Abstract
Aim:
Caffeic acid (3,4-dihydroxycinnamic acid) phenethyl ester (CAPE), the major constituent of propolis, is able to increase the survival of the nematode Caenorhabditis elegans after infection with the fungal pathogen Candida albicans.
Results:
CAPE increases the expression of several antimicrobial proteins involved in the immune response to C. albicans. Structural derivatives of CAPE were synthesized to identify structure–activity relationships and decrease metabolic liability, ultimately leading to a compound that has similar efficacy, but increased in vivo stability. The CED-10(Rac-1)/PAK1 pathway was essential for immunomodulation by CAPE and was a critical component involved in the immune response to fungal pathogens.
Conclusion:
Caenorhabditis elegans is an efficient heterologous host to evaluate immunomodulatory compounds and identify components of the pathway(s) involved in the mode of action of compounds.
Keywords: : caffeic acid phenethyl ester, Candida, innate immune response, p21 kinase, propolis
Caffeic acid (3,4-dihydroxycinnamic acid) phenethyl ester (CAPE) is a flavonoid-like compound derived from honeybee propolis, a wax-like resinous substance that has been used as a folk medicine since at least 300 BC [1–5]. Since its identification as a major constituent of propolis [6], CAPE has been shown to have a wide range of biological activity, including antiviral, antifungal, antioxidative, antitumor and anti-inflammatory properties [2,7]. The immunomodulatory activity of CAPE has been documented on multiple types of immune cells and, in particular, the peripheral blood mononuclear cells (lymphocytes, monocytes and macrophages) [3]. Overall, the molecular mechanism by which CAPE modulates the immune system remains unknown, although nuclear factor of activated T cells (NFAT) and NF-κB have been proposed as critical mediators of this immune response [8].
CAPE inhibits two key transcription factors involved in T-cell activation, NFAT and NF-κB [8,9]. Inhibition of either of these two transcription factors could lead to the decreased production of cytokines associated with CAPE treatment [3,9,10]. Cytokines produced by Th1 (IL-2) and Th2 (IL-4) type lymphocytes, as well as cytokines produced by of macrophages, monocytes and dendritic cells (IL-1β and IL-12), are transcriptionally repressed in the presence of CAPE [3,9,10]. In contrast to these studies, production of the cytokine TGF-β1 was stimulated in the presence of CAPE, and in vivo studies using a murine model indicate CD4+ T-cell populations are increased [3,11]. Moreover, studies using primary splenic lymphocytes exposed indicate that secreted IL-2 and IL-4 cytokine concentrations are significantly increased in the presence of CAPE [11].
Caenorhabditis elegans has served as a useful in vivo heterologous host to identify and evaluate potential therapeutic compounds against several microbes [12] including the clinically relevant yeasts Candida albicans [13] and Cryptococcus neoformans [14]. Of systemic bloodstream infections, Candida spp. are the fourth most common etiological agents responsible for the infection, and within this group of fungi C. albicans is the most prevalent [15,16]. In a multistate point-prevalence survey of healthcare-associated infections, Candida spp. infections represented 6% of all hospital-acquired infections in the USA in 2011, and ranked first among causes of primary bloodstream infections [17].
Several screens for compounds with efficacy against C. albicans using C. elegans as a host have been conducted, and have collectively identified 39 compounds with uncharacterized antifungal activity against Candida spp. [7,18–20]. One of the identified compounds was CAPE, which was able to significantly increase C. albicans-infected nematode survival and disrupt fungal biofilm formation [7]. Importantly, CAPE was able to prolong survival of mice in a candidiasis model of infection [7]. The minimum inhibitory concentration (MIC) of CAPE against C. albicans was 64 μg/ml; however, an increase in nematode survival was observed at lower concentrations (4–8 μg/ml) suggesting the survival may also be due to immunomodulatory activity of the compound in the nematode [7].
The innate immune response of C. elegans to an intestinal infection of C. albicans has indicated that the PMK-1/p38 MAPK cascade is a key component [7,13]. Inactivation of this kinase cascade significantly decreases C. albicans-infected nematode survival [7,13]. Global expression studies revealed that 124 and 189 C. elegans genes were up- or down-regulated twofold or greater, respectively, after 4 h of infection with the fungus [13]. These genes were enriched for confirmed and putative antimicrobial proteins, and genes encoding proteins involved in detoxification and stress response [13]. Interestingly, unlike mammalian cells, C. elegans lacks homologs to NFAT and NF-κB [12] and, therefore, if the increased C. albicans-infected nematode survival is due to the immunomodulatory activity of CAPE, the mechanism responsible for conferring the efficacy must be due to other factors.
We demonstrate herein that CAPE demonstrates immunomodulatory activity in C. elegans and exploited the use of the nematode as a model host to assess the efficacy of 14 structural analogs of CAPE. A divergent and distinct structure–activity relationship (SAR) within CAPE was developed for the observed antifungal and immunomodulatory activities while, at the same time, reducing the inherent metabolic liability of the parent compound. Further studies of the immunomodulatory activity revealed that the efficacy conferred by the immunomodulating-specific compounds required the CED-10/PAK1 pathway in C. elegans. These components were found to be involved in the innate immune response pathway of the nematode against the fungal pathogens C. albicans and C. neoformans.
Materials & methods/experimental
Caenorhabditis elegans strains & culture conditions
Caenorhabditis elegans N2, glp-4(bn2); sek-1(km4), DH26 fer-15(b26)II and RB689 pak-1(ok448)X were obtained from the Caenorhabditis Genetics Center, University of Minnesota. The fer-15;pak-1 double mutant was generated through crosses and is described in detail below. Nematodes were maintained at 15°C, unless otherwise noted and propagated on nematode growth medium seeded with Escherichia coli HB101 according to standard techniques [21].
Caenorhabditis elegans-killing assays
To assess the efficacy of compounds to confer an increase in survival to C. albicans-infected nematodes, the preinfection liquid killing assay was performed using 30–40 glp-4(bn2); sek-1(km4) L4 worms as previously described [22]. Worms that died as a result of adhering to the wall of the plate were not included in the analysis and censored. Each assay was carried out in duplicate and repeated at least twice, and compounds conferring a significant increase in nematode survival were confirmed by repeating the assay using fer-15(b26) nematodes. Nematode survival was calculated by the Kaplan–Meier method, and survival differences were tested for significance using the log–rank test. The level of significance was set at p < 0.05, as indicated. Representative survival assays are indicated in the figures. Survival was monitored once daily (at ∼24-h intervals) and the time points are presented in hours, for accuracy.
