Abstract
Background
Helicobacter pylori (H. pylori) infection is associated with remodeling of gastric microbiota. However, comprehensive analyses of the impact of H. pylori infection, eradication therapy and probiotic supplementation on gut microbiota are still lacking. We aimed to provide evidence for clinical decision making.
Methods
Seventy H. pylori-positive and 35 H. pylori-negative patients (group C) were enrolled. H. pylori-positive patients were randomly assigned to group A (14-day bismuth-containing quadruple therapy) and group B (quadruple therapy supplemented with Clostridium butyricum). Stool samples of group A and B were collected on day 0, 14 and 56 while stool samples of group C were collected on day 0. Gut microbiota was investigated by 16S rRNA sequencing.
Findings
The Sobs index (richness estimator) was significantly higher in H. pylori-positive samples than H. pylori-negative samples (p < .05). Several metabolic pathways were more abundant in H. pylori-positive communities while some disease-associated pathways had higher potential in H. pylori-negative community through KEGG pathway analysis. Abundances of most butyrate-producing bacteria significantly decreased, while several detrimental bacteria increased after eradication therapy. Probiotic supplementation was associated with improved gastrointestinal symptoms as well as increased Bacteroidetes:Firmicutes ratio.
Interpretation
While H. pylori infection may not be necessarily detrimental in all patients, eradication of H. pylori was associated with widespread changes in gut microbial ecology and structure. Probiotic supplementation could relieve more gastrointestinal symptoms by inducing alterations in gut microbiota and host immune responses. As such, the decision to eradicate H. pylori should be based on comprehensive analysis of individual patients.
Keywords: Helicobacter pylori, Gut microbiota, 16S rRNA sequencing, Clostridium butyricum, Bismuth-containing quadruple therapy
Research in context.
Evidence before this study
Infection with H. pylori can lead to widespread changes in gastric microbial community. Animal studies have indicated that H. pylori results in distinct shifts in gut microbiota in uninflamed distal parts of the gastrointestinal tract but not in the stomach. Similarly, in human studies using fluorescence in situ hybridization, fecal samples from H. pylori-infected individuals showed a decrease in abundance of clostridia as well as total anaerobes compared with H. pylori-negative individuals.
Approximately 20% of H. pylori-infected individuals go on to develop complications. A study in Japan reported an increase in Bacteroidetes, Firmicutes, and B:F ratio together with a decrease in Proteobacteria immediately after eradication treatment.
Preclinical data suggest that a butyrate-producing probiotic, Clostridium butyricum, shows promise in treating H. pylori. However, data from human studies were mixed. In addition, these studies were conducted with standard triple eradication therapy, which is likely inadequate in most Asian populations.
Added value of this study
Our results suggest that the nitrate-nitrite-NO pathway may play an important regulatory mechanism in pathologic conditions and may be protective against H. pylori. Relative abundance of 19 disease-associated or metabolic pathways were significantly different between H. pylori-negative and H. pylori-positive patients by KEGG pathway analyses.
Abundances of most butyrate-producing bacteria significantly decreased, and several detrimental bacteria increased immediately after therapy. Longitudinal studies are required to determine the consequences of antibiotic-induced disruption of the gut microbiota.
Supplementation with C. butyricum could relieve gastrointestinal symptoms by inducing alterations in gut microbiota and host immune responses.
Implications of all the available evidence
The interactions between H. pylori, gut microbiota, and host function are complex. The role H. pylori plays in human disease and gut microenvironment homeostasis may not be necessarily detrimental.
The decision to eradicate and application of probiotics should be based on comprehensive analyses of individual patients.
Alt-text: Unlabelled Box
1. Introduction
An estimated 4.4 billion individuals were infected with Helicobacter pylori (H. pylori) worldwide in 2015 [1]. In China, the prevalence of H. pylori infection was 66% among rural populations and 47% in urban settings [2]. In infected patients, H. pylori is the dominant bacterial species in the gastric microbiota [3]. Infection with H. pylori can inhibit gastric acid secretion, induce chronic inflammation of gastric mucosa, and thereby change the gastric microenvironment leading to widespread changes in gastric microbial community [4,5]. In addition, alterations in gut microbiota are associated with a range of gastrointestinal and systemic diseases [6]. Although the stomach has been reported as the exclusive habitat for H. pylori [7], it has been detected through 16S rRNA sequencing in stool samples albeit with low relative abundance [8]. Moreover, animal studies have indicated that H. pylori results in distinct shifts in gut microbiota in uninflamed distal parts of the gastrointestinal tract but not in the stomach [9]. Similarly in human studies using fluorescence in situ hybridization, fecal samples from H. pylori-infected individuals showed a decrease in abundance of clostridia as well as total anaerobes compared with H. pylori-negative individuals [10]. These observations suggest that infection with H. pylori leads to widespread changes in host microbial structure and function likely through alterations in the gastric microenvironment. Despite its widespread prevalence, potential for inducing chronic sequelae, and impact on host-microbe interactions, the interactions between H. pylori, gut microbiota, and host function are still largely unknown.
According to the Kyoto global consensus report, patients diagnosed with H. pylori infection should receive eradication therapy to minimize risk of long-term sequelae, including peptic ulcer disease, gastric adenocarcinoma, and gastric mucosa-associated lymphoid tissue (MALT) lymphoma [11]. However, current evidence indicates that eradication with H. pylori is associated with major disturbances of the intestinal microbiota. This included a decrease in bacterial diversity as well as reductions in number of Bifidobacteria, Lactobacilli, and butyrate producers, such as Faecalibacterium prausnitzii [10]. Another study documented that these changes may persist for up to four years after antibiotic treatment is completed [12]. Given that only approximately 20% of H. pylori-infected individuals go on to develop complications [13] and the accumulating evidence showing the potential harm of antibiotic administration, there is a pressing need to more fully elucidate the complex H. pylori-gut microbiota-host interactions to identify those subjects most at risk for long-term sequelae.
Moreover, supplementation of certain probiotics may have a positive effect on H. pylori eradication by immunological and non-immunological mechanisms [14]. A meta-analysis of 14 randomized trials demonstrated that probiotics increased eradication rate (OR 1.84, 95% CI 1.34–2.54) while decreasing adverse events (OR 0.44, 95% CI 0.30–0.66) [15]. Butyrate is a major short chain fatty acid produced by bacterial fermentation of dietary fibers. In addition to being a major energy source for colonocytes, butyrate has been shown to promote mucosal homeostasis likely through beneficial effects on innate and adaptive immune cells as well as epithelial barrier function [16,17]. Butyrate may also have bactericidal effects against H. pylori. In vitro studies using butyrate as well as supernatants from butyrate-producing bacteria inhibited growth and were associated with destructive effects on the cell envelope of H. pylori [18]. Preclinical data suggest that a butyrate-producing probiotic, Clostridium butyricum (C. butyricum), shows promise in treating H. pylori [19]. However, data from human studies were mixed [20,21]. This may be partially explained by prior studies emphasizing the effect of C. butyricum on eradication rate and side effects. In addition, these studies were conducted with standard triple eradication therapy, which is likely inadequate in most Asian populations. Meanwhile, studies of gut microbial alterations were performed using culture-based rather than with more modern metagenomic approaches.
As such, we aimed to comprehensively investigate the impact of H. pylori infection, 14-day bismuth-containing quadruple therapy (BQT) and probiotic supplementation on gut microbial homeostasis.
