements. Pileipellis a cutis, made up of repent hyphae 3–9 µm wide with the terminal elements 30–80 µm long. Stipitipellis a cutis, with thin-walled hyphae (5–7 μm wide). Clamp-connections present in all tissues.
Habitat and known distribution.
Gregarious, caespitose, or scattered in broad-leaf forest in subtropical temperate transition zone, so far known only from Sichuan and Hunan Provinces in China.
Additional specimens examined.
CHINA, Sichuan Province, Panzhihua City, Yanbian County, Gesala Eco-Tourism Area, at 27°16'N, 101°26'E, alt. 3100 m, 24 Aug 2013, Ming Zhang (GDGM43347), Chao-Qun Wang (GDGM43352); Hunan Province, Zhuzhou City, Yanling County, Taoyuandong National Nature Reserve, at 26°19'N, 114°00'E, alt. 1534 m, 23 Nov 2013, Chao-Qun Wang (GDGM50075 and GDGM50149).
Gloioxanthomyces nitidus
(Berk. & M.A. Curtis) Lodge, Vizzini, Ercole & Boertm., Fungal Diversity 64: 50 (2014)
Figure 5.
Basidiomata of Gloioxanthomycesnitidus (a–b GDGM41710 c–d GDGM42150 e–f GDGM42151).
Figure 6.
Line drawings of Gloioxanthomycesnitidus (GDGM41710). a basidiomata b basidiospores c basidia.
= Hygrophorusnitidus Berk. & M.A. Curtis, Ann. Mag. nat. Hist., Ser. 2 12: 424 (1853).
Description.
Pileus 1.5–3.5 cm wide, convex to nearly plane with a slightly depressed disc, strongly glutinous, yellow, light orange yellow to apricot yellow, even whitish-yellow when mature, clearly striate at margin; pileus margin usually slightly undulating, slightly incurved when young, expanded to flat or partially uplifted when mature. Context thin, yellow to nearly concolorous with pileus, hygrophanous and translucent. Lamellae arcuate-decurrent, narrow at both ends, bright yellow or slightly orange yellow, waxy and fragile, subdistant, usually having 1–3 unequal lamellulae between two lamellae; lamellar edge even, usually gelatinised and sometimes translucent. Stipe 2.5–6 × 0.2–0.5 cm, cylindrical, hollow, yellow to slightly greenish-yellow, smooth, sticky or glutinous with a layer of viscid and translucent material when wet, nearly equal mostly but usually tapering at base.
Basidiospores 7–9(11) × 5–6.5(7.5) μm, Q=1.25–1.7, Qm=1.48, ellipsoid, not constricted, smooth, hyaline, thin-walled. Basidia 29–39 × 7.5–10 μm, clavate, 4-spored; sterigmata up to 5 μm. Lamellar trama subregular, with hyphal elements 10–20 μm wide. Pileipellis an ixotrichoderm. Clamp connections present.
Habitat and known distribution.
Solitary or scattered, on moist ground in a mixed forest with mosses in North-eastern China, so far known in North America and East Asia.
Material examined.
CHINA. Jilin Province, Antu County, Changbaishan Mountains, 20 August 2012, Ming Zhang, Jiang Xu, Chao-Qun Wang (GDGM41710, GDGM42150 and GDGM42151).
Discussion
Phylogenetically, the distinction of the three subfamilies (Lodge et al. 2014) within Hygrophoraceae has very convincing support in the multi-locus tree of this study (Fig. 1). In addition, the establishment of the three well-defined monophyletic tribes in subfamily Hygrocyboideae is supported in this phylogenetic frame where the tribe Hygrocybeae with 73% support values and the tribe Humidicuteae with low support value are sister clades, while the tribe Chromosereae with 76% support values is located at their base. However, the cuphophylloid grade appears not to be monophyletic, thus more studies are still needed to understand the phylogenetic positions of Ampulloclitocybe, Cantharocybe H.E. Bigelow & A.H. Sm. and Cuphophyllus (Donk) Bon.
In the multi-gene analyses, Sinohygrocybe is placed together with two other genera in Chromoserae. Chromosera and Gloioxanthomyces are sister genera under the monophyletic tribe Chromosereae, while Sinohygrocybe is an independent generic lineage; and the distances between Sinohygrocybe and Chromosera or Gloioxanthomyces are further than the distance between Chromosera and Gloioxanthomyces. Such results are confirmed in the ITS phylogenetic tree (Fig. 2). According to the Blastn results, the ITS and LSU sequence identities of the new species to the known taxa are not more than 76% and 96%, respectively, with the Chromosera and Gloioxanthomyces sequences in GenBank. Thus, it is clear the new genus is independent of those two existed genera.
