Abstract
Porcine circovirus associated diseases (PCVAD) have multiple manifestations that have been attributed to porcine circovirus type 2 (PCV2). Recently, a novel porcine circovirus, PCV type 3 (PCV3), was identified in pigs with systemic inflammation of unknown etiology. In this study, we tried to detect the PCV3 genome in tissue samples collected from Japanese pig herds in 2016. The PCV3 genome was detected by PCR in 7 of 73 samples. The homology between each Japanese strain was 99.5% for the full-length sequence and 98.9 to 99.2% for the open reading frame 2. These results suggest that PCV3 has already invaded Japanese pig farms.
Keywords: porcine circovirus type 3 (PCV3), prevalence, swine
Circoviruses belong to the genus Circovirus of the family Circoviridae [6]. Three species of circovirus are known to infect pigs: porcine circovirus type 1 (PCV1), type 2 (PCV2), and type 3 (PCV3). While PCV1 has no pathogenicity in pigs [16], PCV2 causes pathological conditions in pigs that are referred to as porcine circovirus-associated disease (PCVAD), inducing variety of symptoms, which include post-weaning multisystemic wasting syndrome (PMWS), respiratory symptoms, breeding disorders, and porcine dermatitis and nephropathy syndrome (PDNS) [12]. However, PCV2 by itself is limited in its ability to induce the full spectrum of clinical signs and lesions associated with advanced cases of PCVAD [12]. In 2016, a novel circovirus, PCV3, was reported in the US, suggesting that PCV3 might be involved in PDNS and PMWS, which have been regarded as forms of PCVAD [13]. Phan et al. (2016) conducted a metagenomic analysis of the genomes of pigs with systemic inflammation and cardiac pathology of undetermined etiology and detected PCV3 [14].
After the report of PCV3 in the US, it has been reported in many countries of the world [2,3,4,5, 7, 8, 10, 15, 17, 18]. Collins et al. (2017) recently conducted a retrospective study of PCV3 in the UK and showed that PCV3 appeared there at least 10 years earlier than in other countries [2]. However, prior to the current study, it was unclear whether PCV3 is present in Japan. This study aimed to detect the PCV3 genome by PCR in tissue samples collected from Japanese pig herds in 2016 for the diagnosis of pig disease. We report the first identification of PCV3 in Japan.
Tissue samples, collected from 73 pigs at 27 farms in 2016, were used to detect the PCV3 genome. These samples were submitted to the Nippon Institute for Biological Science for routine diagnosis in 2016. The tissue samples originated from 9 prefectures in Japan, and the pigs ranged from fetuses to adults. The clinical history of the pigs included subclinical (n=2), sudden death (n=20), abortion (n=15), diarrhea (n=11), respiratory signs or lung abscesses (n=9), depression (n=6), and neurological symptoms (n=4). No clinical history was provided for the remaining 6 cases. Each sample was collected according to the clinical symptom. For example, five major organs (heart, lung, liver, kidney, and spleen) and lymph nodes were collected from pigs with depression or sudden death; placenta or fetal organs, from pigs with abortion; and intestine or mesenteric lymph node, from pigs with diarrhea. In total, 217 samples were collected: fetal organs (n=55), lung (n=47), liver (n=32), pulmonary lymph nodes (n=25), spleen (n=23), intestine (n=17), kidney (n=14), brain (n=12), tonsil (n=11), mesenteric lymph nodes (n=4), mandibular lymph nodes (n=3), inguinal lymph nodes (n=2), and placenta (n=2). Samples were stored in a −80°C freezer until used in the experiment.
The specimens were used to produce a 10% (w/v) homogenate in Eagle’s minimum essential medium (Thermo Fisher Scientific, Waltham, MA, U.S.A.). The homogenate was centrifuged at 1,500 × g for 15 min at 4°C. The supernatant was filtered through a filter with a pore size of 0.45 µm. For DNA extraction, proteinase K (Thermo Fisher Scientific) was added to the tissue homogenate to achieve a final concentration of 200 µg/ml, and the solution was incubated at 55°C for 2 hr. For RNA extraction, the TRIzol™ Reagent (Thermo Fisher Scientific) was used according to the manufacturer’s instructions.