2. Materials and methods
2.1. Patients
This open-label, prospective clinical trial was performed at Sir Run Run Shaw Hospital, Zhejiang Province, China from December 2016 to August 2017. Patients aged between 18 and 70 years were enrolled. Patients were randomly assigned to treatment groups if they were diagnosed as H. pylori-positive gastritis in the past month by esophagogastroduodenoscopy and histological examination. Subjects who tested negative for H. pylori by esophagogastroduodenoscopy as well as 13C urea breath test were recruited as controls. Exclusion criteria included prior history of treatment for H. pylori infection; confirmed or suspected upper gastrointestinal malignant tumor; peptic ulcer or other upper gastrointestinal lesions; the use of antacids or gastric mucosal protective agents in the past two weeks; the use of antibiotics or probiotics in the past month; known allergy to drugs in this study; other known gastrointestinal diseases; history of gastrointestinal surgery; decompensated cardiac, liver, renal, or pulmonary illness; thyroid disease or diabetes mellitus; pregnant and lactating women; and subjects who could not provide informed consent. This study was approved by the Ethics Committee of Sir Run Shaw Hospital, College of Medicine, Zhejiang University (20161206-21) and registered at Chinese Clinical Trial Registry (Chictr.org.cn, ChiCTR-IPR-16010286). Written informed consent was obtained from all participants before enrollment.
2.2. Study design
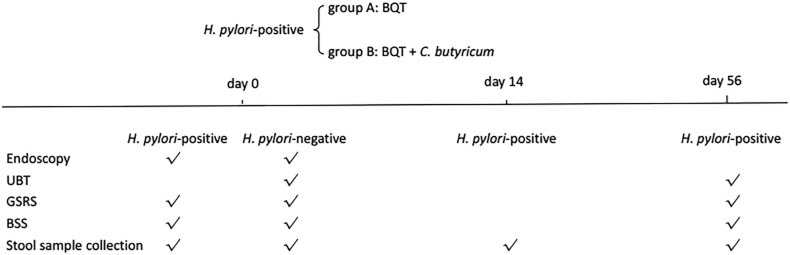
A total of 70 H. pylori-positive patients and 35 H. pylori-negative patients were enrolled. H. pylori-positive patients were randomly assigned to group A and group B. Randomization sequence was generated by a computer algorithm. Patients in group A received 14-day BQT consisting of pantoprazole 40 mg, amoxicillin 1000 mg, furazolidone 100 mg, colloidal bismuth pectin 0.4 g, all twice a day. This eradication therapy was chosen due to low rates of antibiotic resistance to amoxicillin (0.1%) and furazolidone (0.1%) in southeast China [22]. Patients in group B received BQT supplemented with C. butyricum 40 mg three times a day (CBM588, MIYA-BM® tablets, Miyarisan Pharmaceutical, Co., Ltd., Tokyo, Japan) for 14 days. Gastrointestinal symptoms and stool form/frequency were assessed by the Gastrointestinal Symptom Rating Scale (GSRS) [23] and Bristol Stool Scale (BSS) [24] at baseline for all subjects and on day 56 for H. pylori-positive patients. H. pylori-positive patients also underwent 13C urea breath test on day 56 to assess eradication (Fig. 1).
Fig. 1.
Study design. BQT, bismuth-containing quadruple therapy; UBT, 13C urea breath test; GSRS, gastrointestinal symptom rating scale; BSS, Bristol stool scale.
2.3. Stool sample collection
Fresh stool samples were collected from all subjects at baseline. Additional fresh stool samples were collected prospectively from H. pylori-positive patients on day 14 and day 56. All stool samples were immediately frozen and stored at −80 °C.
2.4. DNA isolation, PCR amplification and sequencing
Microbial DNA from stool samples was isolated with TIANamp Stool DNA Kit (TIANGEN BIOTECH, cat. #DP328-02, Beijing, China) according to the manufacturer's instructions. DNA concentration and purification were measured by Nanodrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was checked by 1% agarose gel electrophoresis. The V3-V4 hypervariable regions of 16S rRNA gene were amplified using barcoded primers 338F 5′-ACTCCTACGGGAGGCAGCAG-3′ and 806R 5′-GGACTACHVGGGTWTCTAAT-3′ by thermocycler PCR system (GeneAmp 9700, ABI, USA). The PCR reactions were conducted using the following program: 3 min of denaturation at 95 °C, 27 cycles of 30s at 95 °C, 30s for annealing at 55 °C, and 45 s for elongation at 72 °C, and a final extension at 72 °C for 10 min. PCR reactions were performed in triplicate 20 μL mixture containing 4 μL of 5 × FastPfu Buffer, 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase and 10 ng of template DNA. The amplicons were then extracted from 2% agarose gels, purified by AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA), and quantified by QuantiFluor™ -ST (Promega, USA).
Purified amplicons were pooled in equimolar and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, USA) according to the standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China).
2.5. Taxonomy determination
Raw fastq files were demultiplexed and quality-filtered by Trimmomatic and merged by FLASH (Fast Length Adjustment of Short Reads to Improve Genome Assemblies). A sliding window size of 50 base pair (bp) was set and reads with quality score < 20 were truncated from the 3′ end. Next, paired-end reads were merged with a minimum overlap length of 10 bp and a maximum mismatch rate of 20%. No barcode mismatches were allowed, and primers were exactly matched allowing 2 nucleotide mismatching. Based on the barcodes and primers, samples were identified, and the orientations of sequences were adjusted. Sequences were then de-replicated, and singletons were discarded. Operational Taxonomic Units (OTUs) were clustered with 97% similarity cutoff using UPARSE (version 7.1 http://drive5.com/uparse/). Chimeric sequences were identified and removed using UCHIME. The taxonomy of 16S rRNA gene sequence was determined by Ribosomal Database Project(RDP) Classifier algorithm (version 2.2 http://rdp.cme.msu.edu/) with a confidence threshold of 70%, using Silva (Release128 http://www.arb-silva.de) as the taxonomy reference database. Data were analyzed using the software Quantitative Insights Into Microbial Ecology (QIIME, version 1.9.1).
2.6. Functional predictions
The 16S rRNA functional prediction is to normalize the OTU abundance table through PICRUSt (Phylogenetic Investigation of Communities by Reconstruction of Unobserved States) [25], that is, to remove the influence of 16S marker gene copy number. Then OTUs were categorized into Clusters of Orthologous Groups (COG) [26] and into Kyoto Encyclopedia of Genes and Genome (KEGG) orthology (KO) [27].
According to the COG database, the descriptive information of each COG and its functional information were parsed from the eggNOG database to obtain the functional abundance spectrum. KO, Pathway and Enzyme (EC) information were obtained according to the KEGG database while the abundance of each functional category was calculated according to OTU abundance.
2.7. Statistical analysis
Baseline continuous data were presented as mean ± standard deviation (SD) and were analyzed by one-way ANOVA. Categorical data were described in percentage and compared by χ2 test or Fisher's exact test. All efficacy analyses were performed on an intention-to-treat (ITT) population where patients who dropped out were considered as treatment failures. Secondary per-protocol (PP) analyses were performed which excluded patients lost to follow-up or prematurely withdrew before completion of the study. Differences of GSRS scores in H. pylori-positive groups between day 0 and day 56 were presented as the mean ± standard error (SE) and analyzed by Wilcoxon signed rank test. Both day 0 and day 56 BSS scores were presented as absolute values after subtracting 4 from each value. Wilcoxon signed rank test was subsequently conducted to analyze stool alteration after H. pylori eradication treatment. Differences in relative abundance of bacterial taxa between H. pylori-positive and H. pylori-negative subjects and relative abundance alterations after eradication treatment between group A and group B were compared by Wilcoxon rank sum test. Differences among group A, group B and group C were compared by Kruskal-Wallis test.
Mothur (v.1.30.1) was used to calculate indices of alpha diversity (Shannon index and Simpson index), richness (Sobs index and Ace index) and evenness (Shannoneven index and Heip index). Differences in alpha diversity and predicted pathway abundances between H. pylori-positive and H. pylori-negative subjects were analyzed with Wilcoxon rank sum test. Changes in alpha diversity from day 0 to day 14 and day 56 in group A and group B were compared by Wilcoxon signed rank test. Differences in microbial communities between groups were analyzed using LEfSe (linear discriminant analysis [LDA] coupled with effect size measurements) to avoid high false discovery rates [28]. A p value threshold of 0.05 (Wilcoxon rank sum test) and an effect size threshold of 2 were used for all bacterial taxa.
