Abstract
The ETS family of transcription factors is involved in several normal remodeling events and pathological processes including tumor progression. ETS transcription factors are divided into subfamilies based on the sequence and location of the ETS domain. ETV5 (Ets variant gene 5; also known as ERM) is a member of the PEA3 subfamily. Our meta-analysis of normal, benign, and malignant thyroid samples demonstrated that ETV5 expression is upregulated in papillary thyroid cancer and was predominantly associated with BRAF V600E or RAS mutations. However, the precise role of ETV5 in these lesions is unknown. In this study, we used the KTC1 cell line as a model for human advanced papillary thyroid cancer (PTC) because the cells harbor the heterozygous BRAF (V600E) mutation together with the C250T TERT promoter mutation. The role of ETV5 in PTC proliferation was tested using RNAi followed by high-throughput screening. Signaling pathways driving ETV5 expression were identified using specific pharmacological inhibitors. To determine if ETV5 influences the expression of epithelial-to-mesenchymal (EMT) markers in these cells, an EMT PCR array was used, and data were confirmed by qPCR and ChIP-qPCR. We found that ETV5 is critical for PTC cell growth, is expressed downstream of the MAPK pathway, and directly upregulates the transcription factor TWIST1, a known marker of intravasation and metastasis. Increased ETV5 expression could therefore be considered as a marker for advanced PTCs and a possible future therapeutic target.
Abbreviations: AKT, protein kinase B (or PKB); ATC, anaplastic thyroid carcinoma; BRAF, v-Raf murine sarcoma viral oncogene homolog B (or B-RAF); EMT, epithelial-mesenchymal transition; ERK, extracellular signal-regulated kinase (or mitogen-activated protein kinase 1); ETS, E26 transformation-specific (or E-twenty-six); ETV5, ETS translocation variant 5 (or ERM); GRB2, growth factor receptor-bound protein 2; hTERT, human telomerase reverse transcriptase; MAPK pathway, mitogen-activated protein kinase pathway; MEK, mitogen-activated protein kinase kinase (or MAP2K, MAPKK); mTOR, mammalian target of rapamycin; PDTC, poorly differentiated papillary thyroid carcinoma; PEA3, polyoma enhancer activator 3; PI3K, phosphatidylinositol-4, 5-bisphosphate 3-kinase; PTC, papillary thyroid carcinoma; RAS, rat-sarcoma (gene family); RAF, rapidly accelerated fibrosarcoma (gene family); SOS, son of sevenless; TERT, telomerase reverse transcriptase; TGFBR, transforming growth factor receptor; TSH, thyroid stimulating hormone
Introduction
Mutations of components of the MAPK pathway have been implicated in human oncogenesis [1]. During normal development, this pathway is activated by growth factors that bind to their cognate transmembrane receptor tyrosine kinases. Ligand binding leads to dimerization of the receptors and phosphorylation of their cytoplasmic domains, which enable interaction with the adaptor protein GRB2, binding of SOS, and activation of the small GTPase RAS. In turn, RAS activates members of the RAF family of protein kinases. RAF phosphorylates MEK kinase, which itself phosphorylates and activates the third and final enzyme in the cascade, ERK. Activated ERK phosphorylates a number of substrates, including other kinases and transcription factors, which will induce cell cycle progression [2]. The RAF family contains three serine/threonine kinases, A-RAF, B-RAF, and C-RAF, encoded by the ARAF, BRAF, and CRAF genes respectively. BRAF mutations have been detected in a high proportion of cancers, including melanoma, colorectal carcinoma, carcinoma of the biliary tract, ovarian cancer, and papillary thyroid carcinoma (PTC) [3], [4]. In melanoma and PTCs, the most common mutation affects amino acid position 600 and is characterized by the exchange of valine by glutamate (BRAF (V600E)), which leads to constitutive activation of the pathway [5]. The consequences of this mutation in melanoma have been investigated to a large extent, but less information is available on downstream targets of the activated MAPK pathway in BRAF (V600E) PTCs. Thyroid cancer is the most frequently diagnosed endocrine malignancy especially among women where it is the fifth most common cancer [6]. Thyroid cancers are divided into several forms, with PTC being the most frequent (~80% of cases). Among genetic alterations observed in PTCs, the BRAF (V600E) point mutation is the most common, with a reported frequency of 44%-70%. This mutation is associated with poorer prognosis and aggressive clinical outcome [7], [8], [9], [10], [11], [12]. The BRAF (V600E) inhibitors vemurafenib and dabrafenib have demonstrated promising efficacy in PTCs [13], [14]; however, recent studies show that patients treated with these compounds develop resistance over time [15]. While multiple mechanisms have been proposed to explain how the tumors escape the inhibitory control [16], [17], [18], [19], little is known about downstream effectors (direct or indirect) of mutant BRAF that specifically drive proliferation and metastasis in advanced PTCs.
Transcription factors belonging to the ETS family of proteins were identified as substrates for ERK1/2 and regulate expression of matrix metalloproteases, BCL2 family members, and D-type cyclins, thus mediating cellular invasion and migration, cell survival, and entry into the S phase of the cell cycle [20]. ETS transcription factors are divided into subfamilies based on the sequence and location of the ETS DNA binding domain. ETV5 (Ets variant gene 5; also known as ERM) is a member of the PEA3 subfamily, which has been found to promote metastatic progression in several types of human cancers [21], [22], [23].
In the present study, we demonstrate that ETV5 expression is significantly upregulated in PTC patient samples and a thyroid cancer cell line, KTC1. Expression of this transcription factor solely depends on the activity of the MAPK pathway and mediates PTC cell proliferation. It is also associated with expression of TWIST1 and SNAI1 but only binds to the promoter of TWIST1 to regulate its transcription. Therefore, through TWIST1, ETV5 might play a direct role in the development of more aggressive tumors, and increased levels of ETV5/TWIST1 expression might be considered additional markers for advanced PTC.
Materials and Methods
Immunohistochemistry
Expression of ETV5 in PTC samples was analyzed using a human PTC tissue array (# OD-CT-EdThy03-002, US Biomax, Inc., Rockville, MD) containing 31 patient samples with paired normal tissues. Tumors were staged I-IV according to the TNM grading system. Array slides containing formalin-fixed paraffin-embedded sections were deparaffinized with xylene and rehydrated with decreasing concentrations of ethanol in PBS. Immunohistochemistry was performed using an ABC kit from Abcam (Abcam, Cambridge, MA). Briefly, endogenous peroxidase activity was blocked using 3% H2O2 in PBS. Nonspecific protein binding was blocked with 5% goat serum for 2 hours at room temperature. One array was incubated with a primary rabbit anti-human ETV5 antibody (1: 500, Abcam, Cambridge, MA) overnight at 4°C, followed by a 15-minute wash in PBS. Next, the sections were incubated with the HRP-conjugated goat anti-rabbit antibody (1:1000) included in the kit for 45 minutes at room temperature. After washing with PBS, the chromogen diaminobenzidine was applied according to the ABC kit manufacturer. Sections were mounted using Cytoseal 60, a toluene-based mounting medium (Richard-Allan Scientific, Thermo Fisher Scientific, Waltham, MA). As negative control, another array was stained as above, but the primary antibody was replaced with PBS. Bright-field images were captured using an Olympus BX41 bright-field/fluorescence microscope at magnifications 20× and 40× (Olympus, Waltham, MA). Antibodies are listed in Supplemental Table 1.
Cell Cultures
Immortalized human thyrocytes Nthy-ori 3-1 (Sigma, St Louis, MO) [24] and the papillary thyroid cancer cell lines KTC1 and KTC1-VA7 [19], [25] were used. We have chosen KTC1 over other unique PTC cell lines [26] because it harbors only a BRAF V600E and a TERT promoter mutation as oncogenic drivers (per COSMIC and [27]). The cell line KTC1-VA7, derived from the original KTC1 line and harbors an additional KRAS G12D mutation [19], was also used in transwell experiments. The BCPAP cell line, which is commonly used, is homo/hemizygous for V600E [26] and therefore not clinically relevant. Similarly, the K1 cell line is uniquely tetraploid [28]. The KTC1-VA7 cells harbor an additional KRAS G12D mutation [19]. Cells were maintained and cultured in Dulbecco's modified Eagle's medium (DMEM)/F12 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA). All cell lines were incubated at 37°C in a humidified incubator with 5% CO2. Cells were cultured in microtiter plates or 100-mm–diameter dishes depending on the experimental settings described below. Culture plates and dishes (Corning brand) were purchased from Thermo Fisher Scientific (Waltham, MA).
Cytochemistry
Cells were cultured on glass coverslips for 48 hours, then rinsed with PBS and fixed with 4.0% (w/v) paraformaldehyde diluted in PBS for 30 minutes at room temperature. Cells were then blocked with 0.2% Triton X-100, 0.05% Tween 20, 1.0% BSA, and 10% goat serum in PBS for 1 hour at room temperature. To reveal ACTB (β-actin), fixed cells were exposed to DyLight 488–conjugated phalloidin (Cell Signaling, Danvers, MA) in 0.2% Triton X-100, 0.05% Tween 20, and 1.0% BSA in PBS for 1 hour at room temperature. Nuclei were counterstained with DAPI, and cells were imaged using a Nikon Eclipse Ti confocal microscope.