Assessment of lifespan in liquid media
Synchronized wild-type N2 nematodes were prepared by egg prep and allowed to hatch overnight. Nematodes were transferred into 2-ml S-basal media with cholesterol in six well plates and fed daily with HB101. Nematode survival was monitored daily throughout the duration of the experiment, and nematodes were moved to new assay plates every other day during egg-laying period. The assay was carried out twice with 35 nematodes per assay condition.
Expression studies
Quantitative reverse transcription PCR analysis of genes was carried out using a Bio-Rad CFX96 real-time PCR system. RNA was obtained from nematodes using a previously described procedure [13] with slight modifications. Briefly, approximately 200 synchronized L4 nematodes were added to 1.9-ml SC-complete media with 2× additives in six-well assay plates. For each well, CAPE was added to a final concentration of 8 μg/ml in 1% DMSO as well as 50 μl of a 5× overnight concentrate of HB101 for the nematodes to feed. Nematodes from a single well were collected at 2 and 4 h post-treatment and total RNA extracted using TriReagent (Molecular Research Center, Inc., OH, USA) and reverse transcribed into cDNA using the Ambion RETROscript reverse transcription kit (Life Technologies, NY, USA). Quantitation was carried using iTaq Universal SYBR green supermix (Bio-Rad, CA, USA) and primers listed in Supplementary Table 2. Previously established primers for F08G5.8, F35E12.5, irg-1, irg-2, irg-3, clec-60, T24B8.5, C17H12.8, C32H11.1 and cyp35B1 were kindly provided by the Ausubel laboratory (Massachusetts General Hospital, MA, USA). Ct-values were normalized using snb-1 as the reference and calculated using the 2-ΔΔCt method [23].
Assessment of MIC of compounds
The MIC of the compounds was determined using the established procedure in the Clinical and Laboratory Standards Institute M27-A3 guideline [24].
NF-κB luciferase reporter assay with CAPE derivatives
To evaluate the inhibitory effect of the compounds on NF-κB activity, GloResponse™ NF-κB-RE-luc2P HEK293 Cell Line (Promega Cat# E8520) was used. All compounds were tested in 12 dilutions with assay concentrations ranging from 33.33 to 0.0163 μM in duplicate plates. A similar dose–response curve was generated using parthenolide and oridonin as positive controls. A total of 5000 NF-κB-RE-luc2P HEK293 cells were plated in 384-well plates in duplicate and the next day preincubated with the compounds for 2 h and then stimulated with 20 ng/ml of TNF-α for 5 h and performed Steady-Glo Luciferase assay. Percent of NF-κB activity was calculated by normalizing to treatment with TNFα and solvent alone (DMSO) as 100%.
Phosphate-buffered saline solubility
Solubility was determined in phosphate-buffered saline (PBS) pH 7.4 with 1% DMSO. Each compound was prepared in triplicate at 100 μM in both 100% DMSO and PBS with 1% DMSO. Compounds were allowed to equilibrate at room temperature with a 750-r.p.m. vortex shake for 18 h. After equilibration, samples were analyzed by ultra-performance liquid chromatography–mass spectrometry (Waters, MA, USA) with compounds detected by SIR detection on a single quadrupole mass spectrometer. The DMSO samples were used to create a two-point calibration curve to which the response in PBS was fit.
Plasma stability
Plasma stability was determined at 37°C at 5 h in both human and mouse plasma. Each compound was prepared in duplicate at 5 μM in plasma diluted 50/50 (v/v) with PBS pH 7.4 (0.95% acetonitrile and 0.05% DMSO). Compounds were incubated at 37°C for 5 h with a 350-r.p.m. orbital shake with time points taken at 0 and 5 h. Samples were analyzed by ultra-performance liquid chromatography–mass spectrometry (Waters) with compounds detected by SIR detection on a single quadrupole mass spectrometer.
Generation of fer-15(b26);pak-1(ok448) nematodes
Nematodes carrying the single mutations (DH26 fer-15(b26)II and RB689 pak-1(ok448)X) were crossed generating an F1 population of heterozygous nematodes. These worms were allowed to self-fertilize generating an F2 population that includes the desired homozygous fer-15;pak-1-mutant nematodes. The resulting F2 progeny were initially screened for the homozygous fer-15 mutation by assessment of the temperature-sensitive sterile phenotype conferred by the homozygous fer-15 mutation. Progeny that were temperature sensitive sterile at 25°C (homozygous fer-15) were then subsequently screened for the homozygous pak-1 mutation by two single-worm PCR assays. As the ok448 pak-1 mutation is due to a 1425 bp deletion within the coding region, primers that flank the deletion site (5′-GGA CAG AAT GGG AGA AAT TG-3′ and 5′-ATG GTG AAA CTC CTG CTG AT-3′) will generate PCR products of 2033 and 609 bp indicating the pak-1 wild-type and ok448 alleles, respectively. A second PCR reaction was carried out on the nematodes that indicated the ok448 allele was present using primers within the deletion region (5′-TAC CCA GCT TTC GGT AGT TC-3′ and 5′-ACT GAT GTT GTC ACG GAG TG-3′) to confirm the homozygous pak-1 mutation.
RNA interference
The ced-10 RNAi bacterial clone was obtained from the C. elegans RNAi library of Source Bioscience (Nottingham, UK). Bacteria were grown in Luria broth (LB) with 100 µg/ml ampicillin and spread on 10 cm RNAi plates (nematode growth medium + 50 µg/ml ampicillin + 1 mM Isopropyl-β-d-thiogalactopyranoside). Synchronized L1 larvae were placed on each RNAi plate and incubated at 15°C. Their progeny L1 worms were transferred to new RNAi plates and incubated at 25°C for 2 days. Young adult worms were used in killing assays after washing multiple times in M9.
Results & discussion
Efficacy of CAPE to increase survival of Candida albicans-infected nematodes is due to immunomodulatory activity
In a previous study, CAPE was shown to have direct antifungal activity with an MIC of 225 μM (64 μg/ml in 2% DMSO); a concentration that was higher than necessary to confer an increase in nematode survival (14–28 μM; 4–8 μg/ml) leading to the speculation that the efficacy may be due to immunomodulatory activity [7]. The alternate possibility for the increased survival of C. elegans is that CAPE is able to prolong the overall lifespan of the nematode. In order to assess if the increased survival is due to alteration in life expectancy, we evaluated the nematode lifespan under various conditions in the presence and absence of CAPE to determine if the compound has an influence. Contrary to the increase in survival observed for C. albicans-infected nematodes, there was a significant decrease in survival (p = 0.0015) when the nematodes were assayed in a liquid assay in the presence of 35 μM (10 μg/ml) of CAPE at 20°C (Supplementary Figure 1A). When the temperature for the lifespan assay was increased to the same temperature used in the C. albicans-killing assays (25°C), there was no significant difference in the lifespan of the nematodes (p = 0.2333; Supplementary Figure 1B). The inability of CAPE to alter the nematode lifespan under the same environmental conditions used as the C. albicans survival assay supports the hypothesis that the increased C. albicans-nematode survival could be due to immunomodulatory activity of the compound.
