Abstract
Background
Despite vigorous and ongoing efforts, active immunizations have yet to induce broadly neutralizing antibodies (bNAbs) against HIV-1. An alternative approach is to achieve prophylaxis with long-term expression of potent biologic HIV-1 inhibitors with Adeno-associated Virus (AAV), which could however be limited by hosts' humoral and cellular responses. An approach that facilitates in vivo production of these complex molecules independent of viral-vectored delivery will be a major advantage.
Methods
We used synthetic DNA and electroporation (DNA/EP) to deliver an anti-HIV-1 immunoadhesin eCD4-Ig in vivo. In addition, we engineered a TPST2 enzyme variant (IgE-TPST2), characterized its intracellular trafficking patterns and determined its ability to post-translationally sulfate eCD4-Ig in vivo.
Findings
With a single round of DNA injection, a peak expression level of 80-100μg/mL was observed in mice 14 days post injection (d.p.i). The engineered IgE-TPST2 enzyme trafficked efficiently to the Trans-Golgi Network (TGN). Co-administrating low dose of plasmid IgE-TPST2 with plasmid eCD4-Ig enhanced the potency of eCD4-Ig by three-fold in the ex vivo neutralization assay against the global panel of HIV-1 pseudoviruses.
Interpretation
This work provides a proof-of-concept for delivering anti-HIV-1 immunoadhesins by advanced nucleic acid technology and modulating protein functions in vivo with targeted enzyme-mediated post-translational modifications.
Funding
This work is supported by NIH IPCAVD Grant U19 Al109646-04, Martin Delaney Collaboration for HIV Cure Research and W.W. Smith Charitable Trust.
Keywords: HIV, Immunoadhesin, Post-translational modification, Enzyme trafficking, DNA, Electroporation
Graphical abstract
Highlights
-
•
Use of DNA/electroporation technology to promote in vivo expression of anti-HIV-1 immunoadhesin eCD4-Ig for 6 months
-
•
Engineered a Tyrosylprotein Sulfotransferase 2 variant (IgE-TPST2) that efficiently sulfate eCD4-Ig in vivo at low dose
-
•
IgE-TPST2 mediated sulfation enhanced potency of eCD4-Ig by 3-fold in the neutralizaiton of HIV global panel isolates
Research in context.
Evidence before this study
Despite ongoing efforts, active vaccination has yet to elicit broadly neutralizing antibodies against HIV-1 in humans. Technologies that enable long-term in vivo expression of potent biologics against HIV-1, such as eCD4-Ig, have protected Rhesus Macaques from SHIV and SIV challenges and shown remarkable promises.
Added value of this study
Prior approach to deliver eCD4-Ig in vivo involved the use of Adeno-associated Virus, which can potentially be limited by hosts' immune responses. Here, we use advanced DNA electroporation (DNA/EP) technology to deliver eCD4-Ig in vivo and achieve 6 months of robust expression. Additionally, we engineered an enzyme IgE-TPST2 that improve functionality of eCD4-Ig through post-translational modifications (PTM).
Implications of all the available evidence
Our work demonstrates a proof-of-concept for using DNA/EP for in vivo delivery of complex biologics and modulating their functions through PTMs, and highlights the translational potential of such approach.
Alt-text: Unlabelled Box
1. Introduction
There are currently 37 million people living with HIV/AIDS worldwide, and two million people are newly infected each year [1]. 15–50% of patients chronically infected with HIV have developed antibodies that are considered broadly neutralizing (bNAbs) [2]. However, to date, active vaccination with HIV envelope immunogens have failed to elicit bNAbs in non-human primates (NHPs) and humans [3]. In contrast, passive transfer of bNAbs have protected NHPs from SHIV challenges [[4], [5], [6]]. Additionally, bNAb 10-1074 could transiently suppress viremia in HIV viremic patients [7], while bNAb 3BNC117 can delay viral rebound in HIV patients on analytic interruptions of ART (ATI) [8]. Viral rebound in these patients, typically occurring seven to ten weeks after ATI, is potentially driven by the emergence of HIV viruses resistant to the bNAb.
Recently, AAV-delivery of a potent immunoadhesin construct eCD4-Ig demonstrated protection of Rhesus Macaques (RhM) from repeated challenges of SHIV-AD8 and SIV-Mac239 [9]. eCD4-Ig is a fusion protein consisting of (from N to C-terminus) extracellular D1-D2 domains of CD4, IgG-Fc, and a 15-amino acid CCR5-mimetic peptide. As eCD4-Ig targets the conserved receptor and co-receptor binding sites on HIV envelope, it has demonstrated extreme breadth and potency, neutralizing all isolates tested with IC50 < 5μg/mL [9]. In addition, mutations that allow HIV-1 to escape from eCD4-Ig potentially come at a fitness cost to the virus by lowering the affinity of Env to CD4 and CCR5. To further enhance the potency of eCD4-Ig, AAV-encoded TPST2 was co-administered with AAV-encoded eCD4-Ig because TPST2 can specifically sulfate tyrosine residues in the CCR5 mimetic peptide of eCD4-Ig [9].
While the utility of AAV gene delivery is well-established [10], its successful use in targeting non-immuno-privileged tissues (livers or skeletal muscles) is frequently hindered by pre-existing neutralizing antibodies against the capsid [11], which are extremely prevalent in the human population. For example, neutralizing antibodies to AAV2 are found in 30%–60% of human sera analyzed across different studies [12, 13]. In addition, delivery by AAV induces seroconversion, which precludes the possibility of redosing the biologics with the same vector [14]. Even after successful transduction of target tissues, cytotoxic T-lymphocytic response against the viral capsids may result in premature loss of the transgenes [15].
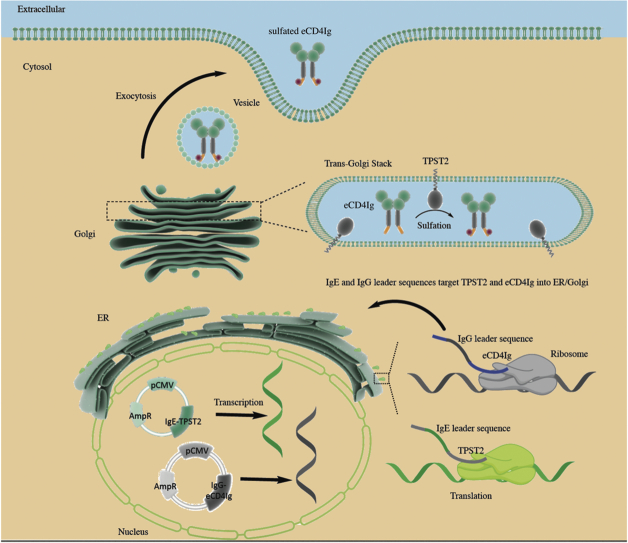
Advances in electroporation technologies (EP) have increased in vivo DNA plasmid driven expression by 100 fold or more [16], which has recently been translated with success in the vaccine space in the clinic [17, 18]. As plasmid delivery is serologically independent, DNA/EP represents a more reproducible approach for gene deliveries [19]. Our group has recently demonstrated the ability of DNA/EP to deliver high levels of plasmid-encoded monoclonal antibodies (DMAbs) against pseudomonas and influenza that can protect mice from lethal challenges [20, 21]. Here, we built on this technology and demonstrated that DNA/EP could express a non-native, highly structured molecule, eCD4-Ig, in vivo. We also showed, for the first time, DNA/EP-mediated in vivo delivery of enzyme IgE-TPST2 that can colocalize with and post-translationally sulfate eCD4-Ig to improve its functionality.