Modulation of ETV5 Expression
Downregulation
Transient ETV5 knockdown experiments in KTC1 cells were carried out using ETV5 siRNA (Accell Human ETV5 siRNA-SMARTpool, GE Dharmacon, Lafayette, CO) or scrambled siRNA as control (GE Dharmacon, Lafayette, CO). To increase efficiency, ETV5 downregulation was also carried out with shRNAs cloned into GIPZ lentiviral vectors expressing TurboGFP (V3LHS_641158, GE Dharmacon, Lafayette, CO). They were obtained from the shRNA and ORFeome Core Facility at MD Anderson Cancer Center. HEK293T cells were grown in T75 flasks in 10 ml of DMEM (10% FBS) and transfected (1 day after plating) with second-generation lentiviral packaging plasmids. pCMV-VSV-G (3 μg) (Addgene, Cambridge, MA), pCMV-dR8.2 dvpr (15 μg) (Addgene, Cambridge, MA), and the shRNA lentiviral vector (15 μg) were combined at a ratio of 0.2:1.0:1.0, using Lipofectamine 2000 per manufacturer instructions (Thermo Fisher Scientific, Waltham, MA). Eighteen hours after transfection, fresh medium was added, and cells were cultured for another 48 hours before culture supernatant containing lentiviruses was collected and filtered. One day after plating, KTC1 cells were transduced in T75 flasks with 5 ml of the lentivirus-containing supernatant mixed with 5 ml DMEM/F12 (10% FBS) and polybrene (#TR-1003-G, Millipore, Billerica, MA) at a concentration of 8 μg/ml for 18 hours. Medium was then changed, and the cells were cultured for an additional 48 hours before cells positive for the shRNA/TurboGFP insertion were selected by FACS. Downregulation of ETV5 expression was verified by qPCR and Western blotting, and cell clones with a reduction of >80% ETV5 expression were utilized for further experiments.
Overexpression
To transiently overexpress ETV5, KTC1 and Nthy-ori 3-1 cells were transfected with pCMV6 expressing both ETV5 and GFP (#RG200366, Origene, Rockville, MD), or the empty expression vector pCMV6-GFP (#PS100010) as control. Alternatively, to stably overexpress ETV5, the human ETV5 lentiviral vector pLenti-GIII-CMV-RFP-2A-Puro was used (Applied Biological Materials, Richmond, BC, Canada).
Cell Proliferation Assays
KTC1 cells were plated into 96-well culture plates (glass bottom, 1000 cells/well) containing DMEM/F12 supplemented with 10% serum and grown overnight. One plate was then collected and fixed in 4% paraformaldehyde prior to treatment (day 0) to be used to determine the starting number of cells. The remaining plates were treated with the pharmacological inhibitors described below, or siRNA, for 3-4 days and then fixed. Plates were stained with the nuclear dye DAPI (1:5000) for 30 minutes, and entire wells were tile-imaged using a high-throughput IN Cell Analyzer 6000 Cell Imaging System (GE Healthcare Life Sciences) kindly made available by the High-Throughput Screening Core Lab, Institute for Biosciences and Technology, Texas A&M Health Science Center (Houston, TX). Images were stitched together, and total cells per well were directly counted using the system's image analysis software. Day 4/5 cell numbers were averaged from the values obtained from the mean of at least eight wells and normalized to the starting cell number (average day 0 values). Relative fold growth was calculated by normalizing to fold growth of control-treated cells. Each data point represents the mean ± SEM from a total of three independent experiments.
Kinase Inhibitors
Pharmacological inhibitors were used to study the signaling molecules and pathways that drive ETV5 expression. All inhibitors were obtained from Selleckchem (Selleck Chemicals, Houston, TX) and their final concentrations used according to the provider. MK2206, a pan-AKT inhibitor, was used at a final concentration of 10 μM. LY294002, a pan-PI3K inhibitor, was used at a final concentration of 20 μM. Rapamycin, an mTOR inhibitor, was used at final concentrations of 20 and 40 μM. GDC0994 a pan-ERK inhibitor was used at a final concentration of 10 μM, and LY210976, a TGFB receptor inhibitor, was used at a final concentration of 10 μM. Vemurafenib, a specific BRAF (V600E) inhibitor, was used at a concentration of 2 μM. All inhibitors were dissolved in DMSO, which was used at a final concentration of 0.1% v/v in D-MEM/F12 culture media. Control groups were cultured in 0.1% v/v DMSO in D-MEM/F12.
qRT-PCR Analysis
Total RNA isolation was carried out using the Qiagen RNeasy kit (Germantown, MD) and cDNA obtained with MuLV reverse transcriptase (Thermo Fisher Scientific, Waltham, MA). Quantitative real-time PCR (qRT-PCR) was performed using the TaqMan Gene Expression Mastermixes (Thermo Fisher Scientific, Waltham, MA). Alternatively, RNA isolation, reverse transcription, and quantitative real-time PCR (qRT-PCR) were carried out using the 2-step Ambion TaqMan Gene Expression Cells-to-CT kit (Thermo Fisher Scientific, Waltham, MA). TaqMan Gene Expression Assays are listed in Supplemental Table 2. Relative quantitative fold change was determined using the ΔΔCt method. In all analyses, the expression value of each gene was normalized to the amount of an internal control gene cDNA (Actb) to calculate a relative amount of RNA in each sample. Quantitative relative fold changes in comparison to controls were calculated. qRT-PCR was carried out with at least four technical replicates per sample and experiment, and at least three independent experiments. P values ≤.05 were denoted significant.
RNA Profiling
The Qiagen RT2 Profiler PCR array EMT PAHS-090Z (Qiagen, Germantown, MD) was used to probe human epithelial-to-mesenchymal transition gene expression. Briefly, total RNA was isolated from KTC1 cells after siRNA ETV5 downregulation and from control KTC1 cells (scrambled siRNA) using the Qiagen RNeasy kit (Qiagen, Germantown, MD). Template cDNAs were prepared from total RNA (1 μg) using the Qiagen RT2 PreAMP cDNA Synthesis Kit and were characterized in triplicates using the PCR array with the RT2 SYBR Green/Fluorescein PCR master mix using a CFX384 BioRad thermocycler.
Western blot analysis
After treatments, cells were rinsed with PBS and lysed for 20 minutes at 4°C with RIPA buffer supplemented with three cocktails of proteases and phosphatases inhibitors, 1:100 dilution (#P8340, #P0044, #P5726, Sigma, St Louis, MO). Lysed cells were collected with a cell scraper, collected in microfuge tubes and sonicated at 30% power on ice for 15 seconds. Samples were centrifuged for 10 minutes at 2000×g and 4 °C, and supernatants were collected. Protein concentration was estimated using the Pierce Bradford Colorimetric Assay (BCA) (Thermo Fisher Scientific, Waltham, MA) with absorbance at 562 nm measured with an Epoch 2 microplate spectrophotometer (BioTek, Winooski, VT). Samples were run on 4%-12% sodium dodecyl sulfate (SDS) polyacrylamide gels (ThermoFisher Scientific, Waltham, MA) and transferred to 0.4-μm nitrocellulose membranes according to standard protocols. Following transfer, the membranes were washed and blocked with 5% powdered milk in Tris-buffered saline (TBS) for 1 hour at room temperature. The blots were incubated in primary antibody solutions (Supplemental Table 1) overnight at 4°C and then washed with 1× TBS-0.1% Tween and probed with appropriate secondary antibodies for 2 hours at room temperature. The secondary antibodies used were LI-COR infrared fluorophore-conjugated secondary antibodies (1:2000, LI-COR Biosciences, Lincoln, NE, Supplemental Table 1) diluted in 5% powdered milk in TBS with 0.1% Tween 20. The blots were visualized, and band intensities were quantified after normalization over ACTB using an Odyssey Fc Imaging system (LI-COR Biotechnology, Lincoln, NE). The band intensities were compared to controls for relative quantification. Each data point represents at least two technical replicates for each of two or three independent experiments.
Scratch/Wound Healing Assay
Untreated KTC1 cells, ETV5 shRNA-transduced KTC1 cells, and control-transduced KTC1 cells were seeded into 96-well plates at a density of ~20,000 cells/well in DMEM/F12 medium containing 10% FBS. Cells were allowed to adhere for 24 hours, and the WoundMaker-96 tool from Essen Bioscience (Ann Arbor, MI) was used to remove cells, creating identical wounds in each well. Images were acquired every 1.5 hours for 72 hours using the IncuCyte ZOOM System microscope (Essen Bioscience). The system's cell migration software module was used for image analysis, and relative wound density metric was used to calculate the rate of migration. Linear regression was used to assess the differences in percent fill between each treatment group and the control group over the period of linear wound closure (initial 36 hours). In the regression model, percent fill was included as the dependent variable and treatment group as the independent variable, while adjusting for time. We calculated 95% confidence intervals (CI) and P values, and statistical significance was defined as P < .05. Stata version 13.1 (College Station, TX) was used for statistical analyses.