In order to confirm that CAPE elicits immunomodulatory activity in C. elegans, we evaluated the induction of genes known to be involved in the immune response. As noted above, our group has investigated the immune response of C. elegans challenged with C. albicans and identified several proteins with putative antimicrobial and pathogen-related functions [13]. The transcription of 8 genes (from a total of 33 genes significantly upregulated ≥3.0-fold) that were induced upon feeding on C. albicans was monitored after treatment with 28 μM (8 μg/ml) CAPE by quantitative PCR (qPCR). These genes were selected on the basis of their increased expression in response to C. albicans infection and their presumed function, and include the antimicrobial proteins abf-2, fipr-22,23, cnc-4 and cnc-7; the chitinases cht-1 and T19H5.1; the ferritin homolog ftn-1; and the predicted lipase Y46H3A.4. The expression of another 11 genes that have differential expression in the presence of bacterial pathogens and are believed to act as part of the immune response or detoxification was also monitored after treatment with CAPE. Of the 19 total genes that were analyzed, 3 were induced by ≥3.0-fold in 4 h after treatment with 28 μM (8 μg/ml) of CAPE (Table 1). These three genes included the antimicrobial proteins encoded by fipr-22,23 and cnc-4 and the chitinase encoded by cht-1. All three of these genes were also increased in expression after treatment with C. albicans, suggesting they may be partially responsible for the increased survival of CAPE-treated nematodes when challenged with the fungus. Of note, the expression of the putative antimicrobial proteins, encoded by fipr-22,23 and cnc-4, with CAPE was at a higher level when compared at the same time point (4 h) after feeding on C. albicans. None of the genes included in the assay were found to be repressed by exposure to CAPE.
Table 1. . Genes that were expressed threefold or greater after treatment with 28 μM (8 μg/ml) caffeic acid (3,4-dihydroxycinnamic acid) phenethyl ester.
| Gene | Function | Average fold increase after 2 h | Average fold increase after 4 h |
|---|---|---|---|
| fipr-22,23 | Putative antimicrobial peptide | 5.26 ± 1.94 | 22.98 ± 1.80 |
| cnc-4 | Putative caenacin antimicrobial peptide | 2.90 ± 2.68 | 31.19 ± 1.30 |
| cht-1 | Chitinase | 1.26 ± 0.66 | 3.50 ± 0.21 |
Increased Candida albicans-infected nematode survival following CAPE treatment is not observed with its metabolite caffeic acid
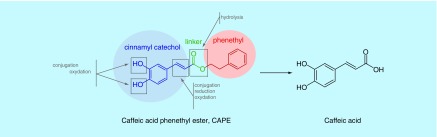
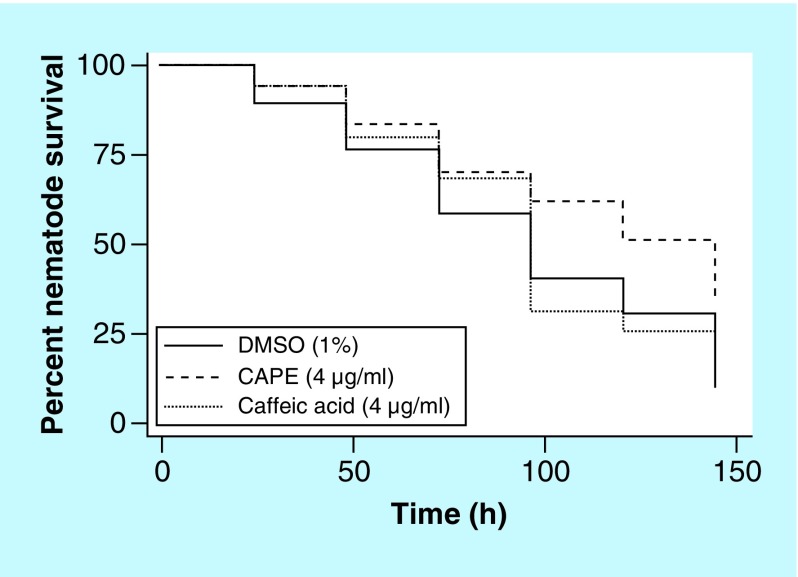
Metabolism studies have shown that CAPE can be readily hydrolyzed into the major metabolite, caffeic acid (Supplementary Material & Figure 1) [25]. Accordingly, the ability of caffeic acid to confer an increase in survival to C. albicans-infected nematodes was assessed (Figure 2). At a concentration of 14 μM (4 μg/ml), CAPE was able to confer a significant increase in survival to nematodes infected with C. albicans (p = 0.0158), while an increase in survival was not observed when treated with concentrations of caffeic acid ranging from 22–177 μM (4–32 μg/ml; p = 0.5625 at 22 μM). The inability of caffeic acid to confer an increase in C. albicans-infected nematode survival demonstrates that the intact parent compound is necessary for the immunomodulatory activity and the activity is not due to active CAPE metabolites. However, it should be noted that it is possible that other metabolites are produced, but their quantity would most likely be negligible.
Figure 1. . Key pharmacophoric elements of caffeic acid (3,4-dihydroxycinnamic acid) phenethyl ester and associated metabolites.
CAPE: Caffeic acid (3,4-dihydroxycinnamic acid) phenethyl ester.
Figure 2. . Survival assay of Caenorhabditis elegans infected with Candida albicans and treated with caffeic acid (3,4-dihydroxycinnamic acid) phenethyl ester or caffeic acid.
Infected nematodes live significantly longer when treated with CAPE (p = 0.0158) while there is no significant difference in survival when infected nematodes are treated with caffeic acid (p = 0.5625).
CAPE: Caffeic acid (3,4-dihydroxycinnamic acid) phenethyl ester.