Beside the molecular analyses, morphological data also support its recognition within tribe Chromosereae. Sinohygrocybe shares a bright pileus colour and decurrent lamellae with the other genera Chromosera and Gloioxanthomyces (Table 2). However, the genus Chromosera, typified by C.cyanophylla (Fr.) Redhead, Ammirati & Norvell, differs from Sinohygrocybe in having omphaloid basidiomata, ephemeral dextrinoid reactions in the context, ratio of basidiospore to basidium length <5, ephemeral pigment bodies in the pileipellis and lilac pigments sometimes present (Redhead et al. 1995, Candusso 1997, Lodge et al. 2014); while Gloioxanthomyces differs from Sinohygrocybe by having weaker/delicate basidiomata, viscid pileus and stipe surface, gelatinised lamellar edge and cheilocystidia, shorter basidia (Boertmann 1990, 2012) with a length ratio of basidium to basidiospore 4–5. Sinohygrocybe shares some macroscopic characters with Hygrocybe, typified by H.conica, including bright colour of basidiomata and the distant lamellae, but Hygrocybe differs from Sinohygrocybe by having more fragile lamellae, more glabrous stipe (at least at the upper portion), often constricted spores and shorter basidia.
Table 2.
Type location, basidiospores and basidia dimensions of species of the tribe Chromosereae.
| Species name | Type location | Basidiospores (μm) | Basidia (μm) | Reference |
|---|---|---|---|---|
| Gloioxanthomyces nitidus | USA, South Carolina | 6.5–9(11) × 4–6.5(7.5) | 29–39 × 7.5–10 | Bessette et al. 2010, this study |
| Gloioxanthomyces vitellinus | Sweden | (6.5)7–9(9) × (5)5.5–7(7.5) | 30–45 × 7–10 | Boertmann 2010 |
| Chromosera citrinopallida | USA, Washington | 7–9(10) × 4.5–5 | 10–45 × 6–8 | Smith and Hesler 1954 |
| Chromosera cyanophylla | Sweden | (6.8)7.2–8.0(8.8) × (3.2)3.6–4.4 | 24–28 × 5.5–6.5 | Holec et al. 2015 |
| Chromosera lilacina | northern Fennoscandia | 7–8.5(10) × (4)5–6(6.5) | 30–45 × 7–9 | Candusso 1997 |
| Chromosera viola | Belgium, Namur Province | 6.5–10.5(11) × 5–7(7.5) | 36–61 × 8–11 | Candusso 1997 |
| Chromosera xanthochroa | Scotland | (5.5)6–8.5(10) × (3.8)4–5.2(5.5) | 25–32 × 6.5–7.5(8.5) | Candusso 1997 |
| Sinohygrocybe tomentosipes | China, Sichuan & Hunan Province | 8–10(10.5) × (4.5)5–7(7.5) | 41–80 × 4–10 | This study |
Sinohygrocybe samples were collected in both late summer (August) and winter (November), showing that they likely have a quite long fruiting season. It should be noted, however, that they are more abundant at times with lower temperature and higher humidity. Therefore, their fruiting in summer may occur only at higher altitude (with the elevation above 1500 m).
As to the Chinese new Gloioxanthomycesnitidus record: 1) phylogenetically, the Chinese samples are nested in the Gloioxanthomyces clade as a sister branch to the North American branch (Fig. 2); 2) morphologically, it shares these characters with the North American G.nitidus: deep yellow basidiomata fading to whitish with age, viscid, hygrophanous surface, central concave pileus and decurrent lamellae (Bessette et al. 2012); 3) geographically, G.nitidus and G.vitellinus are distributed in North America and Asia and Europe, respectively, indicating that Gloioxanthomyces is a Holarctic genus. It is assumed that both North American and East Asian G.nitidus were separated from the same ancestor because of geographical isolation, thus they are very similar at present; however, they may continue to diverge, eventually becoming separate species in the future since they live on detached continents.
Supplementary Material
Acknowledgments
Sincere acknowledgements are expressed to Dr. Xiao-Lan He and Dr. Egon Horak, Mr. Ye-Wei Xia and members of Dr. Wen-Bo Liao’s laboratory, Dr. Jiang Xu and Mrs. Xin Zhang for their help during the field trips in Gesala, Taoyuandong and Changbai Mountains, respectively; to Dr. Tolger Bau for trying to find some additional samples in HMJAU; to Dr. David Boertmann, Dr. Md. Iqbal Hosen and Dr. Wang-Qiu Deng for their improvements and constructive comments on an earlier version of this paper; to Dr. David Hibbett and the International Exchange Scholarship of the University of Chinese Academy of Sciences for providing an opportunity to the first author to learn molecular data analyses. This study was financed by the Ministry of Science and Technology of China (Nos. 2013FY111500, 2013FY111200), the Science and Technology Program of Guangzhou, China (No. 201607020017), the GDAS’ Special Project of Science and Technology Development (2018GDASCX-0907) and the Science and Technology Project of Guangdong Province (2017A030303050).