All statistical tests were two-tailed; p < .05 was considered statistically significant. Statistical analyses were performed using SPSS 19.0 for Windows (SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Baseline demographics and study follow-up
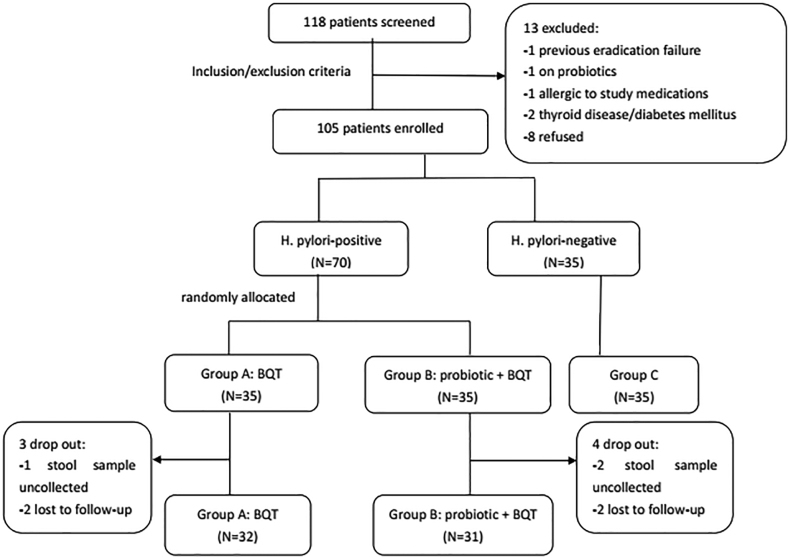
A total of 118 patients were initially screened for the study and 13 patients were excluded (Fig. 2). Therefore, 105 patients were enrolled, including 70 H. pylori-positive patients and 35 H. pylori-negative patients. Sixty-three H. pylori-positive patients (90%) completed therapy, while 3 patients (8.6%) in group A and 4 patients (11.4%) in group B withdrew from the study. A total of 224 stool samples were collected from 63 H. pylori-positive patients and 35 H. pylori-negative patients. Baseline characteristics were similar among the three groups (Table 1).
Fig. 2.
Patient flowchart. BQT, bismuth-containing quadruple therapy.
Table 1.
Baseline characteristics of study patients.
| Characteristics | Group A(n = 35) | Group B(n = 35) | Group C(n = 35) | P |
|---|---|---|---|---|
| Age(years)a | 43.20 ± 12.45 | 43.89 ± 12.50 | 40.89 ± 13.80 | 0.598 |
| BMIa | 22.66 ± 3.25 | 22.33 ± 3.04 | 22.06 ± 2.52 | 0.690 |
| Gender | 0.885 | |||
| Maleb | 12 (34.3%) | 13 (37.1%) | 14 (40.0%) | |
| Femaleb | 23 (65.7%) | 22 (62.9%) | 21 (60.0%) | |
| Marital status | 0.384 | |||
| Marriedb | 28 (80.0%) | 29 (82.9%) | 26 (74.3%) | |
| Unmarriedb | 6 (17.1%) | 4 (11.4%) | 9 (25.7%) | |
| Divorced/Widowedb | 1 (2.9%) | 2 (5.7%) | 0 (0.0%) | |
| Smokingb | 4 (11.4%) | 6 (17.1%) | 6 (17.1%) | 0.745 |
| Alcoholb | 6 (17.1%) | 6 (17.1%) | 4 (11.4%) | 0.745 |
Group A, H. pylori-infected patients treated with bismuth-containing quadruple therapy; Group B, H. pylori-infected patients treated with bismuth-containing quadruple therapy and supplemented with C. butyricum; Group C, H. pylori-negative participants; BMI, body mass index.
Data are presented as mean ± standard deviation (SD).
Data are presented as n (%).
3.2. Differences of gut microbiota between H. pylori-positive and H. pylori-negative populations
We observed a non-statistically significant increase in diversity of H. pylori-positive fecal samples compared to H. pylori-negative subjects based on number and abundance of OTUs, Shannon index and Simpson index. The observed richness of the community as measured by the Sobs index in H. pylori-positive samples was significantly higher relative to H. pylori-negative subjects (p < .05). However, there were no differences in evenness of the microbial communities by Shannoneven or Heip indices between H. pylori-positive and H. pylori-negative groups (Supplementary Fig. 1).
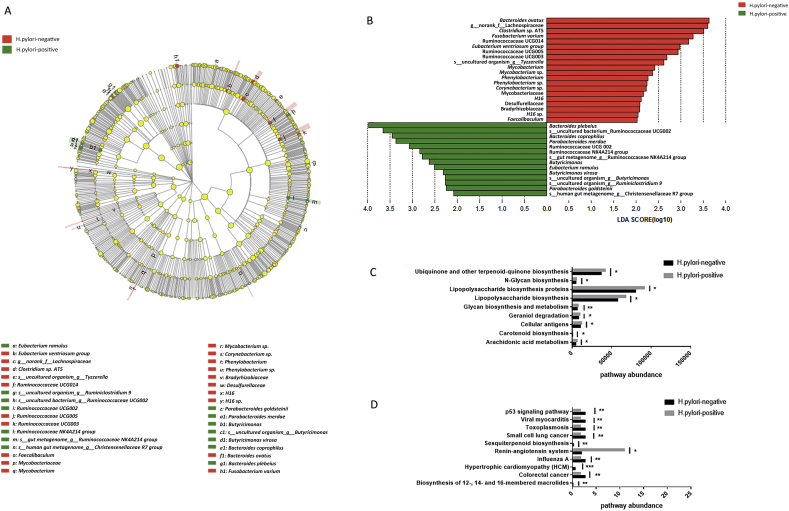
Phylum Nitrospirae was observed exclusively in H. pylori-negative samples (p = .020). The Bacteroidetes:Firmicutes (B:F) ratios were 0.94 and 0.84 in H. pylori-positive and H. pylori-negative communities, respectively. Abundances of 22 bacterial genera and 38 bacterial species (Supplementary Table 1) were significantly different between H. pylori-positive population samples and that in H. pylori-negative population (p < .05). There were no significantly different abundances of several putative beneficial taxa, including Bifidobacterium, Lactobacillus, Clostridium butyricum, Faecalibacterium prausnitzii and Akkermansia muciniphila, between H. pylori-positive and H. pylori-negative samples (data not shown). Neither genus Helicobacter nor species Helicobacter pylori were detected in fecal samples from all patients. There were significant differences in the abundance of different taxa ranging from phyla to species level between these two groups shown by cladogram (Fig. 3A) and LEfSe (Fig. 3B). The relative abundance of 19 pathways were significantly different between H. pylori-negative and H. pylori-positive patients by KEGG pathway analyses. Several disease-associated pathways were predicted to be more active in H. pylori-negative community, such as p53 and colorectal cancer signaling pathways. Many important metabolic pathways were modeled to be more active in patients with H. pylori, such as arachidonic acid metabolism, biosynthesis of 12-, 14- and 16-membered macrolides, carotenoid biosynthesis, glycan biosynthesis and metabolism, lipopolysaccharide biosynthesis, ubiquinone and other terpenoid-quinone biosynthesis (Fig. 3C, D).
Fig. 3.