After centrifugation, the supernatant, obtained as a DNA solution, was subjected to PCR for amplification of the PCV3 open reading frame (ORF) 2. PCR was performed using GoTaq® Green Master Mix (Promega, Madison, WI, U.S.A.) according to the manufacturer’s instructions. Forward and reverse primers were designed according to a previous report [13].
Concerning clinical signs, the general viral pathogens for pigs, which include PCV2, suid herpesvirus 1 (SuHV-1), porcine parvovirus (PPV), porcine reproductive and respiratory syndrome virus (PRRSV), Japanese encephalitis virus (JEV), porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), porcine rotavirus A (PoRV-A), and Getah virus (GETV), were subjected to RT-PCR or simple PCR analysis [11]. Isolation of general bacterial pathogens was attempted on blood agar plates and chocolate agar plates. Colonies were picked, and the 16S rRNA gene was amplified by PCR using Ex Taq® (TAKARA BIO INC., Kusatsu, Japan) according to the manufacturer’s instructions. Isolates were identified by 16S rRNA gene sequencing.
The full-length sequence of the genome was determined for the samples that were found to be positive for the PCV3 ORF2 by PCR. Primer sets were designed according to a previous report, and their respective annealing temperatures were determined [13] (Table 1). The full-length PCV3 genomic sequence was assembled from sequences of each of the amplified products. Each nucleotide sequence was aligned with previously reported sequences of the members of the genus Circovirus and PCV3 ORF2, and was input into the Clustal W multiple sequence alignment program to produce the phylogenetic tree. Bootstrap values were calculated on 1,000 replicates of the alignment, using MEGA ver. 7.0 software [9].
Table 1. Primer sets for sequencing of PCV3 genome.
| Primer sets | Primer name | Position | Primer sequences (5′→3′) | Tm (°C) | Annealing temperature |
Amplification size | Intended purpose |
Reference |
|---|---|---|---|---|---|---|---|---|
| 1 | Full F1 | 74–93 | CACCGTGTGAGTGGATATAC | 57.8 | 55 | 854 | To obtain PCV3 genome sequences |
[13] |
| Full R1 | 927–910 | CAAACCCACCCTTAACAG | 57.9 | |||||
| 2 | Full F2 | 1609–1627 | GTCGTCTTGGAGCCAAGTG | 63.5 | 60 | 825 | [13] | |
| Full R2 | 433–415 | CGACCAAATCCGGGTAAGC | 66.4 | |||||
| 3 | Full F3 | 594–611 | GAAGTTGCGGAGAAGATG | 58.4 | 55 | 550 | original | |
| Full R3 | 1143–1122 | CGCAATAATTGTACACAAGTGC | 61.9 | |||||
| 4 | Full F4 | 1004–1024 | ATATTCGTGGAAAGTTGGAGG | 61.6 | 60 | 778 | original | |
| Full R4 | 1781–1763 | CCACTTCATTACCCGCCTA | 62.4 | |||||
The details of the PCV3-positive samples are shown in Table 2. The PCV3-positive rate at the farm level was 22.2% (6/27). The positive rate of PCV3 in all the samples was 9.6% (7/73). The detected PCV3 ORF2 genomes were designated as TG1923-2/2016, KGS1945-2/2016, KGS1945-3/2016, MZ1950-1/2016, KGS2065-4/2016, KGS2098-1/2016, and GM2124-2/2016 strains. The positive rates of PCV3 by prefecture were 0/5 (0%) in Toyama, 0/5 (0%) in Chiba, 1/6 (16.7%) in Gunma, 1/3 (33.3%) in Tochigi, 0/2 (0%) in Ehime, 0/5 (0%) in Kochi, 0/2 (0%) in Kumamoto, 1/17 (5.9%) in Miyazaki, and 4/28 (14.2%) in Kagoshima. The detection rate of PCV3 by age group was 6.7% (1/15) for fetuses, 13.3% (2/15) for suckling piglets (3–4 weeks of age), 9.0% (1/11) for weaned pigs (5–8 weeks of age), and 9.4% (3/32) for fattening pigs (9 weeks of age and older). The positive rates of PCV3 by organ were 1/55 (1.8%) in fetal organs, 3/47 (6.4%) in lung, 