2. Materials and methods
2.1. DNA design and plasmid synthesis
Protein sequence for ReCD4-Ig was as previously reported [9]. Protein sequences for human TPST2 and HS3SA were obtained from UniProt (accession numbers: O60704 and Q9Y663). Protein sequence for SIVmac239 was obtained from GenBank (accession number M33262). DNA encoding protein sequences were codon and RNA optimized as previously described [20, 21]. The optimized transgenes were synthesized de novo (GenScript, Piscataway, NJ) and cloned into a modified pVAX-1 backbone under the control of the human CMV promoter and bovine growth hormone poly-adenylation signal. Plasmids that encode HIV envelope gp160 for TRO11, 25,710, 398F1, CNE8, X2278, BJOX2000, X1632, CE1176, 246F3, CH119, CE0217 and CNE55 were obtained from NIH-AIDS reagent.
2.2. Cell lines, transfection and ReCD4-Ig purification
HEK293T cells (ATCC Cat# CRL-3216, RRID:CVCL_0063) and TZM-bl cells (NIH-ARP Cat# 8129-442, RRID:CVCL_B478) were maintained in DMEM (Corning Cat# 10-013CV) supplemented with 10% fetal bovine serum (Atlas Biologicals Cat# EF-0500-A) and grown at 37 °C and 5% CO2. Expi293F cells (ThermoFisher Cat # A14527, RRID:CVCL_D615) were maintained in Expi293 expression medium (ThermoFisher Cat# A1435101) at 37 °C and 8% CO2. To determine in vitro sulfation of ReCD4-Ig, cells were seeded at a density of 0.5 × 106 cells/mL in a 6-well plate and transfected with 1.0 μg of p-ReCD4-Ig and varying doses of plasmid encoded enzymes with GeneJammer (Agilent Cat# A204130). Forty-eight hours after transfection, supernatants were collected and centrifuged at 1500g for 5 min to remove cellular debris. Adherent cells were lysed with cell lysis buffer (Cell Signaling Cat# A204130) modified with protease inhibitor cocktail (Roche Cat# 26733200). To obtain ReCD4-Ig standards for quantitative ELISA, Expi 293F cells were plated at a density of 2.5 × 106 cells/mL in Expi293 expression medium, rested overnight and transfected with p-ReCD4-Ig and Expifectamine™ (ThermoFisher Cat# A14525) in OPTI-MEM (ThermoFisher Cat# 31985070). Transfection enhancers were added 20 h after transfection, and supernatant was harvested 5 days after transfection. Magnetic protein G beads (GenScript Cat# L00673S) were used for purification of ReCD4-Ig, and purity was confirmed with Commassie staining of the SDS-Page gels (data not shown) (ThermoFisher Cat# NP0321).
2.3. ELISA
For ELISA-based quantification of ReCD4-Ig, MaxiSorp plates (ThermoFisher Cat# 44-2404-21) were coated with 1μg/mL of JR-FL gp140 (Immune Technology Cat# IT-001-0024∆TMp) overnight at 4 °C. Plates were washed 4 times with Phosphate Buffered Saline +0.1% Tween 20 (BioRad Cat# 1706531) (PBS-T) and blocked with 10% FBS in PBS for 1 h at room temperature. Plates were subsequently washed and incubated with serum samples diluted in PBS-T for one hour at room temperature. Plates were washed again and incubated with secondary goat anti-human Fc HRP (Jackson ImmunoResearch Labs Cat# 109-035-008, RRID:AB_2337579) at 1:5000 dilution for 1 h. The plates were subsequently developed with SigmaFast OPD (SigmaAldrich Cat# P9187) for 10 min before OD450 measurements were performed with Biotek Synergy2 plate reader.
To detect sulfation of ReCD4-Ig in transfection supernatants or sera, MaxiSorp plate were coated at 4 °C overnight with 5μg/mL JR-FL gp140. Plates were washed and blocked with 10% FBS/PBS for 3 h at room temperature. Plates were washed, and samples diluted in PBS-T were added for 1 h incubation at room temperature. Plates were washed again and incubated with 1:250 dilution of mouse anti-sulfotyrosine antibody (Millipore Cat# 05-1100, RRID:AB_11213996) for 1 h at room temperature. Finally, the plates were washed and incubated with 1:5000 dilution of anti-mouse IgG2a HRP secondary antibody (Bethyl Cat# A90-107P, RRID:AB_67155) for 1 h at room temperature. The plates were developed with SigmaFast OPD for 10 min and OD450 signals were measured.
2.4. Western blot
For detection of ReCD4-Ig, 10uL of transfection supernatant was loaded onto pre-cast 4–12% Bis-Tris gels under non-reducing condition and transferred to an Immobilon-FL PVDF membrane (EMD Millipore Cat# IPFL10100) with wet transfer. ReCD4-Ig was identified with IRDye 800CW goat anti-human IgG (LI-COR Biosciences, Lincoln, NE) (which cross-reacts with Rhesus IgG2 Fc) at 1: 10,000 dilution. For detection of sulfated tyrosine in ReCD4-Ig (Fig. 1e), the membrane was incubated overnight with 1:1000 mouse anti-sulfotyrosine (1C-A2) at 4 °C and developed with IRDye 680RD goat anti-mouse IgG (LI-COR Biosciences Cat# 926-32232, RRID:AB_10806644). As anti-mouse and anti-human antibodies are conjugated to dyes with different colors, it is possible to visualize ReCD4-Ig and sulfotyrosine bands simultaneously in a single membrane. For detection of sulfation enzymes expression in vivo, mice were sacrificed at indicated time points after DNA injections/IM-EP. Tibialis anterior (TA) muscles were harvested and homogenized in T-PER extraction buffer (ThermoFisher Cat# 78510) and protease inhibitor. Fifty micrograms of muscle homogenates were loaded onto 4–12% Bis-Tris gel under reducing condition and transferred to a PVDF membrane with wet transfer. The membrane was incubated overnight with polyclonal rabbit anti-TPST2 antibodies (Abcam Cat# ab87407, RRID:AB_10672926), and monoclonal mouse anti-GAPDH antibody (Cell Signaling Cat# 97166S) at 4 °C. The membrane was subsequently developed with IRDye 680RD goat anti-mouse IgG (LI-COR Biosciences Cat# 925–68,070, RRID:AB_2651128) and IRDye 800CW goat anti-rabbit IgG. All membranes were scanned with LI-COR Odyssey CLx (LI-COR Biosciences Cat# 926-32211, RRID:AB_621843).