Cell Migration and Invasion (Transwell Plates)
ETV5 expression was downregulated in KTC1 and KTC1-VA7 cells through shRNA. Transduced control and ETV5 shRNA cells were prepared in serum-free DMEM/F12 medium. Twenty five thousand cells were plated (100 μl volume/well) onto 8-μm pore inserts (24-well plates) that were either tissue culture treated (Corning #3422) or Matrigel coated (Corning #354480) for migration and invasion assays, respectively. Each cell dilution was also aliquoted (50 μl volume/well) into 6 wells of a 96-well tissue culture treated plate to be used for cell number loading normalization. Bottom chambers (receiving chambers) of the 24-well plates were filled with DMEM/F12 medium supplemented with 10% FBS (650 μl volume/well). Following 24-hour incubation at 37°C and 5% CO2, upper and receiving chambers were rinsed with PBS. TrypLE (ThermoFisher #12605036) containing 5 μM Calcein Blue AM (ThermoFisher # C1429) was added to the receiving chambers (350 μl/well), and cells that had migrated through the pores or invaded through Matrigel-coated pores were allowed to dissociate from the bottom surface of the inserts for 1 hour at 37°C. Wells containing dissociated cells were then imaged at 360-nm excitation/449-nm emission using a high-throughput IN Cell Analyzer 6000 Cell Imaging System (GE Healthcare Life Sciences) kindly made available by the High-Throughput Screening Core Lab, Institute for Biosciences and Technology, Texas A&M Health Science Center (Houston, TX). Cell loading normalization plates (96-well) were fixed with 4% paraformaldehyde in PBS and then stained with the nuclear dye DAPI (1:5000) for 30 minutes and also imaged using the IN Cell Analyzer. Images were stitched together, and total cells per well were directly counted using the system's image analysis software. Total migrated/invaded cell numbers for each well were normalized using the average cell loading densities.
Chromatin Immunoprecipitation-Quantitative PCR (ChIP-qPCR)
KTC1 cells were grown to 95% confluence in DMEM:F12 serum-free culture media supplemented with 100 U/ml penicillin and 100 U/ml streptomycin. A minimum of 1×106 cells were used. After washing twice with PBS, protein-DNA were cross-linked with 1% formaldehyde for 15 minutes at room temperature; then formaldehyde was quenched by addition of 0.125 M glycine (Sigma, St Louis, MO) for 5 minutes. Cells were washed twice with ice-cold PBS containing 0.1 M PMSF (Sigma, St Louis, MO) and 1× of a standard protease inhibitor cocktail (Halt, Thermofisher Scientific, Grand Island, NY). Cells were scraped and resuspended in 1 ml PBS containing 0.25% Triton X, 10 mM EDTA, and 10 mM HEPES, with a final pH of 7.9. Samples were incubated for 15 minutes and centrifuged at 4°C and 1000×g to obtain pellets. Pellets were resuspended in lysis buffer containing 0.1% SDS, 10 mM EDTA, and 1× Halt protease inhibitor cocktail in 50 mM Tris-HCl, pH 8.1, and sonicated on ice 10 times for 30 seconds each, with a 30-second interval at the maximum setting (Bioruptor, Diagenode, Denville, NJ). Sonicated chromatin was diluted in lysis buffer without SDS to bring the final concentration of SDS to 0.05%. A chromatin aliquot (10 μl) was run on a 1% agarose gel to confirm presence of desired fragments (300-600 bp). Five micrograms of chromatin was incubated overnight with an ETV5 antibody (Abcam, Cambridge, MA) (Supplemental Table 1) at 4°C with rotation. As negative controls, chromatin was incubated with polyclonal rabbit IgG (Jackson ImmunoResearch, West Grove, PA) or protein G-agarose beads alone. Immunoprecipitation was performed by adding protein G-agarose beads (EZ ChIP kit, Millipore Sigma Cat. 17-295) for 1 hour, then immune-complexes were precipitated by centrifugation as per manufacturer instructions. The beads were washed sequentially for 10 minutes each in a low-salt buffer containing 0.1% SDS, 1% Triton X-100, 2 mM EDTA, 150 mM NaCl in 20 mM Tris–HCl, pH 8.1, followed by a high-salt buffer with 0.1% SDS, 1% Triton X-100, 2 mM EDTA, and 500 mM NaCl in 20 mM Tris–HCl, pH 8.1, and finally a LiCl wash buffer containing 0.25 M LiCl, 1% NP-40, 1% deoxycholate, and 1 mM EDTA in 10 mM Tris–HCl, pH 8.1. The beads were then washed three times with TE buffer. Eluates were reverse-crosslinked in 50 μl TE buffer by heating at 95°C for 15 minutes. The beads were removed, and the released DNA fragments were purified with QIAquick DNA purification Kit (Qiagen, Valencia, CA). qPCR was performed with 1 μl final extract for 45 cycles of amplification with TaKaRa Taq polymerase (TaKaRa Bio USA, Mountain View, CA) and human primers flanking ETV5 binding sites in the TWIST1 and SNAI1 proximal promoters (Supplemental Table 3). As a negative control, primers were chosen for amplification of a DNA sequence without ETV5 binding sites at least 6000 bp away from the binding site-rich sequences. The PCR products were visualized on 1% agarose gel. Triplicate qPCR reactions were performed with 500 nM of primers (Supplemental Table 3), 10 μl of 2× SYBR green I PCR master mix (Applied Biosystems), and 2 μl DNA in a total of 20-μl reaction volume. Results were computed as percent antibody bound per input DNA and then normalized to IgG controls.
Statistical Analysis
Datasets are shown as mean +/− SEM (standard error of mean). Statistical significance between two sample groups was calculated using the two-tailed Student's t test or using the Mann-Whitney test. For comparisons between three or more groups for two independent variables, two-way ANOVA was used, followed by Tukey HSD or Bonferroni post hoc tests. Statistical analysis was performed using the GraphPad Prism software (GraphPad Software, Inc., La Jolla, CA). P values < .05 were denoted significant. For all experimental endpoints, analysis was done with at least three separate repeats and two to three independent experiments.
Results
Overexpression of ETV5 in PTCs
A meta-analysis of ETV5 mRNA expression in PTCs using datasets from NCBI-GEO [29], [30], [31], [32] confirmed a strong association of ETV5 mRNA expression with thyroid tumors known to harbor a BRAF or RAS mutation (Figure 1, A to E) (Supplemental Table 4). Further, integrated analysis of ETS transcription factors mRNA expression in PTCs harboring a RAS or a BRAF mutation in the context of TCGA [33] confirmed that ETV5 mRNA was significantly upregulated in these tumors in comparison to normal thyroid samples (P = 1.0843E-18) (Figure 1F). Other ETS transcription factors (ETV4 and ELF3) were also significantly overexpressed (Supplemental Data 1). Additional ETV5 expression data were obtained from mining NCBI GEO GSE48953 to compare BRAF-mutated to BRAF-WT PTC samples [34]. The data confirmed that ETV5 expression is upregulated in BRAF-mutated PTC samples (Supplemental Fig. 1). We chose to characterize ETV5 over ETV4 and ETV1 because of its strong association with PTC in the NCBI GEO datasets. To confirm the NCBI-GEO mRNA data at the protein level, we next tested ETV5 protein expression in a tissue microarray containing thyroid tissue sections from 31 patients and paired normal thyroid tissues. ETV5 protein expression was markedly increased in all staged (I-IV) tumor samples (Figure 2, C and D) in comparison with expression in follicular cells of normal tissue, which was weak or negative (Figure 2, A and B). In PTCs, the cytoplasmic expression of ETV5 was strong, as has been shown for other ETS transcription factors [35], [36]. ETV5 was detected in most tumor cell nuclei where it adopted a uniformly punctate/speckled pattern, as is often the case for transcription factors [37]. It is noteworthy that normal thyroid C-cells, located between the follicles, expressed ETV5 (arrows, Figure 2B), which might be related to the calcitonin expression of these cells [38]. Control experiments without primary antibody are shown in Supplemental Figure 2.
Figure 1.
Meta-analysis of ETV5 expression in cohorts of PTC patients showing its upregulation in comparison to normal tissues samples. (A-D) Analysis of ETV5 expression in the thyroid of patients harboring a BRAF (V600E) mutation in comparison to normal individuals using the NCBI GEO database (29-32). (E) Analysis of ETV5 expression in thyroid lesions harboring a BRAF (V600E) or a RAS mutation in comparison to normal samples using the NCBI GEO database (29, 32). (F) Analysis of ETV5 expression in BRAF (V600E) or RAS-mutated thyroid cancer samples in comparison to normal contralateral tissue using TCGA (33).
Figure 2.
Immunohistochemistry of ETV5 expression in normal and PTC samples. (A) ETV5 expression in a normal thyroid sample. Most follicular cells are negative or weakly positive for ETV5 expression, which is confined to the nucleus. (B) Magnification of A. Parafollicular cells (C cells) appear positive (arrows). (C and D) ETV5 is strongly upregulated in two different PTC samples, with expression in both nucleus and cytoplasm.
