Abstract
Nuclear lamins are type V intermediate filament proteins. Lamins, including LA, LB1, LB2, and LC, are the major protein components forming the nuclear lamina to support the mechanical stability of the mammalian cell nucleus. Increasing evidence has shown that LA participates in homologous recombination (HR) repair of DNA double-strand breaks (DSBs) . However, the mechanisms underlying this process are incompletely understood. We recently identified the first lamin-binding ligand 1 (LBL1) that directly binds LA and inhibited cancer cell growth. We provided here further mechanistic investigations of LBL1 and revealed that LA interacts with the HR recombinase Rad51 to protect Rad51 from degradation. LBL1 inhibits LA–Rad51 interaction leading to accelerated proteasome-mediated degradation of Rad51, culminating in inhibition of HR repair of DSBs. These results uncover a novel post-translational regulation of Rad51 by LA and suggest that targeting the LA–Rad51 axis may represent a promising strategy to develop cancer therapeutics.
Short abstract
Using the lamin-binding ligand LBL1 as a tool, we discovered that lamin A regulates Rad51 protein stability. LBL1 promotes Rad51 degradation by proteasome to inhibit homologous recombination repair.
Introduction
Nuclear lamins are the major component of nuclear lamina, a meshwork of proteins that lie underneath the inner nuclear membrane.1,2 Lamins are type V intermediate filament (IF) proteins.1 In mammals including humans, there are three lamin genes (LA, LB1, and LB2) encoding four major, highly homologous proteins (LA, LC, LB1, and LB2). LA and LC are alternative splicing products of a single LA gene.3,4LB1 and LB2 are two different genes.4 All the lamin proteins are highly homologous in sequence and predicted secondary structure. These lamin proteins share the same domain structural organization. Similar to other cytosolic IF proteins (e.g., vimentin5), lamins contain a long central, α-helical coiled-coil rod domain,6 which is flanked by a non-α-helical N-terminal head and a C-terminal tail.4 Unlike other cytosolic IF proteins, lamins contain a nuclear localization sequence (NLS) and an immunoglobulin-like (Ig-like) domain with a two-layered sandwich composed of antiparallel β-strands.7,8 This unique structural organization enables lamins to function not only as nuclear scaffold proteins to maintain nuclear mechanical stability, but also as signaling molecules by interacting with other proteins.9−12
Besides the traditional view of lamins as scaffold proteins to maintain the mechanical stability of the nucleus, LA has also been shown to participate in DNA double-strand break (DSB) repair processes. However, the molecular mechanisms underlying this pathway remain incompletely understood.1 For example, a genetic mutation in LA (1824, C → T) activates a cryptic splicing site near the C-terminus of LA resulting in deletion of 50 aa.13,14 Carriers of this mutation develop Hutchinson–Gilford progeria syndrome (HGPS), manifested by accelerated aging in children. Fibroblasts from HGPS patients have been shown to present increased basal level of DSBs,15 increased chromosomal instability,15,16 and defective repair of DSBs.15 Similar results were also observed in mouse and human cells with LA–/– genotype.15,17−19 These results support that LA plays important roles in regulating DSB repair. While LA has been implicated in DSB repair, the precise roles of LA in DSB repair remain to be elucidated.19−21
In mammalian cells, DSBs are repaired by either error-free homologous recombination (HR) or the error-prone nonhomologous end joining (NHEJ) process.22 Dysregulation of DSB repair machinery is frequently observed in various cancer cells,23 which is in part driven by genomic instability, one of the key enabling characteristics of cancer cells.24 Indeed, transformed cancer cells often display increased basal levels of endogenous DSBs25,26 and thus heavily rely on the DSB repair pathways for sustained survival.27 Therefore, targeting the DSB repair pathways is a promising strategy to develop novel cancer therapeutics.28
Previously, we discovered a novel acylated pyrroloquinazoline called LBL1 (lamin-binding ligand 1, Figure 1A) that selectively inhibited breast cancer cell growth.29 Recently, we discovered that LBL1 targets nuclear lamins.30 However, the mechanism of action of LBL1 remains to be established. Here we present our mechanistic studies to reveal that LA interacts with the key HR recombinase Rad51 to protect Rad51 from proteasome-mediated degradation. Binding of LBL1 to LA disrupted the LA–Rad51 interaction and decreased Rad51 protein stability leading to impaired HR efficiency and DSB accumulation inside the cancer cells. Using LBL1 as a chemical tool, we also uncovered a previously unrecognized post-translational regulation of Rad51 by LA. These results suggest that targeting the LA–Rad51 axis may represent a novel strategy to develop cancer therapeutics that inhibit DSB repair.
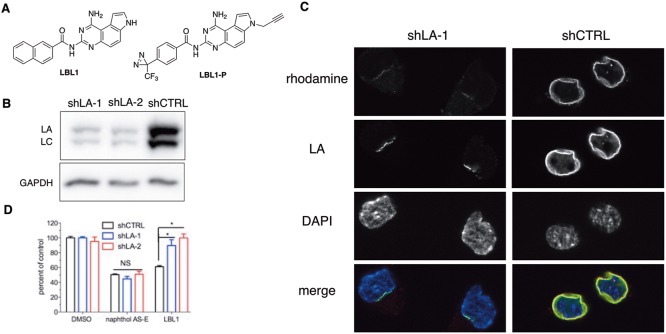
Figure 1.
Lamins were the efficacy targets of LBL1. (A) Chemical structures of LBL1 and LBL1-P. (B) LA expression was silenced by two independent shRNA constructs in DKO MEFs. (C) LBL1-P specifically labeled LA in DKO MEFs cells. The cells from part B were treated with LBL1-P and then subjected to the protocol of photo-cross-linking followed by click reaction with a rhodamine-N3. After click reaction, the cells were stained with anti-LA, and the cells were then analyzed by fluorescence microscopy. (D) DKO MEFs with silenced LA expression were resistant to LBL1. The cells from part B were treated with the indicated drug for 48 h. Then the viable cells were quantified by MTT assay. Data are presented as mean ± SEM (n = 5). * denotes P < 0.05.
Results
Lamins are the Efficacy Targets of LBL1
Using a clickable photoaffinity probe LBL1-P (Figure 1A), we discovered that LBL1 directly targets nuclear lamins.30 To determine if lamins are the efficacy targets of LBL1 in cells, we knocked down LA expression with two independent shRNAs in SV40-immortalized mouse embryonic fibroblasts (MEFs) with LB1–/–LB2–/– (DKO) genetic background.31,32 Significant LA protein knockdown was observed with both shRNAs (Figure 1B). DKO MEFs expressing shLA displayed significantly attenuated growth rate (Figure S1). With the shLA DKO MEF cells, we further investigated the cellular labeling by the clickable photo-cross-linker LBL1-P (Figure 1A) by in-cell click reaction with a rhodamine-azide using a protocol we recently described30 to support its specificity. Similar to what was observed in MDA-MB-231 cells,30LBL1-P colocalized very well with LA in shCTRL DKO MEF cells (Figure 1C, right panels). In the DKO MEF cells with shLA expression, LA protein was significantly knocked down as made evident by a reduced anti-LA labeling signal (Figure 1C, left panels), which is consistent with the Western blot results shown in Figure 1B. Importantly, these residual LA signals were also labeled by LBL1-P (Figure 1C, left panels). No other significant labeling was observed. These results further support the specificity of LA labeling by LBL1-P in living cells.
In a cell growth inhibition assay using the MTT reagent, the lamin-deficient cells were found to be significantly more resistant to LBL1 (Figure 1D). This effect is specific because these cells showed equal sensitivity to an independent compound naphthol AS-E (Figure 1D), which is a cAMP-response element binding protein (CREB) inhibitor.33 Because LBL1 is a fused planar aromatic tricyclic compound, we also tested if LBL1 could intercalate into DNA. Using a gel shift assay, we observed no evidence of intercalation of LBL1 into DNA up to 100 μM concentration (Figure S2). On the other hand, ethidium bromide (EtBr), a known DNA intercalator, exhibited clear intercalation. Altogether, these results demonstrate that lamins are the efficacy targets of LBL1 in living cells.