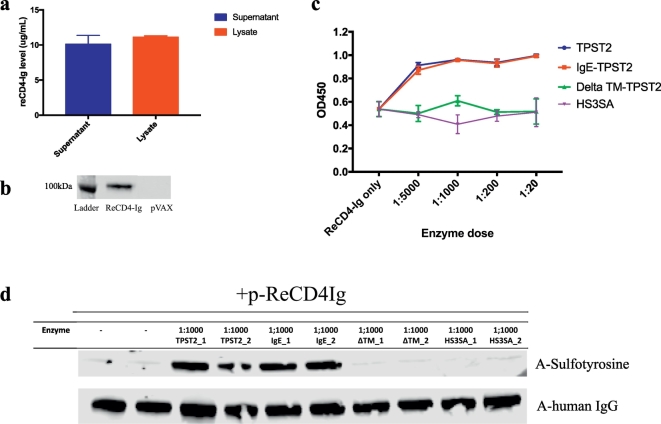
Fig. 1.
-
(A)Expression of ReCD4-Ig in transfection lysate and supernatant of HEK293T cells (n = 4).
-
(B)Western blot of supernatants of HEK293T cells transfected with either p-ReCD4-Ig or pVAX-1.
-
(C)Binding ELISA to detect tyrosine sulfation of ReCD4-Ig in transfection supernatant. HEK293 T cells were transfected with p-ReCD4-Ig or p-ReCD4-Ig with varying doses of plasmid enzyme constructs at (normalized to p-ReCD4-Ig, n = 3).
-
(D)Western blot of supernatants of HEK293T cells transfected with either p-ReCD4-Ig alone or p-ReCD4-Ig and 1:1000 plasmid enzymes. The lower and upper panels show the total amount of ReCD4-Ig and sulfotyrosine respectively.
2.5. Fluorescence microscopy
Eight-well chamber slides (Nunc Cat# 154534) were pre-coated with poly-l-lysine solution (SigmaAldrich Cat# P8920) before HEK293T cells were seeded at a density of 2 × 105 per well overnight. The cells were then transfected with 1.0μg of p-reCD4-Ig and 0.05μg of p-TPST2, p-IgE-TPST2 or p-ΔTM-TPST2 with GeneJammer. Forty-eight hours after transfection, the cells were fixed and permeabilized with 4% formaldehyde in PBS and 0.5% Triton-X-100 (Image-It-Fixation/Permeation Kit, ThermoFisher Cat# R37602) and blocked with 3% BSA in PBS at room temperature for 1 h. The cells were then stained overnight at 4 °C with 1:200 dilution of anti-Golgin 97 antibody (LI-COR Biosciences Cat# 926-32211, RRID:AB_621843) in 1% BSA/PBS-T, and 1:200 dilution of polyclonal rabbit anti-TPST2 antibody (Abcam Cat# ab87407, RRID:AB_10672926). The cells were then washed with PBS-T and stained with 1:500 dilution of Goat anti-Rabbit Alexa Fluor 594 (Thermo Fisher Scientific Cat# A-11037, RRID:AB_2534095), and goat anti-mouse Alexa Fluor 488 (Thermo Fisher Scientific Cat# A32723, RRID:AB_2633275). For nuclear staining, the cells were incubated with 0.5μg/mL of DAPI (SigmaAldrich Cat # D9542) in PBS-T and mounted with cover slips using Prolonged Diamond AntiFade Mountant (ThermoFisher Cat# P36970). Z-stack images were then acquired with Leica TCS SP5 II Scanning Confocal Microscope with a 64× objective. Maximal projections of the Z-stacks, deconvolution, and regions of interest analyses were performed with Leica LASX software to obtain Pearson correlation coefficients.
2.6. Animals
All animal experiments were carried out in accordance with animal protocol 112776 approved by the Wistar Institute Institutional Animal Care and Use Committee (IACUC) (Philadelphia, PA). Six-eight week old female BALB/c (Charles River Cat# 028) and B6.Cg-Foxn1nuJ (Jackson laboratory Cat# 000819, RRID:IMSR_JAX:000819) were housed in the animal facility. For transient immune-modulation, mice were given single intraperitoneal injection of 500μg of anti-mouse CD40L (Bio X Cell Cat# BE0017-1, RRID:AB_1107601). Mice were then given 160μg (2 injections) or 320μg (4 injections) intramuscular injections of DNA co-formulated with hyaluronidase (SigmaAldrich Cat# H4272). One minute after injections, IM-EP was performed at each injection site with the CELLECTRA® 3P device (Inovio Pharmaceutical, Plymouth Meeting, PA).
2.7. Ex vivo neutralization assay
Synthesis of HIV Env pseudotyped viruses and TZM-bl assays were performed as previously described [22]. Briefly, HEK293 T cells were transfected with 4μg of plasmid encoding HIV envelope and 8μg of plasmid encoding HIV backbone pSG3Δenv (NIH-ARP Cat# 11051) with GeneJammer. Forty-eight hours after transfection, the supernatants were filtered with Steriflip (MiliporeSigma Cat# SCGP00525) and stored at −80 °C. Pseudoviruses were titrated with a TZM-bl luciferase reporter assay using Britelight Plus (PerkinElmer Cat# 6066769) to determine a titer that corresponds to at least 150,000 RLU. Mice sera were heat inactivated at 56 °C for 10 min for the TZM-bl neutralization assays to determine serum concentration/titer that would result in 50% virus neutralization (IC50).
2.8. Statistics
One-way ANOVA analysis and pair-wise t-tests (with Holm-Bonferroni Adjustments in the case of multiple comparisons) were performed with GraphPad Prism 7.0. IC50 values were computed with a non-linear regression model of percentage neutralization versus log (reciprocal serum dilution) using Prism 7.0. P-values <0.05 were considered as statistically significant.
3. Results
3.1. Transfection of HEK293T cells enables expression and secretion of ReCD4-Ig in vitro
Since AAV-delivered Rhesus eCD4-Ig (ReCD4-Ig) protected RhMs against SHIV challenges [9], we had focused our current work on ReCD4-Ig for downstream comparisons. An optimized transgene encoding ReCD4-Ig with an N-terminal IgG kappa-leader sequence was designed and synthesized de novo and cloned into a modified pVAX-1 plasmid backbone. Expression of this engineered p-ReCD4-Ig DNA construct was studied in vitro by transfection of HEK293T cells followed by ELISA quantification. In both the transfection supernatant and cell lysate, robust expression of ReCD4-Ig was detected (Fig. 1a). Western blot of the transfection supernatant with anti-human IgG confirmed secretion of ReCD4-Ig (Fig. 1b).
3.2. Co-transfection of p-ReCD4-Ig and p-TPST2 variants allows in vitro sulfation of ReCD4-Ig
Previous studies have demonstrated that tyrosine sulfation of CCR5 mimetic peptide by trans-Golgi resident enzymes, TPST2, enhances potency of ReCD4-Ig [9, 23]. As ReCD4-Ig is targeted to the secretory pathway early in the translation process by the IgG leader sequence, tyrosine sulfation of ReCD4-Ig could only occur if the TPST2 variant is also expressed in the secretory compartment.