SAR assessment of CAPE analogs to identify moieties involved in immunomodulatory activity & metabolic liabilities
The studies detailed in the previous sections established the impact of CAPE in the C. elegans–C. albicans model and provided us with a unique whole-animal model system to study the immune effects of this compound. In the next series of studies, we leveraged the C. elegans–C. albicans system in a series of SAR studies of CAPE. In order to assess potential sites of metabolic liability in CAPE, while tuning the immunomodulatory activity and/or antifungal activity of its analogs, modifications to four structural elements of CAPE were designed and tested: the catechol moiety and the cinnamyl linkage, which are both potential sites of conjugative (glucuronidation) and/or oxidative metabolism; the ester linkage, which undergoes hydrolysis to generate caffeic acid as a metabolite; and the phenethyl ring, which is susceptible to oxidative processes.
First, modifications to the catechol moiety (compounds 1–6; Table 2) were assessed. The phenolic moieties of CAPE were modified via alkylation in a benzodioxole (compound 1) or benzodioxane (compound 2) ring system. By incorporating the phenols into these constrained ring systems, we evaluated the necessity of hydrogen-bond donors at these sites while maintaining the electronic nature of the catechol moiety and mitigating potential conjugative metabolism. In a nematode survival assay, compound 1 showed no change in host survival when challenged with C. albicans. However, compound 2 retained efficacy at a concentration ranging from 13 to 51 μM (4–16 μg/ml), which is similar to the range observed for CAPE (14–28 μM; 4–8 μg/ml). Both compounds had no antifungal activity as measured by MIC against C. albicans. The electronic and/or hydrogen-bond donor/acceptor effects of the individual phenols were further probed by removing one or the other (compound 3) or examining regioisomeric effects (compound 4). Additionally, fused-ring system isosteres for either the electronic (compound 5) or hydrogen-bond acceptor properties (compound 6) were also examined. Compounds 3 and 4 possess a single phenol at the meta or ortho position, respectively. Both compounds showed complete loss of antifungal activity (MIC >256 µg/ml) and had no effect in the nematode survival assay. Similarly, loss of activity was observed in both assays when the catechol moiety was replaced with either an electron-rich indole (compound 5) or an electron-poor benzoxazole (compound 6) ring. The ability of the phenols or the benzodioxane group to act as hydrogen-bond acceptors appears to play a key role in the activity of the compound in a host survival assay. On the other hand, the electronics of the ring system, whether electron-rich or electron-poor does not seem to have an effect.
Table 2. . Structure of caffeic acid (3,4-dihydroxycinnamic acid) phenethyl ester derivatives and selected physiochemical and biological properties.
| Compound | Structure | Nematode survival (μg/ml) | NF-κB inhibition† | MIC (μg/ml) | Solubility in PBS (μM) | Plasma stability (% remaining) | ||
|---|---|---|---|---|---|---|---|---|
| Human | Mouse | Rat | ||||||
| CAPE | 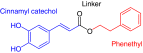 |
4–8 | 66 | 32–64 | 18 | 97 | 2 | ND |
| Caffeic acid | 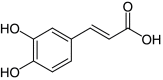 |
>32 | ND | >256 | ||||
| 1 | None | 86 | >256 | |||||
| 2 | 4–16 | 44 | >256 | 2 | 93 | 1 | ND | |
| 3 | None | ND | 128–256 | |||||
| 4 | None | ND | >256 | |||||
| 5 | 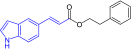 |
32 | 75 | >256 | ||||
| 6 | >256 | 143 | >256 | |||||
| 7 | 4–8 | 49 | 16 | 246 | 100 | 95 | ND | |
| 8 | 64 | 68 | 32 | |||||
| 9 | 64 | 99 | >256 | |||||
| 10 | 4–16 | 113 | >256 | |||||
| 11 | >256 | 42 | 32–64 | |||||
| 12 | >256 | 94 | 32–64 | |||||
| 13 | 4–32 | 50 | 32–64 | 78 | 93 | 91 | ND | |
| 14 | 2–16 | 92 | >256 | 236 | 97 | 99 | 102 | |
†Percent of NF-κB activity was calculated by normalizing to treatment with TNF-α and solvent alone (DMSO) as 100%.
CAPE: Caffeic acid (3,4-dihydroxycinnamic acid) phenethyl ester; MIC: Minimum inhibitory concentration; PBS: Phosphate-buffered saline.
The catechol group (free phenols) appears critical for the minimal direct antifungal activity of CAPE, but was not necessary for the immunomodulatory effects of the compounds as measured via the nematode survival assay (Table 2). MIC studies with other commercially available flavonoids (resveratrol, apigenin and myricetin) demonstrated that highly hydroxylated aromatics also display antifungal activity suggesting a general mechanism, probably driven by oxidative potentials (Supplementary Table 1). Overall, modifications to the catechol moiety led to compound 2 which retained similar efficacy to CAPE in a host survival assay, but demonstrated no inherent antifungal activity. However, these catecholic capping modifications did not lead to improved metabolic stability in rodents and displayed poor solubility (Table 2); therefore, this compound was not tested further.
In order to improve the metabolic stability of compound 2, we examined the cinnamyl moiety within CAPE which may be susceptible to reductive, oxidative and/or conjugative metabolic processes. The reduction of the conjugated double bond to a saturated ethylene linker would eliminate this potential metabolic liability, but also drastically change the conformational flexibility of the small molecule by increasing the number of rotatable bonds. The reduction of the cinnamyl moiety in CAPE provided compound 7 which displayed improved antifungal activity (MIC: 56 μM [16 µg/ml]) and similar C. albicans infected-nematode survival efficacy (14–28 μM; 4–8 μg/ml) compared with its parent CAPE. Additionally, compound 7 demonstrated improved physicochemical properties, as well improved metabolic properties (a PBS solubility of 246 µM and a mouse plasma stability of 100%; Table 2). Notably, the difference between MIC and survival efficacy is only about twofold, and, although this difference was consistent in numerous assays, no final conclusions in regard to antifungal activity or nematode survival are based on this marginal difference.
Next, a series of simple analogs modifying the phenethyl moiety were designed (compounds 8–10; Table 2) to establish and define the SARs for this portion of the molecule. Compounds with a shorter linker length (benzyl, compound 8), as well as more hydrophilic substitutions (the 4-pyridyl, compound 9; the morpholine, compound 10), were evaluated for their antifungal and nematode survival activities. With the exception of compound 10, which displayed moderate activity in the nematode survival assay, these modifications did not provide improved activity relative to the parent compound CAPE. For this reason, we chose to retain the phenethyl moiety in subsequent analogs exploring alternative linking motifs.