Pathways Required for PTC Cell Proliferation
In order to understand the function of ETV5 in the context of PTC/PDTC, we used the KTC1 cell line, which harbors a heterozygous BRAF (V600E) mutation and is originally derived from the pleural effusion of a 68-year-old male PTC patient [25]. We first sought to define which pathways were used by KTC1 cells to proliferate by treating the cells with different pharmacological inhibitors (Figure 3). Our data indicate that these cells can use both PI3K-AKT and RAS/MAPK/ERK pathways since treating them with ERK1/2, pan PI3K, or pan-AKT inhibitors decreases their rates of proliferation over 4 days of culture (Figure 3, A and B). However, inhibition of TGFBR1/2, whose loss of function is found in colon cancers [39], [40] and in some poorly differentiated and anaplastic thyroid cancers [41], did not increase cell proliferation (Figure 3A). Further, inhibition of mTOR downstream of PI3K/AKT also reduced rates of proliferation (Figure 3B). We next assessed the influence of ETV5 on the rate of proliferation of KTC1 cells using a 4-day ETV5 RNAi treatment. Downregulation of ETV5 expression resulted in a significant decrease of the rate of proliferation of KTC1 cells in comparison to controls (Figure 3C). In addition, downregulation of ETV5 was associated with inhibition of CCND1 and CCND2 expression, confirming a possible role of ETV5 in KTC1 cell proliferation (Figure 3D).
Figure 3.
Rates of proliferation of KTC1 cells after different treatments. (A) Inhibition of the MAPK pathway using the pan-ERK inhibitor GDC0994 (10 μM) will decrease the rate of proliferation of KTC1 cells in comparison to cells treated with DMSO alone, while the TGFbeta receptor inhibitor LY210976 (10 μM) has less effect. (B) Inhibition of the PI3K-AKT pathway will also decrease the rate of proliferation of KTC1 cells in comparison to DMSO alone. MK2206-2HCL, a pan-AKT inhibitor, was used at final concentrations of 10 μM. LY294002, a pan-PI3K inhibitor, was used at a final concentration of 20 μM; mTOR inhibition using rapamycin (20 nM) confirms that the PI3K-AKT pathway is used for KTC1 cell proliferation. (C) Downregulation of ETV5 expression by RNAi causes significant reduction of the rate of proliferation of KTC1 cells in comparison to control cultured (mock and scrambled conditions). (D) Downregulation of ETV5 expression with siRNA causes simultaneous downregulation of CCND1 and CCND2 expression in KTC1 cells. **P < .005, ***P < .001.
ETV5 is a Downstream Target of the MAPK Pathway
In order to determine whether PI3K/AKT or MAPK pathway, or both, activate ETV5, we abolished these pathways with ERK1/2 or PI3K pharmacological inhibitors and measured ETV5 mRNA and ETV5 protein expression levels. Figure 4A demonstrates that ETV5 mRNA expression is strongly downregulated after ERK1/2 inhibition. It is not significantly altered by PI3K inhibition. Western blots confirmed that ETV5 protein expression parallels that of activated ERK1/2 (pERK1/2) and is not affected when the PI3K/AKT pathway is inhibited (Figure 4B). The antibody used in this figure (Thermo Fisher) recognizes both ETV5 isoforms produced by alternative splicing at kDa 57.8 and kDa 62.4 (UNIPROT ID P41161-1 and P41161-2). Further, exposure to 1.0 μM vemurafenib, a potent and specific inhibitor of BRAF (V600E), caused a significant downregulation of ETV5 expression at the mRNA level (Figure 4C) and a similar 55% reduction at the protein level (Figure 4D), indicating that ETV5 is expressed downstream of BRAF (V600E). The antibody used in Figure 4Db recognizes only one isoform (Genentech).
Figure 4.
ETV5 is expressed downstream of the MAPK pathway. (A) ETV5 mRNA expression is strongly downregulated by MAPK pathway inhibition but not by PI3K/AKT pathway inhibition. (B) Western blot analysis confirming that ETV5 protein expression parallels expression/phosphorylation of ERK1/2. The chosen antibody (Thermo Fisher) detects the two predicted alternative spliced isoforms of human ETV5 (Uniprot kDa 57.8 and kDa 62.4). ERKi = ERK1/2 inhibitor (GDC0994); PI3Ki = PI3K inhibitor (LY294002). (C) Significant downregulation of ETV5 mRNA expression in KTC1 cells after 48-hour exposure to 1 μM vemurafenib, a specific BRAF (V600E) inhibitor. (D) Significant downregulation of ETV5 protein expression in KTC1 cells after 48-hour exposure to 1 μM vemurafenib. (a) Quantitation of ETV5 protein expression after standardization with ACTB, showing a 55% reduction after vemurafenib treatment. (b) Representative Western blot showing ETV5 protein expression without (A) or with 1 μM vemurafenib (B). The antibody used (Genentech) distinguishes only one isoform. ****P < .0001, **P < .005, n ≥ 3.
Correlation Between ETV5 and EMT Markers Expression in PTC Cells
Because of the known pleiotropic activity of ETV5, we sought to investigate whether ETV5, a transcription factor, is influencing expression of genes important for EMT in PTCs. We first performed an EMT marker expression profile using PCR array gene analysis and compared KTC1 cells after downregulation of ETV5 expression with siRNA to control cells. After 24-hour downregulation of ETV5, the PCR array analysis identified significant decreases in expression of several EMT-related genes such as SNAI1, TWIST1, MMP3, and MMP9, among others (Supplemental Data 2). We confirmed by qPCR that downregulation of SNAI1 expression is associated with downregulation of ETV5 in both KTC1 cells and immortalized primary thyrocytes (Nthy-ori 3-1 cells [24]) (Figure 5, A and B). Further, additional ETV5 downregulation experiments using shRNA identified TWIST1, SNAI2, CDH2, and TGFBR1 as possible ETV5 targets in KTC1 cells (Figure 5C).
Figure 5.
Expression of some EMT markers correlates with ETV5. (A) SNAI1 expression is downregulated in KTC1 cells following knockdown of ETV5 by RNAi. (B) SNAI1 expression in immortalized thyrocytes is downregulated after knockdown of ETV5 by RNAi. (C) Downregulation of the EMT markers TWIST1, SNAI2, CDH2, and TGFBR1 is observed after ETV5 knockdown with ETV5 shRNA.
Correlation Between ETV5 Expression and Cell Invasion in PTC Cells
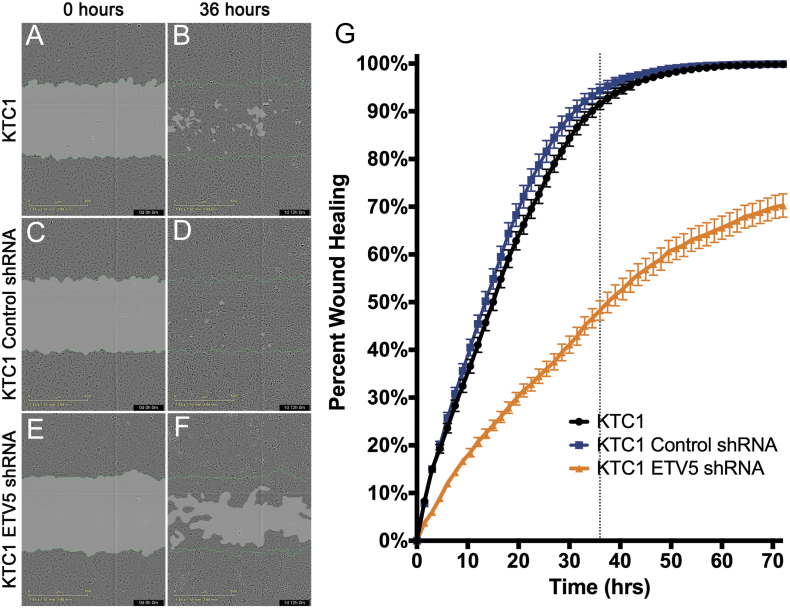
Because some ETV5-target genes in KTC1 cells are markers of EMT and because this process often coexists with increased cell motility, we tested the effects of ETV5 downregulation on migration and invasion. A scratch assay demonstrated that the rate of gap closure significantly decreased when ETV5 expression is downregulated in comparison to native KTC1 cells or the shRNA control cells (Figure 6, A-F). Closure was near completion after 36 hours in native (91.5%) and control-transduced cells (94.3%), producing, on average, a 3.05% (95% CI: −7.26, 1.15) lesser wound fill compared to the native KTC1 cells during the 36-hour period, which was not statistically significant (P = .152). However, wound closure was profoundly delayed for the ETV5-downregulated cells (~48%) compared to the native KTC1 cells at 36 hours, producing, on average, a 35.5% (95 CI lesser wound fill compared to the control treatment during the 36-hour period, a statistically significant difference (P < .001) (Figure 6G).
Figure 6.
ETV5 downregulation impairs gap closure in a scratch/wound healing assay. (A + B) Gap closure of wound healing by control nontreated KTC1 cells (representative sample). (C + D) Gap closure of control KTC1 cells after transduction of an shRNA empty vector showing no significant difference with nontreated cells. Gap nears closure at approximately 36 hours (dotted line) after scratches were made. (E + F) Delay in gap closure of KTC1 cells after downregulation of ETV5 expression through shRNA vector transduction. (G) Quantitation of gap closure over time. The rate of closure is significantly lower when ETV5 expression is knocked down.