LBL1 Induces DSB Formation in Cancer Cells
During our initial investigation of the cell cycle profiles of MDA-MB-231 cells treated with LBL1, we found that the cells were dose-dependently arrested at G2/M phase with a concomitant decrease of G1 and S phase cells (Figure 2A and Figure S3A). In addition, we found that the coefficient of variance (CV%) of the G1 peak in LBL1-treated cells was much bigger than that in DMSO-treated cells (8.8 ± 0.03 versus 12.82 ± 1.98 for 2.5 μM LBL1, P < 0.05) (Figure 2B). This G1 peak broadening is indicative of formation of DSBs inside the cells.15,34 Among the lamin isoforms, LA has been mostly implicated in regulating DSB repair and genomic instability with incompletely understood mechanisms.15−17,35 Therefore, we focused our remaining studies on LA.
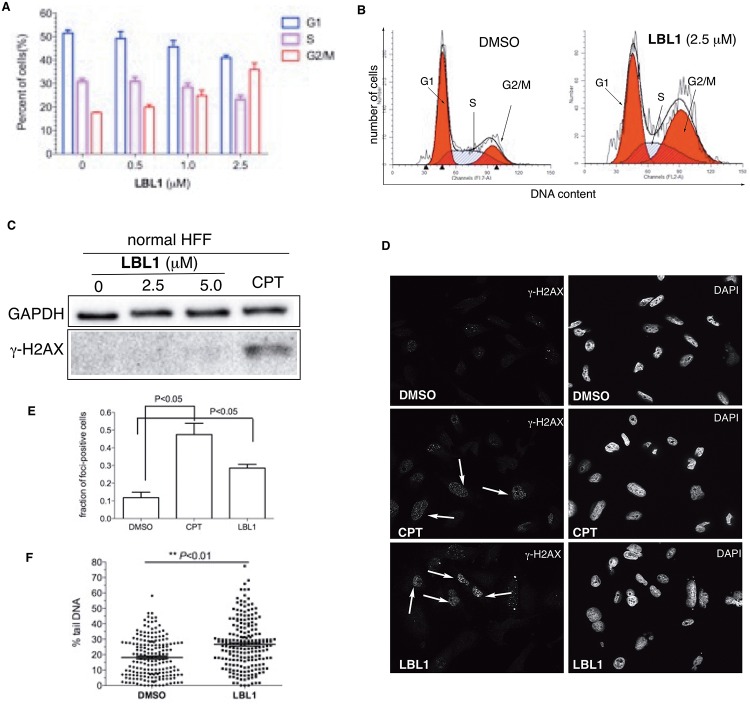
Figure 2.
LBL1 induced DSBs in cancer cells. (A) LBL1 arrested the cells at G2/M phase. The cell cycle distribution of MDA-MB-231 cells treated with different concentrations of LBL1 for 48 h. Then the cell cycle profile was analyzed by flow cytometry after the cells were fixed and stained with PI. (B) LBL1 induced G2/M arrest and G1 peak broadening in MDA-MB-231 cells. The cells were treated with LBL1 for 48 h. The different cell cycle stages (G1, S, G2/M) are indicated. (C) LBL1 did not induce phosphorylation of H2AX in normal primary HFF. The cells were treated with indicated drugs for 24 h. Then the cells were collected, and the lysates were prepared for Western blot analysis with indicated antibodies. CPT was used as a positive control. (D, E) Quantification of γ-H2AX-foci-positive MDA-MB-231 cells. Data are presented as mean ± SD (n = 3, ∼100 cells were analyzed for each experimental condition). Representative fluorescence micrographs are shown in part D, and quantification is shown in part E. (F) Quantification of tail DNA from the neutral COMET assay in MDA-MB-231 cells treated with LBL1. Around 200 cells were analyzed for each condition (n = 193 for DMSO and n = 203 for LBL1-treated cells). The representative micrographs are shown in Figure S3B.
Because LA has been implicated in DSB repair,15,17 we hypothesized that LBL1 might interfere with the function of LA in DSB repair leading to accumulation of DSBs in the cancer cells. Our previous results showed that histone variant H2AX was phosphorylated (referred to as γ-H2AX)36 in MDA-MB-231 cells treated with LBL1.30 In contrast to the breast cancer cells, normal primary human foreskin fibroblasts (HFFs) did not present phosphorylation of H2AX upon LBL1 treatment, although a robust γ-H2AX signal was induced in these rapidly proliferating cells by a topoisomerase I inhibitor camptothecin (CPT) (Figure 2C).37 We further assessed DSB formation through immunofluorescence analysis of LBL1-treated cells using an antibody against γ-H2AX. This analysis showed that a significantly larger fraction of cells became γ-H2AX-foci-positive (Figure 2D,E). To more directly measure DSB formation upon LBL1 treatment in MDA-MB-231 cells, we employed a neutral COMET assay to quantify the amount of DSB formed.38 Significantly more tail DNA and larger comet tails were observed in LBL1-treated cells (Figure 2F and Figure S3B,C). Taken together, these results demonstrate that cytotoxic DSBs are formed selectively in the cancer cells upon LBL1 treatment, which could be the potential basis for LBL1’s selective toxicity in cancer cells.
LBL1 Inhibits HR Repair of DSBs
Our finding that LBL1 induced DSB formation suggested that LBL1 might inhibit DSB repair. Due to activation of various oncogenes, cancer cells are known to present high DNA replication stress leading to unusual amounts of DSB formation,23 a salient feature that has led to development of inhibitors of DSB repair as novel cancer therapeutics.28 The replication associated DSBs are often repaired by the HR mediated by DNA recombinase Rad51.39−41 During HR repair of DSBs, Rad51 undergoes rapid redistribution to accumulate at the site of DSBs to form subnuclear foci.42,43 Therefore, we investigated if Rad51 subnuclear foci formation was inhibited by LBL1. We treated MDA-MB-231 cells with CPT to stimulate Rad51 foci formation (Figure 3A,B). Consistent with our hypothesis, cotreatment of the cells with CPT and LBL1 significantly reduced the efficiency of Rad51 foci formation (Figure 3A,B), and this inhibition of Rad51 foci formation effect was dose-dependent (Figure S4A). Since HR primarily occurs in the S phase of the cell cycle where the homologous DNA template is available for repair, we investigated if the reduced Rad51 foci formation efficiency was due to a change of cell cycle distribution upon LBL1 cotreatment with CPT. As previously reported,44,45 CPT arrested the cells at S phase (Figure S4B). When the cells were cotreated with different concentrations of LBL1 and CPT, the cell cycle profiles were not significantly different from the cells treated with CPT alone (Figure S4B), demonstrating that the inhibition of Rad51 foci formation by LBL1 was not due to lack of sister chromatin template in the S phase for HR repair of DSBs.