Differences of bacterial taxa and predicted functional pathways between H. pylori-positive and H. pylori-negative groups. (A) Cladogram representation of gut microbiota taxa differences between H. pylori-positive and H. pylori-negative groups. (B) Differences of specific bacterial taxa between H. pylori-positive group and H. pylori-negative group by linear discriminant analysis (LDA) effect size (LEfSe). Red indicates taxa enriched in H. pylori-negative group and green indicates taxa enriched in H. pylori-positive group. (C, D) Pathways predicted to show significant different abundances between H. pylori-positive group and H. pylori-negative group according to Kyoto Encyclopedia of Genes and Genome (KEGG) pathway analysis. *, p < .05; **, p < .01.
3.3. Efficacy of H. pylori eradication treatment
The eradication rates of group A and group B were 88.6% and 85.7% by ITT analysis. By PP analysis, the efficacies were 96.9% and 96.8%, respectively. No significant differences in eradication rates between the two groups were observed by ITT or PP analyses (Table 2). After 14-day eradication treatment, there was a significant improvement in overall GI symptoms by GSRS in both group A and group B compared with baseline scores (p < .05). Individual symptom scores also improved, including abdominal distention, feeling of incomplete evacuation, eructation, acid regurgitation and heartburn in group A. Meanwhile, subjects in group B who received probiotics showed improvement in additional symptoms, particularly defecatory function (Supplementary Table 2). This was demonstrated by improvement in BSS scores after treatment indicating an increase in stool frequency and improvement in stool consistency (Supplementary Table 3). There were no significant differences in adverse events or patient compliance between group A and group B (Supplementary Table 4).
Table 2.
Efficacy of different eradication regimens.
| Analysis | Group A | 95% CI | Group B | 95% CI | P |
|---|---|---|---|---|---|
| ITT | 88.6%(31/35) | 73.3%–96.8% | 85.7%(30/35) | 69.7%–95.2% | 1.000 |
| PP | 96.9%(31/32) | 83.8%–99.9% | 96.8%(30/31) | 83.3%–99.9% | 1.000 |
Group A, H. pylori-infected patients treated with bismuth-containing quadruple therapy; Group B, H. pylori-infected patients treated with bismuth-containing quadruple therapy and supplemented with C. butyricum; ITT, intention to treat; PP, per protocol.
3.4. Influence of H. pylori eradication treatment on gut microbiota
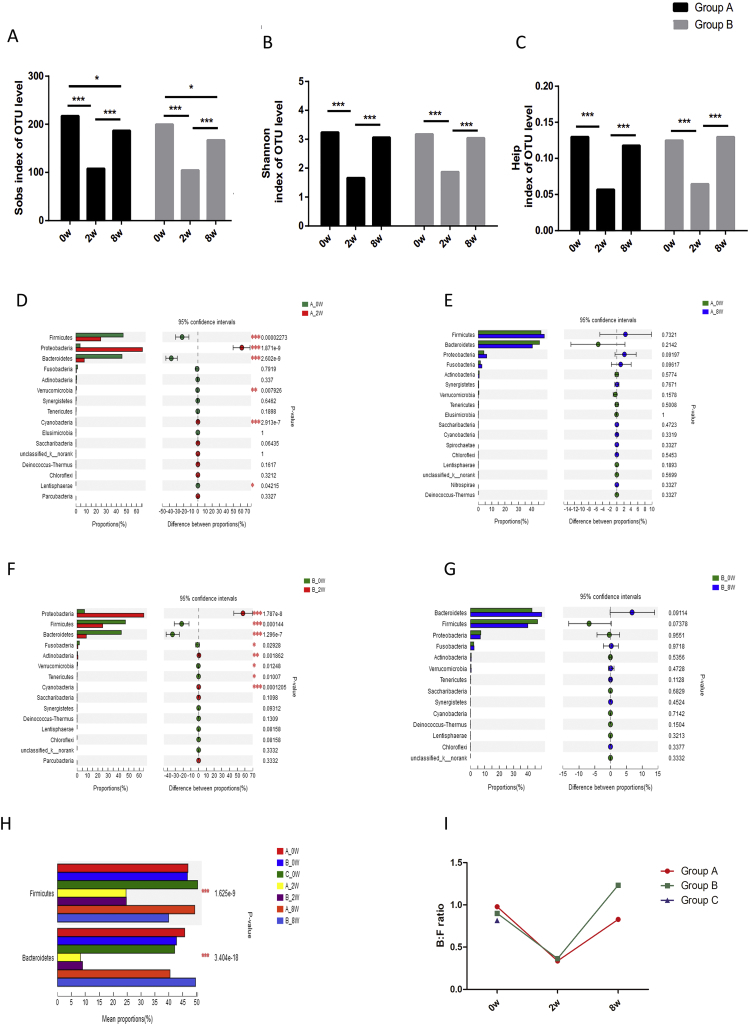
There were significant changes in the composition of the fecal microbiota in both group A and group B after 14-day eradication therapy compared with baseline. The alpha diversity indices were strikingly decreased after treatment (p < .001). This reduction in alpha diversity, as demonstrated by Sobs index, persisted for up to 6 weeks after completion of treatment (p < .05). Although other indices representing richness and evenness were not completely recovered on day 56, no statistical significance was observed compared with baseline (Fig. 4A–C).
Fig. 4.
Impact of eradication therapy and probiotic supplementation on gut microbiota. (A–I) Alpha diversity indices alterations among day 0, day 14 and day 56. Sobs index (A) represents richness of community; Shannon index (B) represents diversity of community; Heip index (C) represents evenness of community. (D-G) Phyla alterations between day 0 and day 14 in group A (D) and group B (F); phyla alterations between day 0 and day 56 in group A (E) and group B (G). Bacteroidetes and Firmicutes proportions (H) and Bacteroidetes:Firmicutes (B:F) ratios (I) within different groups in different time points. *, p < .05; **, p < .01; ***, p < .001.
There were transient changes at the phyla level after 14-day BQT. This included a decrease in relative abundances of Firmicutes, Bacteroidetes, Verrucomicrobia and Lentisphaerae as well as an increase in relative abundances of Proteobacteria and Cyanobacteria on day 14 compared with baseline (Fig. 4D). However, these changes in phyla normalized by day 56 such that no significant difference was detected among these phyla between day 56 and baseline (Fig. 4E). Similarly, there was a decrease in the B:F ratio from 0.98 to 0.34 on day 14 which increased to 0.83 on day 56 (Fig. 4H–I).
There were also marked changes in bacterial community structure at the family, genus and species level. Some of these changes were transient, such as a decrease in the relative abundance of Lachnospiraceae and Ruminococcaceae on day 14 which recovered by day 56. Others, however, were longer lasting, such as an increase in relative abundance of Enterobacteriaceae and Leuconostocaceae, together with a decrease of Rikenellaceae, Christensenellaceae, Peptococcaceae, Clostridiales Family XI, Victivallaceae on day 14 which had not normalized by day 56. There were alterations in the relative abundance of several beneficial anaerobes and some detrimental bacterial taxa on day 14, most of which tended to recover by day 56 (Table 3).
Table 3.