Changes to the ester linkage in CAPE were next assessed (Table 2) in order to modify its propensity for hydrolysis. Although the carbon replacement keto analog (compound 11) and NH–amide analog (compound 12) displayed a complete loss of efficacy in the nematode survival assay, both compounds retained antifungal activity on par with CAPE. Methylation of the amide in compound 12 provided compound 13 which demonstrated good efficacy in both the nematode survival assay and the antifungal assay (13–107 μM; 4–32 µg/ml), demonstrating that the ester linkage in CAPE does not contribute to either of these biological activities. Compound 13 also displayed improved solubility (78 µM) and plasma stability (91% remaining) relative to CAPE, providing more stable linkage alternatives.
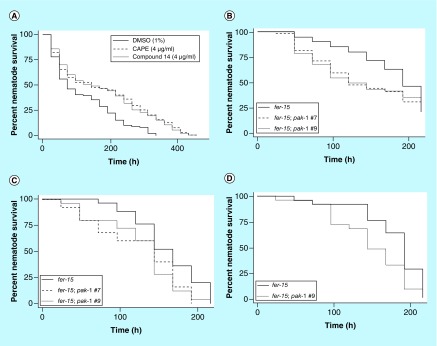
Based on the combined SARs defined above and the observed effects on stability, compound 14 was designed to incorporate the optimal modifications of each CAPE substructure into a single hybrid molecule. We predicted this hybrid molecule would retain the ability of CAPE to enhance nematode survival exclusively through immunomodulatory activity and demonstrate improved metabolic stability. The hybrid compound 14 possesses the benzodioxane moiety of compound 2, the saturated ethyl linkage in compound 7 and the N-methyl amide linker of compound 13. These combined structural modifications in compound 14 conferred an increase in survival to C. albicans-infected nematodes with effective concentration ranges of 6–49 μM (2–16 μg/ml; Figure 3A & Table 2). Moreover, compound 14 displayed improved solubility (236 μM) and mouse plasma stability (99% remaining after 1 h), compared with its parent compound CAPE (2% remaining after 1 h; Table 2) and also displayed complete stability in rat plasma (102%).
Figure 3. . Caenorhabditis elegans survival assays after infection with fungal pathogens.
(A) CAPE and compound 14 are capable of significantly increasing Candida albicans-infected nematode survival. (B) pak-1 is involved in the immune response to C. albicans in a liquid-killing assay. Two independently generated fer-15;pak-1 nematodes were significantly more susceptible to C. albicans infection when compared with fer-15 nematodes. (C) pak-1 is involved in the immune response to C. albicans in a solid killing assay. Two independently generated fer-15;pak-1 nematodes were significantly more susceptible to C. albicans infection when compared with fer-15 nematodes. (D) pak-1 is involved in the immune response to Cryptococcus neoformans. fer-15;pak-1 nematodes were significantly more susceptible to C. neoformans infection when compared with fer-15 nematodes.
CAPE: Caffeic acid (3,4-dihydroxycinnamic acid) phenethyl ester.
As observed with CAPE, at a concentration of 31 μM (10 μg/ml) of compound 14 in a liquid assay at 20°C, the lifespan of C. elegans was decreased (p = 0.0773; Supplementary Figure 2A); whereas when the lifespan was assessed at an assay temperature of 25°C, there was no significant difference in lifespan when compared with solvent alone (p = 0.8544; Supplementary Figure 2B), indicating that the increase in C. albicans-infected nematode survival is due to immunomodulatory activity of compound 14. A correlation between the efficacy of compound 14 to confer nematode survival and expression of fungal immune response genes was found. Compound 14 was able to induce higher expression of fipr-22,23 and cnc-4 in less time than CAPE (Table 3).
Table 3. . Genes that were expressed threefold or greater after treatment with 8 μg/ml of compound 14.
| Gene | Function | Average fold increase after 2 h | Average fold increase after 4 h |
|---|---|---|---|
| fipr-22,23 | Putative antimicrobial peptide | 23.02 ± 12.86 | 36.02 ± 1.76 |
| cnc-4 | Putative caenacin antimicrobial peptide | 6.70 ± 3.16 | 24.57 ± 4.37 |
In mammalian cells, CAPE has previously been demonstrated to have inhibitory activity against the NF-κB transcriptional protein complex [8,9]. As C. elegans lacks a homolog of NF-κB, we evaluated the inhibitory activity of the synthesized CAPE derivatives against this regulator of the immune response. A luciferase reporter assay was used to assess the ability of the CAPE analogs to inhibit NF-κB activity. Of the 14 CAPE analogs, 4 compounds were able to inhibit NF-κB activity at a concentration of 33 μM to a greater extent than CAPE (65.9% NF-κB activity compared with solvent alone; Table 2). The four compounds that were able to inhibit NF-κB were compound 2 (44.4%), compound 7 (49.0%), compound 11 (42.1%) and compound 13 (50.2%; Table 2). Although there was a correlation between NF-κB inhibitory activity and C. albicans-infected C. elegans survival, the only exception was compound 14 which had no inhibitory activity on NF-κB (Table 2).
Overall, as structural modifications were made to CAPE and evaluated in C. albicans-infected C. elegans, an SAR between antifungal and immunomodulatory activity became apparent. Modification of the free phenolic groups on the catechol moiety abrogated antifungal activity (compounds 1–6; Table 2), but retained enhanced nematode survivability (compounds 2, 5 and 14; Table 2) through immunomodulatory effects. Systematic structural modifications allowed us to dissect the multiple biological effects exhibited by CAPE and resolve the multiple inherent metabolic liabilities in the parent compound. Modifications to each of the metabolic hot spots of CAPE, such as saturation of the cinnamyl motif and modification of the ester linkage, greatly increased plasma stability of the compounds across multiple species and improved physicochemical properties (Table 2).
The increased Candida albicans-infected nematode survival of CAPE-treated Caenorhabditis elegans requires CED-10 & p21-activated kinase-1
Previous reports have suggested that the GTPase Rac-1 is a molecular target of CAPE in mammalian cells [26]; however, this hypothesis is based on the structural similarity to caffeic acid [27] which, as we have shown above, is unable to promote survival of infected nematodes (Figure 2). In mammalian cells, Rac-1 activates the p21-activated kinase-1 (PAK1), which is involved in a number of immune responses, including the p38 MAPK cascade and NF-κB [28–31]. These reports prompted investigation into the potential of PAK1 involvement in the C. elegans immune response.
A C. elegans strain harboring the pak-1 mutation in the temperature sterile fer-15 background was generated to aid in these studies to limit progeny production during the killing assays. Two independently generated progeny were obtained that had the desired genotype of fer-15(b26);pak-1(ok448). When both progeny of the fer-15;pak-1 nematodes were challenged with C. albicans in a liquid killing assay, both strains were more susceptible to the fungal infection and had a significant reduction in survival (Figure 3B). A similar correlation was found when the mutants were challenged with C. albicans in a solid media killing assay (Figure 3C). There was no significant difference in survival in both the liquid and solid killing assays between the two fer-15;pak-1 nematode strains (p = 0.7515 for liquid assay and p = 0.8752 for solid assay), and therefore only one strain was used in subsequent experiments.
