Abstract
Mutations in tumors can create a state of increased cellular plasticity that promotes resistance to treatment. Thus, there is an urgent need to develop novel strategies for identifying key factors that regulate cellular plasticity in order to combat resistance to chemotherapy and radiation treatment. Here we report that prostate epithelial cell reprogramming could be exploited to identify key factors required for promoting prostate cancer tumorigenesis and cellular plasticity. Deletion of phosphatase and tensin homolog (Pten) and transforming growth factor-beta receptor type 2 (Tgfbr2) may increase prostate epithelial cell reprogramming efficiency in vitro and cause rapid tumor development and early mortality in vivo. Tgfbr2 ablation abolished TGF-β signaling but increased the bone morphogenetic protein (BMP) signaling pathway through the negative regulator Tmeff1. Furthermore, increased BMP signaling promotes expression of the tumor marker genes ID1, Oct4, Nanog, and Sox2; ID1/STAT3/NANOG expression was inversely correlated with patient survival. Thus, our findings provide information about the molecular mechanisms by which BMP signaling pathways render stemness capacity to prostate tumor cells.
Keywords: prostate cancer, TGF-β, Pten, cancer cell plasticity, epigenetic reprogramming
Introduction
Cancer development is a multistep process, in which an epigenetically defined cell of origin progresses from a normal state to a malignant one through genetic and epigenetic somatic cell alterations (Timp and Feinberg, 2013). Although recent large-scale sequencing studies of major cancer types have identified mutations in many transcription and epigenetic factors (Lawrence et al., 2014), the contribution of these loss-of-function or gain-of-function mutations in cancer development and progression remains unknown. Furthermore, while the order of occurrence and combination of somatic alterations affects cancer cellular plasticity and progression, the mechanism remains unclear.
Previous groundbreaking studies have demonstrated reprogramming of somatic cells to a pluripotent state, meaning that the cell has become stem cell-like and can give rise to the three germ layers. This can occur through ectopic expression of four defined factors [Oct4, Sox2, Klf4, and Myc (OSKM)] in vitro and in vivo. Like tumorigenesis, induced pluripotent stem cell (iPSC) reprogramming is a multistep process associated with striking changes in gene expression, cell proliferation, mesenchymal–epithelial transition (MET), chromatin remodeling, histone modification, and DNA methylation. Similar to cancer cells, iPSCs have unlimited growth potential and premature termination of cellular reprogramming which leads to Wilms tumor-like neoplasia in the kidney (Ohnishi et al., 2014), providing further evidence of the intrinsic link between cellular reprogramming and cancer (Suva et al., 2013). Importantly, iPSCs have epigenetic features that may be reciprocal to those in cancer cells. In addition, many key epigenetic factors that regulate cellular reprogramming, are important in malignant transformation and tumor progression (Apostolou and Hochedlinger, 2013). The transient activation or inactivation of such factors during reprogramming to pluripotency can thus mirror the progression to the cancer phenotype seen in vivo. Moreover, iPSC lines obtained from normal and malignant cells provide models for studying the mechanisms of tumor progression. Differentiated Li–Fraumeni Syndrome (LFS)-derived iPSC mesenchymal stem cells (MSC) were able to generate osteosarcoma after their injection into immunodeficient mice (Lee et al., 2015). We hypothesize that prostate epithelial cell reprogramming could be exploited as a model for not only identifying key factors contributing to tumorigenesis but also for dissecting the molecular mechanisms of cancer cell lineage switching. To test our hypothesis, we used the phosphatase and tensin homolog (PTEN)-null mouse prostate tumor model in our experimental system.
Prostate-specific Pten deletion (PtenPKO) in mice results in prostatic intraepithelial neoplasia, and after a long latency, it results in high-grade adenocarcinoma, with minimally invasive and metastatic features. These Pten-null mice have an 80% survival rate, even after 12 months (Wang et al., 2003). In an effort to understand the long latency and the slow progression of Pten-deficient tumors, both Pten and p53 were knocked out in mice. These mice developed invasive prostate cancer, and died by 7 months of age (Chen et al., 2005). This suggests that p53-dependent cellular senescence plays a critical role in suppression of Pten-deficient tumorigenesis. More recently, it was shown that the prostate-specific deletion of Smad4 (Smad4PKO) accelerates the growth and metastatic progression of Pten-deficient tumors (Ding et al., 2011). These PtenPKO: Smad4PKO mice die by 32 weeks of age, probably due to bladder outlet obstruction. Although the expression of p53, p21, and p27 was comparable in tumor cells derived from PtenPKO and PtenPKO:Smad4PKO mice, PtenPKO:Smad4PKO prostate tumor cells express high levels of Cyclin D1 and SPP1 compared with PtenPKO tumors, suggesting that tumor cell proliferation promotes the generation of a fully-penetrant, invasive, and metastatic prostate cancer phenotype. In addition, p53 mutations are independent predictors of tumor recurrence in low- and intermediate-grade cancers. Thus, loss of Pten and p53 is implicated in aggressive forms of human prostate cancer (Schlomm et al., 2008).
The role of cancer stem cells (CSCs) in tumor progression to an aggressive phenotype is still poorly understood. Several recent studies suggest that not all tumor cells conform to the unidirectional hierarchical CSC model, thus promoting the idea of cancer cell plasticity. Cancer cell plasticity refers to the dynamic ability of shifting from a non-CSC state to a CSC state, and vice versa, under certain conditions (Friedmann-Morvinski and Verma, 2014). The similarities between acquired pluripotency and dedifferentiating tumor cells into CSCs suggests that understanding the mechanisms governing induced pluripotency may aid in deciphering tumorigenesis and the aggressive cancer phenotype. In the present study, we screened a set of genetic and epigenetic factors for their ability to promote efficient reprogramming and found that the combined loss of Pten and Tgfbr2 markedly increased efficiency of prostate epithelial cell reprogramming. Ablation of Pten and Tgfbr2 led to accelerated tumor development in vivo. Importantly, our mechanistic studies identified bone morphogenetic protein 4/inhibitor of differentiation 1 (BMP4/ID1) and interleukin-6/signal transducer and activator of transcription 3 (IL-6/Stat3) pathways as key to pluripotency reactivation and epithelial–mesenchymal transition (EMT)-related gene expression. Expression signatures of ID1, Stat3, and Nanog were identified as useful prognostic markers that were inversely correlated with patient survival. Thus, our study has identified the key molecular mechanisms by which Tgfbr2 ablation accelerates tumorigenesis and cancer lineage plasticity in Pten-deficient prostate cells
Results
Identification of key factors required for prostate epithelial cell reprogramming
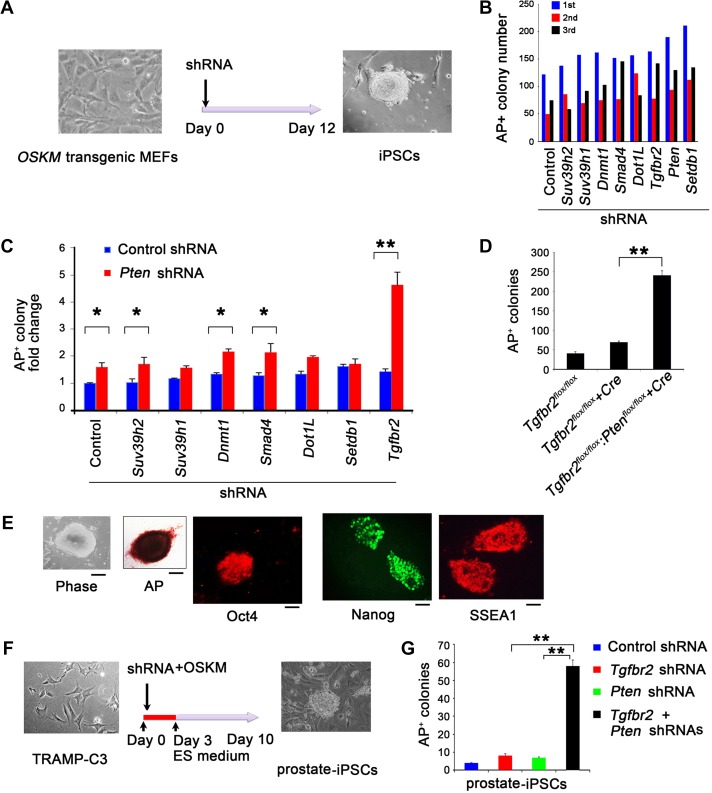
Because of the striking similarities between acquired pluripotency and tumorigenesis, we reasoned that cellular reprogramming could be exploited as a model to identify how key factors interact and contribute to cancer reprogramming and progression. To this end, we first utilized our previously developed Tet-O-OSKM reprogramming screening system (Zhao et al., 2013) to screen a panel of short hairpin RNA (shRNA) that target the genes, Suv39h2, Suv39h1, Dnmt1, Smad4, Dot1L, Tgfbr2, Pten, and Setdb1 (Figure 1A and Supplementary Figure S1A). After three rounds of screening, we found that the gene knockdowns (KD), with the exception of Suv39h2, markedly increased reprogramming efficiency (Figure 1B). Importantly, the combined KD of Pten and Tgfbr2 resulted in the highest increase in reprogramming efficiency (>4-fold change in alkaline phosphatase [AP]+ colony formation) when compared to the Pten KD alone or the KD of other genes (Figure 1C). In addition, Pten and Tgfbr2 gene expression were decreased in embryonic stem cells (ESCs) and wild-type (WT) MEF-derived iPSCs (Supplementary Figure S1B). Consistent with these observations, we found that Pten and Tgfbr2 double KO (DKO, Ptenflox/flox:Tgfbr2flox/flox:Cre) MEFs yielded iPSC AP+ colonies at 3- to 4-fold higher levels than Tgfbr2 KO MEFs (Figure 1D). These Pten and Tgfbr2 DKO MEF-derived iPSCs stained positive for AP, Oct4, Nanog, and SSEA1 (Figure 1E). These results suggest that the combined KD or KO of Pten and Tgfbr2 produces a >4-fold increased cellular reprogramming efficiency.