To highlight our ability to target TPST2 to the right subcellular compartment, we co-transfected HEK293T cells with p-ReCD4-Ig and plasmids encoding TPST2 enzyme variants. The p-IgE-TPST2, a construct with an IgE leader sequence upstream of TPST2, was predicted to sulfate ReCD4-Ig since the IgE leader sequence facilitates trafficking of TPST2 into the endoplasmic reticulum. A second TPST2 construct with a deletion in the transmembrane (TM) motif, p-ΔTM-TPST2, was not expected to sulfate ReCD4-Ig since the TM deletion removes the signal anchor sequence required for Trans-Golgi Network (TGN) targeting of TPST2. Finally, a control plasmid, p-HS3SA, was tested. HS3SA is a Golgi-resident enzyme which can transfer sulfate groups to heparin sulfate and has a similar catalytic site as compared to TPST2 [24]. Varying doses of DNA-encoded enzymes (1:5000 to 1:20, enzyme: ReCD4-Ig) were used to determine the minimal dose required to maximize sulfation of ReCD4-Ig. Using an anti-sulfotyrosine binding ELISA on cell supernatant, higher sulfation was observed for both TPST2 and IgE-TPST2 groups, even at the lowest enzyme dose of 1:5000, as compared to the baseline ReCD4-Ig only group (Fig. 1c). Furthermore, for both TPST2 and IgE-TPST2, sulfation signals were saturated at a remarkable 1:1000 enzyme dose, and higher dose of DNA did not increase sulfation. In comparison, RecCD4-Ig sulfation for both the ΔTM-TPST2 and HS3SA groups were not higher than the baseline. The lack of sulfation with the HS3SA group indicates remarkable specificity of sulfotransferases. To further confirm enzyme-mediated sulfation of ReCD4-Ig, the supernatants were analyzed with Western blots (Fig. 1d). Again, stronger sulfotyrosine bands were observed for the 1:1000 TPST2 and IgE-TPST2 groups than for the ReCD4-Ig only, ΔTM, and HS3SA groups. Taken together, these results suggest that DNA-encoded TPST2, and IgE-TPST2 can mediate in vitro sulfation of ReCD4-Ig at a remarkably low dose.
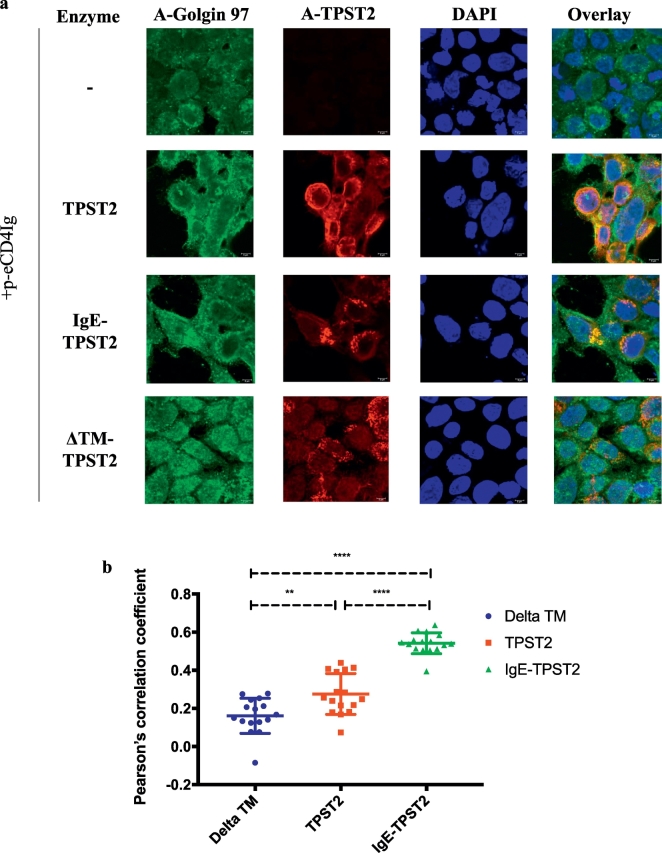
3.3. Incorporation of the N-terminal IgE leader sequence enhances targeting of TPST2 to TGN
We used fluorescence microscopy to determine whether IgE leader sequence can improve trafficking of DNA-encoded IgE-TPST2 to the cellular secretory compartment. HEK293T cells were transfected with either p-ReCD4-Ig only, or p-ReCD4-Ig in combination with p-TPST2, p-IgE-TPST2, or p-ΔTM-TPST2. 48 h after transfections, cells were harvested and stained with DAPI (blue), anti-TPST2 (red), and anti-Golgin 97 (green). Golgin-97 is a trans-Golgi resident protein commonly used to determine the expression of a protein of interest in TGN [25]. Confocal microscopy images of harvested cells illustrate robust expression of TPST2, IgE-TPST2 and ΔTM-TPST2 upon transfection (Fig. 2a). More importantly, IgE-TPST2 appears to co-localize with Golgin 97 to a greater extent than TPST2, whereas ΔTM-TPST2 does not co-localize with Golgin 97. To quantify the extent of co-localization between Golgin 97 and TPST2 variants, we analyzed the Pearson correlation coefficients (PCC) between red and green channels for 16 regions of interests for each group (Fig. 2b). The global one-way ANOVA analysis yields a p-value<0.0001 and post-hoc pairwise t-test shows PCC for the IgE-TPST2 group (PCC = 0.542) is significantly higher than that for the TPST2 group (PCC = 0.275, p < 0.0001). To ensure co-transfection with ReCD4-Ig did not change enzyme localization, we also co-transfected HEK293T cells with plasmid enzymes and pVAX1 backbone, and observed similar colocalization patterns (Supplemental Fig. 1). Taken together, the results support that while both TPST2 and IgE-TPST2 can traffic to TGN, IgE-TPST2 can be targeted to the secretory compartment more efficiently than TPST2, potentially because the N-terminal IgE leader sequence is recognized by signal recognition particle (SRP) more efficiently than the internal signal anchorage sequence of TPST2.
Fig. 2.
-
(A)Confocal microscopy to determine colocalization between TPST2 variant (red) and Golgin 97 (green). Nuclei are stained with DAPI (blue).
-
(B)Quantification of colocalization between TPST2 variants and Golgin 97 with PCC and regions of interest analyses (n = 16). P-values from posthoc pairwise t-tests are indicated: **, p < 0.005, ****, p < 0.0005.