Relative abundance alterations on day 14 and day 56 with different eradication regimens.
| Bacterial taxa |
Group A |
Group B |
||||||
|---|---|---|---|---|---|---|---|---|
| ∆1 | P | ∆2 | P | ∆3 | P | ∆4 | P | |
| Family | ||||||||
| Lachnospiraceae | −16.06000 | *** | 3.13000 | ns | −17.80500 | *** | −2.53000 | ns |
| Ruminococcaceae | −12.71400 | *** | −1.08000 | ns | −10.62300 | *** | −1.49000 | ns |
| Neisseriaceae | 0.00576 | *** | 0.00034 | ns | 0.00348 | ns | −0.00125 | * |
| Bacteroidales S24–7 group | −0.24065 | ** | −0.15133 | ns | −0.50841 | ** | −0.29640 | ns |
| Genus | ||||||||
| Alistipes | −2.31950 | *** | −1.38800 | ** | −1.49970 | *** | −0.05800 | ns |
| Coprococcus 1 | −0.08053 | *** | −0.01773 | ns | −0.05083 | *** | −0.00654 | ns |
| Coprococcus 2 | −0.29260 | ** | −0.37186 | * | −0.22313 | ns | −0.20766 | ns |
| Coprococcus 3 | −0.17960 | *** | −0.11822 | * | −0.33450 | *** | −0.27498 | ** |
| Desulfovibrio | −0.31434 | *** | −0.10010 | ns | −0.19677 | *** | −0.00910 | ns |
| Subdoligranulum | −2.20130 | *** | −1.11500 | ** | −2.81630 | *** | −1.52800 | * |
| Parasutterella | −0.49886 | *** | −0.10090 | ns | −1.16053 | *** | −0.03500 | ns |
| Klebsiella | 16.90610 | *** | −0.26660 | * | 25.67100 | *** | −0.68300 | ns |
| Haemophilus | 0.07340 | * | −0.12320 | ns | −0.11694 | * | 0.00280 | ns |
| Fusobacterium | −1.06650 | ns | 1.09700 | ns | −1.49460 | * | 0.23900 | ns |
| Neisseria | 0.00409 | *** | 0.00020 | ns | 0.00052 | ns | −0.00083 | ns |
| Species | ||||||||
| Bifidobacterium adolescentis | −0.03811 | ** | 0.07002 | ns | −0.04817 | ** | −0.03470 | ns |
| Faecalibacterium prausnitzii | −0.02807 | *** | −0.01711 | ns | −0.02461 | *** | 0.00366 | ns |
| Akkermansia muciniphila | −0.42931 | ** | −0.39807 | ns | −0.22083 | * | 0.08520 | ns |
| Intestinimonas butyriciproducens | −0.02137 | ** | −0.01702 | ns | −0.00113 | * | 0.00679 | ns |
| Roseburia inulinivoran | −0.00566 | *** | −0.00318 | ns | −0.01097 | *** | −0.00657 | ns |
| Butyricimonas virosa | −0.07121 | *** | 0.00188 | ns | −0.08477 | *** | −0.02939 | ns |
| Dorea formicigenerans ATCC27755 | −0.08485 | *** | −0.02717 | * | −0.10362 | *** | −0.07200 | ** |
| Eubacterium rectale | −3.21350 | *** | 0.11800 | ns | −2.73848 | *** | 0.32700 | ns |
| Eubacterium eligens | −0.83190 | *** | 0.02850 | ns | −0.14570 | *** | 0.86860 | ns |
| Eubacterium coprostanoligenes | −0.41991 | *** | 0.05220 | ns | −0.49050 | *** | −0.09010 | * |
| Eubacterium ruminantium | −0.26003 | *** | 0.05180 | ns | −0.16166 | ** | −0.15800 | *** |
| Eubacterium ventriosum | −0.10208 | *** | −0.01447 | ns | −0.09636 | *** | 0.01289 | ns |
| Eubacterium xylanophilum | −0.01384 | ** | 0.02452 | ns | −0.06623 | ** | 0.00642 | ns |
| Eubacterium hallii | −0.00939 | *** | −0.00293 | ns | −0.00699 | ** | −0.00193 | ** |
| Eubacterium fissicatena | −0.00294 | *** | 0.00038 | ns | −0.00588 | ** | −0.00197 | ns |
| Clostridium butyricum | 0.00278 | ns | 0.00097 | ns | 1.23592 | ns | 0.11712 | ns |
| Lactococcus raffinolactis | 0.00176 | * | – | – | −0.00039 | ns | −0.00028 | ns |
| Lactobacillus sakei | 0.00898 | * | – | – | 0.00026 | ns | – | – |
| Acinetobacter baumannii NIPH60 | 0.00039 | * | 0.00027 | ns | 0.00007 | ns | −0.00041 | ns |
Group A, H. pylori-infected patients treated with bismuth-containing quadruple therapy; Group B, H. pylori-infected patients treated with bismuth-containing quadruple therapy and supplemented with C. butyricum; ∆1, relative abundance difference of Group A between day 0 and day 14; ∆2, relative abundance difference of Group A between day 0 and day 56; ∆3, relative abundance difference of Group B between day 0 and day 14; ∆4, relative abundance difference of Group B between day 0 and day 56; p value was to examine the relative abundance differences between day 0 and day 14 in group A and group B; *, p value < .05; **, p value < .01; ***, p value < .001; ns, no significance; −, not detected in day 0 and day 56.
3.5. Impact of H. pylori eradication with probiotic supplementation on gut microbiota
Changes in alpha diversity indices with probiotic supplementation in group B were similar to group A (Fig. 4A–C). The Sobs index had not completely recovered by day 56 (p < .05). The B:F ratio decreased from 0.90 to 0.36 on day 14 and increased to 1.23 on day 56 (Fig. 4H–I).
There were significant changes in phyla at day 14 compared with baseline, including reduction in Firmicutes, Bacteroidetes, and Verrucomicrobia as well as an increased abundance of Proteobacteria and Cyanobacteria. There were also specific changes in phyla seen only in group B, including a significant decrease in Fusobacteria and Tenericutes as well as an increase in Actinobacteria on day 14 (Fig. 4F). All phylum level changes normalized by day 56 in group B (Fig. 4G).
Overall, subjects in group B demonstrated similar changes in relative abundance of specific taxa compared with group A (Table 3). There was a significant decrease of Fusobaterium detected only in group B on day 14 while Lactococcus raffinolactis, Lactobacillus sakei and Acinetobacter baumannii NIPH60 were significantly changed only in group A. Most significant alterations tended to recover by day 56 except some bacteria belonging to Ruminococcaceae, Lachnospiraceae and Eubacterium. The relative abundance of C. butyricum after treatment in group B was significantly higher compared to group A on day 14 as well as H. pylori-negative subjects (p = .001) (Table 4). There was also an increased abundance of C. butyricum on day 14 compared to baseline although it did not meet statistical significance.
Table 4.
Relative abundance alterations of C. butyricum after eradication treatment.
| Relative abundance (%) |
|||||
|---|---|---|---|---|---|
| Group A | Group B | Group C | P1 | P2 | |
| Day 0 | 0.000257 | 0.005076 | 0.000106 | 0.179 | 0.070 |
| Day 14 | 0.003032 | 1.241000 | 0.025 | 0.001 | |
| Day 56 | 0.001224 | 0.122200 | 0.371 | 0.168 | |
| P3 | 0.612 | 0.096 | |||
| P4 | 0.958 | 0.801 | |||
Group A, H. pylori-infected patients treated with bismuth-containing quadruple therapy; Group B, H. pylori-infected patients treated with bismuth-containing quadruple therapy and supplemented with C. butyricum; Group C, H. pylori-negative participants.
P1, p values of difference between group A and group B.
P2, p values of difference among group A, group B and group C.
P3, p values of difference between day 0 and day 14.
P4, p values of difference between day 0 and day 56.
4. Discussion
We have demonstrated that H. pylori infection is associated with widespread alterations in the fecal microbiota compared with H. pylori-negative individuals and highlights the intricate and complex interactions between the host and gut microbiota. H. pylori has developed mechanisms to co-exist in the harsh gastric microenvironment, whereby it induces mucosal inflammation, immune activation, hypergastrinemia as well as variable effects on gastric acid production. Although the exact mechanisms by which H. pylori colonization in the stomach may lead to changes in fecal microbiota are not clearly defined, there is accumulating evidence that interplay between the bacteria and host responses may shape commensal microbiota composition. H. pylori has been associated with changes in the gastric microbiota, particularly in patients with advanced premalignant lesions or gastric cancer [29]. Furthermore, this may influence the gut microbiota distally since changes in luminal pH as well as end products of bacterial fermentation are key regulators in driving the community structure of the gut microbiota [30]. H. pylori may further induce other host responses, including inflammatory cytokines or neuroendocrine mediators, which could also conceivably modulate the gut microbiota in non-inflamed locations.