Citation
Wang C-Q, Zhang M, Li T-H, Liang X-S, Shen Y-H (2018) Additions to tribe Chromosereae (Basidiomycota, Hygrophoraceae) from China, including Sinohygrocybe gen. nov. and a first report of Gloioxanthomyces nitidus. MycoKeys 38: 59–76. https://doi.org/10.3897/mycokeys.38.25427
References
- Bessette AE, Roody WC, Sturgeon WE, Bessette AR. (2012) Waxcaps mushrooms of eastern North America. Syracuse University Press, New York.
- Boertmann D. (1990) The identity of Hygrocybevitellina and related species. Nordic Journal of Botany 10(3): 311–317. 10.1111/j.1756-1051.1990.tb01775.x [DOI] [Google Scholar]
- Boertmann D. (2010) The genus Hygrocybe, 2nd revised edition (Fungi of Northern Europe – vol. 1). Danish Mycological Society, Copenhagen.
- Boertmann D. (2011) Relationship of Hygrocybevitellina and H.nitida–Preliminary Report. Omphalina 2(1): 4–5. [Google Scholar]
- Boertmann D. (2012) Update on Hygrocybenitida. Omphalina 3(1): 13–15. [Google Scholar]
- Candusso M. (1997) Fungi Europaei 6. Hygrophorus s.l. Libreria Basso, Alassio.
- Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G. (2004) MycoBank: an online initiative to launch mycology into the 21st century. Studies in Mycology 50: 19–22. [Google Scholar]
- Dentinger BTM, Gaya E, O’Brien H, Suz LM, Lachlan R, Díaz-Valderrama JR, Koch RA, Aime MC. (2016) Tales from the crypt: Genome mining from fungarium specimens improves resolution of the mushroom tree of life. Biological Journal of the Linnean Society 117(1): 11–32. 10.1111/bij.12553 [DOI] [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. (2017) MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefings in Bioinformatics: 1–7. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed]
- Kornerup A, Wanscher JH. (1978) Methuen handbook of colour (3rd edn). Eyre Methuen, London, 252 pp. [Google Scholar]
- Lawrey JD, Lücking R, Sipman HJM, Chaves JL, Redhead SA, Bungartz F, Sikaroodi M, Gillevet PM. (2009) High concentration of basidiolichens in a single family of agaricoid mushrooms (Basidiomycota, Agaricales, Hygrophoraceae). Mycological Research 113: 1154–1171. 10.1016/j.mycres.2009.07.016 [DOI] [PubMed] [Google Scholar]
- Lodge DJ, Padamsee M, Matheny PB, Aime MC, Cantrell SA, Boertmann D, Kovalenko A, Vizzini A, Dentinger BTM, Kirk PM, Ainsworth AM, Moncalvo JM, Vilgalys R, Larsson E, Lücking R, Griffith GW, Smith ME, Norvell LL, Desjardin DE, Redhead SA, Ovrebo CL, Lickey EB, Ercole E, Hughes KW, Courtecuisse R, Young A, Binder M, Minnis AM, Lindner DL, Ortiz-Santana B, Haight J, Læssøe T, Baroni TJ, Geml J, Hattori T. (2014) Molecular phylogeny, morphology, pigment chemistry and ecology in Hygrophoraceae (Agaricales). Fungal Diversity 64: 1–99. 10.1007/s13225-013-0259-0 [DOI] [Google Scholar]
- Matheny PB. (2005) Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe, Agaricales). Molecular Phylogenetics and Evolution 35: 1–20. 10.1016/j.ympev.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees, Gateway Computing Environments Workshop (GCE), 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Redhead SA, Lutzoni F, Moncalvo JM, Vilgalys R. (2002) Phylogeny of agarics: partial systematics solutions for core omphalinoid genera in the Agaricales (Euagarics). Mycotaxon 83: 19–57. [Google Scholar]
- Redhead SA, Ammirati JF, Norvell LL. (1995) Omphalina sensu lato in North America 3: Chromosera gen. nov. Beihefte Sydowia 10: 155–167. [Google Scholar]
- Redhead SA, Ammirati JF, Norvell LL, Vizzini A, Contu M. (2012) Validation of combinations with basionyms published by Fries in 1861. Mycotaxon 118: 455–458. 10.5248/118.455 [DOI] [Google Scholar]
- Smith AH, Hesler LR. (1954) Additional North American Hygrophori. Sydowia 8: 304–333. [Google Scholar]
- Stamatakis A. (2014) RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9): 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR protocols: a guide to methods and applications.Academic Press, San Diego, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Yang ZL, Feng B, Hao YJ. (2013) Pseudoarmillariellabacillaris, a new species with bacilliform basidiospores in Asia. Mycosystema 32: 127–132. [Google Scholar]
- Young AM. (2005) Fungi of Australia: Hygrophoraceae. ABRS, Canberra; CSIRO Publishing, Melbourne.
- Young AM, Wood AE. (1997) Studies on the Hygrophoraceae (Fungi: Homobasidiomycetes: Agaricales) of Australia. Australian Systematic Botany 10: 911–1030. 10.1071/SB96005 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.