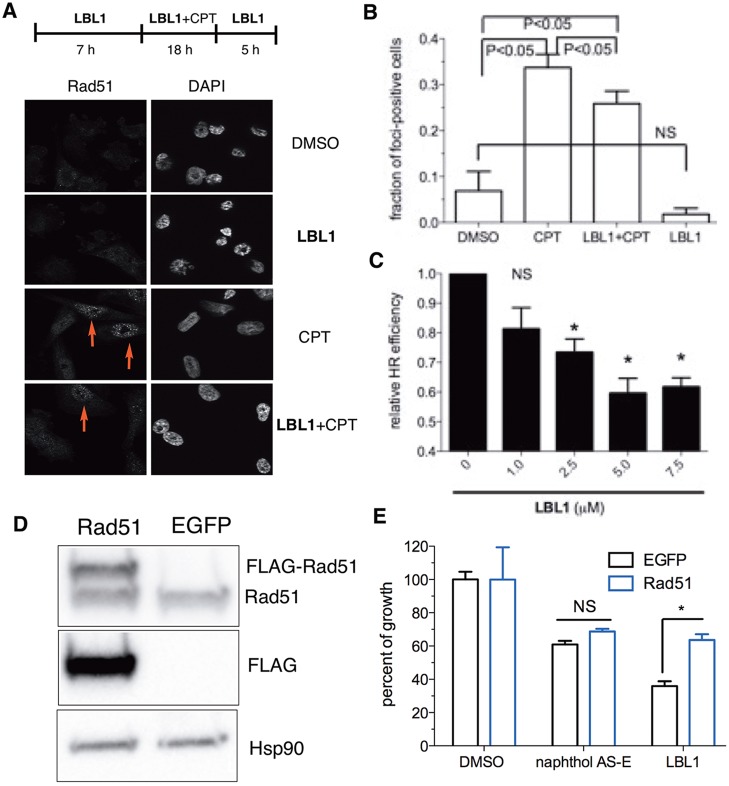
Figure 3.
LBL1 inhibited HR. (A) LBL1 inhibited Rad51 subnuclear foci formation stimulated by CPT. MDA-MB-231 cells were treated as described at the top. Then the cells were analyzed by immunofluorescence analysis. Representative images are shown. (B) Quantification of Rad51-foci-positive cells from part A. Data are presented as mean ± SD (n = 3, ∼100 cells were analyzed for each experimental condition). (C) LBL1 inhibited HR as assessed in a GFP-based reporter assay in MDA-MB-231 cells. The cells were transfected with a GFP-based HR reporter and DsRed as described in the Experimental Section. DSBs were induced by expressing I-SceI. Then the cells were treated with indicated concentrations of LBL1. The cells were then analyzed by flow cytometry. The ratio of GFP+/DsRed+ was registered as relative HR efficiency with vehicle-treated cells defined as 1.0 (n = 3). (D, E) Overexpression of Rad51 rescued LBL1’s antiproliferative activity in MDA-MB-231 cells. FLAG-tagged Rad51 was overexpressed in MDA-MB-231 cells, and the cell lysates were analyzed by Western blot with indicated antibodies. (E) The cells were treated with indicated drugs for 48 h. The cellular growth was quantified by the MTT assay. Data are presented as mean ± SEM (n = 3). * denotes P < 0.05.
To more directly assess the effect of LBL1 on HR repair, we employed a green-fluorescent-protein-based (GFP-based) functional HR reporter assay by flow cytometry.46 The HR reporter plasmid contains two defective GFP alleles and would not provide functional GFP in the absence of DSB and HR (Figure S4C).46 Successful HR to repair the DSB induced by the expression of a rare endonuclease I-SceI would lead to restore functional GFP that could be detected by flow cytometry. Therefore, MDA-MB-231 cells were transfected with the HR reporter and an I-SceI-expressing plasmid. A constitutively expressed DsRed was also included as a transfection control.46 Then the cells were treated with different concentrations of LBL1 for 48 h. The HR efficiency was calculated by the ratio of GFP+/DsRed+. As shown in Figure 3C, LBL1 treatment decreased the HR efficiency in a dose-dependent manner, consistent with the results from the Rad51 foci formation assay. Using a related GFP-based NHEJ reporter,46 we did not observe a decrease of NHEJ efficiency upon LBL1 treatment (Figure S4D), indicating LBL1’s selectivity in the HR pathway.
To further test if Rad51 is a key downstream target of LBL1’s interaction with LA, we overexpressed FLAG-tagged Rad51 in MDA-MB-231 cells to see if this overexpression could rescue LBL1’s effect on growth inhibition. FLAG-tagged Rad51 could be readily detected by Western blot, and it was migrated slightly slower with a higher molecular weight than endogenous Rad51 (Figure 3D). Importantly, the ectopically expressed FLAG-Rad51 was functional in forming subnuclear foci upon CPT treatment (Figure S4E). As presented in Figure 3E, cells with Rad51 overexpression were significantly more resistant to LBL1 than cells with EGFP overexpression. As a control, these cells maintained equal sensitivity to the CREB inhibitor naphthol AS-E. These results indicate that Rad51 is a key downstream mediator of LBL1’s effect.
LBL1 Disrupts LA–Rad51 Interaction Leading to Decreased Rad51 Protein Stability
LA is known to be implicated in DSB repair, and our results showed that LBL1 binds LA and inhibits HR. Therefore, we asked if LBL1 could modulate the level of Rad51, an essential DNA recombinase in the HR pathway. Treating MDA-MB-231 cells with LBL1 resulted in a decreased level of Rad51 protein (Figure 4A). Previous studies using shRNA to knockdown LA showed that the Rad51 mRNA level was decreased.17 To discern if a transcription mechanism was involved in Rad51 modulation by LBL1, we investigated the Rad51 transcript level by qRT-PCR. Our acute LBL1 treatment in MDA-MB-231 cells did not result in a decrease of the mRNA level of Rad51 (Figure 4B), suggesting another previously unrecognized post-translational regulation of Rad51 by LA was occurring. To investigate if a proteasome-mediated degradation mechanism was involved, we treated the cells with a combination of proteasome inhibitor MG132 and LBL1. Inhibition of the proteasome activity effectively rescued Rad51 downregulation induced by LBL1 (Figure 4A), suggesting that Rad51 might be ubiquitylated, and LBL1 could facilitate proteasome-mediated Rad51 degradation. To directly test if Rad51 was ubiquitylated, we employed an in vivo ubiquitylation assay47 by transfecting HEK 293T cells with FLAG-tagged ubiquitin (FLAG-Ub). Then the cells were treated with MG132 along with or without LBL1. Immunoprecipitation using anti-Rad51 under denaturing conditions showed that Rad51 was indeed polyubiquitylated (Figure 4C). Furthermore, LBL1 treatment presented more polyubiquitylated Rad51, likely driving Rad51 toward degradation. To quantitatively evaluate the stability of Rad51 in cells, we treated MDA-MB-231 cells with a protein synthesis inhibitor cycloheximide (CHX) along with or without LBL1. In the absence of LBL1, the half-life (t1/2) of Rad51 was 3.2 h (Figure 4D,E). With LBL1 treatment, the t1/2 was significantly decreased to 0.67 h.
Figure 4.