Although H. pylori is present in over half of the global populations, sequelae of infection occur in 20% while severe complications, such as gastric cancer, occur in <3% of infected individuals. Indeed, some studies have suggested that humans have developed a symbiotic relationship with H. pylori. As H. pylori has co-evolved with humans for over 60,000 years [31], there is speculation that this infection may enhance mucosal and systemic immunity to provide a survival benefit to the host. Perry et al. showed H. pylori seropositive subjects with latent tuberculosis were more likely to mount an enhanced Th1 response compared with H. pylori-negative subjects [32]. Meanwhile, global consensus statements have recommended eradication therapy in all individuals infected with H. pylori. However, the consequences of eradicating H. pylori and disrupting the gut microbiota on host function are still unknown. There is an inverse relationship between rates of H. pylori infection and prevalence of gastroesophageal reflux disease, Barrett's esophagus, and esophageal adenocarcinoma [33]. In addition, there is accumulating evidence linking decreased prevalence of H. pylori with increasing rates of asthma and atopic disorders [34]. Taken collectively, H. pylori colonization may have beneficial actions on host immunity while eradication may have deleterious consequences in some individuals.
Animal models suggest that the interplay between H. pylori infection, host immune response, and commensal microbiota may be associated with development of H. pylori-associated diseases. For example, infection with wild-type only, but not mutant strains, of H. pylori led to downstream changes in colonic and fecal microbiota in gerbils [9]. Meanwhile, only wild-type strains of H. pylori contain the cytotoxin-associated antigen A (cagA) virulence factor. Importantly, H. pylori strains containing cagA genes have been implicated as important prerequisites for long-term sequelae in humans [35]. As such, dynamic and ongoing crosstalk between H. pylori and commensal microbiota may influence the host response and susceptibility to complications of H. pylori infection.
There were intriguing phyla level changes in fecal microbiota with only H. pylori-negative subjects demonstrating presence of Nitrospirae. Nitrospirae are the most abundant and diverse nitrite-oxidizing bacteria which converts nitrite to nitrate [36]. While risk for H. pylori-associated gastric cancer has been associated with increased nitrites and formation of N-nitroso compounds in the stomach [37], there is no data available regarding the relationship between nitrite-oxidizing bacteria in the colon, such as Nitrospirae, and H. pylori. However, recent evidence suggests that nitrate and nitrite are storage pools for nitric oxide (NO)-like bioactivity which may play important regulatory mechanisms in humans. However, bioactivation of these molecules require presence of commensal bacteria as mammals lack the specific reductase enzymes [38]. Generation of NO is greatly enhanced during periods of hypoxia or acidosis, which allows for vasodilatation and other protective modulatory responses. Indeed, gastric ulcers are common adverse events from taking non-steroidal anti-inflammatory drugs which block generation of NO while administration of dietary nitrate prevented NSAID-induced ulcers in rats [39]. Furthermore, nitrite was shown to have bactericidal effects against H. pylori [40]. Thus, the nitrate-nitrite-NO pathway may play an important regulatory mechanism in pathologic conditions and our results suggest may be protective against H. pylori.
According to 16S rRNA functional predictions, pathways including lipopolysaccharide biosynthesis were higher in H. pylori positive group than in H. pylori negative group. As a complex glycolipid of outer membrane of gram-negative bacteria, lipopolysaccharide plays vital roles in outer-membrane integrity and is essential for the viability of bacteria. The capacity to synthesize and export lipopolysaccharide efficiently accounts for rapid growth and cell division [41]. Sobs index, representing richness of community, was significantly higher in H. pylori-positive community than H. pylori-negative community in our study. This phenomenon may explain the higher lipopolysaccharide biosynthesis pathway abundance in H. pylori-positive group. In addition, as a potent endotoxin responsible for triggering mucosal inflammation, lipopolysaccharide of H. pylori may interfere with DNA repair system [42]. Interestingly, colorectal cancer pathways were predicted to be higher in H. pylori-negative group in our study, which contrasts from several retrospective and prospective studies reporting that H. pylori infection was associated with increased risk of colorectal cancer or serrated colonic polyps [[43], [44], [45]]. Further studies are warranted to clarify the relationship between H. pylori infection, intestinal inflammation and colorectal carcinogenesis.
We observed striking changes in the gut microbiota after completion of 14-day BQT. This included changes at the phyla level, such as decrease in Firmicutes, Bacteroidetes, Verrucomicrobia, and Lentispaerae as well as an increase in Proteobacteria and Cyanobacteria, which was in consistent with findings by Ping-I Hsu et al. [46]. Changes in the gut microbial communities, such as decreased B:F ratio and increased abundance of Proteobacteria, have been associated with obesity and the metabolic syndrome [47]. In contrast, Yanagi et al. reported an increase in Bacteroidetes, Firmicutes, and B:F ratio while Proteobacteria decreased immediately after treatment [48]. These discrepant observations may be explained at least partially by different eradication regimens, drug doses and treatment duration lengths between the two studies. Furthermore, many of these changes persisted even up to 3 months after completion of antibiotics. Similar observations were made in a study from Malaysia which showed a decreased B:F ratio in H. pylori patients for up to 18 months after treatment [8], indicating long-lasting perturbations in gut microbiota that may have deleterious effects on the host.
There were also important taxonomic changes at the family level after treatment. BQT treatment was associated with decreased relative abundance of Lachnospiraceae and Ruminococcaceae, which are known to have beneficial effects, such as producing the short chain fatty acid butyrate. Decreases in these bacterial taxa are common after antibiotic exposure, associated with increased risk for Clostridium difficile infection, and are restored after fecal microbiota transplantation [49]. Similarly, we also observed reductions in major butyrate-producing bacteria at the genus and species level, including Eubacterium, Coprococcus, Faecalibacterium prausnitzii, Intestinimonas butyriciproducens, Roseburia inulinivoran and Butyricimonas virosa immediately after treatment. Other putative beneficial microbes, including Bifidobacterium adolescentis, Akkermansia muciniphila, Dorea formicigenerans ATCC27755 and Subdoligranulum also decreased after BQT [50,51]. On the other hand, there was an increase in relative abundances of detrimental bacteria, such as Acinetobacter baumannii NIPH60, Klebsiella and Haemophilus. Longitudinal studies are required to determine the consequences of antibiotic-induced disruption of the gut microbiota.
No significant difference in eradication rates were observed with the supplementation of BQT with probiotics (Table 2). This may be related to high eradication rates with both therapeutic regimens (>95% eradication) as well as limited sample size. BQT significantly improved several gastrointestinal symptoms including abdominal distention, feeling of incomplete evacuation, eructation, acid regurgitation and heartburn in H. pylori-infected patients. Supplementation with C. butyricum also led to improvement of other gastrointestinal symptoms, particularly defecatory function as evidenced by improved BSS scores, such as increased flatus, increased passage of stools and hard stools. Although a statistically significant difference was not observed, supplementation with C. butyricum showed a trend towards improving adverse events with antibiotic treatment. Prior systematic reviews and meta-analyses have demonstrated decreased antibiotic-associated adverse events with probiotic supplementation [52,53]. However, most studies reported the effects of Lactobacillus strains while our research studied the effect of C. butyricum. It is possible that different probiotic strains as well as a larger sample study may explain differences in our results. The relative abundance of C. butyricum in group B was strikingly higher when compared to that in group A and H. pylori-negative community on day 14. Furthermore, the B:F ratio on day 56 in the probiotic group was significantly higher compared with BQT alone which has previously been shown to correlate with serum IgG and IgM levels [54]. In contrast to group A, relative abundances of Neisseria and Acinetobacter baumannii NIPH60 in group B were not significantly increased while relative abundances of Haemophilus and Fusobacterium were reduced on day 14. It is possible that probiotic supplementation induced alterations in gut microbiota proportion alteration, leading to changes in host immune response and improved gastrointestinal symptoms. Taken together, eradication of H. pylori was associated with significant alterations in the gut microbiota which did not completely recover even 6 weeks after completion of treatment. Administration of a probiotic containing C. butyricum improved gastrointestinal symptoms with increased B:F ratio.