Considering gap closure could be dependent on cell proliferation only and not on active migration, we performed cell migration and invasion assays in Boyden chambers. Figure 7A shows that ETV5 does not promote migration of KTC1 or KTC1-VA7 (BRAF/KRAS clone) cells. However, as demonstrated in Figure 7B, downregulation of ETV5 expression significantly reduced cell invasion through collagen-coated pores by approximately 33%. Cells with reduced ETV5 expression kept the same morphology as the control cells (Figure 7, C and D), while forced overexpression of ETV5 induced a striking phenotypic difference as all cells increase in size and exhibit prominent actin stress fibers (Figure 7E). However, they were prone to lose adherence and died quickly, which precluded further experimentation.
Figure 7.
ETV5 levels of expression influence cell invasion and cell morphology. (A + B) KTC1 and KTC1-VA7 cell migration versus invasion. ETV5 downregulation does not significantly impair cell migration in a Boyden chamber assay (A). Cell invasion through a collagen layer is significantly reduced when ETV5 expression is downregulated (B).
(C-E) Levels of ETV5 expression significantly alter KTC1 cell morphology in culture. Control cells (C) and cells with a downregulated ETV5 expression (D) retain a cuboidal or fusiform shape, while cells that overexpress ETV5 (E) increase in size and exhibit prominent actin stress fibers (stars). Cells were stained with DAPI (nuclear staining) and DyLight 488 Phalloidin (ACTB, β-actin).
Direct Control of TWIST1 Expression by ETV5 in PTC Cells
In order to confirm that ETV5 expression impacts cell invasion or markers of metastasis in BRAF-mutated PTC cells, we undertook promoter analysis of TWIST1 and SNAI1. TWIST1 is a pleiotropic transcription factor involved in intra- and extravasation and protects cells from apoptosis [42]. TWIST1 is also a recognized marker of the dedifferentiated anaplastic thyroid cancer phenotype [43], [44] and plays a role in cancer drug resistance [45]. In association with SNAI1, it is a major inducer of EMT [46]. Correlation between ETV5 and TWIST1 expression was confirmed by siRNA and qPCR analysis (Figure 8A). Proximal promoter analysis using the Eukaryotic Promoter Database indicated that six ETV5 consensus binding sequences (C/G)CGGAA(G/A) are present in the TWIST1 proximal promoter (Figure 8B). ChIP-PCR analysis using an ETV5 antibody and primer sets recognizing binding regions 1-3 (Figure 8B) indicated that, in KTC1 cells, ETV5 significantly interacts with region 3 of the human TWIST1 proximal promoter (primer pair for region 3, PP3) (Figure 8C). Interaction is strong and significant compared with binding of IgG only (black bars) or when using control primers outside of the proximal promoter (PPC, primer pair control). There is an 11-fold enrichment using the ETV5 antibody in comparison with IgG in binding region 3 that contains ETV5 binding sequences at bp −126 to −119 (5′-GCGGAAA-3′) and bp −40 to −36 (5′-CGGA-3′). One consensus sequence is also present in the SNAI1 proximal promoter at bp −402 to bp −397 from the TSS, but no significant enrichment was observed after ChIP-qPCR (data not shown). Altogether, our results indicate that ETV5 directly controls expression of TWIST1 but not that of SNAI1. Increased ETV5 expression levels will therefore increase PTC cells proliferation and their invasive potential but will only indirectly influence SNAI1 and events downstream of this transcription factor.
Figure 8.
ETV5 binds to the TWIST1 promoter. (A) Downregulation of TWIST1 expression after ETV5 shRNA vector transduction. (B) Analysis of the human TWIST1 promoter with respect to ETV5 putative binding sites. The human TWIST1 proximal promoter contains six ETV5 consensus sequences for possible binding of ETV5 to DNA. Regions amplified with three different primer pairs (PP1, PP2, PP3) are represented, which were used after ChIP (Supplemental Table 3). (C) ChIP-qPCR analysis of the human TWIST1 promoter showing binding of ETV5 in the region bp −179 to −87 probed with primer pair 3 (PP3). Primer pair 1 (PP1) and primer pair 2 (PP2) did not amplify, indicating no chromatin immunoprecipitation. Similarly, no amplification was obtained with PPC = control primer pair (~−6000 bp from TSS). Results show fold enrichment in comparison to immunoprecipitation using IgG instead of ETV5 antibody and are given as means ± standard error of the mean. **P < .01, ***P < .001.
Discussion
The recent discovery of translocations involving ETS proteins has increased interest in the function of these transcriptional activators in carcinogenesis [47], [48], [49]. Among ETS proteins from the PEA3 family, ETV4 and ETV5 upregulation (with or without translocation) has often been correlated to cell proliferation, migration, and invasiveness [21], [22], [50], [51], [52]. Occasionally, all three behavioral characteristics can be observed in the same cell type, making PEA3 proteins the most versatile of these pleiotropic factors [53]. In the present study, we demonstrate that ETV5 overexpression specifically depends on activation of the MAPK pathway and drives papillary thyroid cancer cell proliferation. ETV5 also directly regulates the expression of TWIST1, which itself plays an important role in processes such as metastasis in several cancers [45].
ETS transcription factors are controlled by a variety of signaling pathways that include MAPK, PI3K, and Ca2+ signaling [54]. However, PEA3 protein activation, in particular ETV5, is more restricted downstream of MAPK and PKA signaling [55], [56] and has been linked to RET/MAPK activation in spermatogonial stem cell self-renewal [57], [58]. Although PTC tumors and KTC1 cell proliferation following BRAF inhibition depend on both MAPK pathway and reactivated PI3K pathway [16], [17], [19], we now demonstrate that ETV5 expression in PTC cells mainly depends on the MAPK pathway and ERK1/2 phosphorylation. It is presently not known if ERK1/2, a kinase with DNA binding ability [59], directly associates with ETV5 or is inducing ETV5 expression through activation/phosphorylation of intermediates. Patients with advanced BRAF (V600E) PTCs respond well to vemurafenib and dabrafenib, which has been studied in clinical trials [60]. However, like in BRAF (V600E) melanoma or colon carcinoma patients, responses are incomplete and the disease progresses within 1 year [13], [14], [61], [62]. In an attempt to overcome this problem, BRAF (V600E) inhibitors have been combined with the MEK inhibitor trametinib; however, incidences of acquired resistance have already been reported [63], [64]. Therefore, targeting PEA3 transcription factors such as ETV5 alone or in combination therapies might be an alternative approach. However, potential issues have already been invoked in ETS-targeted therapies, such as redundancy between these transcription factors and inhibition of negative feedback loops that might cause resistance to cell death [65]. Further investigations are therefore necessary to overcome these obstacles.
Our examination of potential downstream targets of ETV5 in KTC1 cells indicates that CCND1 and CCND2 might be mediators of ETV5-dependent proliferation. A recent study demonstrated that CCND1 was upregulated together with PEA3 transcription factors in a subset of gliomas [66], and CCND2 was recently identified as a new ETV4 target gene in proliferating and migrating mouse mammary cancer cells [67], [68]. Expression of CCND2 in thyroid malignancies is controversial as its downregulation was demonstrated in follicular thyroid carcinomas [69], while a subsequent study demonstrated that CCND2 expression is upregulated in all thyroid cancers in comparison to benign neoplasms [70]. Nonetheless, ETV5 and its putative targets CCND1/2 might represent markers of proliferation specifically in advanced PTCs. As CCND1/2 complex with CDK4 and CDK6 to allow G1/S transition of the cell cycle, targeted CDK4/6 inhibitors such as palbociclib and LEE011 could become promising therapy options for this disease [71].
Acquisition of invasiveness and metastatic abilities by epithelial tumor cells is associated with morphological and functional changes linked to de novo expression of genes regulating EMT [72]. Few studies have investigated the molecular mechanisms driving cell migration and invasion in advanced papillary thyroid cancer. In the present study, we show that changes in ETV5 expression do not affect PTC cell migration, but its downregulation affects cell invasion and matrix metalloproteases MMP3 and MMP9 (Figure 7 and Supplemental Data 2). In addition, ETV5 directly drives the expression of TWIST1, a known determinant of intravasation, metastasis, and chemoresistance in a variety of solid tumors [45]. Interestingly, TWIST1 expression is increased in anaplastic thyroid cancers in comparison to PTCs and PDTC [43], and expression of another member of the TWIST family, TWIST2, has been recently reported as a key effector downstream of BRAF (V600E) in a zebrafish thyroid carcinoma model [73]. While TWIST1 together with SNAI1 in humans is a recognized enabler of EMT in certain cancer cells [46], TWIST2 is also responsible for maintaining preosteoblasts in an undifferentiated state [74]. Along with SNAI1, TWIST1 is a known driver of N-Cadherin (CDH2) expression, and we demonstrated in the present study significant downregulation of CDH2 expression following ETV5 downregulation, indicating possible dependence of CDH2 on the BRAF(V600E)/ETV5/TWIST1 axis. As TWIST1 is phosphorylated by ERK1/2, itself activated by BRAF (V600E), it is likely that cooperation between ETV5 and ERK1/2 contributes to increased TWIST1 expression and activation in these cells.