Figure 1.
Cellular reprogramming to pluripotency in DKD of Pten and Tgfbr2. (A) Microscopic image of MEFs on Day 0 of reprogramming (left). Experimental scheme for iPSC generation from Tet-O-OSKM MEFs. Microscopic image of iPSC clone on Day 12 after reprogramming with OSKM. (B) Number of alkaline phosphatase (AP)+ colonies in three rounds of shRNA screening, on Day 12 after doxycycline (Dox) treatment, generated from Tet-O-4F (OSKM) MEFs transduced with specific shRNAs compared with control shRNA. (C) Fold change in number of AP+ colonies derived from Tet-O-OSKM MEFs transduced with control or Pten-specific shRNA in combination with other gene-specific shRNAs as indicated. (D) Number of AP+ colonies on Day 14 after OSKM-mediated reprogramming in Tgfbr2flox/flox (control), Tgfbr2flox/flox + Cre (Tgfbr2-deficient), and Tgfbr2flox/flox:Ptenflox/flox + Cre (Pten and Tgfbr2 double-deficient) MEF cells. (E) Representative phase-contrast, brightfield (AP staining) images and immunofluorescence (Oct4, Nanog, and SSEA1 staining) images in Pten and Tgfbr2 double-deficient iPSC colonies. Scale bar, 50 μm. (F) Microscopic image of TRAMP-C3s on Day 0 of reprogramming (left). Experimental scheme for iPSC cell generation from the mouse prostate epithelial cell line, TRAMP-C3. Microscopic image of prostate iPSC clone on Day 10 after reprogramming with OSKM. (G) Number of AP+ colonies from prostate epithelial cell-derived stem cells on Day 10 after reprogramming and after transduction with control or specific shRNAs, as indicated. In B, C, D, and G, data are plotted as mean ± SD and are representative of three independent experiments. *P < 0.05, **P < 0.01 vs. corresponding control.
To ensure that prostate epithelial cells could also undergo the process of reprogramming into a pluripotent state, we tested whether mouse prostate epithelial cells (TRAMP-C3, a SV40 large T antigen-transformed cell line) could be reprogrammed to iPSCs, and whether reprogramming efficiency could be enhanced by KD of Pten and Tgfbr2. Using OSKM, we reprogrammed TRAMP-C3 cells and found iPSC colonies 10 days after transduction (Figure 1F). Combined KD of Pten and Tgfbr2 in TRAMP-C3 cells using shRNAs strikingly increased the efficiency of OSKM-mediated reprogramming compared to KD of either Pten or Tgfbr2 alone (Figure 1G). Furthermore, Pten and Tgfbr2 double KD (DKD) in TRAMP-C3 cells increased the number of prostate spheres cultured in 3D Matrigel by 1.5-fold when compared to Pten or Tgfbr2 KD alone (Supplementary Figure S1C). Taken together, these findings suggest that KD of both Tgfbr2 and Pten augments somatic cell reprogramming into a pluripotent state.
Tgfbr2 ablation promotes Pten-null prostate cancer growth and invasiveness, and enhances pluripotency markers
To determine whether the increased reprogramming efficiency by Pten and Tgfbr2 DKD or DKO could be translated into in vivo prostate tumor development, we generated mice with prostate-specific deletion of either Pten (PtenPKO), Tgfbr2 (Tgfbr2PKO), or both (PtenPKO:TgfbrPKO) (Supplementary Figure S2A). As expected, WT, PtenPKO, and Tgfbr2PKO mice had no early mortality, but all PtenPKO:TgfbrPKO male mice died by 13 weeks of age (median survival of 11 weeks) (Supplementary Figure S3A). This increased mortality in PtenPKO:TgfbrPKO male mice was most likely due to bladder outlet obstruction due to large tumors (Supplementary Figure S3B), suggesting that deletion of Tgfbr2 in PtenPKO mice markedly accelerates prostate tumor initiation and growth observed in PtenPKO mice. Histopathological analysis of prostate tissues by hematoxylin and eosin staining revealed normal morphology in the WT and Tgfbr2PKO mice, hyperplasia and prostate intraepithelial neoplastic lesions in PtenPKO mice, and invasive adenocarcinoma in PtenPKO:Tgfbr2PKO mice (Supplementary Figure S3C). Immunohistochemical staining of prostate tissues showed robust Pten and TGFβR2 expression in WT mouse prostate epithelium. As expected, Pten expression was absent in PtenPKO and in PtenPKO:Tgfbr2PKO prostate tissue, while TGFβR2 expression was absent in Tgfbr2PKO and PtenPKO:Tgfbr2PKO prostate tissue (Supplementary Figure S3D). To assess the downstream signaling effects of Pten deletion, such as phosphorylation of Akt, which augments cell survival, proliferation, and senescence, we stained prostate tissues with phosphorylated Akt (p-Akt) antibody. Minimal or no p-Akt signal was observed in the WT and Tgfbr2PKO prostate tissues. However, Akt activation was clearly present in prostate tissues from PtenPKO (Chen et al., 2005) and PtenPKO:Tgfbr2PKO mice (Supplementary Figure S3D), which was confirmed by Western blot (WB) analysis (Supplementary Figure S2B). During the course of our work, another group reported similar findings (Bjerke et al., 2014), indicating that prostate-specific ablation of Pten and Tgfb2 resulted in large, invasive prostate cancer and metastatic progression. We found that PtenPKO:Tgfbr2PKO tumors had increased Cyclin D1, SPP1, and Ki-67 expression (Supplementary Figures S2C and S3E). This is consistent with previous results obtained with Pten:Smad4 DKO tumors (Ding et al., 2011), further suggesting that TGFβR2 plays a role in Pten-deficient mediated prostate cancer cell proliferation.
Because TGF-β signaling was reported to induce cellular senescence, we stained WT, Tgfbr2PKO, PtenPKO, and PtenPKO:Tgfbr2PKO prostate tissue sections for β-galactosidase activity and p27 (biomarkers of senescence). Both β-galactosidase activity and p27 expression were increased in PtenPKO prostate tissues, but were markedly reduced in PtenPKO:Tgfbr2PKO prostate tissues (Supplementary Figures S2D and S3E). Increased expression of the apoptotic inhibitors (i.e. Skp2, Bcl-2, and Bcl-xL) were also observed in the PtenPKO:Tgfbr2PKO prostate tissues compared with Tgfbr2PKO or PtenPKOprostate tissues (Supplementary Figure S2D). However, we did not find any appreciable differences in the expression of p53, Ink4a, Arf, or p21 between PtenPKO and PtenPKO:Tgfbr2PKO prostate samples (data not shown). Furthermore, we detected CK18+ cells in lung tissues from 12-week-old PtenPKO:Tgfbr2PKO mice, indicating prostate cancer metastasis (Supplementary Figure S3F). These data suggest that Tgfbr2 deficiency markedly increases prostate cell proliferation and survival, decreases cell senescence, and accelerates tumor development and metastasis in PtenPKO mice. These data may partially explain the increased prostate cancer growth observed in PtenPKO:Tgfbr2PKO mice. However, this does not explain why Tgfbr2 ablation has a more dramatic effect on prostate cancer development and progression in PtenPKO mice than the previously reported Smad4 ablation in PtenPKO mice.
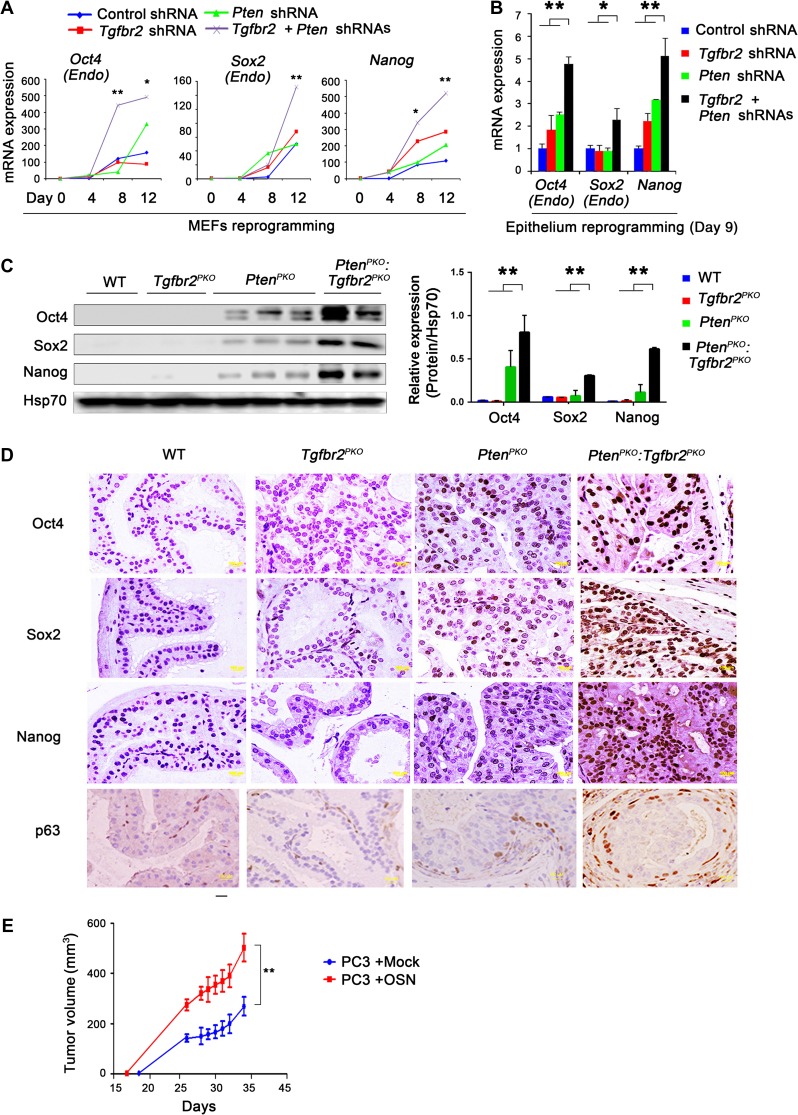
To understand whether accelerated prostate cancer growth observed in PtenPKO:Tgfbr2PKO mice is associated with increased cell stemness, we first reprogrammed MEFs into a pluripotent state. When Pten and Tgfbr2 were simultaneously knocked down, endogenous Oct4, Sox2, Nanog, Rex1, and Cripto gene expression increased in a time-dependent manner (Figure 2A and Supplementary Figure S4A). Next, we performed iPSC reprogramming using TRAMP-C3 cells and found that KD of Pten and Tgfbr2 in prostate epithelial cells simultaneously undergoing reprogramming also had dramatically increased expression of endogenous Oct4, Sox2, Nanog, Rex1, and Cripto (Figure 2B and Supplementary Figure S4B). We also performed WB analysis of pluripotent stem cell markers in WT, Tgfbr2PKO, PtenPKO, and PtenPKO:Tgfbr2PKO prostate tissue. We found strikingly increased expression of Oct4, Sox2, and Nanog in PtenPKO:Tgfbr2PKO tumors compared to PtenPKO tissues (Figure 2C). This observation was confirmed by tissue staining with antibodies specific to Oct4, Sox2, and Nanog (Figure 2D). The prostate stem cell marker, p63, was also highly expressed in PtenPKO:Tgfbr2PKO tumors compared to PtenPKO prostate tissue (Figure 2D). To provide direct evidence that ectopic expression of Oct4, Sox2, and Nanog affects prostate cancer progression, we transduced the human prostate cancer cell line PC3 with three genes. Xenograft assays indicated that ectopic expression of Oct4, Sox2, and Nanog (OSN) augmented PC3 growth in vivo (Figure 2E). Taken together, these results suggest that core cellular pluripotency markers are strongly upregulated in PtenPKO:Tgfbr2PKO tumor cells when compared to WT, Tgfbr2PKO, and PtenPKO prostate cells.
Figure 2.
Induction of pluripotency factors in prostate tumor cells upon ablation of Pten and Tgfbr2. (A) Tet-O-OSKM MEF cells were transduced with the indicated shRNAs, with subsequent OSKM-mediated reprogramming for 12 days. RNA samples were collected at the indicated time points. Expression of endogenous Oct4, Sox2, and Nanog was analyzed by quantitative real-time RT-PCR (qRT-PCR). (B) TRAMP-C3 cells were transduced with the indicated shRNAs, with subsequent OSKM-mediated reprogramming for 9 days. Endogenous Oct4, Sox2, and Nanog expression levels were analyzed by qRT-PCR. (C) WB analysis demonstrating elevated Oct4, Sox2, and Nanog levels in prostates of PtenPKO:Tgfbr2PKO mice. (D) Immunohistochemical analysis of Oct4, Sox2, Nanog, and p63, a marker of prostate epithelial stem cells, expression in prostate tissues of mice with the indicated genotypes. Scale bar, 10 μm. (E) Human PC3 cells transduced with OSN (Oct4, Sox2, and Nanog). Next, 106 cells were subcutaneously injected into male NOD/SCID mice. Results are expressed as the mean tumor size ± SD of the size (n = 10 mice/group). Each mouse was inoculated bilaterally with cells at both rear axillas (two injections per mouse). In A, B, and E, data are plotted as mean ± SD and are representative of three independent experiments. *P < 0.05, **P < 0.01 vs. corresponding control.
BMP/SMAD/ID1/STAT signaling promotes prostate tumorigenesis and progression
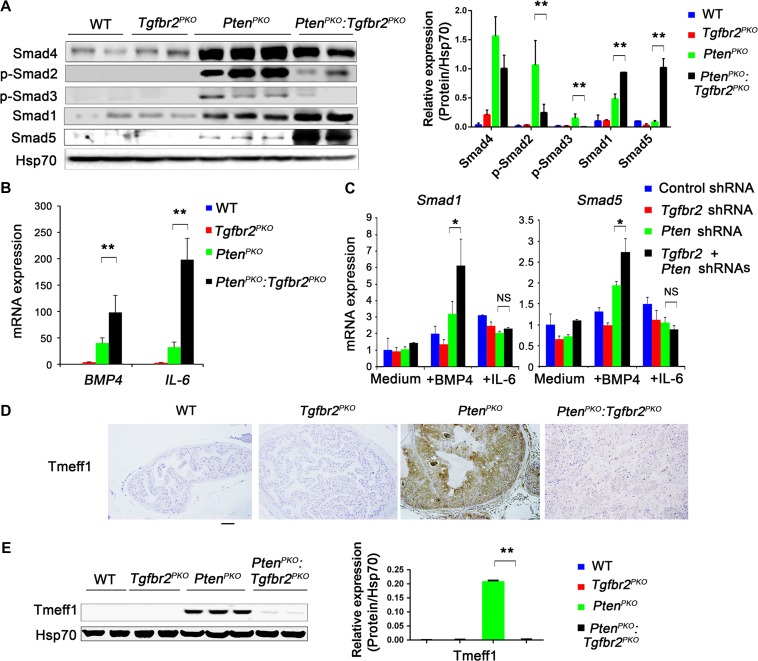
Because the TGF-β family of proteins and their signaling pathways play an important role in maintaining pluripotency, we determined the effects of Tgfbr2 deletion on Smad signaling in prostate tumor cells. We did not detect any significant differences between the Smad-independent signaling pathway or the NF-κB signaling pathway (phosphorylated p70 S6, p65, JNK, ERK, and p38) and PtenPKO or PtenPKO:Tgfbr2PKO prostate tissues (Supplementary Figure S5B). However, WB analysis revealed sustained expression of Smad4 but little or no phosphorylation of Smad2 (p-Smad2) and Smad3 (p-Smad3) in PtenPKO:Tgfbr2PKO prostate tumors; when compared to PtenPKO prostate tumors, suggesting that downstream signaling from TGFβR2 is abolished (Figure 3A). Furthermore, this was not due to gene expression changes in the TGFβR2 agonists, TGF-β1, TGF-β2, TGF-β3, or changes in BMP receptors and BMP antagonists (Gremlin1/2 and Noggin) (Supplementary Figure S5C−E). In contrast, when compared to PtenPKO, expression of Smad1 and Smad5 were increased 2-fold and 12-fold, respectively, in PtenPKO:Tgfbr2PKO prostate tumors, suggesting that the bone morphogenetic pathway (BMP) is enhanced in PtenPKO:Tgfbr2PKO prostate tumors (Figure 3A and Supplementary Figure S5A). Although no appreciable differences in the BMP7 levels were observed (Supplementary Figure S5C), we detected increased BMP4 and IL-6 mRNA levels in PtenPKO:Tgfbr2PKO prostate tumors (Figure 3B). Previous studies have demonstrated that BMP4/ID1 and IL-6/Stat3 signaling pathways promote prostate tumorigenesis and progression (Ling et al., 2003; Abdulghani et al., 2008). We found that exogenous BMP4, but not IL-6, increased gene expression of Smad1 and Smad5 in Pten and Tgfbr2 DKD TRAMP-C3 cells (Figure 3C). The increased downstream Smad1 and Smad5 signaling in response to BMP4 and the increased BMP4 expression upon depletion of Pten and Tgfbr2 suggests a positive feedback loop for BMP signaling. Although mRNA change in Smad5 in PtenPKO:Tgfbr2PKO prostate tumors or Pten and Tgfbr2 DKD TRAMP-C3 cells were significant, striking changes in Smad5 protein in PtenPKO:Tgfbr2PKO prostate tumors, compared to PtenPKO prostate tissue (Figure 3A), may suggest an alternative regulatory mechanism for the Smad5 protein.
Figure 3.
BMP/SMAD/ID1/STAT signaling upon ablation of Pten and Tgfbr2. (A) WB analysis of Smad4, pSmad2, pSmad3, Smad1, and Smad5 expression in prostates from the indicated mice. (B) Expression levels of BMP4 and IL-6 were analyzed by qRT-PCR in prostate tissues from the indicated mice. (C) TRAMP-C3 cells were transduced with the indicated shRNAs for 48 h, and subsequently treated with 50 ng/ml BMP4 or 100 ng/ml IL-6 for an additional 24 h. Expression of Smad1 or Smad5 was analyzed by qRT-PCR. (D) Immunohistochemical analysis of Tmeff1 expression in prostate tissues of mice with the indicated genotypes. Scale bar, 100 μm. (E) WB analysis of Tmeff1 expression in prostate tissues from mice with the indicated genotypes. In A, B, C, and E, data are plotted as mean ± SD and are representative of three independent experiments. *P < 0.05, **P < 0.01 vs. corresponding control.
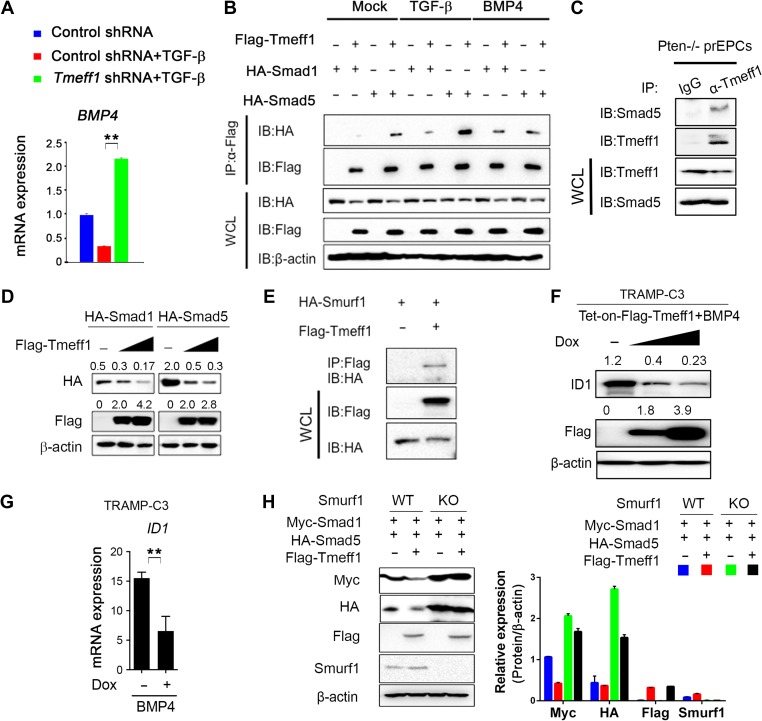
To gain further insight into how depletion of Tgfbr2 and Pten leads to BMP signaling and upregulation of Smad1 and Smad5 proteins, we first determined the expression pattern of Tmeff1, a BMP pathway inhibitor in hair follicle stem cells (Oshimori and Fuchs, 2012). WB and quantitative real-time RT-PCR (qRT-PCR) analyses demonstrated decreased Tmeff1 expression in PtenPKO:Tgfbr2PKO prostate tumors compared to PtenPKO prostate tissue (Figure 3D and E, Supplementary Figure S5F). We further showed that Tmeff1 expression was induced by TGF-β but not by BMP4 (Supplementary Figure S5G). In addition, KD of Tmeff1 led to an increase of BMP4 expression in response to TGF-β and BMP4 (Figure 4A, Supplementary Figure S5H and I). Further co-immunoprecipitation (co-IP) and immunoblot analyses showed that Tmeff1 interacted with both Smad1 and Smad5 in HEK293T cells with or without BMP4 and TGF-β stimulation (Figure 4B). More importantly, we found endogenous interaction of Tmeff1 with Smad5 in PtenPKO prostate cells (Figure 4C). Tmeff1 induced dose-dependent degradation of Smad1 and Smad5 through interaction with Smurf1, the E3 ligase of Smad1 and Smad5 (Figure 4D and E). Consistent with these observations, we found that doxycycline induced overexpression of Flag-tagged Tmeff1 in TRAMP-C3 cells and reduced ID1 expression in a dose-dependent manner (Figure 4F and G). To confirm that Tmeff1-induced Smad1 and Smad5 degradation depends on Smurf1, we found that deletion of Smurf1 abolished Tmeff1-induced degradation of Smad1 and Smad5 in HEK293T cells (Figure 4H). Taken together, these results suggest that Tmeff1 induced by TGF-β signaling interacts with Smad1 and Smad5, causing them to degrade, and thus inhibiting BMP signaling in prostate tumors. Depletion of Tgfbr2 abolishes Tmeff1-mediated inhibition of BMP signaling for accelerated prostate cancer growth.
Figure 4.
Tmeff1 interacts with Smad1/5 and causes their degradation. (A) TRAMP-C3 cells were transduced with control shRNA or Tmeff1 shRNA, and treated with or without 25 ng/ml TGF-β. BMP4 expression was analyzed by qRT-PCR after 24 h. (B) HEK293T cells were co-transfected with vectors for Flag-Tmeff1 and HA-Smad1 or Flag-Tmeff1 and HA-Smad5, followed by the treatment with 25 ng/ml TGF-β or 50 ng/ml BMP4. Cell lysates were used for immunoprecipitation with anti-Flag beads and immunoblot analysis with anti-HA. WCL, whole-cell lysates. (C) Extracts of prostate epithelial cells (prEPCs) isolated from PtenPKO mice were subjected to IP with anti-Tmeff1 or IgG and immunoblotting with anti-Smad5 antibody. (D) WB analysis of HA-Smad1 or HA-Smad5 expression in HEK293T cells transfected with increasing amount of Flag-Tmeff1 (0, 200, and 400 ng). (E) Co-IP of Flag-Tmeff1 and HA-Smurf1 in HEK293T cells transfected with the indicated plasmids. (F) WB analysis of ID1 expression in Tet-on-Flag-Tmeff1 TRAMP-C3 cells treated with 50 ng/ml BMP4 and doxycycline (0, 500 ng/ml, and 1 μg/ml). (G) ID1 mRNA level in Tet-on-Flag-Tmeff1 TRAMP-C3 cells treated with 50 ng/ml BMP4 and doxycyline (0 and 500 ng/ml). (H) WB analysis of Myc-Smad1 and HA-Smad5 in WT and Smurf1-deleted HEK293T cells transfected with Flag-Tmeff1 along with Myc-Smad1 or HA-Smad5. In A, G, and H, data are plotted as mean ± SD and are representative of three independent experiments. *P < 0.05, **P < 0.01 vs. corresponding control.
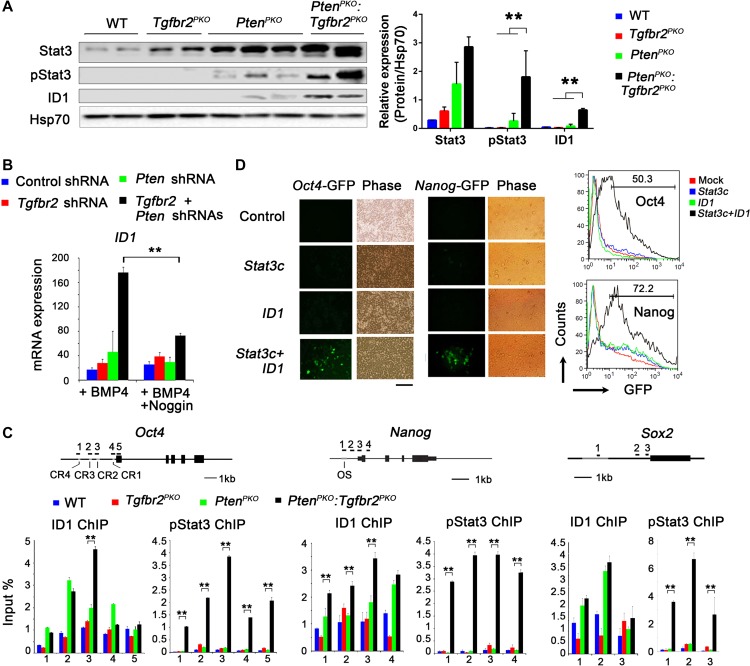
To investigate whether Stat3 and ID1 play a role in PtenPKO:Tgfbr2PKO prostate tumor cell growth, we measured Stat3 and ID1 expression levels. We found increased levels of ID1 protein and phosphorylated Stat3 (pStat3) protein in PtenPKO:Tgfbr2PKO prostate tumors when compared to PtenPKO tumor samples (Figure 5A). Further experiments using TRAMP-C3 cells showed that BMP4 augmented ID1 expression in Pten- and Tgfbr2-DKD cells compared to Pten- or Tgfbr2-single KD cells (Figure 5B and Supplementary Figure S6A). This BMP4-mediated increase in ID1 expression could be blocked by the addition of Noggin, a BMP antagonist (Figure 5B). Taken together, these data indicate that the ID1 and Stat3 signaling pathways, in the absence of Pten and Tgfrb2, may be involved in tumorigenesis.
Figure 5.
Increased pluripotency and ID1 and pStat3 binding to pluripotency gene promoters upon ablation of Pten and Tgfr2. (A) WB analysis of Stat3, pStat3, and ID1 expression in prostates from mice with the indicated genotypes. (B) TRAMP-C3 cells were transduced with the indicated shRNA for 48 h, and subsequently treated with 50 ng/ml BMP4 or a combination of 50 ng/ml BMP4 and 10 ng/ml Noggin for an additional 24 h. Expression of ID1 was analyzed by qRT-PCR. (C) ChIP–qPCR analysis to determine ID1 and pStat3 binding to the promoter regions of Oct4, Nanog, and Sox2 in prostate tissue from mice with the indicated genotypes. Numbered black bars indicate primer locations (top panel). Values are reported as fold enrichment relative to input DNA. (D) Human 293T cells harboring Oct4 or Nanog promoter-fused-GFP plasmids were transfected with activated Stat3 (Stat3c) or ID1. Fluorescence images (left) and fluorescence-activated cell sorting (FACS) results of GFP expression in 293T cells at 2 days after transfection. Scale bar, 50 μm. In A, B, and C, data are plotted as mean ± SD and are representative of three independent experiments. *P < 0.05, **P < 0.01 vs. corresponding control.
ID1 and Stat3 bind to the promoter regions of pluripotent marker genes
ID1 and Stat3 have been identified as critical regulators in the maintenance of ESC self-renewal (Ying et al., 2003). To examine whether ID1 and pStat3 bind directly to the Oct4, Sox2, and Nanog genes, we performed chromatin immunoprecipitation (ChIP) assays on prostate tumors using the pY705-Stat3 (pStat3)-specific and ID1-specific antibodies. In PtenPKO:Tgfbr2PKO prostate tumors, the highest enrichment of ID1 in the Oct4 promoter was found in a distal enhancer region (Conserved Region 2, CR2) (Figure 5C). PtenPKO prostate tissues showed increased ID1-binding sites in the CR1 to CR4 regions, compared to WT tissues. In addition, the enrichment of pStat3 was found in all CR1 to CR4 regions in PtenPKO:Tgfbr2PKO prostate tumors. Conversely, the ChIP assay did not detect any pStat3-binding sites in the Oct4 enhancer regions in WT or PtenPKO prostate tissues. pStat3 enrichment sites were also found in the Nanog and Sox2 promoter regions in PtenPKO:Tgfbr2PKO prostate tumors. In contrast, ID1 enrichment sites were only found in the Nanog promoter, and not in the Sox2 promoter of PtenPKO:Tgfbr2PKO prostate tumors. To confirm that pStat3 and ID1 bind to the promoter regions of pluripotency genes upon ablation of Pten and Tgfrb2, a series of ChIP–qPCR experiments were performed using TRAMP-C3 cells, with occupancy sites selected from the previous experiment. ID1 and pStat3 were both enriched in and occupied the regulatory regions of Oct4 and Nanog in Tgfbr2 and Pten DKD TRAMP-C3 cells (Supplementary Figure S6B). After silencing Pten and Tgfbr2, we performed ChIP–qPCR assays to determine if the modifications of Oct4 and Nanog created a more open chromatin structure, affecting H3K4me3 and DNA 5hmC levels. We found that KD Pten and Tgfbr2 led to H3K4me3 and 5hmC enriched regions of Oct4 and Nanog loci, dependent on BMP4 signaling (Supplementary Figure S6C and D). In addition, ID1 and Stat3 inhibition decreased prostate epithelial cell growth (Supplementary Figure S6E and F). Consistently, overexpressing ID1 and Stat3 in prostate cancer cells (PC3 and LNCap) significantly increased prostate sphere formation (Supplementary Figure S6G). To determine whether Oct4 and Nanog promoter binding of ID1 and pStat3 led to transcriptional activation, Oct4-GFP and Nanog-GFP reporter constructs containing the 4.6 kb and 2.5 kb upstream promoter regions of Oct4 and Nanog, respectively, were used. Following 24 h of transfection, 293T cells expressing ID1 alone or Stat3 alone showed no GFP expression in Oct4-GFP or Nanog-GFP reporter-transfected cells. In contrast, cells transfected with both ID1 and Stat3 together exhibited a dramatic increase in GFP expression compared to control cells, or cells transfected with ID1 alone or Stat3 alone (>50% in Oct4-GFP and >70% in Nanog-GFP) (Figure 5D). These data suggest that ID1 and Stat3 together enhance the expression of Oct4 and Nanog.
EMT in Pten- and Tgfbr2-deficient prostate tumor progression
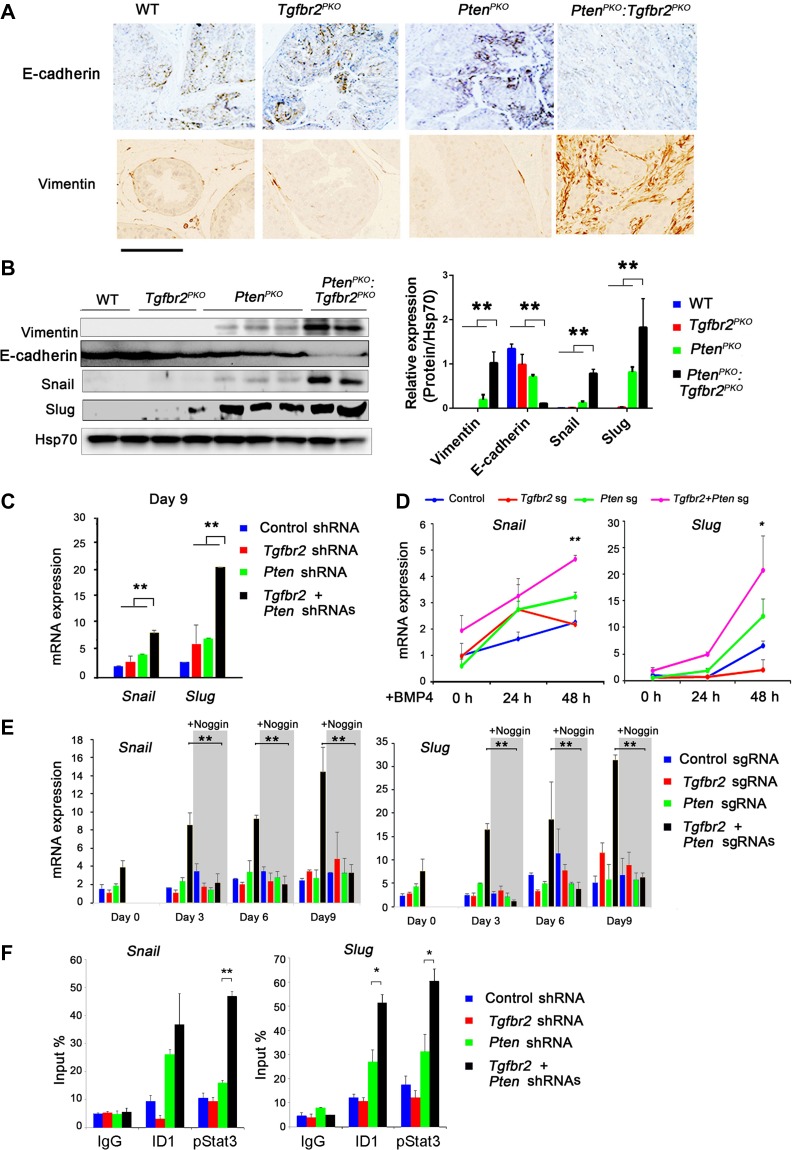
Previous studies have demonstrated that the key characteristics of EMT are closely associated with CSCs signatures (Scheel and Weinberg, 2012) and that TGF-β and BMP signaling plays an important role in EMT (Li et al., 2010). BMP4 has also been shown to induce EMT in many types of cancer (Theriault et al., 2007). Therefore, we investigated whether EMT might contribute to accelerated prostate cancer growth observed in PtenPKO:Tgfbr2PKO mice. We found that expression of the EMT marker, Vimentin, was increased in PtenPKO:Tgfbr2PKO tumors, whereas the expression of the MET marker, E-cadherin, was decreased (Figure 6A). In addition, WB analysis showed increased expression of the EMT-related proteins, Vimentin, Snail, and Slug, in PtenPKO:Tgfbr2PKO tumors (Figure 6B). Previous studies have shown that MET is critical in cellular reprogramming of MEF into a pluripotent state (Li et al., 2010). We asked whether EMT or MET is required for OSKM reprogramming in TRAMP-C3 epithelial cells. We knocked down either Tgfbr2, Pten, or both genes in TRAMP-C3 cells. The EMT genes, Snail and Slug, were upregulated on Day 9 during reprogramming of TRAMP-C3 cells with KD of Tgfbr2 and Pten (Figure 6C). Next, we generated Pten KO, Tgfbr2 KO, and DKO TRAMP-C3 cells using the CRISPR/Cas9 KO system (Supplementary Figure S7A and B). Forty-eight hours after the addition of BMP4, DKO cells had a 2-fold and 4-fold increase in Snail and Slug gene expression, respectively, when compared to control cells (Figure 6D). This BMP4-induced Snail and Slug expression during reprogramming could be completely blocked by addition of the BMP4 antagonist, Noggin (Figure 6E). To determine whether the BMP4 downstream regulator, ID1, binds to the promoter regions of Snail and Slug, we performed ChIP–qPCR experiments using TRAMP-C3 cells. ID1, as well as pStat3, were enriched in the promoter regions of Snail and Slug in Tgfbr2 and Pten DKD TRAMP-C3 cells (Figure 6F). These data suggest that ablation of Pten and Tgfbr2 or exogenous BMP4 promotes EMT-related gene expression, which may contribute, in part, to the accelerated prostate cancer growth and progression in PtenPKO:Tgfbr2PKO mice.
Figure 6.
EMT-related gene expression upon ablation of Pten and Tgfbr2. (A) Immunohistochemical staining of E-cadherin and Vimentin in prostate tissues from mice with the indicated genotypes. Scale bar, 100 μm. (B) WB analysis of Vimentin, Snail, Slug, and Hsp70 (loading control) levels in prostate tissues from mice with the indicated genotypes. (C) TRAMP-C3 cells were transduced with the indicated shRNAs, and subsequently OSKM-reprogrammed for 9 days. Expression of Snail and Slug was analyzed by qRT-PCR. (D) The indicated KO TRAMP-C3 cells were generated using the CRISPR/Cas9 shRNA. Next, the cells were treated with 50 ng/ml BMP4 for 48 h. Expression of Slug and Snail was analyzed by qRT-PCR. (E) The indicated KO TRAMP-C3 cells were OSKM-reprogrammed for 9 days with or without 10 ng/ml Noggin treatment. The RNA was collected at different time points. Expression of Snail and Slug was analyzed by qRT-PCR. (F) Determination of ID1 and pStat3 binding to the promoter regions of Snail and Slug in TRAMP-C3 cells transduced with the indicated shRNAs by ChIP−qPCR analysis. Values are reported as fold enrichment relative to input DNA. In B, C, D, E, and F, data are plotted as mean ± SD and are representative of three independent experiments. *P < 0.05, **P < 0.01 vs. corresponding control.
Expression signature of ID1, STAT3, and Nanog is a prognostic marker for aggressive human prostate cancer
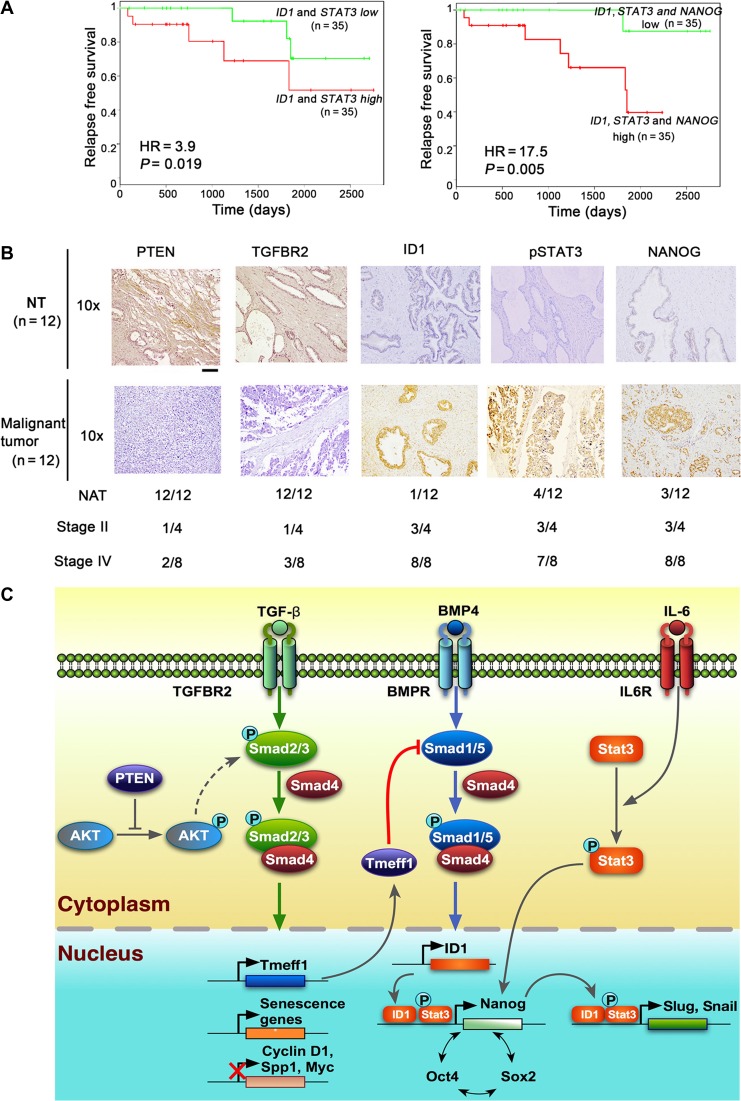
Many prostate cancer patients develop recurrent cancer that progresses to lethal metastatic disease. Therefore, identifying prognostic factors in early-stage prostate cancer to predict tumor progression or recurrence is important. Since CSCs have been linked to tumor recurrence, we assessed whether our identified gene regulators of prostate cancer stemness could predict the risk of recurrence in prostate cancer patients. We performed Kaplan–Meier relapse survival analysis by comparing two combinations of stage II prostate cancer patients obtained from a published microarray data set based on their gene expression levels: (i) low expression of both ID1 and STAT3 vs. high expression of both ID1 and STAT3; and (ii) low expression of ID1, STAT3, and NANOG vs. high expression of ID1, STAT3, and NANOG. Kaplan–Meier survival analysis revealed that patients with elevated expression of ID1, STAT3, and NANOG had the shortest time to prostate cancer recurrence (P = 0.005, HR = 17.5) (Figure 7A). Notably, high expression of ID1 and STAT3 in prostate tumors was also associated with worse outcomes for tumor recurrence (P = 0.019, HR = 3.93) (Figure 7A). However, elevated expression of ID1 alone, STAT3 alone, or NANOG alone did not significantly correlate with the time to recurrence (P > 0.05) (Supplementary Figure S8A). Next, we performed immunohistochemical analysis of PTEN, TGFBR2, ID1, pSTAT3, and NANOG in a tumor tissue microarray (Chen et al., 2005) consisting of 24 patient specimens (Figure 7B and Supplementary Figure S8B). Examination of clinical prostate specimens revealed higher ID1 and NANOG expression levels in prostate tumor cells in comparison to normal prostate epithelium (Figure 7B), and ID1, STAT3, and NANOG were expressed in 75%, 75%, and 75% of stage II cases, respectively, with a trend toward increased expression in stage IV tumors. In addition, ID1/STAT3/NANOG expression was negatively correlated with PTEN/TGFBR2 expression, which was confirmed in prostate tumor tissues collected at Houston Methodist Hospital (Supplementary Figure S8C). These results demonstrate that the three-gene signature of ID1, STAT3, and NANOG is a robust prognostic indicator, predicting indolent versus aggressive prostate cancer after primary treatment.
Figure 7.
Prognostic potential of a three-gene signature in human prostate cancer. (A) Kaplan–Meier plot of the recurrence based on the expression of a three-gene set (ID1/STAT3/NANOG) in prostate cancer patients. HR, hazard ratio. (B) Representative immunohistochemical staining with specific antibodies against PTEN, TGFBR2, ID1, STAT3, and NANOG in human prostate cancer tissue array (n = 24). Scale bar, 200 μm. The numbers of PTEN, TGFBR2, ID1, STAT3, and NANOG-positive graded tumors or normal tissues are depicted. NT, normal tissue. (C) Schematic illustration of our working model by which Tgfbr2 ablation abolishes TGF-β signaling for derepression of Cyclin D and SPP1, and activation of BMP signaling through downregulation of a negative regulator, Tmeff1.
Discussion
In this study, we demonstrated that prostate epithelial cell reprogramming is a key process contributing to cancer cellular plasticity and progression. We also dissected the mechanisms which lead to gene mutations or alterations in stemness acquisition and drug resistance. Our initial screening identified Tgfbr2 as a key negative regulator based on its ability to increase cellular reprogramming. We showed that the combined deletions of Pten and Tgfbr2 markedly increased reprogramming of prostate epithelial cells to pluripotency in vitro, and resulted in aggressive prostate cancer development and progression in vivo. More importantly, we have provided insight into the molecular mechanisms by which Pten and Tgfbr2 gene ablation disrupts TGF-β signaling and activates BMP and IL-6 signaling, this in turn promotes ID1 expression and Stat3 activation; and induces core pluripotency gene expression and EMT, thus accelerating tumor development and progression. Although Pten is frequently mutated in prostate and other cancers, its mutation or deletion results in prostatic intraepithelial neoplasia, with a long latency of minimally invasive and metastatic state before converting to a high-grade aggressive cancer. This suggests that there are multiple barriers that inhibit tumor progression of Pten-null tumor cells until the necessary number of mutations occur. Our results show that TGFβRII-mediated signaling is a major negative regulator that inhibits tumor progression in Pten-null tumor cells.