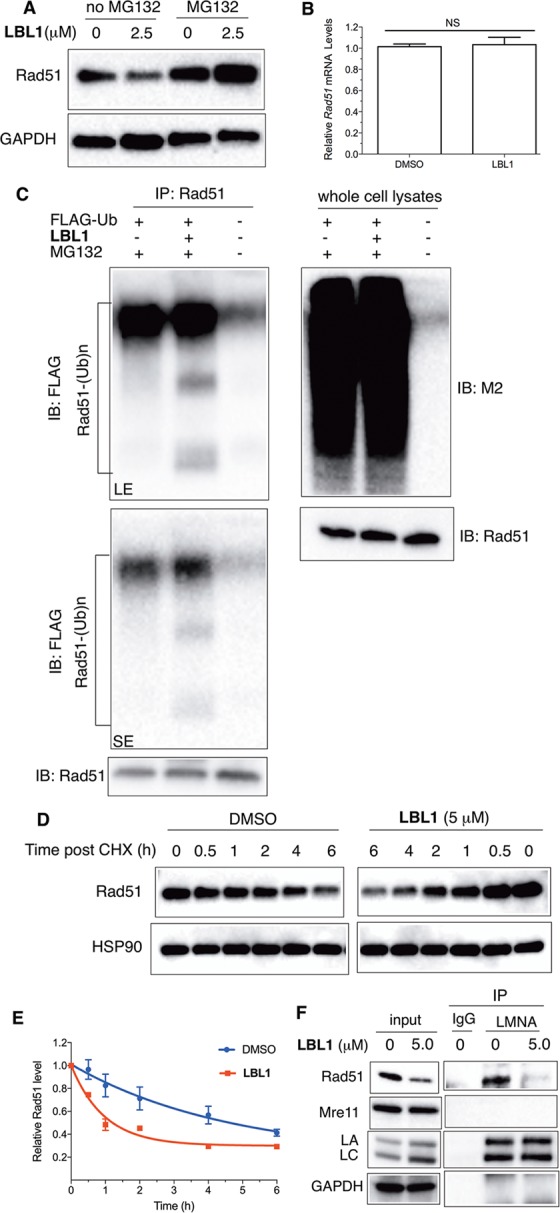
LBL1 induced proteasome-mediated degradation of Rad51. (A) LBL1-induced downregulation of Rad51 was mediated by proteasome. MDA-MB-231 cells were treated with the indicated concentration of LBL1 in the presence or absence of MG132 for 24 h. Then the cells were collected, and the lysates were prepared for Western blot analysis with indicated antibodies. (B) LBL1 did not decrease the mRNA level of Rad51. MDA-MB-231 cells were treated with LBL1 (0 or 2.5 μM) for 24 h. Then the total RNA was isolated for qRT-PCR analysis using HPRT as a reference gene (n = 3). (C) LBL1 promoted Rad51 ubiquitylation. HEK 293T cells were transfected with FLAG-Ub. Then the cells were treated with MG132 (20 μM) along with or without LBL1 (5 μM) for 6 h. The cells were then subjected to denaturing immunoprecipitation, and the precipitated proteins were analyzed by Western blot with indicated antibodies. SE, short exposure; LE, long exposure. (D) LBL1 decreased Rad51 protein stability in MDA-MB-231 cells. The cells were treated with LBL1 (0 or 5.0 μM) along with CHX for indicated time periods. The cells were collected, and the lysates were prepared for Western blot analysis with indicated antibodies. (E) Quantification of data shown in panel D (n = 2). (F) LA physically interacted with Rad51, and LBL1 disrupted this interaction in MDA-MB-231 cells. The cells were treated with indicated drugs for 24 h. Then the lysates were prepared as described in the Experimental Section for co-IP with anti-LA or IgG control. The precipitated proteins were analyzed by Western blot with indicated antibodies.
The proteasome-mediated regulation of Rad51 by LA is a novel mechanism independent of the reported transcriptional regulation. To investigate how LA might protect Rad51 from proteasome-mediated degradation, we found that LA interacted with Rad51 as made evident by a coimmunoprecipitation (co-IP) assay with anti-LA (Figure 4F). Furthermore, LBL1 inhibited the interaction between LA and Rad51 (Figure 4F). The inhibition of LA–Rad51 interaction by LBL1 was dose-dependent (Figure S5A). Another HR protein Mre11 didn’t interact with LA, nor did its level decrease upon LBL1 treatment (Figure 4F), suggesting LA’s specificity in regulating Rad51. We further studied other upstream proteins participating in the HR pathway including Nijmegen breakage syndrome 1 (NBS1), CtBP-interacting protein (CtIP), and breast and ovarian cancer susceptibility protein 1 (BRCA1). The protein level of these proteins was unchanged after the cells were treated with LBL1 (Figure S5B), further suggesting LBL1’s specificity in modulating Rad51. This newly discovered Rad51 post-translational regulation by LA and LA–Rad51 interaction was also observed in nonsmall cell lung cancer A549 cells (Figure S5C,D). Taken as a whole, these results suggest that LA is involved in protecting Rad51 from proteasome-mediated degradation. LBL1 was able to disrupt this LA–Rad51 interaction leading to accelerated Rad51 degradation and inhibition of HR repair of DSBs.
Discussion and Conclusions
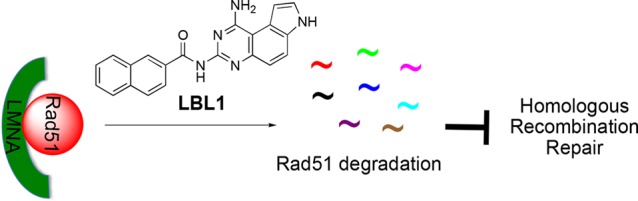
LBL1 is a novel compound we previously discovered from a phenotypical screening to possess selective toxicity in cancer cells with an unknown mechanism of action.29 In a recent study, we identified nuclear lamins as the direct molecular targets of LBL1 through an unbiased chemoproteomics strategy using a clickable photo-cross-linker LBL1-P.30 In this study, using LBL1 as a chemical tool, we found that LA interacted with Rad51 and that LBL1 disrupted this interaction leading to accelerated Rad51 interaction to interfere with HR repair of DSBs. This in turn led to accumulation of DSBs and G2/M cell cycle arrest and cellular apoptosis (Figure S6).
Increasingly, lamins’ functions have been expanded beyond their role as nucleoskeletal proteins.1 LA has been implicated in DSB repair pathways and genomic stability.15,17,48 More than 300 mutations in human LA have been discovered, and these mutations cause a wide spectrum of diseases collectively called laminopathies.49 Among these, HGPS associated LA mutation activates a cryptic splicing site resulting in a truncated LA mutant called progerin that is permanently farnesylated.13 Among the many phenotypes of HGPS cells are their elevated basal level of DSBs and impaired capability in HR repair of DSBs.15 Similar phenotypes have also been observed in other mouse models of HGPS and LA–/– cells.17,35 However, the underlying mechanisms are incompletely understood. Previously studies using long-term shRNA to knockdown LA showed that LA had a positive effect on transcriptional regulation of Rad51 to affect Rad51 protein level.17 In our current study using LBL1 as a unique chemical tool for acute treatment, we found that while the Rad51 protein level was decreased upon LBL1 treatment, the Rad51 transcript level was unchanged, suggesting the presence of a novel mechanism of Rad51 regulation by LA. In this regard, we found that the proteasome-mediated degradation pathway is involved in Rad51 regulation by LA. This discovery adds a new layer of post-translational regulation of Rad51 by LA besides previously described transcriptional regulation,17 highlighting the complexity of LA involvement in DSB repair. Supporting that Rad51 is a critical mediator of LBL1’s effect, ectopic expression of Rad51 in MDA-MB-231 cells could at least partially rescue the effect of LBL1 (see Figure 3E). However, complete rescue was not achieved. This could be due to the following nonmutually exclusive possibilities. The first is that there exist other downstream mediators besides Rad51. The other possibility is due to the relatively low expression level of FLAG-Rad51 (see Figure 3D). It was difficult for higher expression clones to survive as demonstrated previously.50
Rad51 protein level in the cells has to be tightly regulated to maintain genomic integrity. Upon DSB formation, Rad51 accumulates at the DSB sites forming distinct subnuclear foci.51 Our finding that LA physically interacts with Rad51 suggests that LA might play an important role in providing a reservoir for Rad51 at basal state (i.e., in the absence of DSBs). Through elegant FRAP (fluorescence recovery after photobleach) studies, Rad51 was previously shown to be diffusing at a much slower rate than its expected molecular weight inside the nucleus.52 However, the identities of proteins other than BRCA2 holding Rad51 remain unknown. The results shown here suggest that LA is perhaps one such protein to sequester Rad51 from proteasome-mediated degradation. We speculate that Rad51 might undergo a post-translational modification (e.g., phosphorylation) upon DSB formation to mobilize to the DSB sites to initiate HR repair. In the case of LBL1 treatment, while robust DSBs were generated inside the cancer cells, Rad51 did not accumulate at the DSB sites to form Rad51 foci (see Figure 3A), suggesting there was a Rad51 localization defect as well as Rad51 degradation upon LBL1 treatment, both of which could contribute to the decreased HR efficiency. Taken altogether, our discovery sheds new insights into the function of LA in regulating DSB repair using LBL1 as a chemical tool.