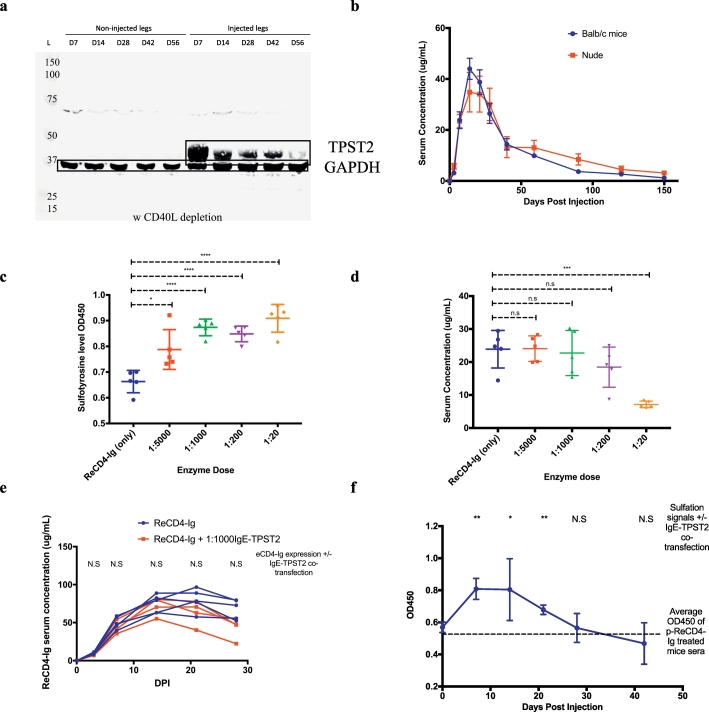
3.4. DNA/EP mediates in vivo expression of ReCD4-Ig and sulfation enzymes
We next determined whether we can express ReCD4-Ig and the sulfation enzyme TPST2 in vivo with DNA/EP. We observed strong TPST2 expression in the injected as compared to the contralateral legs of mice for at least 56 days (Fig. 3a). DNA/EP also mediated robust in vivo expression of IgE-TPST2 (Supplemental Fig. 2). Since ReCD4-Ig sequence is RhM based, strong anti-drug antibodies could develop in immune competent mice and influence the expression profile of ReCD4-Ig. Thus, immunodeficient B6.Cg-Foxn1nu/J (nude) mice were used initially. We observed a high level of expression of ReCD4-Ig, which peaked at an average of 35μg/mL 14 days post injection (d.p.i) (Fig. 3b). Remarkably, expression was detected as early as 3 d.p.i (average 5.7μg/ml) and lasted for at least 150 days (average 3.1μg/ml). In addition, ReCD4-Ig expression profile in transiently immune-depleted BALB/c mice demonstrated a similar pattern.
Fig. 3.
-
(A)Western blot of muscle homogenates 7–56 d.p.i demonstrates expression of TPST2 (43 kDa) in the injected legs as compared to the contralateral legs. GAPDH (37 kDa) serves as loading controls.
-
(B)Serum expression of ReCD4-Ig in B6.Cg-Foxn1nu/J and transiently immune-depleted BALB/c from a single round of DNA injection (160μg). N = 5 for each group.
-
(C)Binding ELISA to determine ReCD4-Ig tyrosine sulfation in sera of BALB/c mice injected with p-ReCD4-Ig alone or p-ReCD4-Ig with varying doses of p-IgE-TPST2.
-
(D)Serum expression level of ReCD4-Ig in mice cotreated with 320μg p-ReCD4-Ig and varying plasmid IgE-TPST2 doses 7 d.p.i.
-
(E)Serum expression level of ReCD4-Ig at different timepoints in BALB/c treated with either 320μg of p-ReCD4-Ig or 320μg of p-ReCD4-Ig and 0.32μg of p-IgE-TPST2. Each line represents an individual mouse.
-
(F)Average sulfation signals of ReCD4-Ig in the sera of BALB/c treated with 320μg of p-ReCD4- and 0.32μg of p-IgE-TPST2. N = 5 for the group. Dotted line represents average sulfation signals of ReCD4-Ig in the sera of BALB/c treated with 320μg of p-ReCD4 alone 7 d.p.i.
3.5. Low dose of p-IgE-TPST2 leads to in vivo sulfation of ReCD4-Ig without decreasing its expression
We next tested the ability of DNA-encoded IgE-TPST2 to sulfate ReCD4-Ig in vivo in transiently immune-depleted BALB/c mice. Remarkably, we observed that 1:1000 dose of IgE-TPST2 can saturate the sulfation OD450 signals detected in the mice sera 7 d.p.i, as compared to 1:20 IgE-TPST2 dose group (Fig. 3c). Additionally, sulfation of ReCD4-Ig was significantly higher, even at a lower 1:5000 dose of the enzyme, as compared to the baseline ReCD4-Ig only group. Previous studies describe that co-transfection of a high dose of TPST2 with its target proteins in vitro could lead to decreased secretion of the target proteins [26]. We observed a similar phenomenon both in vitro and in vivo (Fig. 3d and Supplemental Fig. 3a). To study if we could find an effective dose that would not limit ReCD4-Ig expression, we titrated the amount of IgE-TPST2 co-administered in vivo. A high dose (1:20) of IgE-TPST2 co-transfected with ReCD4-Ig resulted in 67% and 70% decreases in the expression of ReCD4-Ig in transfection supernatants and mice sera, respectively, as compared to ReCD4-Ig only groups. Additionally, suppression of ReCD4-Ig secretion is not directly driven by IgE-TPST2-mediated sulfation since co-administration of p-ReCD4-Ig and 1:20 p-HS3SA also reduces ReCD4-Ig expression by 52% (Supplemental Fig. 3b). Importantly, at the minimal 1:1000 dose of p-IgE-TPST2 required for optimal sulfation of ReCD4-Ig, we did not observe a difference in ReCD4-Ig expression 7 d.p.i between ReCD4-Ig only and ReCD4-Ig + 1:1000 IgE-TPST2 groups (Fig. 3d). Such pattern was again observed when the mice injected with either pReCD4-Ig alone or pReCD4-Ig + 1:1000 p-IgE-TPST2 were followed over time (Fig. 3e). In addition, using binding ELISA to assess the durability of sulfation of ReCD4-Ig by IgE-TPST2 in vivo, we observed peak levels of sulfation in the co-treated mice between 7 and 14 d.p.i (Fig. 3f). Sulfation of ReCD4-Ig in these co-treated mice declines over time but remains significantly higher than the mice which did not receive plasmid enzyme co-treatment for at least three weeks. Taken together, these results illustrate that plasmid encoded enzymes delivered by electroporation can enable in vivo sulfation of ReCD4-Ig at a remarkably low dose without decreasing the expression of ReCD4-Ig.