1/32 (3.1%) in liver, 2/25 (8.0%) in pulmonary lymph nodes, 1/23 (4.3%) in spleen, 2/17 (11.8%) in intestine, 4/14 (28.6%) in kidney, 1/12 (8.3%) in brain, 0/11 (0%) in tonsil, 1/4 (25.0%) in mesenteric lymph nodes, 1/3 (33.3%) in mandibular lymph nodes, 0/2 (0%) in inguinal lymph nodes, and 0/2 (0%) in placenta. Although the PCV3 genome was detected in various organs of the whole body, we could not confirm a specific target organ for PCV3 infection. Similar to our results, Ku et al. (2017) reported that the PCV3 genome was detected in various organs of the whole body [8]. The PCV3 genome was also detected in fetal organs, suggesting that PCV3 may be transmitted vertically from dams to fetuses. Previous epidemiological studies in the US and China reported that the PCV3 genome was also detected in fetal organs, and the authors refer to the possibility that PCV3 is transmitted vertically, and is involved in abortion [8, 13]. Pigs with the PCV3 genome had shown some clinical symptoms, such as sudden death (n=4), abortion (n=1), depression (n=1), and neurological symptoms (n=1). Co-infection with other pathogens, such as PPV, PRRSV, Streptococcus suis, Escherichia coli, and Staphylococcus haemolyticus, was detected in the PCV3-positive samples.
Table 2. Details of samples detected PCV3 genome.
| Viral strain | Days of age | Clinical symptomsa) | Other detected pathogensb) | Organsc) | Prefecture | GenBank accession No. |
|---|---|---|---|---|---|---|
| KGS2065-4/2016 | Abortus | Abo | PPV, Ec | Lu | Kagoshima | |
| KGS1945-2/2016 | 14–21 | SD | PPV | Ki | Kagoshima | |
| KGS1945-3/2016 | 14–21 | SD | PPV, Sh | Ki | Kagoshima | |
| GM2124-2/2016 | 50 | NS | PRRSV, Ec | Lu, HLN, BI | Gunma | LC269727 |
| KGS2098-1/2016 | 72 | SD | Ss | He, Lu, Li, Ki, Sp, MLN, JI, Co | Kagoshima | LC383841 |
| TG1923-2/2016 | 130 | Dep, AB | PRRSV | Lu, HLN, ILN | Tochigi | LC383840 |
| MZ1950-1/2016 | 180 | SD | Ss | Ki | Miyazaki |
a) Dep: Depression, AB: Abdominal breathing, SD: Sudden death, Abo: abortion, NS: Neurological symptoms. b) Ec: Escherichia coli, Sh: Staphylococcus haemolyticus, Ss: Streptococcus suis, PPV: porcine parvovirus, PRRSV: porcine reproductive and respiratory syndrome virus. c) Lu: Lung, He: Heart, Li: Liver, Sp: Spleen, Ki: Kidney, JI: Jejunum Ileum, Co: Colon, Bl: Blain, MLN: Mesenteric lymph nodes, ILN: Inguinal lymph nodes, HLN: Hilar lymph nodes.
Previous epidemiological studies in the US, China, and Poland showed that the positive rates of PCV3 in individual samples were 12.5% (34/271), 34.7% (77/222), and from 0 to 65.0% (the median was 22.2%), respectively [8, 13, 15]. Kwon et al. (2017) reported that the positive rates of PCV3 in pen-based oral fluid samples and at the farm level were 44.2% (159/360) and 72.6% (53/73), respectively, in South Korea [10]. The PCV3 genome-positive rate in this study was within the range of detection rate reported in other countries.
In previous reports, the PCV3 genome was detected in samples from young pigs aged 14–21 days and 63–70 days, and in aborted fetuses [13, 14]. Stadejek et al. (2017) reported that PCV3 was most common among weaned pigs, finishers, and sows (26.1, 28.0, and 29% of serum pools, respectively); however, only 5% of serum pools from suckling piglets were PCV3 genome-positive [15]. The authors suggested that the difference in the detection rate among the age groups is due to the protective effect of maternal immunity. However, in this study, the PCV3 genome was detected in all age groups.