The results described here provide a potential explanation for the selective toxicity of LBL1 in cancer cells over normal cells. Activation of various oncogenes in cancer cells results in aberrantly high DNA replication stress leading to formation of lethal DSBs.23 These DSBs must be efficiently repaired by DSB repair pathways including HR in order for the cells to survive in the presence of activated oncogenes.23 Therefore, the evolved cancer cells are addicted to the DSB repair mechanisms for survival. On the other hand, normal cells do not endure high levels of DNA replication stress and are therefore less dependent on the DSB machinery. This is consistent with our results that normal cells did not produce γ-H2AX signal upon LBL1 treatment. Given the challenges in developing small molecules to directly target HR,53 extensive efforts are ongoing to identify other druggable regulatory proteins in the HR pathway.54,55 Our finding that the LA–Rad51 interaction is amenable for modulation by small molecules like LBL1 support that the LA–Rad51 axis represents a novel avenue for developing cancer therapeutics that inhibit DSB repair.
Experimental Section
Plasmids
All lentiviral shLA plasmids were purchased from Open Biosystems. The lentiviral shRNA packaging vectors and FLAG-Ub were gifts from Dr. Mushui Dai (Oregon Health & Science University). The lentiviral EGFP plasmid and its packaging vectors were gifts from Dr. Michael Cohen (Oregon Health & Science University). Rad51 overexpression plasmid was constructed using lentiviral EGFP plasmid as a backbone. All the plasmids were sequence-verified by Sanger sequencing. The primers used are available upon request. The NHEJ and HR reporter plasmids were generous gifts from Dr. Gorbunova (University of Rochester). The plasmid expressing I-SceI (pCBASceI) and the plasmid expressing DsRed (DsRed2-N1) were from Addgene.
Cell Lines and Culture
MDA-MB-231 was purchased from Developmental Therapeutics Program at the National Cancer Institute. A549, HFF, and HEK293T were purchased from American Type Culture Collection (ATCC). DKO MEFs were described before.56 The cells were tested for mycoplasma contamination regularly by PCR. Cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies) supplemented with 10% FBS (Hyclone) and 10% nonessential amino acids (Life Technologies) at 37 °C with 5% CO2. All the cells were used within 50 passages.
Chemicals
MG132 was from Calbiochem. S-(+)-Camptothecin, cycloheximide, and naphthol AS-E were from Sigma. LBL1 and LBL1-P were synthesized as previously described.29,30
Lentiviruses
HEK293T cells were transfected with lentiviral expression plasmids along with packaging vectors using the calcium-phosphate method. The supernatants containing lentiviral particles were collected, passed through a 0.45 μM filter, and stored at −80 °C prior to use. For lentiviral transduction, DKO MEF cells were plated in 24-well plates and infected with lentiviruses for 3 days with hexadimethrine bromide (8 μg/mL) and selected with puromycin (0.5 μg/mL). MDA-MB-231 cells were plated in 6-well plates and infected with lentiviruses for 3 days without hexadimethrine bromide and puromycin.
Growth Inhibition Assay
The cellular growth inhibition assay was carried out as described before.57
Immunofluorescence
For imaging γ-H2AX and Rad51 foci, the cells growing on the coverslips were treated as indicated. Then the cells were fixed with 3.7% formaldehyde and permeabilized with 0.5% triton X-100. The cells were washed with 1× PBS and blocked with 3% BSA before incubating with primary antibodies. The following primary antibodies were used: anti-phospho-H2AX (rabbit, Cell Signaling Technology, catalogue no. 9718, 1:400); anti-Rad51 (rabbit, Santa Cruz Biotechnology, catalogue no. sc-8349, 1:100); anti-LA (mouse, Sigma, catalogue no. SAB4200236, 1:1000); anti-FLAG M2 (mouse, Sigma, catalogue no. F3165, 1:400). The cells were then incubated with secondary antibodies for 1 h at room temperature. Cy-3-conjugated donkey antirabbit secondary antibody (Jackson Immunoresearch) was used at 1:1000 dilution. Coverslips were mounted in ProLong Gold AntiFade reagent with DAPI (Life Technologies), and images were acquired with a fluorescence microscope ApoTome (Zeiss). The cells were considered foci-positive if >10 foci were observed per nucleus. DAPI was used to count cell nuclei. Around 100 cell nuclei were analyzed for each experiment, and results are shown as the average of three independent cell preparations. For colocalization analysis, the Z-stack of images were reconstructed, and colocalization Pearson correlation coefficients were determined using the Coloc module in the Imaris software package (Bitplane).
Western Blot
For Western blot analysis, the following primary antibodies were used: anti-Rad51 (rabbit, Santa Cruz Biotechnology, catalogue no. sc-8349, 1:1000), anti-phospho-histone H2AX (rabbit, Cell Signaling Technology, catalogue no. 9718, 1:1200), anti-LA/C (mouse, Sigma, catalogue no. SAB4200236, 1:2000), anti-FLAG M2 (mouse, Sigma, catalogue no. F4049, 1:4000), anti-Hsp90 (rabbit, Cell Signaling Technology, catalogue no. 4874, 1:1000), anti-Mre11 (rabbit, Cell Signaling Technology, catalogue no. 4895, 1:1000), anti-GAPDH (mouse, Santa Cruz Biotechnology, catalogue no. sc-32233, 1:4000), anti-NBS1 (rabbit, Cell Signaling Technology, catalogue no. 14956, 1:1000), anti-CtIP (rabbit, Cell Signaling Technology, catalogue no. 9201, 1:1000), anti-BRCA1 (rabbit, Cell Signaling Technology, catalogue no. 9010, 1:1000). The HRP-conjugated secondary antibodies were from Cell Signaling Technology or Jackson ImmunoResearch Laboratories.
Coimmunoprecipitation
The cells were treated as indicated for 24 h. Then the cells were collected by scraping and washed twice with ice-cold PBS. The cell pellets were lysed in a lysis buffer B (50 mM TrisHCl, 5 mM EDTA, 150 mM NaCl, 1 mM DTT, 0.5% Nonidet P-40, pH 8.0) supplemented with 1 mM PMSF and protease inhibitor cocktail (Pierce) on ice for 15 min. The lysates were centrifuged at 14 000× rpm for 15 min at 4 °C. An equal amount of the supernatant was precleared with mouse IgG (Jackson ImmunoResearch) and protein A/G agarose beads (Thermo Scientific). The precleared lysates were then incubated with anti-LA overnight at 4 °C followed by the addition of protein A/G agarose beads for 1 h at 4 °C. The agarose beads were separated from unbound proteins by centrifuging at 3000× rpm for 2 min at 4 °C. The beads were then washed three times with lysis buffer B, and the bound proteins were eluted with 1× SDS-PAGE buffer by heating at 95 °C for 5 min. The eluted proteins were analyzed by SDS-PAGE followed by Western blot.
In-Cell Click Reaction
MDA-MB-231 cells growing on the coverslips were washed once with PBS, and then the cells were covered with PBS. The cells were treated with DMSO or LBL1 for 20 min followed by LBL1-P for 20 min. After being UV-irradiated for 5 min, the cells were fixed and permeabilized as above. After the cells were washed with 3% BSA in PBS, the cells were incubated with Click-iT cell reaction cocktail (Life Technologies) supplemented with rhodamine-N3 for 30 min at room temperature. Then the cells were washed with 3% BSA in PBS before being incubated with anti-LA overnight. The cells were then incubated with Alexa Fluor 488 donkey antimouse secondary antibody for 1 h at room temperature, and the coverslips were mounted as above. The fluorescent micrographs were acquired with an ApoTome fluorescence microscope (Zeiss).