Our study had some limitations. First, although this was a prospective study, longer duration of follow-up is necessary to determine consequences of antibiotic-induced alterations in the gut microbiota. Secondly, fecal samples are not representative of the entire gut microbiota as the gut microbiota differs depending on location. In addition, fecal samples provide a measure of luminal microbes as opposed to mucosa-associated microbiota, which are more intimately associated with the host.
In conclusion, H. pylori infection may not be necessarily detrimental in all patients. After H. pylori eradication treatment gut microenvironment homeostasis may be disturbed with unknown consequences. Our study provided data for effectiveness of probiotic supplementation in improving gastrointestinal symptoms. Collectively, our results argue that the decision to eradicate H. pylori should be based on estimating the risk/benefit ratio for each individual.
Acknowledgments
Acknowledgements
We thank our patients and colleagues at Sir Run Run Shaw Hospital, Zhejiang University.
Funding sources
This study was supported by Natural Science Foundation of Zhejiang Province (LY18H160011), Science and Technology Plan of Medicine and Health of Zhejiang Province (2017KY398), Science Foundation of Zhejiang Traditional Medicine Bureau (2017ZB063), Public Welfare Technology Application Research Plan of Zhejiang Province for Laboratory Animals (2017C37149), and National Institutes of Health (NIH) grant (KL2TR002241). The funders had no role in study design, data collection, data analysis, interpretation and writing of the report.
Conflicts of interests
The authors declare that they have no competing interests.
Author contributions
According to the guidelines of the International Committee of Medical Journal Editors (ICMJE), all authors contributed to the four criteria. Shujie Chen and Jianmin Si conceived and designed the study. Luyi Chen, Wenli Xu, Jiamin He, Wenfang Zheng, Tingting Su, Sanchuan Lai, Yanqin Long, Hua Chu, Yujia Chen, Lan Wang, Kan Wang and Shujie Chen acquired the data. Lan Wang and Bixia Huang analyzed and interpreted the data. Luyi Chen drafted the manuscript. Shujie Chen, Jianmin Si and Allen Lee critically revised the manuscript. All authors read and approved the final manuscript.
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.08.028.
Contributor Information
Jianmin Si, Email: jianmin_si@zju.edu.cn.
Shujie Chen, Email: chenshujie77@zju.edu.cn.
Appendix A. Supplementary data
References
- 1.Hooi J.K.Y., Lai W.Y., Ng W.K. Global prevalence of Helicobacter pylori Infection: systematic review and meta-analysis. Gastroenterology. 2017;153:420–429. doi: 10.1053/j.gastro.2017.04.022. [DOI] [PubMed] [Google Scholar]
- 2.Nagy P., Johansson S., Molloy-Bland M. Systematic review of time trends in the prevalence of Helicobacter pylori infection in China and the USA. Gut Pathog. 2016;8:8. doi: 10.1186/s13099-016-0091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maldonado-Contreras A., Goldfarb K.C., Godoy-Vitorino F. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011;5:574–579. doi: 10.1038/ismej.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process—first American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 5.Ferreira R.M., Pereira-Marques J., Pinto-Ribeiro I. Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut. 2018;67:226–236. doi: 10.1136/gutjnl-2017-314205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng Q., Chen W.D., Wang Y.D. Gut microbiota: an integral moderator in health and disease. Front Microbiol. 2018;9:151. doi: 10.3389/fmicb.2018.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minalyan A., Gabrielyan L., Scott D., Jacobs J., Pisegna J.R. The gastric and intestinal microbiome: role of proton pump inhibitors. Curr Gastroenterol Rep. 2017;19:42. doi: 10.1007/s11894-017-0577-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yap T.W., Gan H.M., Lee Y.P. Helicobacter pylori eradication causes perturbation of the human gut microbiome in young adults. PLoS One. 2016;11 doi: 10.1371/journal.pone.0151893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heimesaat M.M., Fischer A., Plickert R. Helicobacter pylori induced gastric immunopathology is associated with distinct microbiota changes in the large intestines of long-term infected Mongolian gerbils. PLoS One. 2014;9 doi: 10.1371/journal.pone.0100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myllyluoma E., Ahlroos T., Veijola L., Rautelin H., Tynkkynen S., Korpela R. Effects of anti-Helicobacter pylori treatment and probiotic supplementation on intestinal microbiota. Int J Antimicrob Agents. 2007;29:66–72. doi: 10.1016/j.ijantimicag.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 11.Sugano K., Tack J., Kuipers E.J. Kyoto global consensus report on Helicobacter pylori gastritis. Gut. 2015;64:1353–1367. doi: 10.1136/gutjnl-2015-309252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jakobsson H.E., Jernberg C., Andersson A.F., Sjolund-Karlsson M., Jansson J.K., Engstrand L. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang A.Y., Peura D.A. The prevalence and incidence of Helicobacter pylori-associated peptic ulcer disease and upper gastrointestinal bleeding throughout the world. Gastrointest Endosc Clin N Am. 2011;21:613–635. doi: 10.1016/j.giec.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 14.Homan M., Orel R. Are probiotics useful in Helicobacter pylori eradication? World J Gastroenterol. 2015;21:10644–10653. doi: 10.3748/wjg.v21.i37.10644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tong J.L., Ran Z.H., Shen J., Zhang C.X., Sd Xiao. Meta-analysis: the effect of supplementation with probiotics on eradication rates and adverse events during Helicobacter pylori eradication therapy. Aliment Pharmacol Ther. 2007;25:155–168. doi: 10.1111/j.1365-2036.2006.03179.x. [DOI] [PubMed] [Google Scholar]
- 16.Okamoto T., Sasaki M., Tsujikawa T., Fujiyama Y., Bamba T., Kusunoki M. Preventive efficacy of butyrate enemas and oral administration of Clostridium butyricum M588 in dextran sodium sulfate-induced colitis in rats. J Gastroenterol. 2000;35:341–346. doi: 10.1007/s005350050358. [DOI] [PubMed] [Google Scholar]
- 17.Cushing K., Alvarado D.M., Ciorba M.A. Butyrate and mucosal inflammation: new scientific evidence supports clinical observation. Clin Transl Gastroenterol. 2015;6:e108. doi: 10.1038/ctg.2015.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yonezawa H., Osaki T., Hanawa T. Destructive effects of butyrate on the cell envelope of Helicobacter pylori. J Med Microbiol. 2012;61:582–589. doi: 10.1099/jmm.0.039040-0. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi M., Taguchi H., Yamaguchi H., Osaki T., Kamiya S. Studies of the effect of Clostridium butyricum on Helicobacter pylori in several test models including gnotobiotic mice. J Med Microbiol. 2000;49:635–642. doi: 10.1099/0022-1317-49-7-635. [DOI] [PubMed] [Google Scholar]
- 20.Shimbo I., Yamaguchi T., Odaka T. Effect of Clostridium butyricum on fecal flora in Helicobacter pylori eradication therapy. World J Gastroenterol. 2005;11:7520–7524. doi: 10.3748/wjg.v11.i47.7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Imase K., Takahashi M., Tanaka A. Efficacy of Clostridium butyricum preparation concomitantly with Helicobacter pylori eradication therapy in relation to changes in the intestinal microbiota. Microbiol Immunol. 2008;52:156–161. doi: 10.1111/j.1348-0421.2008.00026.x. [DOI] [PubMed] [Google Scholar]
