Abstract
Background and aims
Defective autophagy has been proposed as an important event in a growing number of autoimmune and inflammatory diseases such as rheumatoid arthritis and lupus. However, the precise role of mechanistic target of rapamycin (mTOR)-dependent autophagy and its underlying regulatory mechanisms in the intestinal epithelium in response to inflammation and oxidative stress remain poorly understood.
Methods
The levels of p-mTOR, LC3B, p62 and autophagy in mice and LPS-treated cells were examined by immunoblotting, immunohistochemistry, confocal microscopy and transmission electron microscopy (TEM). We evaluated the expression of IL-1β, IL-8, TNF-α, MDA, SOD and T-AOC by quantitative real time-polymerase chain reaction (qRT-PCR) and commercially available kits after silencing of mTOR and ATG5. In vivo modulation of mTOR and autophagy was achieved by using AZD8055, rapamycin and 3-methyladenine. Finally, to verify the involvement of TLR4 signalling and the NF-κB pathway in cells and active ulcerative colitis (UC) patients, immunofluorescence, qRT-PCR, immunoblotting and TEM were performed to determine TLR4 signalling relevance to autophagy and inflammation.
Results
The mTOR-dependent autophagic flux impairment in a murine model of colitis, human intestinal epithelial cells and active UC patients is probably regulated by TLR4-MyD88-MAPK signalling and the NF-κB pathway. Silencing mTOR remarkably attenuated, whereas inhibiting ATG5 aggravated, LPS-induced inflammation and oxidative injury. Pharmacological administration of mTOR inhibitors and autophagy stimulators markedly ameliorated experimental colitis and oxidative stress in vivo.
Conclusions
Our findings not only shed light on the regulatory mechanism of mTOR-dependent autophagy, but also provided potential therapeutic targets for intestinal inflammatory diseases such as refractory inflammatory bowel disease.
Keywords: Autophagy, mTOR, Inflammation, Oxidative stress
Research in context.
Evidence before this study
mTOR plays a central role in many major cellular processes including growth, metabolism, homeostasis and autophagy/macroautophagy induction. Autophagy is an evolutionarily conserved intracellular degradation process that mediates the triggering of and modulates many aspects of innate and adaptive immune responses, such as antigen presentation, antimicrobial peptide production and cytokines secretion. Cross talk interactions between autophagy and inflammation have been reported in numerous inflammatory and autoimmune diseases such as rheumatoid arthritis and lupus, and we explored its role in the intestinal epithelium and its underlying regulatory mechanism in response to inflammation and oxidative stress.
Added value of this study
The activated mTOR and defective autophagy play essential roles in intestinal inflammatory responses and oxidative stress injury. Silencing mTOR remarkably attenuated, whereas inhibiting ATG5 aggravated, LPS-induced inflammation and oxidative injury. Treatment with mTOR inhibitors and autophagy stimulators remarkably protected the mice from intestinal inflammation and oxidative injury. mTOR-dependent autophagy regulates gut inflammatory responses via its upstream TLR4-MyD88-MAPK signalling and the downstream NF-κB pathway.
Implications of all the available evidence
mTOR-dependent autophagy, along with its regulatory axis TLR4-MyD88-MAPK and NF-κB, may serve as an alternative therapeutic target for patients with gut inflammation. mTOR inhibitors and autophagy stimulators (e.g., rapamycin) are promising candidates for patients with refractory IBD and should be tested in further clinical trials.
Alt-text: Unlabelled Box
1. Introduction
Cytokines and oxidative stress in the gastrointestinal tract in time and space orchestrate the initiation, development, exacerbation and recurrence of the inflammatory process in various intestinal inflammatory diseases [[1], [2], [3]]. The gut epithelium, often considered the primary defence for the host, forms a robust barrier to various infectious or noninfectious stimulations to maintain intestinal mucosal homeostasis [4].
Autophagy is a dynamic intracellular degradation process through which organelles and long-lived proteins undergo lysosome-mediated self-digestion and recycling. The autophagy machinery is initiated under stress conditions such as hypoxia, starvation, exposure to toxic molecules or other reticular stress to maintain cellular homeostasis and adapt to adverse environments [5]. A growing body of evidence has emerged supporting the view that autophagy mediates the crucial functions of triggering and modulating innate and adaptive immune responses such as antigen presentation, cytokines secretion and antimicrobial peptide production [6]. Furthermore, in the intestine, it has been demonstrated that hampered autophagy results in inflammation and contributes to an increased susceptibility to Crohn's Disease (CD) [[7], [8], [9]]. Intense investigations have begun to evaluate the value of autophagy as a novel therapeutic target and a highly needed diagnostic tool. Recent evidence indicates that restoring impaired autophagy can alleviate inflammation in intestinal diseases such as granulomatous diseases and CD both in mice and humans [10]. Therefore, autophagy seems to be an appealing target for regulating and normalizing the imbalanced inflammatory responses in intestinal inflammation.