3.6. In vivo sulfation increases the potency of ReCD4-Ig
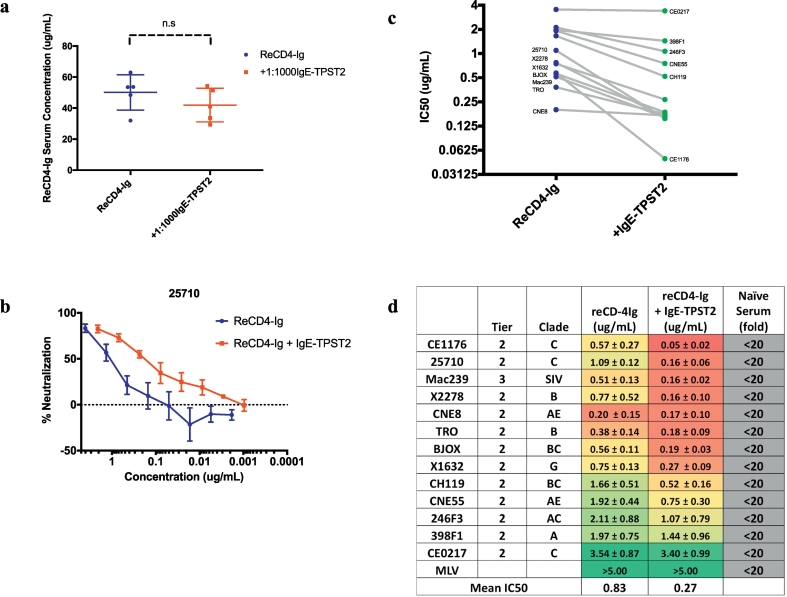
We next determined if in vivo sulfation of ReCD4-Ig can enhance its potency. Similar levels of ReCD4-Ig expression (40μg/mL) was again observed for both p-ReCD4-Ig only and p-ReCD4-Ig + 1:1000 p-IgE-TPST2 groups 7 d.p.i in immune-depleted BALB/c mice (Fig. 4a). We first tested the ability of in vivo produced ReCD4-Ig to neutralize one of the pseudoviruses from the global panel (25,710, Tier 2, clade C) using the standard TZM-bl assay [27]. Sulfation mediated by IgE-TPST2 significantly enhanced the ability of ReCD4-Ig to neutralize this isolate, as evidenced by a right-ward shift in the neutralization curve (Fig. 4b). We then evaluated the effects of ReCD4-Ig sulfation with a panel of 13 pseudoviruses comprising of the global panel and the tier 3 isolate SIVmac239 [27]. We observed that ReCD4-Ig neutralized all 13 viruses in the panel with an IC50 less than 5μg/mL and a mean IC50 of 0.83μg/mL (Fig. 4c and Fig. 4d). Naïve mice sera, in comparison, did not neutralize any of the pesudovirus at a titer of 1:20. Our results validated the remarkable breadth of eCD4-Ig as described in previous studies [9]. In addition, sulfation of ReCD4-Ig enhances its potency in neutralizing 8/12 pseudoviruses in the global panel (CE1176, 25710, X2278, TRO, BJOX, X1632, CH119, CNE55) and Mac239 (Fig. 4c–d and Supplemental Fig. 4). Sulfation exhibits the most drastic effect on the ability of ReCD4-Ig to neutralize CE1176, with a 10-fold drop in IC50 (0.57 ± 0.27 μg/mL to 0.05 ± 0.02 μg/mL). Overall, IgE-TPST2 mediated sulfation led to a decrease in the geometric mean of IC50 against the viral panel from 0.83μg/mL to 0.27μg/mL. Of note, the IC50 of sulfated ReCD4-Ig in neutralization of Mac239 is 0.16 ± 0.06 μg/mL, similar to the IC50 reported for AAV delivered ReCD4-Ig [9]. Taken together, our results validated in vivo sulfation of ReCD4-Ig by plasmid-encoded IgE-TPST2 from a functional standpoint and demonstrated the ability of DNA-encoded enzymes to modulate biological functions of a target protein through post-translational modification (PTM).
Fig. 4.
-
(A)Serum concentrations of ReCD4-Ig at the time of terminal bleed (7 d.p.i) in immune-depleted BALB/c mice injected with 320μg of p-ReCD4-Ig alone or p-ReCD4-Ig + 1:1000 p-IgE-TPST2.
-
(B)Neutralization of 25,710 pseudovirus versus serum concentration of ReCD4-Ig.
-
(C)Comparison of ReCD4-Ig IC50 with or without IgE-TPST2 treatment.
-
(D)IC50 values of ReCD4-Ig in sera of mice with and without IgE-TPST2 treatment. Geometric mean of IC50 across the panel (except for MLV) is also given.
4. Discussion
Sulfation plays an important role in molecular interactions between CCR5, gp120, and CD4 to support HIV infection. Sulfated tyrosine residues on the N-terminus of CCR5 and CD4i antibody (412d) mediate their intermolecular interactions with the V3 loop of Env in the CD4-bound post-fusion state [28]. As such, sulfated CD4i antibodies neutralize primary HIV-1 isolates more potently than non-sulfated prototypical CD4i antibodies 17b and 48d [29]. In addition, the CDRH3s of many V2-apex bNAbs (PGDM1400, PG9) are also tyrosine sulfated [30]. By using DNA/EP to express both ReCD4Ig and IgE-TPST2 in vivo, we have significantly increased potency of the immuneadhesin through post-translational sulfation, validating and extending prior findings [9].
Importantly, this is the first report utlizing DNA to encode an enzyme that can carry out PTM of a target protein in vivo. Through modulating protein function in vivo, DNA/EP provides a platform with diverse applications. For example, by modifying the glycosylation pattern in the Fc portion of immunoglobulin, we can fine-tune its effector functions. Afucosylation of IgG1 Fc with endoglycosidase/fucosidase, for instance, can potentially enhance antibody-dependent cell-mediated cytotoxicity (ADCC) of the modified antibody. Terminal sialylation, in the context of core fucosylation, exhibits an opposite effect [31, 32]. In the area of vaccine design, PTMs of an antigen can create new epitopes for recognition by the immune system. For instance, both germline encoded and somatically mutated antibodies in the CAP256.VRC26 lineage recognize sialic-acid bearing glycans at N160, or N156 positions of the HIV envelope [33]. Co-administration of DNA-encoded HIV Env antigens and sialytransferases could likely elicit such glycan-dependent neutralizing antibodies.
Our results illustrate that a remarkably low dose of 1:1000 p-IgE-TPST2 is required for in vivo sulfation of ReCD4-Ig. We expected this finding since a single molecule of the enzyme should be able to turn over multiple copies of target proteins. Specifically, since TPST2 has a turnover number (kcat) of 5.1 × 10−3 s−1 (for a mono-sulfated CCR8 peptide) and half-life of a Golgi-resident enzyme is about 20 h, a single copy of TPST2 enzyme should be able to turnover at least hundreds of copies of ReCD4-Ig [34, 35]. Of note, the dose required to sulfate ReCD4-Ig is much lower for DNA-encoded IgE-TPST2 (1:1000) than AAV-encoded TPST2 (1:4). This demonstrates the high efficiency of DNA/EP mediated enzyme delivery and that muscle cells have received separate copies of both p-IgE-TPST2 and p-ReCD4-Ig simultaneously. This is likely due to directionally pulsed electric fields that can create transient pores in the plasma membrane, which move polyanionic plasmid DNA directly into the cells to improve transfection efficiency by 100–1000 folds [16]. In comparison, uptake of AAV-encoded genetic materials (ReCD4-Ig and TPST2) into cells requires clathrin-dependent endocytosis or micropinocytosis [36, 37], and transduction of muscles cells by both AAV-TPST2 and AAV-eCD4-Ig can occur in a stochastic fashion.