PCV3 infection might be associated with PDNS, reproductive failure [8, 13], multisystemic inflammation [14], and congenital tremors [1]. Additionally, in this study, PCV3 was detected in an abortive fetus. PCV3 genomes were also identified from subclinical cases [19]. PCV2 likely requires other cofactors to induce the full spectrum of clinical signs and lesions associated with advanced cases of PCVAD [12]. As other pathogens were detected in PCV3-positive samples, it is possible that co-infection with PCV3 and other pathogens induced the complicated symptoms observed, such as PCVAD. To investigate the pathogenicity of PCV3, it is necessary to further investigate the pathogenicity of both PCV3 singular infection cases and co-infection cases.
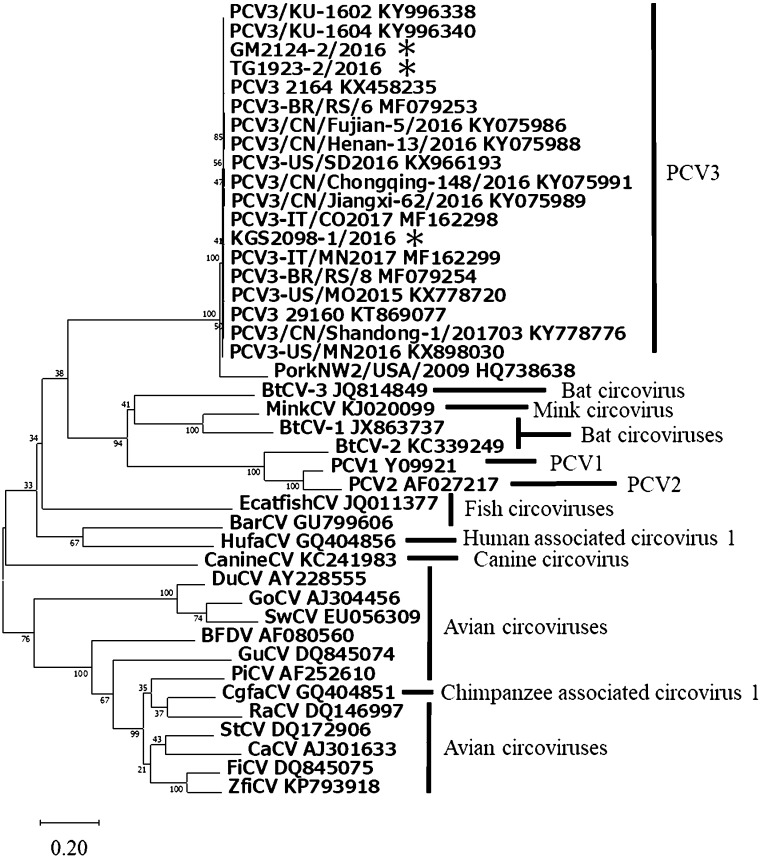
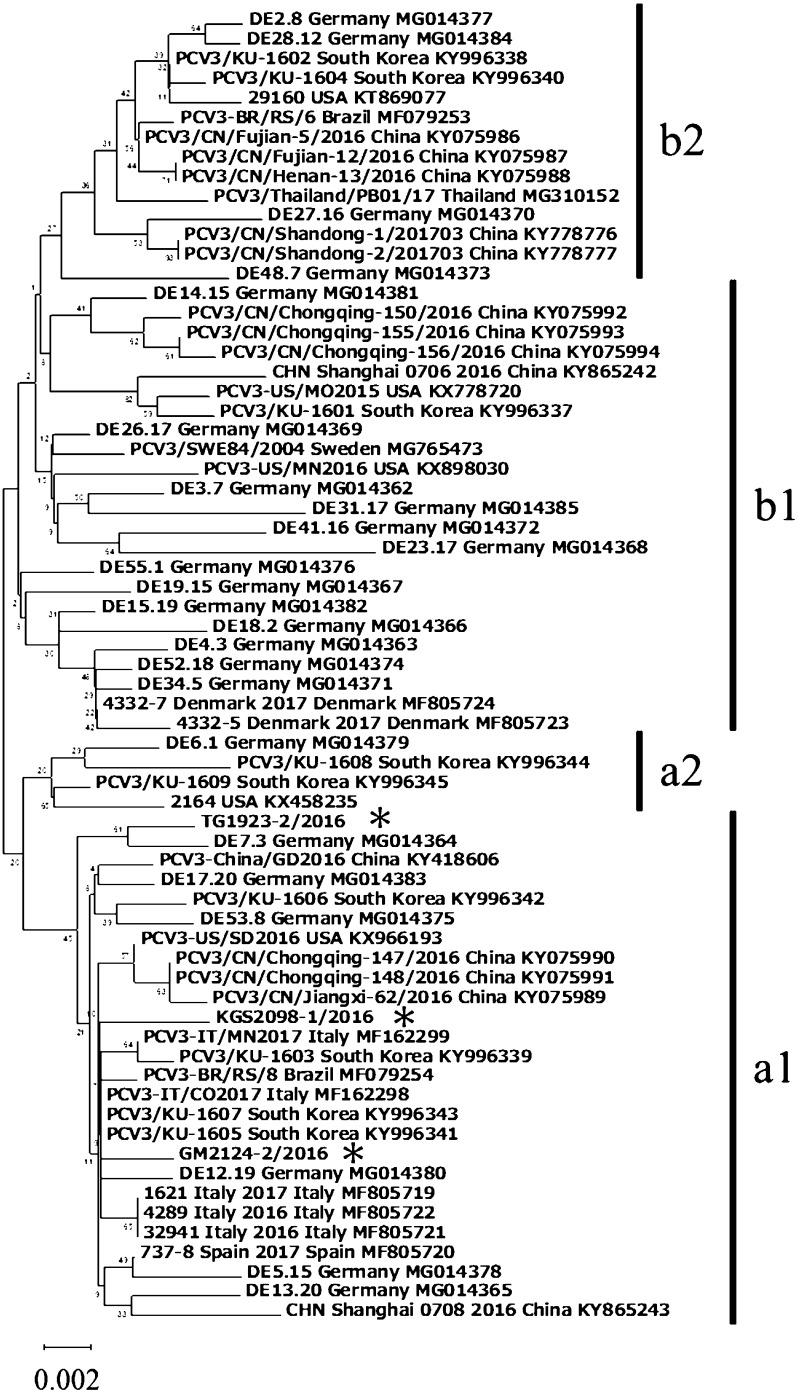
Of the seven PCV3 positive samples, three whole genome sequences were obtained, and the genomes were found to consist of 2 kb single circular DNA. The sequences of the three strains, GM2124-2/2016, KGS2098-1/2016, and TG1923-2/2016, were subjected to phylogenetic analysis. The three strains grouped in a clade previously reported as PCV3, and which was separated from all other members of the genus Circovirus (Fig. 1). The homology between each Japanese strain was 99.5% for the full-length sequence, and 98.9 to 99.2% for the ORF 2. Fux et al. showed that the sequences of PCV3 could be divided into two main groups (a and b) and several subclusters (a1, a2, b1 and b2) [5]. A phylogenetic tree was constructed based on the nucleotide sequence of the ORF2 (Fig. 2). In this study, all the genotypes of PCV3 detected in Japan (GM2124-2/2016, KGS2098-1/2016, and TG1923-2/2016) belonged to the a1 subgroup. However, PCV3 reported from the other countries (Brazil, China, Germany, Italy, and South Korea) belonged to various clusters, and did not present any genotype and geographical correlation. The fact that all three strains obtained in this study classified into the same subcluster could be attributed to the small number of strains examined. Thus, further investigations using a large number of Japanese PCV3 strains are necessary.
Fig. 1.