Except for TWIST1 and SNAI1, our screen did not identify significant changes in expression of other drivers of EMT such as ZEB1 and ZEB2 upon downregulation of ETV5 in KTC1 cells [75], and while SNAI1 expression parallels that of ETV5, we were unable to demonstrate binding of ETV5 to the SNAI1 promoter. Therefore, ETV5 may drive SNAIl expression indirectly. Altogether, our data indicate that ETV5 itself is not a major driver of EMT in KTC1 cells.
Morphologically, only a few native KTC1 cells exhibited a cuboidal shape and expressed cortical actin, which is specific for epithelial cells (Figure 7C) [76]. This might be due to the fact that KTC1 cells derive from a pleural effusion [25] and that increased expression of ETV5 and TWIST1, in the presence of a TERT mutation, might already confer a dedifferentiated phenotype. Forced overexpression of ETV5 led to a dramatic increase in cell size and prominent actin stress fibers (Figure 7E), a hallmark of transition toward the EMT state [76]. Further studies will be crucial to demonstrate whether ETV5 influences hTERT expression and activity in these cells, as mutated hTERT promoters create de novo ETS binding sites.
In conclusion, we have demonstrated that the ETS transcription factor ETV5 is upregulated in BRAF-mutated PTC cells. It is critical for their proliferation and might contribute to cell invasion, possibly through direct upregulation of the transcription factor TWIST1. ETV5 and TWIST1 might therefore serve as useful diagnostic or prognostic factors predicting a more aggressive phenotype in papillary thyroid cancers.
The following are the supplementary data related to this article.
mRNA expression of ETS transcription factors in 53 PTC samples and adjacent/contralateral normal tissues (TCGA).
PCR array profile of upregulated and downregulated genes in KTC1 cells after downregulation of ETV5 by RNAi for 24 hours. Scatter plot of the log base 10 of the hybridization intensity of each gene in the two groups [x axis = control group (scrambled); y axis = treated group with ETV5 siRNA (Group 1)]. The middle line indicates a fold-change (2−ΔCt) of 1. The top and the bottom lines indicate the desired two-fold change in gene expression threshold.
Supplementary figures
Supplementary tables
Declarations of Interest
None.
Author's Contributions
O. R. P.: design, data collection, data analysis and interpretation, manuscript writing. B. P. D.: design, data collection, data analysis and interpretation, manuscript writing. E. M. B.: design, data collection, data analysis and interpretation. D. K. S.: data collection. N. M. H.: data collection. R. T. P.: data analysis. H. D.: statistical analysis. M. E. C.: data interpretation, final approval of manuscript. G. J. C.: data collection, data interpretation, final approval of manuscript. M. C. H.: conception and design, financial support, administrative support, data analysis and interpretation, manuscript writing, final approval of manuscript.
Acknowledgements
The authors thank Dr. Clifford Stephan, Ph.D., Director of the Center for Translational Cancer Research at Texas A&M Health Science Center (Houston, TX), for providing access to the High-Throughput Screening Core Lab. We thank Ms. Mary Sobieski for her assistance with the In Cell Analyzer 6000 Cell Imaging System. The authors also thank Dr. Jun-ichi Abe and Ms. Tamlyn Thomas, Department of Cardiology, Division of Internal Medicine, MD Anderson Cancer Center, for providing training to the Olympus confocal microscope. We also wish to acknowledge the MD Anderson Cancer Center Support CORE Grant NCI 5P30 CA016672.
Footnotes
Funding: This work was funded by NCI1P50 CA168505 Thyroid Cancer SPORE grant (Developmental Research Program to M. C. H.) and by an MD Anderson Cancer Center Startup Package to M. C. H.
Contributor Information
Oorvashi Roy Puli, Email: oorvi.roy@gmail.com.
Brian P Danysh, Email: danysh@gmail.com.
Elena McBeath, Email: emfujiwara@mdanderson.org.
Deepankar K Sinha, Email: dksinha@bcm.edu.
Nguyet M Hoang, Email: nnhoang@wisc.edu.
Reid T Powell, Email: rpowell@ibt.tamhsc.edu.
Heather E Danysh, Email: hdanysh@rti.org.
Maria E Cabanillas, Email: mcabani@mdanderson.org.
Gilbert J Cote, Email: gcote@mdanderson.org.
Marie-Claude Hofmann, Email: mhofmann@mdanderson.org.
References
- 1.Burotto M, Chiou VL, Lee JM, Kohn EC. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120:3446–3456. doi: 10.1002/cncr.28864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M, Tafuri A. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta. 2007;1773:1263–1284. doi: 10.1016/j.bbamcr.2006.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 4.Dankner M, Rose AAN, Rajkumar S, Siegel PM, Watson IR. Classifying BRAF alterations in cancer: new rational therapeutic strategies for actionable mutations. Oncogene. 2018;37:3183–3199. doi: 10.1038/s41388-018-0171-x. [DOI] [PubMed] [Google Scholar]
- 5.Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949–954. doi: 10.1038/nature00766. [DOI] [PubMed] [Google Scholar]
- 6.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 7.Henderson YC, Shellenberger TD, Williams MD, El-Naggar AK, Fredrick MJ, Cieply KM, Clayman GL. High rate of BRAF and RET/PTC dual mutations associated with recurrent papillary thyroid carcinoma. Clin Cancer Res. 2009;15:485–491. doi: 10.1158/1078-0432.CCR-08-0933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cabanillas ME, Patel A, Danysh BP, Dadu R, Kopetz S, Falchook G. BRAF inhibitors: experience in thyroid cancer and general review of toxicity. Horm Cancer. 2015;6:21–36. doi: 10.1007/s12672-014-0207-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakayama H, Yoshida A, Nakamura Y, Hayashi H, Miyagi Y, Wada N, Rino Y, Masuda M, Imada T. Clinical significance of BRAF (V600E) mutation and Ki-67 labeling index in papillary thyroid carcinomas. Anticancer Res. 2007;27:3645–3649. [PubMed] [Google Scholar]
- 10.Xing M, Alzahrani AS, Carson KA, Viola D, Elisei R, Bendlova B, Yip L, Mian C, Vianello F, Tuttle RM. Association between BRAF V600E mutation and mortality in patients with papillary thyroid cancer. JAMA. 2013;309:1493–1501. doi: 10.1001/jama.2013.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen Y, Xing M, Mambo E, Guo Z, Wu G, Trink B, Beller U, Westra WH, Ladenson PW, Sidransky D. BRAF mutation in papillary thyroid carcinoma. J Natl Cancer Inst. 2003;95:625–627. doi: 10.1093/jnci/95.8.625. [DOI] [PubMed] [Google Scholar]
- 12.Liu D, Liu Z, Condouris S, Xing M. BRAF V600E maintains proliferation, transformation, and tumorigenicity of BRAF-mutant papillary thyroid cancer cells. J Clin Endocrinol Metab. 2007;92:2264–2271. doi: 10.1210/jc.2006-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim KB, Cabanillas ME, Lazar AJ, Williams MD, Sanders DL, Ilagan JL, Nolop K, Lee RJ, Sherman SI. Clinical responses to vemurafenib in patients with metastatic papillary thyroid cancer harboring BRAF(V600E) mutation. Thyroid. 2013;23:1277–1283. doi: 10.1089/thy.2013.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dadu R, Shah K, Busaidy NL, Waguespack SG, Habra MA, Ying AK, Hu MI, Bassett R, Jimenez C, Sherman SI. Efficacy and tolerability of vemurafenib in patients with BRAF(V600E)-positive papillary thyroid cancer: M.D. Anderson Cancer Center off label experience. J Clin Endocrinol Metab. 2015;100:E77–E81. doi: 10.1210/jc.2014-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holderfield M, Deuker MM, McCormick F, McMahon M. Targeting RAF kinases for cancer therapy: BRAF-mutated melanoma and beyond. Nat Rev Cancer. 2014;14:455–467. doi: 10.1038/nrc3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montero-Conde C, Ruiz-Llorente S, Dominguez JM, Knauf JA, Viale A, Sherman EJ, Ryder M, Ghossein RA, Rosen N, Fagin JA. Relief of feedback inhibition of HER3 transcription by RAF and MEK inhibitors attenuates their antitumor effects in BRAF-mutant thyroid carcinomas. Cancer Discov. 2013;3:520–533. doi: 10.1158/2159-8290.CD-12-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Byeon HK, Na HJ, Yang YJ, Kwon HJ, Chang JW, Ban MJ, Kim WS, Shin DY, Lee EJ, Koh YW. c-Met-mediated reactivation of PI3K/AKT signaling contributes to insensitivity of BRAF(V600E) mutant thyroid cancer to BRAF inhibition. Mol Carcinog. 2016;55:1678–1687. doi: 10.1002/mc.22418. [DOI] [PubMed] [Google Scholar]