The results also support an approach to target an enzyme to a specific subcellular compartment to maximize its functions. While the efficiency of IgE-TPST2 mediated sulfation appears similar to that of TPST2-mediated sulfation (Fig. 1d), selective targeting of the IgE-TPST2 can potentially reduce cytosolic expression of the enzyme and off-target effects. This approach can be further extended to target proteins to other subcellular compartments for therapeutic and investigational purposes. Specifically, an N-terminal sequence consisting of 10–70 amino acids that forms amphipathic helices could target proteins to the mitochondria; a dileucine motif DXXLL, or a tyrosine-based motif YXXØ, in the cytoplasmic tail of a transmembrane protein could target it to the lysosome; whereas a unit of 5 basic positively charged amino acids could target a protein to the nucleus [38, 39].
Finally, we have demonstrated the ability of DNA/EP to enable robust and long-term in vivo expression of immunoadhesins like ReCD4-Ig. With a single round of injection, a peak expression level of 80-100μg/mL in mice was observed, with levels that remains above 3μg/mL for 150 days. The decline in ReCD4-Ig expression between 28 and 42 d.p.i (Fig. 3b) and in ReCD4-Ig sulfation between 14 and 28 d.p.i (Fig. 3f) is under further investigation. Approaches such as modifications to the plasmid backbone to reduce in vivo promoter silencing and protein engineering to increase enzyme half-lives are worthy of future investigations [40]. The in vivo folding of ReCD4-Ig into its proper conformational state is confirmed by the observation that DNA-encoded ReCD4-Ig demonstrates excellent potency and breadth in neutralizing all isolates from the global panel with an IC50 less than 5μg/mL and a mean IC50 of 0.27μg/mL. Recently, our preliminary studies demonstrate microgram per milliliter levels of expression of other monoclonal antibodies can be achieved in NHPs with Cellectra EP delivered DNA, supporting the potential translation of this approach (work in progress). This study illustrates the utility of DNA/EP for in vivo expression and subcellular targeting of an enzyme and provides a strategy for modifying protein functions in vivo through PTM. Further study to determine whether the DNA/EP can be a tool to deliver and modify immunoadhesins and other important biologics to prevent and control HIV-1 infection and other diseases is likely important.
Acknowledgments
Acknowledgement
We would like to thank Animal Facility staff at the Wistar Institute for providing house and care to the animals. We thank Imaging Facility Core at the Wistar Institute for assistance with confocal microscopy experiments. We thank Michael Farzan for prior work on the development of eCD4-Ig which inspired this work and invaluable feedback provided during preparation of this manuscript. We thank Ronald Collman for providing HIV envelope expression plasmids and NIH AIDS Research and Reference Reagent Program for reagents. We thank the NIH, Martin DeLaney Collaboration and W.W. Smith Charitable Trust for providing funding and support.
Funding sources
This research is supported by NIH IPCAVD Grant U19 Al109646-04 awarded to D.B·W and made possible through generous support of Martin Delaney Collaboration for HIV Cure Research and W.W. Smith Charitable Trust. The funding sources were not involved in the design of this study, collection and analyses of data, decision to submit, and preparation of the manuscript.
Conflicts of interests
Z.X, M.C·W and D.B·W have a pending patent 206193-0006-P1-US.607603. M.C·W is an employee of Inovio Pharmaceuticals and as such receives salary and benefits including ownership of stock and stock options from the company. K.M. receives grants and consulting fees from Inovio related to DNA vaccine development. D.B·W. has received grant funding, participates in industry collaborations, has received speaking honoraria, and has received fees for consulting, including serving on scientific review committees and board series. Remuneration received by D.B·W. includes direct payments, stock or stock options, and in the interest of disclosure he notes potential conflicts associated with his work with Inovio and possible others.
Author contributions
Z.X., M.C.W., and D.B.W planned the experiments. Z.X., M.C.W., H.C., A.P.P., A.P., E.T.R., and J.D.C. conducted the experiments. K.M. contributed crucial reagents. Z.X. analyzed the data. Z.X., M.C.W., and D.B.W wrote the paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.08.027.
Appendix A. Supplementary data
supplementary material
References
- 1.Tetteh R.A., Yankey B.A., Nartey E.T., Lartey M., Leufkens H.G., Dodoo A.N. Pre-exposure prophylaxis for HIV prevention: safety concerns. Drug Saf. 2017;40(4):273–283. doi: 10.1007/s40264-017-0505-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hraber P., Seaman M.S., Bailer R.T., Mascola J.R., Montefiori D.C., Korber B.T. Prevalence of broadly neutralizing antibody responses during chronic HIV-1 infection. AIDS. 2014;28(2):163–169. doi: 10.1097/QAD.0000000000000106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sok D., Le K.M., Vadnais M. Rapid elicitation of broadly neutralizing antibodies to HIV by immunization in cows. Nature. 2017;548(7665):108–111. doi: 10.1038/nature23301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Julg B., Liu P.T., Wagh K. Protection against a mixed SHIV challenge by a broadly neutralizing antibody cocktail. Sci Transl Med. 2017;9(408) doi: 10.1126/scitranslmed.aao4235. eaao4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shingai M., Donau O.K., Plishka R.J. Passive transfer of modest titers of potent and broadly neutralizing anti-HIV monoclonal antibodies block SHIV infection in macaques. J Exp Med. 2014;211(10):2061–2074. doi: 10.1084/jem.20132494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moldt B., Rakasz E.G., Schultz N. Highly potent HIV-specific antibody neutralization in vitro translates into effective protection against mucosal SHIV challenge in vivo. Proc Natl Acad Sci U S A. 2012;109(46):18921–18925. doi: 10.1073/pnas.1214785109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caskey M., Schoofs T., Gruell H. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat Med. 2017;23(2):185–191. doi: 10.1038/nm.4268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scheid J.F., Horwitz J.A., Bar-On Y. HIV-1 antibody 3BNC117 suppresses viral rebound in humans during treatment interruption. Nature. 2016;535(7613):556–560. doi: 10.1038/nature18929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gardner M.R., Kattenhorn L.M., Kondur H.R. AAV-expressed eCD4-Ig provides durable protection from multiple SHIV challenges. Nature. 2015;519(7541):87–91. doi: 10.1038/nature14264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naso M.F., Tomkowicz B., Perry W.L., 3rd, Strohl W.R. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs. 2017;31(4):317–334. doi: 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fitzpatrick Z., Leborgne C., Barbon E. Influence of pre-existing anti-capsid neutralizing and binding antibodies on AAV vector transduction. Mol Ther Methods Clin Dev. 2018;9:119–129. doi: 10.1016/j.omtm.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mimuro J., Mizukami H., Shima M. The prevalence of neutralizing antibodies against adeno-associated virus capsids is reduced in young Japanese individuals. J Med Virol. 2014;86(11):1990–1997. doi: 10.1002/jmv.23818. [DOI] [PubMed] [Google Scholar]