Phylogenetic tree of full-length circovirus genome. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is displayed next to the branches. GenBank accession numbers are written at the end. Previously published sequences used as reference sequences for this phylogenetic analysis included PCV3, KT869077, KX458235, KX778720, KX898030, KX966193, KY075986, KY075988, KY075989, KY075991, KY778776, KY996338, KY996340, MF162298, MF162299, MF079253, and MF079254; Barbel circovirus (BarCV), GU799606; Bat circovirus-1 (BtCV-1), JX863737; BtCV-2, KC339249; BtCV-3, JQ814849; Beak and feather disease virus (BFDV), AF080560; Canary circovirus (CaCV), AJ301633; Canine circovirus (CanineCV), KC241983; Chimpanzee associated circovirus 1 (CgfaCV), GQ404851; Duck circovirus (DuCV), AY228555; European catfish circovirus (EcatfishCV), JQ011377; Finch circovirus (FiCV), DQ845075; Goose circovirus (GoCV), AJ304456; Gull circovirus (GuCV), DQ845074; Human associated circovirus 1 (HufaCV), GQ404856; Mink circovirus (MiCV), KJ020099; Pigeon circovirus (PiCV), AF252610; PCV1, Y09921; PCV2, AF027217; PorkNW2/USA/2009, HQ738638; Raven circovirus (RaCV), DQ146997; Starling circovirus (StCV), DQ172906; Swan circovirus (SwCV), EU056309; and Zebra finch circovirus (ZfiCV), KP793918. The PCV3 strain analyzed in this study was indicated by an asterisk.
Fig. 2.
Phylogenetic tree of PCV3 ORF2 regions. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is displayed next to the branches. The country of origin and GenBank accession numbers are written at the end. Previously published sequences used as reference sequences for this phylogenetic analysis included PCV3 from Brazil, China, Denmark, Germany, Italy, South Korea, Spain, Sweden, Thailand, and the US. The PCV3 strains analyzed in this study were indicated by an asterisk.
In conclusion, this is the first report on the prevalence of PCV3 in Japan. We investigated the occurrence of the PCV3 genome in tissue samples collected from Japanese pig herds in 2016 using PCR. In the examination of the 73 samples collected from 27 farms, the PCV3 genome was detected in 7 samples from 6 farms in the Kanto and Kyusyu regions of Japan. Our results indicate that PCV3 is prevalent in commercial pig herds in Japan. To elucidate the influences of PCV3 on the pig industry, further studies involving virus isolation and elucidation of PCV3 pathogenicity in pigs are needed.
Nucleotide sequence accession numbers: The GenBank accession numbers for the full PCV3 genomes sequence are LC269727, LC383840, and LC383841.
REFERENCES
- 1.Chen G. H., Mai K. J., Zhou L., Wu R. T., Tang X. Y., Wu J. L., He L. L., Lan T., Xie Q. M., Sun Y., Ma J. Y.2017. Detection and genome sequencing of porcine circovirus 3 in neonatal pigs with congenital tremors in South China. Transbound. Emerg. Dis. 64: 1650–1654. doi: 10.1111/tbed.12702 [DOI] [PubMed] [Google Scholar]
- 2.Collins P. J., McKillen J., Allan G.2017. Porcine circovirus type 3 in the UK. Vet. Rec. 181: 599. doi: 10.1136/vr.j5505 [DOI] [PubMed] [Google Scholar]
- 3.Faccini S., Barbieri I., Gilioli A., Sala G., Gibelli L. R., Moreno A., Sacchi C., Rosignoli C., Franzini G., Nigrelli A.2017. Detection and genetic characterization of Porcine circovirus type 3 in Italy. Transbound. Emerg. Dis. 64: 1661–1664. doi: 10.1111/tbed.12714 [DOI] [PubMed] [Google Scholar]
- 4.Franzo G., Legnardi M., Hjulsager C. K., Klaumann F., Larsen L. E., Segales J., Drigo M.2018. Full-genome sequencing of porcine circovirus 3 field strains from Denmark, Italy and Spain demonstrates a high within-Europe genetic heterogeneity. Transbound. Emerg. Dis. 65: 602–606. doi: 10.1111/tbed.12836 [DOI] [PubMed] [Google Scholar]
- 5.Fux R., Söckler C., Link E. K., Renken C., Krejci R., Sutter G., Ritzmann M., Eddicks M.2018. Full genome characterization of porcine circovirus type 3 isolates reveals the existence of two distinct groups of virus strains. Virol. J. 15: 25. doi: 10.1186/s12985-018-0929-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.ICTV. 2017. ICTV Online Report. https://talk.ictvonline.org/taxonomy [accessed on May 7, 2018].