COMET Assay
The neutral COMET assays were performed using a COMET assay reagent kit (Trevigen) according to the manufacturer’s protocol. MDA-MB-231 cells were treated as indicated for 30 h, and then collected and washed with ice-cold PBS. The washed cells were mixed with 1% low-melting agarose (Sigma) and spread on the slides (Trevigen). The slides were incubated with lysis buffer (Trevigen) overnight at 4 °C. After electrophoresis in neutral running buffer, the slides were stained with SYBR gold (Life Technologies) and visualized by an ApoTome fluorescence microscope (Zeiss). The images were analyzed with CASPlab software,58 and the percentage of DNA in the tail and tail moment were used to compare different samples.
Cell Cycle Analysis
MDA-MB-231 cells were treated as indicated, trypsin digested, collected, and washed twice with ice-cold PBS. The cells were resuspended in 1× binding buffer (BD Pharmingen, Annexin V: FITC apoptosis detection kit I). Ethanol was added to a final concentration of 70%, and the cells were left on ice for at least 2 h. After fixation, the cells were washed with ice-cold PBS and resuspended in 1× binding buffer. Then the cells were stained with propidium iodide and RNase A to a final concentration of 100 μg/mL for 30 min at room temperature. The stained cells were then analyzed using a BD FACSCANTO II flow cytometer. The resulting data were analyzed by ModFit (Verity Software House).
qRT-PCR
MDA-MB-231 cells were treated as indicated for 24 h, and the total RNA was extracted using a NucleoSpin RNA kit (Clontech). The first strand cDNA was synthesized using PrimeScript first strand cDNA synthesis kit (Clontech). qPCR was performed using the SYBR Advantage qPCR premix (Clontech), and a QuantStudio 7 Flex system (Life Technologies) was used to collect data. The results were evaluated by the 2–ΔΔCT method using HPRT as a reference gene.
DNA Intercalation Assay
The DNA ladder (250 ng, 1 kb, NEB) was incubated with indicated drugs in PBS for 30 min at room temperature. Then 6× DNA loading dye (NEB) was added, and the DNA ladder was separated on 1% agarose gel electrophoresis without EtBr. After electrophoresis, the gel was stained in the presence of EtBr for 30 min at room temperature.
In Vivo Ubiquitination Assay
HEK 293T cells in a 10 cm plate were transfected with 4 μg of FLAG-ubiquitin plasmid with Lipofectamin2000. Then, 48 h after transfection, the cells were treated with MG132 (20 μM) along with or without LBL1 (5 μM) for 6 h. The cells were collected by scraping and washed twice with cold PBS. Then the cells were lysed in lysis buffer B supplemented with 1% SDS, and the mixture was further sonicated. The lysates were centrifuged at 14 000× rpm for 10 min at room temperature, and the supernatant was diluted 10× with lysis buffer B. About 500 μg of lysates was incubated with 1 μg of anti-Rad51 for overnight at 4 °C, when protein A/G beads were added, and the mixture was tumbled for another 1 h. The agarose beads were separated from unbound proteins by centrifuging at 3000× rpm for 2 min at 4 °C. The beads were then washed three times with lysis buffer B, and the bound proteins were eluted with 1× SDS-PAGE buffer by heating at 95 °C for 5 min. The eluted proteins were analyzed by SDS-PAGE followed by Western blot.
Rad51 Degradation
MDA-MB-231 cells were treated with cycloheximide (100 μg/mL) and LBL1 (5 μM) for the indicated time periods. The cells were collected by scraping and washed twice with ice-cold PBS. The lysates were prepared with lysis buffer B supplemented with 8 M urea.
NHEJ and HR Reporter Assay
MDA-MB-231 cells were first transfected with NheI linearized NHEJ or HR plasmids and selected in the media with G418 (Life Technologies, 3 mg/mL). Then the cells were transfected with a plasmid expressing I-SceI and a plasmid expressing DsRed. The transfected cells were treated with LBL1 for 48 h. The cells were then collected for live cell flow cytometry analysis. The successful NHEJ or HR was measured as the ratio of GFP+/DsRed+.
Statistical Analysis
The statistical analyses were carried out using the Student t-test either in Excel 2011 or Prism 5.0. A P value of <0.05 was denoted as significance. The data are presented as mean ± SD or SEM. Variance was estimated and found to be similar among different groups. The investigators were not blinded to the sample groups.
Acknowledgments
We thank Dr. Stephanie Kaech and Aurelie Snyder for technical help on fluorescence microscopy image acquisition and analysis. We thank Professor Mushui Dai for technical help on the ubiquitination assay, and Mandy Gilchrist and Pamela Canaday for technical support in flow cytometry analysis. Professor Vera Gorbunova (University of Rochester) generously provided the HR and NHEJ reporter plasmids. This work was supported by NIH R01CA211866, and partially by R01GM122820 and R21CA220061. OHSU School of Medicine, OHSU Office of Technology Transfer and Business Development, Oregon Clinical & Translational Research Institute, Oregon Medical Research Foundation, and Lloyd Fund partially supported this research.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.8b00379.
Additional figures including MEF proliferation, DNA intercalation assay, COMET assay, HR assay, Rad51 modulation, and a working model (PDF)
Author Contributions
B.X.L. and X.X. conceived the project, analyzed the data, and wrote the manuscript, which was reviewed and approved by all the authors. B.X.L. performed most of the experiments. J.C. and B.C. synthesized and characterized LBL1 and LBL1-P. Y.Z. contributed SV40-immortalized MEF and experimental design on MEF.
The authors declare no competing financial interest.
Notes
Safety statement: no unexpected or unusually high safety hazards were encountered during the course of the experiments. Caution should be taken when handling ethidium bromide.