- 18.Duquette M, Sadow PM, Husain A, Sims JN, Antonello ZA, Fischer AH, Song C, Castellanos-Rizaldos E, Makrigiorgos GM, Kurebayashi J. Metastasis-associated MCL1 and P16 copy number alterations dictate resistance to vemurafenib in a BRAFV600E patient-derived papillary thyroid carcinoma preclinical model. Oncotarget. 2015;6:42445–42467. doi: 10.18632/oncotarget.6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Danysh BP, Rieger EY, Sinha DK, Evers CV, Cote GJ, Cabanillas ME, Hofmann MC. Long-term vemurafenib treatment drives inhibitor resistance through a spontaneous KRAS G12D mutation in a BRAF V600E papillary thyroid carcinoma model. Oncotarget. 2016;7:30907–30923. doi: 10.18632/oncotarget.9023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sizemore GM, Pitarresi JR, Balakrishnan S, Ostrowski MC. The ETS family of oncogenic transcription factors in solid tumours. Nat Rev Cancer. 2017;17:337–351. doi: 10.1038/nrc.2017.20. [DOI] [PubMed] [Google Scholar]
- 21.Llaurado M, Abal M, Castellvi J, Cabrera S, Gil-Moreno A, Perez-Benavente A, Colas E, Doll A, Dolcet X, Matias-Guiu X. ETV5 transcription factor is overexpressed in ovarian cancer and regulates cell adhesion in ovarian cancer cells. Int J Cancer. 2012;130:1532–1543. doi: 10.1002/ijc.26148. [DOI] [PubMed] [Google Scholar]
- 22.Monge M, Colas E, Doll A, Gonzalez M, Gil-Moreno A, Planaguma J, Quiles M, Arbos MA, Garcia A, Castellvi J. ERM/ETV5 up-regulation plays a role during myometrial infiltration through matrix metalloproteinase-2 activation in endometrial cancer. Cancer Res. 2007;67:6753–6759. doi: 10.1158/0008-5472.CAN-06-4487. [DOI] [PubMed] [Google Scholar]
- 23.Power PF, Mak IW, Singh S, Popovic S, Gladdy R, Ghert M. ETV5 as a regulator of matrix metalloproteinase 2 in human chondrosarcoma. J Orthop Res. 2013;31:493–501. doi: 10.1002/jor.22227. [DOI] [PubMed] [Google Scholar]
- 24.Lemoine NR, Mayall ES, Jones T, Sheer D, McDermid S, Kendall-Taylor P, Wynford-Thomas D. Characterisation of human thyroid epithelial cells immortalised in vitro by simian virus 40 DNA transfection. Br J Cancer. 1989;60:897–903. doi: 10.1038/bjc.1989.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurebayashi J, Tanaka K, Otsuki T, Moriya T, Kunisue H, Uno M, Sonoo H. All-trans-retinoic acid modulates expression levels of thyroglobulin and cytokines in a new human poorly differentiated papillary thyroid carcinoma cell line, KTC-1. J Clin Endocrinol Metab. 2000;85:2889–2896. doi: 10.1210/jcem.85.8.6732. [DOI] [PubMed] [Google Scholar]
- 26.Schweppe RE, Klopper JP, Korch C, Pugazhenthi U, Benezra M, Knauf JA, Fagin JA, Marlow LA, Copland JA, Smallridge RC. Deoxyribonucleic acid profiling analysis of 40 human thyroid cancer cell lines reveals cross-contamination resulting in cell line redundancy and misidentification. J Clin Endocrinol Metab. 2008;93:4331–4341. doi: 10.1210/jc.2008-1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landa I, Ganly I, Chan TA, Mitsutake N, Matsuse M, Ibrahimpasic T, Ghossein RA, Fagin JA. Frequent somatic TERT promoter mutations in thyroid cancer: higher prevalence in advanced forms of the disease. J Clin Endocrinol Metab. 2013;98:E1562–E1566. doi: 10.1210/jc.2013-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribeiro FR, Meireles AM, Rocha AS, Teixeira MR. Conventional and molecular cytogenetics of human non-medullary thyroid carcinoma: characterization of eight cell line models and review of the literature on clinical samples. BMC Cancer. 2008;8:371. doi: 10.1186/1471-2407-8-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Giordano TJ, Kuick R, Thomas DG, Misek DE, Vinco M, Sanders D, Zhu Z, Ciampi R, Roh M, Shedden K. Molecular classification of papillary thyroid carcinoma: distinct BRAF, RAS, and RET/PTC mutation-specific gene expression profiles discovered by DNA microarray analysis. Oncogene. 2005;24:6646–6656. doi: 10.1038/sj.onc.1208822. [DOI] [PubMed] [Google Scholar]
- 30.Pita JM, Banito A, Cavaco BM, Leite V. Gene expression profiling associated with the progression to poorly differentiated thyroid carcinomas. Br J Cancer. 2009;101:1782–1791. doi: 10.1038/sj.bjc.6605340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.He H, Jazdzewski K, Li W, Liyanarachchi S, Nagy R, Volinia S, Calin GA, Liu CG, Franssila K, Suster S. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci U S A. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giordano TJ, Au AY, Kuick R, Thomas DG, Rhodes DR, Wilhelm KG, Jr., Vinco M, Misek DE, Sanders D, Zhu Z. Delineation, functional validation, and bioinformatic evaluation of gene expression in thyroid follicular carcinomas with the PAX8-PPARG translocation. Clin Cancer Res. 2006;12:1983–1993. doi: 10.1158/1078-0432.CCR-05-2039. [DOI] [PubMed] [Google Scholar]
- 33.Cancer Genome Atlas Research N Integrated genomic characterization of papillary thyroid carcinoma. Cell. 2014;159:676–690. doi: 10.1016/j.cell.2014.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smallridge RC, Chindris AM, Asmann YW, Casler JD, Serie DJ, Reddi HV, Cradic KW, Rivera M, Grebe SK, Necela BM. RNA sequencing identifies multiple fusion transcripts, differentially expressed genes, and reduced expression of immune function genes in BRAF (V600E) mutant vs BRAF wild-type papillary thyroid carcinoma. J Clin Endocrinol Metab. 2014;99:E338–E347. doi: 10.1210/jc.2013-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prescott JD, Koto KS, Singh M, Gutierrez-Hartmann A. The ETS transcription factor ESE-1 transforms MCF-12A human mammary epithelial cells via a novel cytoplasmic mechanism. Mol Cell Biol. 2004;24:5548–5564. doi: 10.1128/MCB.24.12.5548-5564.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prescott JD, Poczobutt JM, Tentler JJ, Walker DM, Gutierrez-Hartmann A. Mapping of ESE-1 subdomains required to initiate mammary epithelial cell transformation via a cytoplasmic mechanism. Mol Cancer. 2011;10:103. doi: 10.1186/1476-4598-10-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang HM, Li L, Papadopoulou N, Hodgson G, Evans E, Galbraith M, Dear M, Vougier S, Saxton J, Shaw PE. Mitogen-induced recruitment of ERK and MSK to SRE promoter complexes by ternary complex factor Elk-1. Nucleic Acids Res. 2008;36:2594–2607. doi: 10.1093/nar/gkn099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peleg S. Modified binding of proteins from calcitonin-negative tumor cells to the neuroendocrine-specific CANNTG motif of the calcitonin gene. Nucleic Acids Res. 1993;21:5360–5365. doi: 10.1093/nar/21.23.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brannon AR, Vakiani E, Sylvester BE, Scott SN, McDermott G, Shah RH, Kania K, Viale A, Oschwald DM, Vacic V. Comparative sequencing analysis reveals high genomic concordance between matched primary and metastatic colorectal cancer lesions. Genome Biol. 2014;15:454. doi: 10.1186/s13059-014-0454-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Principe DR, DeCant B, Staudacher J, Vitello D, Mangan RJ, Wayne EA, Mascarinas E, Diaz AM, Bauer J, McKinney RD. Loss of TGFbeta signaling promotes colon cancer progression and tumor-associated inflammation. Oncotarget. 2017;8:3826–3839. doi: 10.18632/oncotarget.9830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Landa I, Ibrahimpasic T, Boucai L, Sinha R, Knauf JA, Shah RH, Dogan S, Ricarte-Filho JC, Krishnamoorthy GP, Xu B. Genomic and transcriptomic hallmarks of poorly differentiated and anaplastic thyroid cancers. J Clin Invest. 2016;126:1052–1066. doi: 10.1172/JCI85271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 43.Salerno P, Garcia-Rostan G, Piccinin S, Bencivenga TC, Di Maro G, Doglioni C, Basolo F, Maestro R, Fusco A, Santoro M. TWIST1 plays a pleiotropic role in determining the anaplastic thyroid cancer phenotype. J Clin Endocrinol Metab. 2011;96:E772–E781. doi: 10.1210/jc.2010-1182. [DOI] [PubMed] [Google Scholar]
- 44.Di Maro G, Orlandella FM, Bencivenga TC, Salerno P, Ugolini C, Basolo F, Maestro R, Salvatore G. Identification of targets of Twist1 transcription factor in thyroid cancer cells. J Clin Endocrinol Metab. 2014;99:E1617–E1626. doi: 10.1210/jc.2013-3799. [DOI] [PubMed] [Google Scholar]