- 13.Li C., Narkbunnam N., Samulski R.J. Neutralizing antibodies against adeno-associated virus examined prospectively in pediatric patients with hemophilia. Gene Ther. 2012;19(3):288–294. doi: 10.1038/gt.2011.90. [DOI] [PubMed] [Google Scholar]
- 14.Tse L.V., Klinc K.A., Madigan V.J. Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion. Proc Natl Acad Sci U S A. 2017;114(24) doi: 10.1073/pnas.1704766114. (E4812-E21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manno C.S., Pierce G.F., Arruda V.R. Successful transduction of liver in hemophilia by AAV-factor IX and limitations imposed by the host immune response. Nat Med. 2006;12(3):342–347. doi: 10.1038/nm1358. [DOI] [PubMed] [Google Scholar]
- 16.Sardesai N.Y., Weiner D.B. Electroporation delivery of DNA vaccines: prospects for success. Curr Opin Immunol. 2011;23(3):421–429. doi: 10.1016/j.coi.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tebas P., Roberts C.C., Muthumani K. Safety and immunogencity of an anti-Zika virus DNA vaccine—preliminary report. N Engl J Med. 2017 doi: 10.1056/NEJMoa1708120. published online Oct 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Trimble C.L., Morrow M.P., Kraynyak K.A. Safety, efficacy, and immunogenicity of VGX-3100, a therapeutic synthetic DNA vaccine targeting human papillomavirus 16 and 18 E6 and E7 proteins for cervical intraepithelial neoplasia 2/3: a randomised, double-blind, placebo-controlled phase 2b trial. Lancet. 2015;386(10008):2078–2088. doi: 10.1016/S0140-6736(15)00239-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Almqvist N., Winkler T.H., Martensson I.L. Autoantibodies: focus on anti-DNA antibodies. Self Nonself. 2011;2(1):11–18. doi: 10.4161/self.2.1.15087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel A., Digiandomenico A., Keller A.E. An engineered bispecific DNA-encoded IgG antibody protects against Pseudomonas aeruginosa in a pneumonia challenge model. Nat Commun. 2017;8(1):637. doi: 10.1038/s41467-017-00576-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elliott S.T.C., Kallewaard N.L., Benjamin E. DMAb inoculation of synthetic cross reactive antibodies protects against lethal influenza a and B infections. NPJ Vaccines. 2017;2:18. doi: 10.1038/s41541-017-0020-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sarzotti-Kelsoe M., Bailer R.T., Turk E. Optimization and validation of the TZM-bl assay for standardized assessments of neutralizing antibodies against HIV-1. J Immunol Methods. 2014;409:131–146. doi: 10.1016/j.jim.2013.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seibert C., Cadene M., Sanfiz A., Chait B.T., Sakmar T.P. Tyrosine sulfation of CCR5 N-terminal peptide by tyrosylprotein sulfotransferases 1 and 2 follows a discrete pattern and temporal sequence. Proc Natl Acad Sci U S A. 2002;99(17):11031–11036. doi: 10.1073/pnas.172380899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teramoto T., Fujikawa Y., Kawaguchi Y. Crystal structure of human tyrosylprotein sulfotransferase-2 reveals the mechanism of protein tyrosine sulfation reaction. Nat Commun. 2013;4:1572. doi: 10.1038/ncomms2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lu L., Tai G., Hong W. Autoantigen Golgin-97, an effector of Arl1 GTPase, participates in traffic from the endosome to the trans-golgi network. Mol Biol Cell. 2004;15(10):4426–4443. doi: 10.1091/mbc.E03-12-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W., Bardhi A., Feng Y. Improving the CH1-CK heterodimerization and pharmacokinetics of 4Dm2m, a novel potent CD4-antibody fusion protein against HIV-1. MAbs. 2016;8(4):761–774. doi: 10.1080/19420862.2016.1160180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Decamp A., Hraber P., Bailer R.T. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol. 2014;88(5):2489–2507. doi: 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farzan M., Mirzabekov T., Kolchinsky P. Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell. 1999;96(5):667–676. doi: 10.1016/s0092-8674(00)80577-2. [DOI] [PubMed] [Google Scholar]
- 29.Choe H., Li W., Wright P.L. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell. 2003;114(2):161–170. doi: 10.1016/s0092-8674(03)00508-7. [DOI] [PubMed] [Google Scholar]
- 30.Moore P.L., Gorman J., Doria-Rose N.A., Morris L. Ontogeny-based immunogens for the induction of V2-directed HIV broadly neutralizing antibodies. Immunol Rev. 2017;275(1):217–229. doi: 10.1111/imr.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold J.N., Wormald M.R., Sim R.B., Rudd P.M., Dwek R.A. The impact of glycosylation on the biological function and structure of human immunoglobulins. Annu Rev Immunol. 2007;25:21–50. doi: 10.1146/annurev.immunol.25.022106.141702. [DOI] [PubMed] [Google Scholar]
- 32.Li T., Dilillo D.J., Bournazos S., Giddens J.P., Ravetch J.V., Wang L.X. Modulating IgG effector function by fc glycan engineering. Proc Natl Acad Sci U S A. 2017;114(13):3485–3490. doi: 10.1073/pnas.1702173114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andrabi R., Su C.Y., Liang C.H. Glycans function as anchors for antibodies and help drive HIV broadly neutralizing antibody development. Immunity. 2017;47(3):524–537. doi: 10.1016/j.immuni.2017.08.006. [e3] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danan L.M., Yu Z., Ludden P.J., Jia W., Moore K.L., Leary J.A. Catalytic mechanism of Golgi-resident human tyrosylprotein sulfotransferase-2: a mass spectrometry approach. J Am Soc Mass Spectrom. 2010;21(9):1633–1642. doi: 10.1016/j.jasms.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strous G.J. Golgi and secreted galactosyltransferase. CRC Crit Rev Biochem. 1986;21(2):119–151. doi: 10.3109/10409238609113610. [DOI] [PubMed] [Google Scholar]
- 36.Stoneham C.A., Hollinshead M., Hajitou A. Clathrin-mediated endocytosis and subsequent endo-lysosomal trafficking of adeno-associated virus/phage. J Biol Chem. 2012;287(43):35849–35859. doi: 10.1074/jbc.M112.369389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinberg M.S., Nicolson S., Bhatt A.P., McLendon M., Li C., Samulski R.J. Recombinant adeno-associated virus utilizes cell-specific infectious entry mechanisms. J Virol. 2014;88(21):12472–12484. doi: 10.1128/JVI.01971-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Regev-Rudzki N., Yogev O., Pines O. The mitochondrial targeting sequence tilts the balance between mitochondrial and cytosolic dual localization. J Cell Sci. 2008;121(Pt 14):2423–2431. doi: 10.1242/jcs.029207. [DOI] [PubMed] [Google Scholar]
- 39.Braulke T., Bonifacino J.S. Sorting of lysosomal proteins. Biochim Biophys Acta. 2009;1793(4):605–614. doi: 10.1016/j.bbamcr.2008.10.016. [DOI] [PubMed] [Google Scholar]
- 40.Chen P., Tian J., Kovesdi I., Bruder J.T. Promoters influence the kinetics of transgene expression following adenovector gene delivery. J Gene Med. 2008;10(2):123–131. doi: 10.1002/jgm.1127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary material