Abstract
A newly isolated bacterial strain SHJ was found to be capable of degrading diethyl phthalate (DEP) very efficiently. Its growth characteristics and 16S rDNA gene sequence were analyzed. Its whole genome was also sequenced. Strain SHJ was identified as Sphingobium yanoikuyae SHJ.
Keywords: Sphingobium yanoikuyae, Gene sequence, Genome, Diethyl phthalate, Biodegradation
Specifications table
| Subject area | Biology |
| More specific subject area | Microbial characterization, identification and phylogenetic analysis |
| Type of data | Table, figure |
| How data was acquired | Microscope, SEM, DNA sequencing, bioinformatics |
| Data format | Raw, analyzed and deposited |
| Experimental factors | Strain SHJ was cultured for observation and 16S rDNA gene sequencing analysis |
| Experimental features | A new microbe was isolated, cultured, observed under a scanning electron microscope. The morphology of its colonies on agar plate was described. Its 16S rDNA gene was sequenced, for which phylogenetic analysis was performed. |
| Data source location | Sample was collected at 30°28′19″N, 113°59′13″E (longitude, latitude), Wuhan, Hubei, China |
| Data accessibility | With this article, GenBank accession number JFFT01000000, DDBJ/EMBL/GenBank under the accession JFFT00000000 |
Value of the data
-
•
The whole genome sequence data of strain SHJ is available by its accession number.
-
•
Characterization and identification of the newly isolated Sphingobium yanoikuyae SHJ.
-
•
Biodiversity with capability of bio-remediating phthalate esters-contaminated aquifer.
1. Data
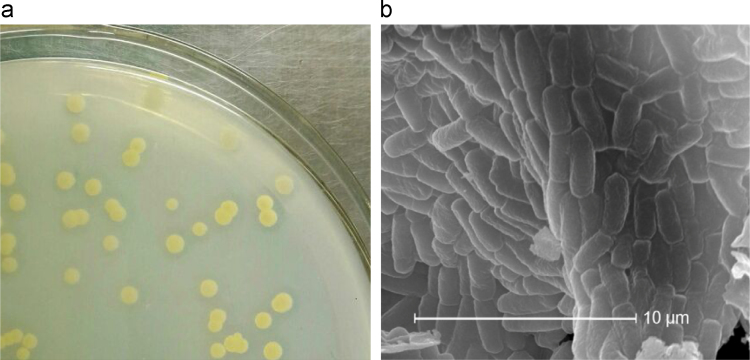
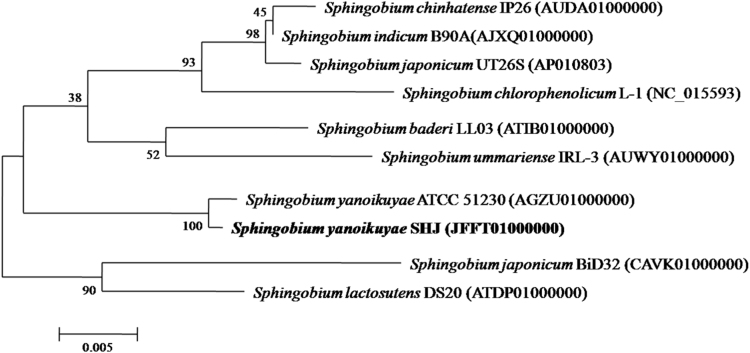
A new bacterium strain SHJ was isolated from the shallow aquifer sediment of Jianghan plain, Hubei, China. It grew on NB agar plate containing 400 mg L−1 DEP as sole carbon source and appeared to be yellow colony (Fig. 1a), and it was observed to be short rod under a scanning electron microscope (Fig. 1b). It was found to be capable of degrading DEP very efficiently under simulated shallow aquifer (SSA) conditions which are dark, oxygen-limited, at pH 7 and 18 °C [1]. However, the most well-known DEP-degrading bacterial isolates that are purely aerobic are listed in Table 1. Classification and general features of the strain SHJ were listed in Table 2. Its 16S rDNA gene sequence (GenBank accession number JFFT01000000) showed the highest similarity with Sphingobium yanoikuyae ATCC 51230 (Fig. 2). Therefore, strain SHJ was classified as Sphingobium yanoikuyae SHJ. The Whole Genome Shotgun project of S. yanoikuyae SHJ has been deposited at DDBJ/EMBL/GenBank under the accession JFFT00000000 and the release date of its GenBank Data is February 28, 2017.
Fig. 1.
The growth of strain SHJ on NB agar plate containing 400 mg L−1 DEP (a) and its cell morphology under a scanning electron microscope (b).
Table 1.
Several DEP-degrading bacterial strains isolated from various environments.
| Species | Isolation | DEP (mg L−1) | Performance | References |
|---|---|---|---|---|
| Bacillus subtilis 3C3 | Soil | 100 | 60% after 24 h | Navacharoen et al. [2] |
| Bacillus thuringiensis | Agricultural soil | 400 | 88% after 80 h | Surhio et al. [3] |
| Rhodococcus sp. L4 | Activated sludge | 100 | 100% after 6 days | Lu et al. [4] |
| Mycobacterium sp YC-RL4 | Petroleum-contaminated soil | 50 | 100% after 5 days | Ren et al. [5] |
| Acinetobacter sp. LMB-5 | Vegetable greenhouse soil | 100 | 95% after 45 h | Fang et al. [6] |
| Acinetobacter sp. JDC-16 | River sludge | 500 | 100% after 27 h | Liang et al. [7] |
| Pseudomonas fluoresences FS1 | Activated sludge at a petrochemical factory | 100 | 100% after 36 h | Zeng et al. [8] |
| Pleurotus ostreatus | Forest soil | 100 | 100% after 8 days | Hwang et al. [9] |
| Gordonia alkanivorans YC-RL2 | Petroleum-contaminated soil | 100 | 100% after 7 days | Nahurira et al. [10] |
| Sphingomonas sp. C28242 | Activated sludge | 450 | 100% after 120 h | Fang et al. [6] |
| Sphigomonas sp. DK4 | River sediment | 100 | 56% after 7 days | Chang et al. [11] |
| Corynebacterium sp.O18 | Petrochemical sludge | 100 | 100% after 7 days | Chang et al. [11] |
Table 2.
Classification and general features of Sphingobium yanoikuyae SHJ according to the MIGS (miRNA-induced gene silencing) recommendation.
| MIGS ID | Property | Term | Evidence codea |
|---|---|---|---|
| Current classification | Domain Bacteria | TAS [12] | |
| Phylum Proteobacteria | TAS [13] | ||
| Class Alphaproteobacteria | TAS [14] | ||
| Order Sphingomonadales | TAS [15] | ||
| Family Sphingomonadaceae | TAS [16] | ||
| Genus Sphingobium | TAS [17] | ||
| Species yanoikuyae | TAS [17] | ||
| Gram stain | Gram-negative | IDA | |
| Cell shape | Short rod-shaped | IDA | |
| Motility | Non-motile | IDA | |
| Sporulation | Non-spore-forming | IDA | |
| Temperature range | 13–30 °C | IDA | |
| Optimum temperature | 28 °C | IDA | |
| pH range; Optimum | 6–9;6.8 | IDA | |
| Carbon source | L-arabinose, D-xylose, galactose, Salicin, mannose, D-turanose, and caprate | TAS [17] | |
| Energy source | Chemoheterotrophic | TAS [17] | |
| MIGS-6 | Habitat | Sediments | IDA |
| MIGS-6.3 | Salinity | Slight Halophilic | IDA |
| MIGS-22 | Oxygen | Facultative aerobe | IDA |
| MIGS-15 | Biotic relationship | Free living | IDA |
| MIGS-14 | Pathogenicity | None | NAS |
| MIGS-4 | Geographic location | Caidian District, Wuhan, Hubei, China | IDA |
| MIGS-5 | Sample collection time | 2008 | IDA |
| MIGS-4.1 | Latitude | 30°28′19″ N | NAS |
| MIGS-4.2 | Longitude | 113°59′13″ E | NAS |
| MIGS-4.3 | Depth | 2.2 m | NAS |
| MIGS-4.4 | Altitude | 24 m | NAS |
Evidence codes - IDA: Inferred from Direct Assay; TAS: Traceable Author Statement (i.e., a direct report exists in the literature); NAS: Non-traceable Author Statement (i.e., not directly observed for the living, isolated sample, but based on a generally accepted property for the species, or anecdotal evidence). These evidence codes are from the Gene Ontology project.
Fig. 2.
Phylogenetic analysis of 16S rDNA sequences. The tree was built using the maximum-likelihood method with the Hasegawa-Kishino-Yano model assuming non-uniformity of evolutionary rates among sites (https://www.megasoftware.net/index.php). Bootstrap analysis with 1000 replicates was performed to assess the support of the clusters. The corresponding GenBank accession numbers are displayed in parentheses.
2. Experimental design, materials and methods
2.1. Chemicals and reagents
DEP was purchased from Tianjin Hengxing Chemical Reagent Co., Ltd., China. DEP standard solutions were prepared at various concentrations in methanol and kept in dark at 4 °C.
2.2. DEP-degrading bacterial strain
The DEP-degrading strain SHJ was isolated from the sediments collected from the quaternary shallow aquifer from a depth of 2.2 m in Jianghan Plain, Hubei, China, with a precise GPS location of 30°28′19″N, 113°59′13″E (longitude, latitude). The strain SHJ was grown using the method described previously [18]. It was pre-grown for 24 h at pH 7.2 and 30 °C in nutrient broth (NB), which contained peptone 5 g L−1, beef extract 3 g L−1, NaCl 5 g L−1. Nutrient agar plates were prepared using NB supplemented with agar (1.5%). NB-DEP agar plate was prepared by diffusing 400 mg L−1 DEP solution into the nutrient agar medium. All media were sterilized for 20 min at 121 °C before inoculation. Detection and identification of DEP degradation intermediates ethyl methyl phthalate (EMP), monoethyl phthalate (MEP), monomethyl phthalate (MMP) and phthalic acid (PA) was carried out as described previously [1].
2.3. Identification of strain SHJ
Colonies of the strain SHJ on NB agar plate were picked for Gram staining, and the morphology of the strain was observed using an optical microscope.
Microbial identification and phylogenetic analysis of strain SHJ were performed by 16S rDNA gene sequencing. One ml overnight culture of bacterium grown in NB media in a rotary shaker (150 rpm) at 30 °C was centrifuged at 6000×g for 10 min. The cells obtained were washed three times using sterile water and re-suspended in sterile water. Genome DNA was extracted from the isolate using UltraClean® Microbial DNA Isolation Kit (MoBio, USA) according to the manufacturer׳s protocol. 16S rDNA gene of the strain SHJ was amplified from its genomic DNA by using PCR procedures [18]. The bacterial universal primers F27 and R1492 were used for amplifying the full length of 16S rRNA gene fragments. The Shanghai Personal Biotechnology Co., Ltd performed the sequencing and assembly of strain SHJ using Illumina MiSeq sequencing platform, and gene prediction and annotation were completed using National Center for Biotechnology Information (NCBI) Prokaryotic Genome Annotation Pipeline (PGAP, https://www.ncbi.nlm.nih.gov/genome/annotation_prok/) [1]. The 16S rDNA gene sequence of strain SHJ was searched against GenBank database under the accession JFFT00000000 using BLASTn at the website of NCBI (http://www.ncbi.nlm.nih.gov/BLAST/). Based on the 16S rDNA gene sequences obtained, phylogenetic analysis of strain SHJ was performed by molecular evolutionary genetics analysis (MEGA 6, https://www.megasoftware.net/index.php) after all sequences alignment by using Clustal W (https://www.ebi.ac.uk/Tools/msa/clustalw2/).
Acknowledgements
This research was supported by the Grant for Innovative Research Groups of the National Natural Science Foundation of China (41521001) and the research project of National Natural Science Foundation of China (Grant No. 41073069). And we would like to thank the Shanghai Personal Biotechnology Co., Ltd for performing the sequencing and assembly of the whole genome sequence.
Footnotes
Transparency data associated with this article can be found in the online version at 10.1016/j.dib.2018.09.033.
Transparency document. Supplementary material
Supplementary material.
.