- 45.Zhao Z, Rahman MA, Chen ZG, Shin DM. Multiple biological functions of Twist1 in various cancers. Oncotarget. 2017;8:20380–20393. doi: 10.18632/oncotarget.14608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 48.De Braekeleer E, Douet-Guilbert N, Morel F, Le Bris MJ, Basinko A, De Braekeleer M. ETV6 fusion genes in hematological malignancies: a review. Leuk Res. 2012;36:945–961. doi: 10.1016/j.leukres.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 49.Seethala RR, Chiosea SI, Liu CZ, Nikiforova M, Nikiforov YE. Clinical and morphologic features of ETV6-NTRK3 translocated papillary thyroid carcinoma in an adult population without radiation exposure. Am J Surg Pathol. 2017;41:446–457. doi: 10.1097/PAS.0000000000000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hollenhorst PC, Paul L, Ferris MW, Graves BJ. The ETS gene ETV4 is required for anchorage-independent growth and a cell proliferation gene expression program in PC3 prostate cells. Genes Cancer. 2011;1:1044–1052. doi: 10.1177/1947601910395578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akagi T, Kuure S, Uranishi K, Koide H, Costantini F, Yokota T. ETS-related transcription factors ETV4 and ETV5 are involved in proliferation and induction of differentiation-associated genes in embryonic stem (ES) cells. J Biol Chem. 2015;290:22460–22473. doi: 10.1074/jbc.M115.675595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mesci A, Taeb S, Huang X, Jairath R, Sivaloganathan D, Liu SK. Pea3 expression promotes the invasive and metastatic potential of colorectal carcinoma. World J Gastroenterol. 2014;20:17376–17387. doi: 10.3748/wjg.v20.i46.17376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pellecchia A, Pescucci C, De Lorenzo E, Luceri C, Passaro N, Sica M, Notaro R, De Angioletti M. Overexpression of ETV4 is oncogenic in prostate cells through promotion of both cell proliferation and epithelial to mesenchymal transition. Oncogenesis. 2012;1:e20. doi: 10.1038/oncsis.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yordy JS, Muise-Helmericks RC. Signal transduction and the Ets family of transcription factors. Oncogene. 2000;19:6503–6513. doi: 10.1038/sj.onc.1204036. [DOI] [PubMed] [Google Scholar]
- 55.Janknecht R, Monte D, Baert JL, de Launoit Y. The ETS-related transcription factor ERM is a nuclear target of signaling cascades involving MAPK and PKA. Oncogene. 1996;13:1745–1754. [PubMed] [Google Scholar]
- 56.Liu D, Liu Z, Liu H, Li H, Pan X, Li Z. Brain-derived neurotrophic factor promotes vesicular glutamate transporter 3 expression and neurite outgrowth of dorsal root ganglion neurons through the activation of the transcription factors Etv4 and Etv5. Brain Res Bull. 2016;121:215–226. doi: 10.1016/j.brainresbull.2016.02.010. [DOI] [PubMed] [Google Scholar]
- 57.Schmidt JA, Avarbock MR, Tobias JW, Brinster RL. Identification of glial cell line-derived neurotrophic factor-regulated genes important for spermatogonial stem cell self-renewal in the rat. Biol Reprod. 2009;81:56–66. doi: 10.1095/biolreprod.108.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tyagi G, Carnes K, Morrow C, Kostereva NV, Ekman GC, Meling DD, Hostetler C, Griswold M, Murphy KM, Hess RA. Loss of Etv5 decreases proliferation and RET levels in neonatal mouse testicular germ cells and causes an abnormal first wave of spermatogenesis. Biol Reprod. 2009;81:258–266. doi: 10.1095/biolreprod.108.075200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McReynolds AC, Karra AS, Li Y, Lopez ED, Turjanski AG, Dioum E, Lorenz K, Zaganjor E, Stippec S, McGlynn K. Phosphorylation or mutation of the ERK2 activation loop alters oligonucleotide binding. Biochemistry. 2016;55:1909–1917. doi: 10.1021/acs.biochem.6b00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brose M.S., Cabanillas M.E., Cohen E.E., Wirth L.J., Riehl T., Yue H., Sherman S.I., Sherman E.J. Vemurafenib in patients with BRAF(V600E)-positive metastatic or unresectable papillary thyroid cancer refractory to radioactive iodine: a non-randomised, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:1272–1282. doi: 10.1016/S1470-2045(16)30166-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, Dummer R, Garbe C, Testori A, Maio M. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sullivan RJ, Flaherty KT. Resistance to BRAF-targeted therapy in melanoma. Eur J Cancer. 2013;49:1297–1304. doi: 10.1016/j.ejca.2012.11.019. [DOI] [PubMed] [Google Scholar]
- 63.Wagle N, Van Allen EM, Treacy DJ, Frederick DT, Cooper ZA, Taylor-Weiner A, Rosenberg M, Goetz EM, Sullivan RJ, Farlow DN. MAP kinase pathway alterations in BRAF-mutant melanoma patients with acquired resistance to combined RAF/MEK inhibition. Cancer Discov. 2014;4:61–68. doi: 10.1158/2159-8290.CD-13-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fofaria NM, Frederick DT, Sullivan RJ, Flaherty KT, Srivastava SK. Overexpression of Mcl-1 confers resistance to BRAFV600E inhibitors alone and in combination with MEK1/2 inhibitors in melanoma. Oncotarget. 2015;6:40535–40556. doi: 10.18632/oncotarget.5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tetsu O, Phuchareon J, Eisele DW, McCormick F. ETS1 inactivation causes innate drug resistance to EGFR inhibitors. Mol Cell Oncol. 2016;3 doi: 10.1080/23723556.2015.1078924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gleize V, Alentorn A, Connen de Kerillis L, Labussiere M, Nadaradjane AA, Mundwiller E, Ottolenghi C, Mangesius S, Rahimian A, Ducray F. CIC inactivating mutations identify aggressive subset of 1p19q codeleted gliomas. Ann Neurol. 2015;78:355–374. doi: 10.1002/ana.24443. [DOI] [PubMed] [Google Scholar]
- 67.Firlej V, Ladam F, Brysbaert G, Dumont P, Fuks F, de Launoit Y, Benecke A, Chotteau-Lelievre A. Reduced tumorigenesis in mouse mammary cancer cells following inhibition of Pea3- or Erm-dependent transcription. J Cell Sci. 2008;121:3393–3402. doi: 10.1242/jcs.027201. [DOI] [PubMed] [Google Scholar]
- 68.Ladam F, Damour I, Dumont P, Kherrouche Z, de Launoit Y, Tulasne D, Chotteau-Lelievre A. Loss of a negative feedback loop involving pea3 and cyclin d2 is required for pea3-induced migration in transformed mammary epithelial cells. Mol Cancer Res. 2013;11:1412–1424. doi: 10.1158/1541-7786.MCR-13-0229. [DOI] [PubMed] [Google Scholar]
- 69.Weber F, Shen L, Aldred MA, Morrison CD, Frilling A, Saji M, Schuppert F, Broelsch CE, Ringel MD, Eng C. Genetic classification of benign and malignant thyroid follicular neoplasia based on a three-gene combination. J Clin Endocrinol Metab. 2005;90:2512–2521. doi: 10.1210/jc.2004-2028. [DOI] [PubMed] [Google Scholar]
- 70.Shibru D, Hwang J, Khanafshar E, Duh QY, Clark OH, Kebebew E. Does the 3-gene diagnostic assay accurately distinguish benign from malignant thyroid neoplasms? Cancer. 2008;113:930–935. doi: 10.1002/cncr.23703. [DOI] [PubMed] [Google Scholar]
- 71.Asghar U, Witkiewicz AK, Turner NC, Knudsen ES. The history and future of targeting cyclin-dependent kinases in cancer therapy. Nat Rev Drug Discov. 2015;14:130–146. doi: 10.1038/nrd4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 73.Anelli V, Villefranc JA, Chhangawala S, Martinez-McFaline R, Riva E, Nguyen A, Verma A, Bareja R, Chen Z, Scognamiglio T. Oncogenic BRAF disrupts thyroid morphogenesis and function via twist expression. Elife. 2017;6 doi: 10.7554/eLife.20728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakamura T, Toita H, Yoshimoto A, Nishimura D, Takagi T, Ogawa T, Takeya T, Ishida-Kitagawa N. Potential involvement of Twist2 and Erk in the regulation of osteoblastogenesis by HB-EGF-EGFR signaling. Cell Struct Funct. 2010;35:53–61. doi: 10.1247/csf.10001. [DOI] [PubMed] [Google Scholar]
- 75.Puisieux A, Brabletz T, Caramel J. Oncogenic roles of EMT-inducing transcription factors. Nat Cell Biol. 2014;16:488–494. doi: 10.1038/ncb2976. [DOI] [PubMed] [Google Scholar]
- 76.Shankar J, Nabi IR. Actin cytoskeleton regulation of epithelial mesenchymal transition in metastatic cancer cells. PLoS One. 2015;10 doi: 10.1371/journal.pone.0119954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
mRNA expression of ETS transcription factors in 53 PTC samples and adjacent/contralateral normal tissues (TCGA).
PCR array profile of upregulated and downregulated genes in KTC1 cells after downregulation of ETV5 by RNAi for 24 hours. Scatter plot of the log base 10 of the hybridization intensity of each gene in the two groups [x axis = control group (scrambled); y axis = treated group with ETV5 siRNA (Group 1)]. The middle line indicates a fold-change (2−ΔCt) of 1. The top and the bottom lines indicate the desired two-fold change in gene expression threshold.
Supplementary figures
Supplementary tables