Pten is one of the most commonly deleted or mutated tumor suppressor genes in human prostate cancer and its deletion promotes reprogramming of MEFs into iPSCs (Liao et al., 2013). TGF-β signaling has a complex, and sometimes opposing, role in cancer. It inhibits tumor development in early stage, but promotes tumor growth and metastasis in later stages (Massague, 2008). Recent studies also show that inhibition of TGF-β signaling enhances both the efficiency and kinetics of OSKM-reprogrammed MEFs (Ichida et al., 2009) and PTEN and TGFBR2 cooperate in the development of mouse prostate cancer (Bjerke et al., 2014). Despite the importance and availability of PTEN and TGF-β signaling studies in stem cells and cancer, the molecular mechanisms responsible for TGF-β signaling-mediated inhibition of cancer development and progression in Pten-null prostate cells remain poorly understood. In this study, we have identified the molecular mechanisms by which Tgfbr2 ablation promotes Pten-deficient prostate tumor progression, leading to mouse death within 13 weeks after birth. Consistent with a previous report (Ding et al., 2011), ablation of Pten increases TGF-β signaling by upregulation of multiple Smad components (Smad4, p-Smad3, and p-Smad2). Ablation of Smad4 in PTEN-null prostate cells leads to inactivation of TGF-β signaling and acceleration of tumor development, causing mice to die within 30 weeks (Ding et al., 2011). By contrast, we show that Tgfbr2 ablation in Pten-null prostate cells results in loss of p-Smad3 and p-Smad2, but a high level of Smad4 still remains. More importantly, we observed high levels of Smad1 and Smad5 in PtenPKO:Tgfbr2PKO prostate cancer cells, probably due to reduced expression of Tmeff1, a negative regulator of BMP signaling that is induced by TGF-β signaling (Oshimori and Fuchs, 2012). To gain additional insight into how Tmeff1 suppresses BMP signaling, we show that Tmeff1 interacts with the Smad1–Smad5 complex and promotes Smad1 and Smad5 protein degradation by recruiting the E3 ligase, Smurf1, resulting in suppression of BMP signaling and ID1 expression. This is consistent with previous findings (Zhu et al., 1999). Thus, deletion of Pten and Tgfbr2 not only inactivates TGF-β signaling, which leads to upregulation of CyclinD1 and Spp1, as observed in Pten and Smad4 DKO cells (Ding et al., 2011), but also activates BMP signaling. It should be noted that both TGF-β and BMP signaling pathways are completely abrogated in Pten and Smad4 double KO prostate cells. This key difference in BMP signaling in Pten and Tgfbr2-deficient mice may, in part, explain the aggressive development and early mortality in prostate cancer in comparison to cancer development in Pten and Smad4 double KO mice.
Reprogramming could be used as a powerful platform to study the interplay between transcription factors and chromatin structure, as well as the epigenetic barriers that resist cell fate changes. Our data indicate that Pten and Tgfbr2 repress pluripotency-associated loci such as Oct4 and Nanog and are dependent on BMP4 signaling. Loss of Pten and Tgfbr2 may allow for OSN-mediated epigenetic reprogramming and cellular plasticity which induce cancer. To understand the molecular mechanisms of how increased BMP signaling accelerates tumor development and cancer cellular plasticity in PtenPKO:Tgfbr2PKO mice, our results show that increased BMP signaling leads to upregulation of pluripotency core genes (Oct4 and Nanog) required for reprogramming. These findings are consistent with a recent study showing that BMP signaling promotes early stages of cellular reprogramming (Samavarchi-Tehrani et al., 2010). It has been known that BMP signaling, in cooperation with STAT3 signaling (Ying et al., 2003), induces ID1 expression, which in turn reactivates Nanog expression (Suzuki et al., 2006). Indeed, we found high levels of ID1, phosphorylated Stat3, Oct4, Sox2, and Nanog in PtenPKO:Tgfbr2PKO prostate cancer, compared to PtenPKO prostate cancer cells. However, expression of these pluripotency core genes has not been reported in prostate cancer in PtenPKO:Smad4PKO mice. Inhibition of TGF-β signaling or increased BMP signaling can facilitate cellular reprogramming by promoting MET in the early phases of reprogramming and increased Oct4, Nanog, and Sall4 expression can facilitate cellular reprogramming by promoting MET in the later stage of reprogramming (Ichida et al., 2009; Li et al., 2010; Samavarchi-Tehrani et al., 2010). However, we show that upregulation of EMT-related genes (Snail and Slug) was observed in reprogramming of prostate epithelial cells to stem cell status and this upregulation was also observed in prostate cancer clinical samples. It is likely that upregulation of BMP4 facilitates the increased plasticity of MET/EMT transitions and allows for greater flexibility in transitioning between states (non-CSC back to the CSC state).
It is well known that human cancers are composed of cells with distinct phenotypes, genotypes, and epigenetics states. This so-called ‘intratumoral heterogeneity’ is further supported by recent single-cell RNA sequencing results (Patel et al., 2014). It is becoming increasingly clear that CSCs can contribute to intratumoral heterogeneity and drive tumor growth (Pattabiraman and Weinberg, 2014). CSCs express many proteins similar to early ESCs, such as Oct4, Nanog, and Sox2 (Orkin and Hochedlinger, 2011). The overexpression of these three proteins occurs in human malignancies, especially in poorly differentiated tumors (Ben-Porath et al., 2008). Transient expression of OSKM establishes an enhanced pro-metastatic state in the colon tumor, which indicates that epigenetic reprogramming of cancer cells may enhance metastases (Ben-Porath et al., 2008; Singovski et al., 2016). Recently, it was demonstrated that local accumulation of BMP-SMAD1 signaling enhances the recovery of LIF/STAT3 responsiveness (Ohnishi et al., 2014). We have demonstrated that BMP4/ID1 and IL-6/STAT3 axes act in concert to reactivate the expression of stem cell core factors in prostate cancer cells, which likely contributes to prostate cancer cellular plasticity. Although it is known that TGF-β signaling promotes EMT, our results show that the BMP signaling may also promote EMT in PtenPKO:Tgfbr2PKO prostate tumors. MET and EMT are two dynamic processes during cancer development and metastasis. In general, EMT has been associated with metastatic cancer cells having stem-like properties (Pattabiraman and Weinberg, 2014). More recent studies have shown that the levels and duration of expression of Twist1 differentially regulate tumor development, stemness and metastasis (Beck et al., 2015). For this reason, we did not observe a marked increase in Twist1 expression in PtenPKO:Tgfbr2PKO prostate tumors (data not shown). By contrast, we showed that there was increased expression of Snail and Slug in PtenPKO:Tgfbr2PKO prostate tumors when comparing to PtenPKO prostate tumor cells, and this was regulated by ID1 and STAT3 through their binding to the promoter regions of the Snail and Slug genes. Furthermore, we showed that BMP4 is essential for activation of Snail and Slug during OSKM-mediated epithelial cell reprogramming. This BMP4-mediated epithelial–mesenchymal plasticity of prostate cancer cells may be harnessed for therapeutic intervention to target CSCs.
Prostate-specific antigen (PSA) has been used as an early prostate cancer biomarker, but it cannot distinguish between aggressive and indolent prostate cancer. The signature set of three genes identified in this study (i.e. ID1/STAT3/NANOG) is significantly associated with the recurrence of aggressive prostate cancer and may be useful as biomarkers for prostate cancer prognosis. Based on our results, we propose a working model to illustrate how Tgfbr2 ablation accelerates tumor development and progression in Pten-deficient prostate cells (Figure 7C). First, Tgfbr2 inactivation reduces Pten deficiency-induced prostate cell senescence and increases cell proliferation (Myc, Cyclin D, and Spp1) due to loss of TGF-β signaling. Second, the reduced expression of Tmeff1 by Tgfbr2 inactivation releases its inhibitory effect on BMP signaling. Then, enhanced BMP signaling promotes ID1 expression, which collaborates with IL-6/Stat3 to activate pluripotent genes Nanog, Oct4, and Sox2. These pluripotency core factors may form a positive feedback loop, facilitating prostate epithelial cell reprogramming. Finally, ID1, Stat3, and Nanog in PtenPKO:Tgfbr2PKO prostate tumors further promote expression of EMT-related genes such as Snail and Slug. The ID1/STAT3/NANOG expression signature may serve as a useful biomarker for prostate cancer prognosis in patients.
In summary, our findings demonstrate that prostate epithelial cell reprogramming can be used to identify key factors that contribute to cancer cellular plasticity and progression in vitro and in vivo. Our results demonstrate a previously unrecognized mechanism by which ablation of Tgfbr2 in Pten-null prostate cells eliminates Smad2/3-mediated TGF-β signaling and Tmeff1 expression and increases BMP signaling pathways, which in turn, initiates a cascade of signaling events that activates multiple signaling pathways, ultimately leading to reactivation of pluripotency genes and EMT-related genes. These signaling pathways in PtenPKO:Tgfbr2PKO prostate cells act in concert to accelerate cancer development and progression. The ID1/STAT3/NANOG expression signature can be used as a useful biomarker for the prognosis of patient survival.