In order to assess if the involvement of pak-1 in the immune response is specific for C. albicans, killing assays were conducted using the pak-1 mutant nematodes and the fungus C. neoformans. As observed when challenged with C. albicans, pak-1 mutant nematodes are more susceptible to infection by C. neoformans and survival is significantly reduced (p = 0.0119; Figure 3D). Collectively the data demonstrate that PAK1 is important in the immune response to pathogenic fungi.
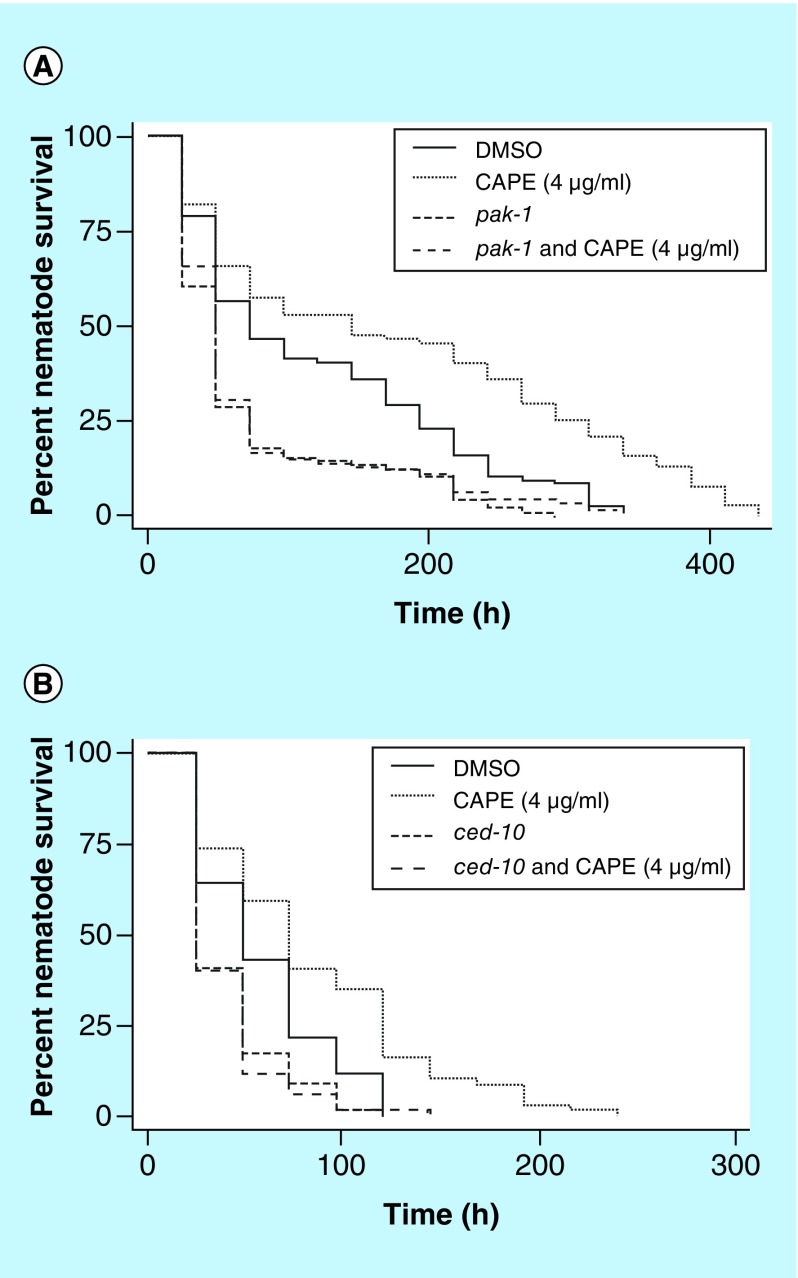
The ability of CAPE to confer an increase in C. albicans-infected nematode survival was evaluated using the pak-1 mutant nematodes. When pak-1 was disrupted, an increase in nematode survival was not observed and the survival was significantly deceased compared with control nematodes (p ≤ 0.0001) and they were not significantly different to untreated CAPE nematodes (pak-1 vs pak-1 and 14 μM [4 μg/ml] CAPE p = 0.2193; ced-10 RNAi vs ced-10 RNAi and 14 μM [4 μg/ml] CAPE p = 0.6607; Figure 4A).
Figure 4. . PAK1 and CED-10 is involved in the immune response to Candida albicans.
(A) pak-1 is involved in conferring the efficacy of CAPE against Candida albicans. pak-1-mutant nematodes do not respond to treatment with CAPE and the survival of CAPE-treated nematodes is not significantly different than nematodes treated with solvent alone. (B) ced-10 is involved in conferring the efficacy of CAPE against C. albicans. Nematodes suppressed by RNAi for ced-10 do not respond to treatment with CAPE and the survival of CAPE-treated nematodes is not significantly different than nematodes treated with solvent alone.
CAPE: Caffeic acid (3,4-dihydroxycinnamic acid) phenethyl ester.
As the Rho GTPase Rac-1 activates PAK1 in mammalian cells, the involvement of the rac-1 ortholog, ced-10, in the C. elegans immune response during infection of C. albicans was investigated. Nematodes that were suppressed in transcription of ced-10 by RNAi were more susceptible to infection by C. albicans and had a significantly shorter survival time when compared with control nematodes (p = 0.0005; Figure 4B). As with the pak-1 mutant nematodes, when the C. albicans-infected ced-10 RNAi nematodes were treated with CAPE, there was no significant increase in survival and was decreased in comparison to control nematodes (p = 0.0001) and they were not significantly different than untreated CAPE nematodes (ced-10 RNAi vs ced-10 RNAi and 14 μM [4 μg/ml] CAPE; p = 0.6607; Figure 4B).
A similar trend was observed when compound 14 was assayed. Nematodes lived significantly longer when treated with 12 μM (4 μg/ml) of compound 14 (fer-15, 2% DMSO vs fer-15, compound 14; p = 3 × 10-6) which was absent in nematodes lacking pak-1 (fer-15;pak-1, 2% DMSO vs fer-15;pak-1, compound 14; p = 0.459). Collectively, these studies support that the immunomodulatory activity of CAPE and compound 14 both have the same mode of action and that this immunomodulatory activity is mediated by the PAK1 kinase.