References
- 1.Wang Y., Liu H., Peng Y.E., Tong L., Feng L., Ma K. New pathways for the biodegradation of diethyl phthalate by Sphingobium yanoikuyae SHJ. Process Biochem. 2018;71C:152–158. [Google Scholar]
- 2.Navacharoen A., Vangnai A.S. Biodegradation of diethyl phthalate by an organic-solvent-tolerant Bacillus subtilis strain 3C3 and effect of phthalate ester coexistence. Int. Biodeterior. Biodegrad. 2011;65(6):818–826. [Google Scholar]
- 3.Surhio M.A., Talpur F.N., Nizamani S.M., Talpur M.K., Amin F., Khaskheli A.A., Bhurgri S., Afridi H.I., Rahman S.U. Effective bioremediation of endocrine-disrupting phthalate esters, mediated by Bacillus strains. Water Air Soil Pollut. 2017;228(10) [Google Scholar]
- 4.Lu Y., Tang F., Wang Y., Zhao J.H., Zeng X., Luo Q.F., Wang L. Biodegradation of dimethyl phthalate, diethyl phthalate and di-n-butyl phthalate by Rhodococcus sp L4 isolated from activated sludge. J Hazard Mater. 2009;168(2–3):938–943. doi: 10.1016/j.jhazmat.2009.02.126. [DOI] [PubMed] [Google Scholar]
- 5.Ren L., Jia Y., Ruth N., Qiao C., Wang J.H., Zhao B.S., Yan Y.C. Biodegradation of phthalic acid esters by a newly isolated Mycobacterium sp YC-RL4 and the bioprocess with environmental samples. Environ. Sci. Pollut. Res. 2016;23(16):16609–16619. doi: 10.1007/s11356-016-6829-4. [DOI] [PubMed] [Google Scholar]
- 6.Fang H.H.P., Liang D.W., Zhang T. Aerobic degradation of diethyl phthalate by Sphingomonas sp. Bioresour. Technol. 2007;98(3):717–720. doi: 10.1016/j.biortech.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Liang R.X., Wu X.L., Wang X.N., Dai Q.Y., Wang Y.Y. Aerobic biodegradation of diethyl phthalate by Acinetobacter sp JDC-16 isolated from river sludge. J. Cent. South Univ. Technol. 2010;17(5):959–966. [Google Scholar]
- 8.Zeng F., Cui K., Li X., Fu J., Sheng G. Biodegradation kinetics of phthalate esters by Pseudomonas fluoresences FS1. Process Biochem. 2004;39(9):1125–1129. [Google Scholar]
- 9.Hwang S.S., Choi H.T., Song H.G. Biodegradation of endocrine-disrupting phthalates by Pleurotus ostreatus. J. Microbiol. Biotechnol. 2008;18(4):767–772. [PubMed] [Google Scholar]
- 10.Nahurira R., Ren L., Song J.L., Jia Y., Wang J.H., Fan S.H., Wang H.S., Yan Y.C. Degradation of Di(2-ethylhexyl) Phthalate by a Novel Gordonia alkanivorans Strain YC-RL2. Curr. Microbiol. 2017;74(3):309–319. doi: 10.1007/s00284-016-1159-9. [DOI] [PubMed] [Google Scholar]
- 11.Chang B., Yang C., Cheng C., Yuan S. Biodegradation of phthalate esters by two bacteria strains. Chemosphere. 2004;55(4):533–538. doi: 10.1016/j.chemosphere.2003.11.057. [DOI] [PubMed] [Google Scholar]
- 12.Woese C.R., Kandler O., Wheelis M.L. Towards a natural system of organisms: proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA. 1990;87(12):4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.G.M. Garrity, T.G. Lilburn, J.R. Cole, The taxonomic outline of bacteria and archaea, 2007.
- 14.G.M. Garrity, J.A. Bell, T. Lilburn, Class I. Alphaproteobacteria class. nov1-574, 2005.
- 15.Garrity G.M., Holt J.G. The road map to the manual. In: Boone David R., Castenholz Richard W., editors. Bergey’s Manual® of Systematic Bacteriology. 2001. pp. 119–166. [Google Scholar]
- 16.Kosako Y., Yabuuchi E., Naka T., Fujiwara N., Kobayashi K. Proposal of Sphingomonadaceae Fam. Nov., Consisting of Sphingomonas Yabuuchi et al. 1990, Erythrobacter Shiba and Shimidu 1982, Erythromicrobium Yurkov et al. 1994, Porphyrobacter Fuerst et al. 1993, Zymomonas Kluyver and van Niel 1936, and Sandaraci. Microbiol. Immunol. 2000;44(7):563–575. doi: 10.1111/j.1348-0421.2000.tb02535.x. [DOI] [PubMed] [Google Scholar]
- 17.Yabuuchi E., Yano I., Oyaizu H., Hashimoto Y., Ezaki T., Yamamoto H. Proposals of Sphingomonas paucimobilis gen. nov. and comb. nov., Sphingomonas parapaucimobilis sp. nov., Sphingomonas yanoikuyae sp. nov., Sphingomonas adhaesiva sp. nov., Sphingomonas capsulata comb. nov., and two genospecies of the genus Sphingomonas. Microbiol. Immunol. 1990;34(2):99–119. doi: 10.1111/j.1348-0421.1990.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 18.Wu Q., Liu H., Ye L.S., Li P., Wang Y.H. Biodegradation of di-n-butyl phthalate esters by Bacillus sp SASHJ under simulated shallow aquifer condition. Int. Biodeterior. Biodegrad. 2013;76:102–107. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.