Materials and methods
Animals
Pten flox/flox mice were originally obtained from Dr Hong Wu (Wang et al., 2003) and crossed with Pb-Cre (PB4-Cre) mice (NCI Mouse Repository) to obtain conditional Pten knockout mice on a C57BL/6 background. Tgfbr2flox/flox mice were obtained from NCI Mouse Repository (Chytil et al., 2002). To generate Ptenflox/flox:Tgfbr2flox/flox:Pb-Cre mouse, Ptenflox/flox:Pb-Cre mice were bred with Tgfbr2flox/flox mice. Tet-O-Sox2 and Tet-O-Klf4 transgenic mice were generated and crossed with Rosa-rtTA, Tet-O-Oct4 (from the Jackson Laboratories), and Tet-O-Myc transgenic mice. Tet-O-4F MEFs that express rtTA and Tet-O-Oct4, Sox2, Klf4, and Myc were established and used for reprogramming. Animal experiments were performed using 8- to 12-week-old male mice.
Generation of iPSCs from Tet-O-OSKM MEFs and TRAMP-C3 cells
Tet-O-OSKM MEFs were used to generate iPSCs by treating MEFs with doxycycline in mouse ESC medium. The efficiency of iPSC formation was calculated based on the number of alkaline phosphatase (AP)+ iPSC colonies and the initial cell number of seeded MEFs. Mouse iPSCs were generated from TRAMP-C3 cells (directly from ATCC in June 2008, where cell lines were authenticated by short tandem repeat profiling) as previously described (Takahashi and Yamanaka, 2006), with minor modifications.
Co-IP and ChIP−PCR analysis
Co-IP was performed as described previously (He et al., 2015). Briefly, cells were lysed with 1× RIPA buffer containing a protease inhibitor cocktail. Extracts were incubated overnight with anti-Flag M2 affinity resin (A2220, Sigma) or anti-Tmeff1 antibody. The mixture was thoroughly washed with 1× RIPA buffer and eluted with 1× SDS sample buffer. The ChIP assay was performed using the Imprint Ultra Chromatin Immunoprecipitation kit (Sigma), as previously described (Zhao et al., 2013). See Supplementary Materials and methods for details.
Statistics
All analyses were performed using GraphPad Prism version 5.0 (GraphPad Software). Data are presented as mean ± SD, unless otherwise stated. Statistical significance of differences between two groups was assessed using an unpaired Student’s t-test and a P-value < 0.05 was considered significant.
Ethics statement
All in vivo experiments in mice were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine and Houston Methodist Research Institute.
Supplementary Material
Acknowledgements
We would like to thank Dr Hong Wu at UCLA for providing Ptenflox/flox mice and Dr Dan Liu at Baylor College of Medicine for assistance with shRNA screening. We also thank Jana Burchfield for her editing. W.L. was supported by Xiangya Hospital, Central South University, China.
Funding
This work was supported by grants from the National Key Research and Development Program of China (2017YFA0103800), the National Natural Science Foundation of China (81572766 and 31771630), Guangdong Innovative and Entrepreneurial Research Team Program (2016ZT06S029), Guangdong Natural Science Foundation (2016A030313215 and 2016A030313238), SYSU Young Teachers Training Program (16YKZD14), the National Cancer Institute (NCI), the National Institutes of Health (NIH) (R01CA090327 and R01CA101795), and the Cancer Prevention and Research Institute of Texas (CPRIT) (RP170537).
Conflict of interest: none declared.
Author contributions: W.Z., Q.Z., and P.T. designed and performed the experiments and wrote the manuscript. A.A. and Q.L. performed the animal experiments. T.L. and W.L. performed IHC staining and immunofluorescence staining. P.L. performed shRNA screening. B.N. and H.Y.W. conducted bioinformatics analysis of patient data. R.W. designed experiments, interpreted data, supervised the project, and wrote the manuscript.
References
- Abdulghani J., Gu L., Dagvadorj A., et al. (2008). Stat3 promotes metastatic progression of prostate cancer. Am. J. Pathol. 172, 1717–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolou E., and Hochedlinger K. (2013). Chromatin dynamics during cellular reprogramming. Nature 502, 462–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck B., Lapouge G., Rorive S., et al. (2015). Different levels of Twist1 regulate skin tumor initiation, stemness, and progression. Cell Stem Cell 16, 67–79. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I., Thomson M.W., Carey V.J., et al. (2008). An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 40, 499–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerke G.A., Yang C.S., Frierson H.F., et al. (2014). Activation of Akt signaling in prostate induces a TGFβ-mediated restraint on cancer progression and metastasis. Oncogene 33, 3660–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Trotman L.C., Shaffer D., et al. (2005). Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 436, 725–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chytil A., Magnuson M.A., Wright C.V., et al. (2002). Conditional inactivation of the TGF-β type II receptor using Cre:Lox. Genesis 32, 73–75. [DOI] [PubMed] [Google Scholar]
- Ding Z., Wu C.J., Chu G.C., et al. (2011). SMAD4-dependent barrier constrains prostate cancer growth and metastatic progression. Nature 470, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedmann-Morvinski D., and Verma I.M. (2014). Dedifferentiation and reprogramming: origins of cancer stem cells. EMBO Rep. 15, 244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L., Zhang Y., Ma G., et al. (2015). Near-infrared photoactivatable control of Ca2+ signaling and optogenetic immunomodulation. eLife 4, e10024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida J.K., Blanchard J., Lam K., et al. (2009). A small-molecule inhibitor of Tgf-β signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 5, 491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence M.S., Stojanov P., Mermel C.H., et al. (2014). Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 505, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.F., Su J., Kim H.S., et al. (2015). Modeling familial cancer with induced pluripotent stem cells. Cell 161, 240–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., et al. (2010). A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63. [DOI] [PubMed] [Google Scholar]
- Liao J., Marumoto T., Yamaguchi S., et al. (2013). Inhibition of PTEN tumor suppressor promotes the generation of induced pluripotent stem cells. Mol. Ther. 21, 1242–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling M.T., Wang X., Ouyang X.S., et al. (2003). Id-1 expression promotes cell survival through activation of NF-κB signalling pathway in prostate cancer cells. Oncogene 22, 4498–4508. [DOI] [PubMed] [Google Scholar]
- Massague J. (2008). TGFβ in cancer. Cell 134, 215–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K., Semi K., Yamamoto T., et al. (2014). Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell 156, 663–677. [DOI] [PubMed] [Google Scholar]
- Orkin S.H., and Hochedlinger K. (2011). Chromatin connections to pluripotency and cellular reprogramming. Cell 145, 835–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimori N., and Fuchs E. (2012). Paracrine TGF-β signaling counterbalances BMP-mediated repression in hair follicle stem cell activation. Cell Stem Cell 10, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.P., Tirosh I., Trombetta J.J., et al. (2014). Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science 344, 1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattabiraman D.R., and Weinberg R.A. (2014). Tackling the cancer stem cells – what challenges do they pose? Nat. Rev. Drug Discov. 13, 497–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P., Golipour A., David L., et al. (2010). Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77. [DOI] [PubMed] [Google Scholar]
- Scheel C., and Weinberg R.A. (2012). Cancer stem cells and epithelial–mesenchymal transition: concepts and molecular links. Semin. Cancer Biol. 22, 396–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlomm T., Iwers L., Kirstein P., et al. (2008). Clinical significance of p53 alterations in surgically treated prostate cancers. Mod. Pathol. 21, 1371–1378. [DOI] [PubMed] [Google Scholar]
- Singovski G., Bernal C., Kuciak M., et al. (2016). In vivo epigenetic reprogramming of primary human colon cancer cells enhances metastases. J. Mol. Cell Biol. 8, 157–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suva M.L., Riggi N., and Bernstein B.E. (2013). Epigenetic reprogramming in cancer. Science 339, 1567–1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki A., Raya A., Kawakami Y., et al. (2006). Nanog binds to Smad1 and blocks bone morphogenetic protein-induced differentiation of embryonic stem cells. Proc. Natl Acad. Sci. USA 103, 10294–10299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., and Yamanaka S. (2006). Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676. [DOI] [PubMed] [Google Scholar]
- Theriault B.L., Shepherd T.G., Mujoomdar M.L., et al. (2007). BMP4 induces EMT and Rho GTPase activation in human ovarian cancer cells. Carcinogenesis 28, 1153–1162. [DOI] [PubMed] [Google Scholar]
- Timp W., and Feinberg A.P. (2013). Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer 13, 497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Gao J., Lei Q., et al. (2003). Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell 4, 209–221. [DOI] [PubMed] [Google Scholar]
- Ying Q.L., Nichols J., Chambers I., et al. (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT3. Cell 115, 281–292. [DOI] [PubMed] [Google Scholar]
- Zhao W., Li Q., Ayers S., et al. (2013). Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell 152, 1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Kavsak P., Abdollah S., et al. (1999). A SMAD ubiquitin ligase targets the BMP pathway and affects embryonic pattern formation. Nature 400, 687–693. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.