Although several pathways have been characterized in C. elegans that are involved in immune response signaling, the involvement of the CED-10/PAK1 pathway in the innate immune response had not been previously known. Importantly, this study demonstrates that by taking advantage of C. elegans as an alternative model host, immunomodulatory compounds can be further developed. In particular, the lack of a homolog to NF-κB in the nematode allowed identification of additional molecular targets that might have been unnoticed in the presence of the transcription factor complex.
The MAPK cascade composed of NSY-1/SEK-1/PMK-1 is an integral signaling component of the innate immune response of C. elegans to several fungal pathogens [7,13,32,33], and the expression of fipr-22,23, cnc-4 and cnc-7 requires PMK-1 [13]. Expression studies with CAPE and compound 14 demonstrated that fipr-22,23 and cnc-4 were significantly upregulated after 4 h of treatment with either of the compounds (Tables 1 & 3), suggesting that CED-10/PAK1 and the PMK-1 MAPK cascade are in the same pathway. In mammalian cells, RAC1 and PAK1 function upstream of several signaling pathways including the p38 MAPK cascade [28,34]. The involvement of PAK1 and PMK-1 in the same pathway in C. elegans suggests that there exists a commonality between the pathways of the nematode and mammalian cells. Additionally, other known PMK-1-regulated genes that are differentially transcribed in response to bacterial pathogens (C17H12.8, C32H11.12, F08G5.6, F35E12.5 and T24B8.5) [35] were not altered in gene expression upon treatment with CAPE, suggesting other factors, which are dependent on the pathogen, may be involved.
In summary, the diverse biological activity of CAPE is mediated by the RAC1/PAK1 pathway. PAK1 is responsible for upstream phosphorylation of several important pathways in mammalian cells. In addition to the p38 MAPK cascade, PAK1 is responsible for activation of pathways involving ERK, c-Jun N-terminal kinase and NF-κB [30,31]. The previously described inhibitory activity of CAPE on NF-κB activity could actually be the result of inhibitory activity on PAK1, as opposed to NF-κB activity. Stimulation of NF-κB activity for various types of immune cells requires PAK1 and a constitutively active PAK1 is capable of stimulating NF-κB activity [30]. Furthermore, PAK1 is implicated in the development of various tumor types and the inhibition of PAK1 could be responsible for conferring the antitumor activity of CAPE [31]. Therefore, the development of the metabolically stable CAPE analogs that retain activity, such as compound 14, could have far reaching implications, as it could potentially be further developed for multiple clinical applications.
Conclusion
The immunomodulatory activity of CAPE is responsible for the increased nematode survival when challenged with the clinically relevant fungus C. albicans. Structural analogs of CAPE have been synthesized that have increased plasma stability, which do not have decreased potency. CAPE and analogs of the compound require the CED-10/PAK1 pathway in C. elegans. This pathway is important for conferring an innate immune response in the nematode.
Future perspective
The further development of immunomodulatory compounds may provide a means for clinical treatment against many pathogenic microbes and potentially autoimmune diseases.
Executive summary.
Immunomodulatory activity
Caffeic acid(3,4-dihydroxycinnamic acid) phenethyl ester (CAPE) has immunomodulatory activity in the nematode Caenorhabditis elegans.
Several genes implicated in the innate immune response to Candida albicans are upregulated after treatment with CAPE including putative antimicrobial peptides.
Structural analogs of CAPE
The level of hydroxylation on the cinnamyl moiety of CAPE is responsible for the antifungal activity of the compound.
Modification of the ester and double bond of CAPE increases the plasma stability of the molecule, without decreasing the immunomodulatory activity.
Immune response in Caenorhabditis elegans
The CED-10(Rac-1)/PAK1 pathway in C. elegans is involved in the innate immune response to the pathogenic fungi C. albicans and Cryptococcus neoformans.
CED-10/PAK1 is required for the immunomodulatory activity of CAPE in the nematode.
Supplementary Material
Acknowledgements
The authors would like to thank R Pukkila-Worley for guidance and oligos regarding the expression studies. The authors would like to thank S Johnston for analytical chemistry support and Sai Life Sciences for their analog synthesis support.
Footnotes
Financial & competing interests disclosure
This research was made possible by NIH funding from the National Institute of Allergy and Infectious Diseases P01 AI083214 to E Mylonakis and National Cancer Institute RC2 CA148399 to AN Koehler, and a grant from the Rhode Island Medical Foundation to JJ Coleman. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
- 1.Banskota AH, Tezuka Y, Kadota S. Recent progress in pharmacological research of propolis. Phytother. Res. 2001;15:561–571. doi: 10.1002/ptr.1029. [DOI] [PubMed] [Google Scholar]
- 2.Sforcin JM. Propolis and the immune system: a review. J. Ethnopharmacol. 2007;113:1–14. doi: 10.1016/j.jep.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Ansorge S, Reinhold D, Lendeckel U. Propolis and some of its constituents down-regulate DNA synthesis and inflammatory cytokine production but induce TGF-beta1 production of human immune cells. Z. Naturforsch. C. 2003;58:580–589. doi: 10.1515/znc-2003-7-823. [DOI] [PubMed] [Google Scholar]
- 4.Russo A, Longo R, Vanella A. Antioxidant activity of propolis: role of caffeic acid phenethyl ester and galangin. Fitoterapia. 2002;73(Suppl. 1):S21–S29. doi: 10.1016/s0367-326x(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 5.Miorin P, Levy N, Jr, Custodio A, Bretz W, Marcucci M. Antibacterial activity of honey and propolis from Apis mellifera and Tetragonisca angustula against Staphylococcus aureus . J. Appl. Microbiol. 2003;95:913–920. doi: 10.1046/j.1365-2672.2003.02050.x. [DOI] [PubMed] [Google Scholar]
- 6.Grunberger D, Banerjee R, Eisinger K, et al. Preferential cytotoxicity on tumor cells by caffeic acid phenethyl ester isolated from propolis. Experientia. 1988;44:230–232. doi: 10.1007/BF01941717. [DOI] [PubMed] [Google Scholar]