- 22.Su P., Li Y., Li H. Antibiotic resistance of Helicobacter pylori isolated in the Southeast Coastal Region of China. Helicobacter. 2013;18:274–279. doi: 10.1111/hel.12046. [DOI] [PubMed] [Google Scholar]
- 23.Svedlund J., Sjodin I., Dotevall G. GSRS—a clinical rating scale for gastrointestinal symptoms in patients with irritable bowel syndrome and peptic ulcer disease. Dig Dis Sci. 1988;33:129–134. doi: 10.1007/BF01535722. [DOI] [PubMed] [Google Scholar]
- 24.Blake M.R., Raker J.M., Whelan K. Validity and reliability of the Bristol Stool Form Scale in healthy adults and patients with diarrhoea-predominant irritable bowel syndrome. Aliment Pharmacol Ther. 2016;44:693–703. doi: 10.1111/apt.13746. [DOI] [PubMed] [Google Scholar]
- 25.Langille M.G., Zaneveld J., Caporaso J.G. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatusov R.L., Koonin E.V., Lipman D.J. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 27.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Segata N., Izard J., Waldron L. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noto J.M., Peek R.M., Jr. The gastric microbiome, its interaction with Helicobacter pylori, and its potential role in the progression to stomach cancer. PLoS Pathog. 2017;13 doi: 10.1371/journal.ppat.1006573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cremer J., Arnoldini M., Hwa T. Effect of water flow and chemical environment on microbiota growth and composition in the human colon. Proc Natl Acad Sci U S A. 2017;114:6438–6443. doi: 10.1073/pnas.1619598114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moodley Y., Linz B., Bond R.P. Age of the association between Helicobacter pylori and man. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perry S., de Jong B.C., Solnick J.V. Infection with Helicobacter pylori is associated with protection against tuberculosis. PLoS One. 2010;5 doi: 10.1371/journal.pone.0008804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peek R.M., Jr., Blaser M.J. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat Rev Cancer. 2002;2:28–37. doi: 10.1038/nrc703. [DOI] [PubMed] [Google Scholar]
- 34.Blaser M.J., Chen Y., Reibman J. Does Helicobacter pylori protect against asthma and allergy? Gut. 2008;57:561–567. doi: 10.1136/gut.2007.133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saha A., Hammond C.E., Beeson C., Peek R.M., Jr., Smolka A.J. Helicobacter pylori represses proton pump expression and inhibits acid secretion in human gastric mucosa. Gut. 2010;59:874–881. doi: 10.1136/gut.2009.194795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Daims H., Nielsen J.L., Nielsen P.H., Schleifer K.H., Wagner M. In situ characterization of Nitrospira-like nitrite-oxidizing bacteria active in wastewater treatment plants. Appl Environ Microbiol. 2001;67:5273–5284. doi: 10.1128/AEM.67.11.5273-5284.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leach S.A., Thompson M., Hill M. Bacterially catalysed N-nitrosation reactions and their relative importance in the human stomach. Carcinogenesis. 1987;8:1907–1912. doi: 10.1093/carcin/8.12.1907. [DOI] [PubMed] [Google Scholar]
- 38.Lundberg J.O., Weitzberg E., Gladwin M.T. The nitrate-nitrite-nitric oxide pathway in physiology and therapeutics. Nat Rev Drug Discov. 2008;7:156–167. doi: 10.1038/nrd2466. [DOI] [PubMed] [Google Scholar]
- 39.Jansson E.A., Petersson J., Reinders C. Protection from nonsteroidal anti-inflammatory drug (NSAID)-induced gastric ulcers by dietary nitrate. Free Radic Biol Med. 2007;42:510–518. doi: 10.1016/j.freeradbiomed.2006.11.018. [DOI] [PubMed] [Google Scholar]
- 40.Dykhuizen R.S., Fraser A., McKenzie H., Golden M., Leifert C., Benjamin N. Helicobacter pylori is killed by nitrite under acidic conditions. Gut. 1998;42:334–337. doi: 10.1136/gut.42.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Whitfield C., Trent M.S. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem. 2014;83:99–128. doi: 10.1146/annurev-biochem-060713-035600. [DOI] [PubMed] [Google Scholar]
- 42.Slomiany B.L., Slomiany A. Role of LPS-elicited signaling in triggering gastric mucosal inflammatory responses to H. pylori: modulatory effect of ghrelin. Inflammopharmacology. 2017;25:415–429. doi: 10.1007/s10787-017-0360-1. [DOI] [PubMed] [Google Scholar]
- 43.Epplein M., Pawlita M., Michel A., Peek R.M., Jr., Cai Q., Blot W.J. Helicobacter pylori protein-specific antibodies and risk of colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2013;22:1964–1974. doi: 10.1158/1055-9965.EPI-13-0702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kountouras J., Polyzos S.A., Doulberis M. Potential impact of Helicobacter pylori-related metabolic syndrome on upper and lower gastrointestinal tract oncogenesis. Metabolism. 2018;87:18–24. doi: 10.1016/j.metabol.2018.06.008. [DOI] [PubMed] [Google Scholar]
- 45.Kumar A., Kim M., Lukin D.J. Helicobacter pylori is associated with increased risk of serrated colonic polyps: Analysis of serrated polyp risk factors. Indian J Gastroenterol. 2018;37:235–242. doi: 10.1007/s12664-018-0855-8. [DOI] [PubMed] [Google Scholar]
- 46.Hsu P.I., Pan C.Y., Kao J.Y. Helicobacter pylori eradication with bismuth quadruple therapy leads to dysbiosis of gut microbiota with an increased relative abundance of Proteobacteria and decreased relative abundances of Bacteroidetes and Actinobacteria. Helicobacter. 2018;23 doi: 10.1111/hel.12498. [DOI] [PubMed] [Google Scholar]
- 47.Compare D., Rocco A., Sanduzzi Zamparelli M., Nardone G. The gut bacteria-driven obesity development. Dig Dis. 2016;34:221–229. doi: 10.1159/000443356. [DOI] [PubMed] [Google Scholar]
- 48.Yanagi H., Tsuda A., Matsushima M. Changes in the gut microbiota composition and the plasma ghrelin level in patients with Helicobacter pylori-infected patients with eradication therapy. BMJ Open Gastroenterol. 2017;4 doi: 10.1136/bmjgast-2017-000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Song Y., Garg S., Girotra M. Microbiota dynamics in patients treated with fecal microbiota transplantation for recurrent Clostridium difficile infection. PLoS One. 2013;8 doi: 10.1371/journal.pone.0081330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanninen A., Toivonen R., Poysti S. Akkermansia muciniphila induces gut microbiota remodelling and controls islet autoimmunity in NOD mice. Gut. 2017;67:1445–1453. doi: 10.1136/gutjnl-2017-314508. [DOI] [PubMed] [Google Scholar]
- 51.Routy B., Le Chatelier E., Derosa L. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 52.Lu M., Yu S., Deng J. Efficacy of probiotic supplementation therapy for Helicobacter pylori eradication: a meta-analysis of randomized controlled trials. PLoS One. 2016;11 doi: 10.1371/journal.pone.0163743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McFarland L.V., Huang Y., Wang L., Malfertheiner P. Systematic review and meta-analysis: multi-strain probiotics as adjunct therapy for Helicobacter pylori eradication and prevention of adverse events. United Eur Gastroenterol J. 2016;4:546–561. doi: 10.1177/2050640615617358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shen X., Miao J., Wan Q. Possible correlation between gut microbiota and immunity among healthy middle-aged and elderly people in Southwest China. Gut Pathog. 2018;10:4. doi: 10.1186/s13099-018-0231-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.