- 7.Kedkovid R., Woonwong Y., Arunorat J., Sirisereewan C., Sangpratum N., Lumyai M., Kesdangsakonwut S., Teankum K., Jittimanee S., Thanawongnuwech R.2018. Porcine circovirus type 3 (PCV3) infection in grower pigs from a Thai farm suffering from porcine respiratory disease complex (PRDC). Vet. Microbiol. 215: 71–76. doi: 10.1016/j.vetmic.2018.01.004 [DOI] [PubMed] [Google Scholar]
- 8.Ku X., Chen F., Li P., Wang Y., Yu X., Fan S., Qian P., Wu M., He Q.2017. Identification and genetic characterization of porcine circovirus type 3 in China. Transbound. Emerg. Dis. 64: 703–708. doi: 10.1111/tbed.12638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kumar S., Stecher G., Tamura K.2016. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 33: 1870–1874. doi: 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwon T., Yoo S. J., Park C. K., Lyoo Y. S.2017. Prevalence of novel porcine circovirus 3 in Korean pig populations. Vet. Microbiol. 207: 178–180. doi: 10.1016/j.vetmic.2017.06.013 [DOI] [PubMed] [Google Scholar]
- 11.Ogawa H., Taira O., Hirai T., Takeuchi H., Nagao A., Ishikawa Y., Tuchiya K., Nunoya T., Ueda S.2009. Multiplex PCR and multiplex RT-PCR for inclusive detection of major swine DNA and RNA viruses in pigs with multiple infections. J. Virol. Methods 160: 210–214. doi: 10.1016/j.jviromet.2009.05.010 [DOI] [PubMed] [Google Scholar]
- 12.Opriessnig T., Meng X. J., Halbur P. G.2007. Porcine circovirus type 2 associated disease: update on current terminology, clinical manifestations, pathogenesis, diagnosis, and intervention strategies. J. Vet. Diagn. Invest. 19: 591–615. doi: 10.1177/104063870701900601 [DOI] [PubMed] [Google Scholar]
- 13.Palinski R., Piñeyro P., Shang P., Yuan F., Guo R., Fang Y., Byers E., Hause B. M.2016. A novel porcine circovirus distantly related to known circoviruses is associated with porcine dermatitis and nephropathy syndrome and reproductive failure. J. Virol. 91: e01879–e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Phan T. G., Giannitti F., Rossow S., Marthaler D., Knutson T. P., Li L., Deng X., Resende T., Vannucci F., Delwart E.2016. Detection of a novel circovirus PCV3 in pigs with cardiac and multi-systemic inflammation. Virol. J. 13: 184. doi: 10.1186/s12985-016-0642-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stadejek T., Woźniak A., Miłek D., Biernacka K.2017. First detection of porcine circovirus type 3 on commercial pig farms in Poland. Transbound. Emerg. Dis. 64: 1350–1353. doi: 10.1111/tbed.12672 [DOI] [PubMed] [Google Scholar]
- 16.Tischer I., Mields W., Wolff D., Vagt M., Griem W.1986. Studies on epidemiology and pathogenicity of porcine circovirus. Arch. Virol. 91: 271–276. doi: 10.1007/BF01314286 [DOI] [PubMed] [Google Scholar]
- 17.Tochetto C., Lima D. A., Varela A. P. M., Loiko M. R., Paim W. P., Scheffer C. M., Herpich J. I., Cerva C., Schmitd C., Cibulski S. P., Santos A. C., Mayer F. Q., Roehe P. M.2018. Full-Genome Sequence of Porcine Circovirus type 3 recovered from serum of sows with stillbirths in Brazil. Transbound. Emerg. Dis. 65: 5–9. doi: 10.1111/tbed.12735 [DOI] [PubMed] [Google Scholar]
- 18.Ye X., Berg M., Fossum C., Wallgren P., Blomström A. L.2018. Detection and genetic characterisation of porcine circovirus 3 from pigs in Sweden. Virus Genes 54: 466–469. doi: 10.1007/s11262-018-1553-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng S., Wu X., Zhang L., Xin C., Liu Y., Shi J., Peng Z., Xu S., Fu F., Yu J., Sun W., Xu S., Li J., Wang J.2017. The occurrence of porcine circovirus 3 without clinical infection signs in Shandong Province. Transbound. Emerg. Dis. 64: 1337–1341. doi: 10.1111/tbed.12667 [DOI] [PMC free article] [PubMed] [Google Scholar]