Supplementary Material
References
- Burke B.; Stewart C. L. The nuclear lamins: Flexibility in function. Nat. Rev. Mol. Cell Biol. 2013, 14, 13–24. 10.1038/nrm3488. [DOI] [PubMed] [Google Scholar]
- Aebi U.; Cohn J.; Buhle L.; Gerace L. The nuclear lamina is a meshwork of intermediate-type filaments. Nature 1986, 323, 560–564. 10.1038/323560a0. [DOI] [PubMed] [Google Scholar]
- Lin F.; Worman H. J. Structural organization of the human gene encoding nuclear lamin a and nuclear lamin c. J. Biol. Chem. 1993, 268, 16321–16326. [PubMed] [Google Scholar]
- Dechat T.; Adam S. A.; Taimen P.; Shimi T.; Goldman R. D. Nuclear lamins. Cold Spring Harbor Perspect. Biol. 2010, 2, a000547. 10.1101/cshperspect.a000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyatina A. A.; Nicolet S.; Aebi U.; Herrmann H.; Strelkov S. V. Atomic structure of the vimentin central alpha-helical domain and its implications for intermediate filament assembly. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 13620–13625. 10.1073/pnas.1206836109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelkov S. V.; Schumacher J.; Burkhard P.; Aebi U.; Herrmann H. Crystal structure of the human lamin a coil 2b dimer: Implications for the head-to-tail association of nuclear lamins. J. Mol. Biol. 2004, 343, 1067–1080. 10.1016/j.jmb.2004.08.093. [DOI] [PubMed] [Google Scholar]
- Dhe-Paganon S.; Werner E. D.; Chi Y. I.; Shoelson S. E. Structure of the globular tail of nuclear lamin. J. Biol. Chem. 2002, 277, 17381–17384. 10.1074/jbc.C200038200. [DOI] [PubMed] [Google Scholar]
- Ruan J.; Xu C.; Bian C.; Lam R.; Wang J. P.; Kania J.; Min J.; Zang J. Crystal structures of the coil 2b fragment and the globular tail domain of human lamin b1. FEBS Lett. 2012, 586, 314–318. 10.1016/j.febslet.2012.01.007. [DOI] [PubMed] [Google Scholar]
- Zastrow M. S.; Vlcek S.; Wilson K. L. Proteins that bind a-type lamins: Integrating isolated clues. J. Cell Sci. 2004, 117, 979–987. 10.1242/jcs.01102. [DOI] [PubMed] [Google Scholar]
- Shumaker D. K.; Solimando L.; Sengupta K.; Shimi T.; Adam S. A.; Grunwald A.; Strelkov S. V.; Aebi U.; Cardoso M. C.; Goldman R. D. The highly conserved nuclear lamin ig-fold binds to pcna: Its role in DNA replication. J. Cell Biol. 2008, 181, 269–280. 10.1083/jcb.200708155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechat T.; Korbei B.; Vaughan O. A.; Vlcek S.; Hutchison C. J.; Foisner R. Lamina-associated polypeptide 2alpha binds intranuclear a-type lamins. J. Cell Sci. 2000, 113, 3473–3484. [DOI] [PubMed] [Google Scholar]
- Zastrow M. S.; Flaherty D. B.; Benian G. M.; Wilson K. L. Nuclear titin interacts with a- and b-type lamins in vitro and in vivo. J. Cell Sci. 2006, 119, 239–249. 10.1242/jcs.02728. [DOI] [PubMed] [Google Scholar]
- Eriksson M.; Brown W. T.; Gordon L. B.; Glynn M. W.; Singer J.; Scott L.; Erdos M. R.; Robbins C. M.; Moses T. Y.; Berglund P.; Dutra A.; Pak E.; Durkin S.; Csoka A. B.; Boehnke M.; Glover T. W.; Collins F. S. Recurrent de novo point mutations in lamin a cause hutchinson-gilford progeria syndrome. Nature 2003, 423, 293–298. 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman R. D.; Shumaker D. K.; Erdos M. R.; Eriksson M.; Goldman A. E.; Gordon L. B.; Gruenbaum Y.; Khuon S.; Mendez M.; Varga R.; Collins F. S. Accumulation of mutant lamin a causes progressive changes in nuclear architecture in hutchinson-gilford progeria syndrome. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 8963–8968. 10.1073/pnas.0402943101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B.; Wang J.; Chan K. M.; Tjia W. M.; Deng W.; Guan X.; Huang J. D.; Li K. M.; Chau P. Y.; Chen D. J.; Pei D.; Pendas A. M.; Cadinanos J.; Lopez-Otin C.; Tse H. F.; Hutchison C.; Chen J.; Cao Y.; Cheah K. S.; Tryggvason K.; Zhou Z. Genomic instability in laminopathy-based premature aging. Nat. Med. 2005, 11, 780–785. 10.1038/nm1266. [DOI] [PubMed] [Google Scholar]
- Mukherjee A. B.; Costello C. Aneuploidy analysis in fibroblasts of human premature aging syndromes by fish during in vitro cellular aging. Mech. Ageing Dev. 1998, 103, 209–222. 10.1016/S0047-6374(98)00041-4. [DOI] [PubMed] [Google Scholar]
- Redwood A. B.; Perkins S. M.; Vanderwaal R. P.; Feng Z.; Biehl K. J.; Gonzalez-Suarez I.; Morgado-Palacin L.; Shi W.; Sage J.; Roti-Roti J. L.; Stewart C. L.; Zhang J.; Gonzalo S. A dual role for a-type lamins in DNA double-strand break repair. Cell Cycle 2011, 10, 2549–2560. 10.4161/cc.10.15.16531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M.; Hunt C. R.; Pandita R. K.; Kumar R.; Yang C. R.; Horikoshi N.; Bachoo R.; Serag S.; Story M. D.; Shay J. W.; Powell S. N.; Gupta A.; Jeffery J.; Pandita S.; Chen B. P.; Deckbar D.; Lobrich M.; Yang Q.; Khanna K. K.; Worman H. J.; Pandita T. K. Lamin a/c depletion enhances DNA damage-induced stalled replication fork arrest. Mol. Cell. Biol. 2013, 33, 1210–1222. 10.1128/MCB.01676-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez I.; Redwood A. B.; Perkins S. M.; Vermolen B.; Lichtensztejin D.; Grotsky D. A.; Morgado-Palacin L.; Gapud E. J.; Sleckman B. P.; Sullivan T.; Sage J.; Stewart C. L.; Mai S.; Gonzalo S. Novel roles for a-type lamins in telomere biology and the DNA damage response pathway. EMBO J. 2009, 28, 2414–2427. 10.1038/emboj.2009.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs-Seymour I.; Markiewicz E.; Bekker-Jensen S.; Mailand N.; Hutchison C. J. Lamin a/c-dependent interaction with 53bp1 promotes cellular responses to DNA damage. Aging Cell 2015, 14, 162–169. 10.1111/acel.12258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S.; Liu B.; Wang Y.; Hao Q.; Zhou Z. Lamin a is an endogenous sirt6 activator and promotes sirt6-mediated DNA repair. Cell Rep. 2015, 13, 1396–1406. 10.1016/j.celrep.2015.10.006. [DOI] [PubMed] [Google Scholar]
- Chapman J. R.; Taylor M. R.; Boulton S. J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell 2012, 47, 497–510. 10.1016/j.molcel.2012.07.029. [DOI] [PubMed] [Google Scholar]
- Halazonetis T. D.; Gorgoulis V. G.; Bartek J. An oncogene-induced DNA damage model for cancer development. Science 2008, 319, 1352–1355. 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hanahan D.; Weinberg R. A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Bartkova J.; Rezaei N.; Liontos M.; Karakaidos P.; Kletsas D.; Issaeva N.; Vassiliou L. V.; Kolettas E.; Niforou K.; Zoumpourlis V. C.; Takaoka M.; Nakagawa H.; Tort F.; Fugger K.; Johansson F.; Sehested M.; Andersen C. L.; Dyrskjot L.; Orntoft T.; Lukas J.; Kittas C.; Helleday T.; Halazonetis T. D.; Bartek J.; Gorgoulis V. G. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 2006, 444, 633–637. 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Di Micco R.; Fumagalli M.; Cicalese A.; Piccinin S.; Gasparini P.; Luise C.; Schurra C.; Garre M.; Nuciforo P. G.; Bensimon A.; Maestro R.; Pelicci P. G.; d'Adda di Fagagna F. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 2006, 444, 638–642. 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Bartkova J.; Horejsi Z.; Koed K.; Kramer A.; Tort F.; Zieger K.; Guldberg P.; Sehested M.; Nesland J. M.; Lukas C.; Orntoft T.; Lukas J.; Bartek J. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 2005, 434, 864–870. 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- Helleday T.; Petermann E.; Lundin C.; Hodgson B.; Sharma R. A. DNA repair pathways as targets for cancer therapy. Nat. Rev. Cancer 2008, 8, 193–204. 10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- Chen J.; Kassenbrock A.; Li B. X.; Xiao X. Discovery of a potent anti-tumor agent through regioselective mono--acylation of 7-pyrrolo[3,2-]quinazoline-1,3-diamine. MedChemComm 2013, 4, 1275–1282. 