- 7.Breger J, Fuchs BB, Aperis G, Moy TI, Ausubel FM, Mylonakis E. Antifungal chemical compounds identified using a C. elegans pathogenicity assay. PLoS Pathog. 2007;3:e18. doi: 10.1371/journal.ppat.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Natarajan K, Singh S, Burke T, Jr, Grunberger D, Aggarwal B. Caffeic acid phenethyl ester is a potent and specific inhibitor of activation of nuclear transcription factor NF-kappa B. Proc. Natl Acad. Sci. USA. 1996;93:9090–9095. doi: 10.1073/pnas.93.17.9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marquez N, Sancho R, Macho A, Calzado MA, Fiebich BL, Munoz E. Caffeic acid phenethyl ester inhibits T-cell activation by targeting both nuclear factor of activated T-cells and NF-κB transcription factors. J. Pharmacol. Exp. Ther. 2004;308:993–1001. doi: 10.1124/jpet.103.060673. [DOI] [PubMed] [Google Scholar]
- 10.Wang LC, Lin YL, Liang YC, et al. The effect of caffeic acid phenethyl ester on the functions of human monocyte-derived dendritic cells. BMC Immunol. 2009;10:39. doi: 10.1186/1471-2172-10-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park JH, Lee JK, Kim HS, et al. Immunomodulatory effect of caffeic acid phenethyl ester in Balb/c mice. Int. Immunopharmacol. 2004;4:429–436. doi: 10.1016/j.intimp.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 12.Irazoqui JE, Urbach JM, Ausubel FM. Evolution of host innate defense: insights from Caenorhabditis elegans and primitive invertebrates. Nat. Rev. Immunol. 2010;10:47–58. doi: 10.1038/nri2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pukkila-Worley R, Ausubel FM, Mylonakis E. Candida albicans infection of Caenorhabditis elegans induces antifungal immune defenses. PLoS Pathog. 2011;7:e1002074. doi: 10.1371/journal.ppat.1002074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Means TK, Mylonakis E, Tampakakis E, et al. Evolutionarily conserved recognition and innate immunity to fungal pathogens by the scavenger receptors SCARF1 and CD36. J. Exp. Med. 2009;206:637–653. doi: 10.1084/jem.20082109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pfaller MA, Diekema DJ. Epidemiology of invasive candidiasis: a persistent public health problem. Clin. Microbiol. Rev. 2007;20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andes D, Safdar N, Baddley J, et al. Impact of treatment strategy on outcomes inpatients with candidemia and other forms of invasive candidiasis: a patient-level quantitative review of randomized trials. Clin. Infect. Dis. 2012;54:1110–1122. doi: 10.1093/cid/cis021. [DOI] [PubMed] [Google Scholar]
- 17.Magill S, Edwards J, Bamberg W, et al. Multistate point-prevalence survey of health care-associated infections. N. Engl. J. Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Okoli I, Coleman JJ, Tampakakis E, et al. Identification of antifungal compounds active against Candida albicans using an improved high-throughput Caenorhabditis elegans assay. PLoS ONE. 2009;4:e7025. doi: 10.1371/journal.pone.0007025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coleman JJ, Okoli I, Tegos GP, et al. Characterization of plant-derived saponin natural products against Candida albicans . ACS Chem. Biol. 2010;5:321–332. doi: 10.1021/cb900243b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coleman JJ, Ghosh S, Okoli I, Mylonakis E. Antifungal activity of microbial secondary metabolites. PLoS ONE. 2011;6:e25321. doi: 10.1371/journal.pone.0025321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brenner S. The genetics of Caenorhabditis elegans . Genetics. 1974;77(1):71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muhammed M, Coleman JJ, Mylonakis E. Caenorhabditis elegans: a nematode infection model for pathogenic fungi. Method Mol. Biol. 2012;845:447–454. doi: 10.1007/978-1-61779-539-8_31. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.National Committee for Clinical Laboratory Standards. Reference Method for Broth Dilution Susceptibility Testing of Yeasts. Tentative Standard M27-A. PA, USA: 1995. [Google Scholar]
- 25.Celli N, Dragani LK, Murzilli S, Pagliani T, Poggi A. In vitro and in vivo stability of caffeic acid phenethyl ester, a bioactive compound of propolis. J. Agric. Food Chem. 2007;55:3398–3407. doi: 10.1021/jf063477o. [DOI] [PubMed] [Google Scholar]
- 26.Demestre M, Messerli S, Celli N, et al. CAPE (caffeic acid phenethyl ester)-based propolis extract (Bio 30) suppresses the growth of neurofibromatosis (NF) tumor xenografts in mice. Phytother. Res. 2009;23:226–230. doi: 10.1002/ptr.2594. [DOI] [PubMed] [Google Scholar]
- 27.Xu JW, Ikeda K, Kobayakawa A, et al. Downregulation of Rac1 activation by caffeic acid in aortic smooth muscle cells. Life Sci. 2005;76:2861–2872. doi: 10.1016/j.lfs.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 28.Zhang S, Han J, Sells MA, et al. Rho family GTPases regulate p38 mitogen-activated protein kinase through the downstream mediator Pak1. J. Biol. Chem. 1995;270:23934–23936. doi: 10.1074/jbc.270.41.23934. [DOI] [PubMed] [Google Scholar]
- 29.Mainiero F, Soriani A, Strippoli R, et al. RAC1/p38 MAPK signaling pathway controls beta1 integrin-induced interleukin-8 production in human natural killer cells. Immunity. 2000;12:7–16. doi: 10.1016/s1074-7613(00)80154-5. [DOI] [PubMed] [Google Scholar]
- 30.Frost JA, Swantek JL, Stippec S, Yin ML, Gaynor R, Cobb MH. Stimulation of NF kappaB activity by multiple signaling pathways requires PAK1. J. Biol. Chem. 2000;275:19693–19699. doi: 10.1074/jbc.M909860199. [DOI] [PubMed] [Google Scholar]
- 31.Kumar R, Gururaj A, Barnes C. p21-activated kinases in cancer. Nat. Rev. Cancer. 2006;6:459–471. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 32.Mylonakis E, Ausubel FM, Perfect JR, Heitman J, Calderwood SB. Killing of Caenorhabditis elegans by Cryptococcus neoformans as a model of yeast pathogenesis. Proc. Natl Acad. Sci. USA. 2002;99:15675–15680. doi: 10.1073/pnas.232568599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muhammed M, Fuchs BB, Wu MP, Breger J, Coleman JJ, Mylonakis E. The role of mycelium production and a MAPK-mediated immune response in the C. elegans-Fusarium moel system. Med. Mycol. 2012;50:488–496. doi: 10.3109/13693786.2011.648217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 2005;15:11–18. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- 35.Troemel E, Chu S, Reinke V, Lee S, Ausubel F, Kim D. p38 MAPK regulates expression of immune response genes and contributes to longevity in C. elegans . PLoS Genet. 2006;10:e183. doi: 10.1371/journal.pgen.0020183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.