10.1039/c3md00134b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. X.; Chen J.; Chao B.; David L. L.; Xiao X. Anticancer pyrroloquinazoline LBL1 targets nuclear lamins. ACS Chem. Biol. 2018, 13, 1380–1387. 10.1021/acschembio.8b00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.; Sharov A. A.; McDole K.; Cheng M.; Hao H.; Fan C. M.; Gaiano N.; Ko M. S.; Zheng Y. Mouse b-type lamins are required for proper organogenesis but not by embryonic stem cells. Science 2011, 334, 1706–1710. 10.1126/science.1211222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y.; Kim Y.; Shimi T.; Goldman R. D.; Zheng Y. Concentration-dependent lamin assembly and its roles in the localization of other nuclear proteins. Mol. Biol. Cell 2014, 25, 1287–1297. 10.1091/mbc.e13-11-0644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B. X.; Yamanaka K.; Xiao X. Structure-activity relationship studies of naphthol as-e and its derivatives as anticancer agents by inhibiting creb-mediated gene transcription. Bioorg. Med. Chem. 2012, 20, 6811–6820. 10.1016/j.bmc.2012.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luzi P.; Bruni A.; Mangiavacchi P.; Cevenini G.; Marini D.; Tosi P. Ploidy pattern and cell cycle in breast cancer as detected by image analysis and flow cytometry. Cytometry 1994, 18, 79–87. 10.1002/cyto.990180205. [DOI] [PubMed] [Google Scholar]
- Varela I.; Cadinanos J.; Pendas A. M.; Gutierrez-Fernandez A.; Folgueras A. R.; Sanchez L. M.; Zhou Z.; Rodriguez F. J.; Stewart C. L.; Vega J. A.; Tryggvason K.; Freije J. M.; Lopez-Otin C. Accelerated ageing in mice deficient in zmpste24 protease is linked to p53 signalling activation. Nature 2005, 437, 564–568. 10.1038/nature04019. [DOI] [PubMed] [Google Scholar]
- Rogakou E. P.; Boon C.; Redon C.; Bonner W. M. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 1999, 146, 905–916. 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y. DNA topoisomerase i inhibitors: Chemistry, biology, and interfacial inhibition. Chem. Rev. 2009, 109, 2894–2902. 10.1021/cr900097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive P. L.; Banath J. P. The comet assay: A method to measure DNA damage in individual cells. Nat. Protoc. 2006, 1, 23–29. 10.1038/nprot.2006.5. [DOI] [PubMed] [Google Scholar]
- Johnson R. D.; Jasin M. Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J. 2000, 19, 3398–3407. 10.1093/emboj/19.13.3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C.; Erixon K.; Arnaudeau C.; Schultz N.; Jenssen D.; Meuth M.; Helleday T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 2002, 22, 5869–5878. 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong L. N.; Li Y.; Sun E.; Ang K.; Hwang P. Y.; Wu X. Homologous recombination is a primary pathway to repair DNA double-strand breaks generated during DNA rereplication. J. Biol. Chem. 2014, 289, 28910–28923. 10.1074/jbc.M114.576488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorensen C. S.; Hansen L. T.; Dziegielewski J.; Syljuasen R. G.; Lundin C.; Bartek J.; Helleday T. The cell-cycle checkpoint kinase chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 2005, 7, 195–201. 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- Gildemeister O. S.; Sage J. M.; Knight K. L. Cellular redistribution of rad51 in response to DNA damage: Novel role for rad51c. J. Biol. Chem. 2009, 284, 31945–31952. 10.1074/jbc.M109.024646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C. B.; Clements M. K.; Wasi S.; Daoud S. S. Sensitivity to camptothecin of human breast carcinoma and normal endothelial cells. Cancer Chemother. Pharmacol. 1997, 40, 475–483. 10.1007/s002800050690. [DOI] [PubMed] [Google Scholar]
- Shao R. G.; Cao C. X.; Shimizu T.; O'Connor P. M.; Kohn K. W.; Pommier Y. Abrogation of an s-phase checkpoint and potentiation of camptothecin cytotoxicity by 7-hydroxystaurosporine (ucn-01) in human cancer cell lines, possibly influenced by p53 function. Cancer Res. 1997, 57, 4029–4035. [PubMed] [Google Scholar]
- Mao Z.; Seluanov A.; Jiang Y.; Gorbunova V. Trf2 is required for repair of nontelomeric DNA double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 2007, 104, 13068–13073. 10.1073/pnas.0702410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M. S.; Zeng S. X.; Jin Y.; Sun X. X.; David L.; Lu H. Ribosomal protein l23 activates p53 by inhibiting mdm2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 2004, 24, 7654–7668. 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Suarez I.; Redwood A. B.; Gonzalo S. Loss of a-type lamins and genomic instability. Cell Cycle 2009, 8, 3860–3865. 10.4161/cc.8.23.10092. [DOI] [PubMed] [Google Scholar]
- Butin-Israeli V.; Adam S. A.; Goldman A. E.; Goldman R. D. Nuclear lamin functions and disease. Trends Genet. 2012, 28, 464–471. 10.1016/j.tig.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flygare J.; Falt S.; Ottervald J.; Castro J.; Dackland A. L.; Hellgren D.; Wennborg A. Effects of hsrad51 overexpression on cell proliferation, cell cycle progression, and apoptosis. Exp. Cell Res. 2001, 268, 61–69. 10.1006/excr.2001.5265. [DOI] [PubMed] [Google Scholar]
- West S. C. Molecular views of recombination proteins and their control. Nat. Rev. Mol. Cell Biol. 2003, 4, 435–445. 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- Yu D. S.; Sonoda E.; Takeda S.; Huang C. L.; Pellegrini L.; Blundell T. L.; Venkitaraman A. R. Dynamic control of rad51 recombinase by self-association and interaction with brca2. Mol. Cell 2003, 12, 1029–1041. 10.1016/S1097-2765(03)00394-0. [DOI] [PubMed] [Google Scholar]
- Sanford-Crane H.; Pejovic T.; Xiao X. Drugging homologous recombination: Back to the future. Future Med. Chem. 2018, 10, 1279–1281. 10.4155/fmc-2018-0096. [DOI] [PubMed] [Google Scholar]
- Sun C.; Yin J.; Fang Y.; Chen J.; Jeong K. J.; Chen X.; Vellano C. P.; Ju Z.; Zhao W.; Zhang D.; Lu Y.; Meric-Bernstam F.; Yap T. A.; Hattersley M.; O′Connor M. J.; Chen H.; Fawell S.; Lin S. Y.; Peng G.; Mills G. B. Brd4 inhibition is synthetic lethal with parp inhibitors through the induction of homologous recombination deficiency. Cancer Cell 2018, 33, 401–416. 10.1016/j.ccell.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. F.; Cruz C.; Greifenberg A. K.; Dust S.; Stover D. G.; Chi D.; Primack B.; Cao S.; Bernhardy A. J.; Coulson R.; Lazaro J. B.; Kochupurakkal B.; Sun H.; Unitt C.; Moreau L. A.; Sarosiek K. A.; Scaltriti M.; Juric D.; Baselga J.; Richardson A. L.; Rodig S. J.; D'Andrea A. D.; Balmana J.; Johnson N.; Geyer M.; Serra V.; Lim E.; Shapiro G. I. Cdk12 inhibition reverses de novo and acquired parp inhibitor resistance in brca wild-type and mutated models of triple-negative breast cancer. Cell Rep. 2016, 17, 2367–2381. 10.1016/j.celrep.2016.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimi T.; Kittisopikul M.; Tran J.; Goldman A. E.; Adam S. A.; Zheng Y.; Jaqaman K.; Goldman R. D. Structural organization of nuclear lamins a, c, b1, and b2 revealed by superresolution microscopy. Mol. Biol. Cell 2015, 26, 4075–4086. 10.1091/mbc.e15-07-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F.; Li B. X.; Kassenbrock A.; Xue C.; Wang X.; Qian D. Z.; Sears R. C.; Xiao X. Identification of a potent inhibitor of creb-mediated gene transcription with efficacious in vivo anticancer activity. J. Med. Chem. 2015, 58, 5075–5087. 10.1021/acs.jmedchem.5b00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konca K.; Lankoff A.; Banasik A.; Lisowska H.; Kuszewski T.; Gozdz S.; Koza Z.; Wojcik A. A cross-platform public domain pc image-analysis program for the comet assay. Mutat. Res., Genet. Toxicol. Environ. Mutagen. 2003, 534, 15–20. 10.1016/S1383-5718(02)00251-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.