In addition, autophagy is negatively regulated by the upstream protein kinase mTOR, and both are central regulators of organism growth and homeostasis [11]. mTOR is a highly conserved serine/threonine protein kinase, which consists of two forms of complexes termed mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). Accumulating evidence has revealed that mTORC1 activation is associated with a wide range of intricate signalling pathways, including its downstream target ribosomal protein S6 (RPS6), as well as inflammatory responses [12]. In a recent study of lupus-prone mice, Oaks et al. observed that mTORC1-dependent mitochondrial dysfunction in the livers of lupus-prone mice contributed to the generation of antiphospholipid antibodies (aPL) and this preceded the development of systemic lupus erythaematosus (SLE) [13]. Moreover, studies have shown that the mTOR signalling pathway is of great importance to Nod-2 mediated tolerance and anti-inflammatory cytokines production, and nutrient modulation of mTOR might directly modulate inflammatory responses in CD [14]. Sirolimus (rapamycin), a drug commonly used in post-transplantation management due to its immunosuppressive and antineoplastic properties, has shown significant efficacy in reversing the activation of mTORC1 in T cells in lupus-prone mice and in patients with SLE [13,15,16]. In addition, as has been recently recognized, rapamycin has demonstrated great clinical efficacy in inducing and maintaining clinical remission in IBD patients who were refractory to conventional anti-tumour necrosis factor alpha (anti-TNF-α) therapies [17,18]. However, the functions and the precise mechanisms by which mTOR and autophagy interact in the pathogenesis of intestinal inflammatory diseases, especially in intestinal epithelial cells, remain inconclusive.
Therefore, in this study, LPS was used to induce experimental colitis and oxidative stress in Balb/c mice, human colon adenocarcinoma cell line Caco-2 and HT-29 cells to investigate the interrelations between mTOR and autophagy and the underlying mechanisms. We demonstrated that the activated mTOR and impaired autophagy play essential roles in the intestinal epithelium in intestinal inflammation and oxidative injury based on our in vivo and in vitro studies. Mechanistically, LPS activates mTOR and inhibits autophagy in experimental colitis via the upstream toll-like receptor 4 (TLR4) signalling pathway, and mTOR-dependent autophagy orchestrates its downstream NF-κB activation thereby leading to the production of pro-inflammatory cytokines and oxidative stress. In addition, the dysregulation of mTOR phosphorylation and autophagy-related proteins was further verified in inflamed colorectal tissues from human patients with Active Ulcerative Colitis (A-UC), and the proposed TLR4-MyD88-MAPK signalling and NF-κB pathways were also involved in such regulation. Therefore, our findings not only reveal a novel mechanism for the mTOR-autophagy axis, but also provide solid evidence for further clinical trials of mTOR inhibitors (e.g., rapamycin) as a candidate therapy for intestinal inflammatory diseases such as refractory IBD.
2. Materials and methods
2.1. Cell culture and reagents
The human colonic adenoma cell line Caco-2 and HT-29 were purchased from the Chinese Academy of Sciences (Shanghai, China), and were cultured in DMEM (HyClone, USA) supplemented with 10% foetal bovine serum (FBS, Gibco, USA), 1% penicillin/streptomycin and 1% non-essential amino acids (NEAA, Gibco, USA) in a humidified, 5% CO2 air atmosphere at 37 °C. The chemical reagents used in this study are shown in Supplementary Table 1.
2.2. Patients and specimens
Normal and inflamed colonic mucosal biopsies were collected from healthy controls (HC, n = 19) and active ulcerative colitis (A-UC, n = 31) patients who underwent endoscopy at the Department of Gastroenterology, Xinhua Hospital of Shanghai Jiao Tong University School of Medicine (Shanghai, China) from June 2015 to February 2018. The clinical characteristics of these patients are shown in the Supplementary Table 2. These tissues samples were used for qRT-PCR and TEM analysis. The colorectal inflamed tissues (I, n = 5) and paired distant normal tissues (N, n = 5) were obtained from patients with A-UC who received curative surgery at the Department of Colorectal Surgery in our hospital from October 2015 to November 2017. These tissue samples were used for Western bolt analysis. Moreover, 8 pairs of qualified formalin-fixed paraffin-embedded surgical specimens derived from A-UC patients who underwent curative surgery at the Department of Colorectal Surgery in our hospital from May 2016 to November 2017 were collected and then subjected to immunofluorescence staining. The study was approved by the Research and Ethics committee of Xinhua Hospital. Written informed consent was also obtained from all subjects before the study protocol.
2.3. Western blotting
Tissues and cells were rinsed with pre-chilled PBS twice and then lysed using ice-cold RIPA buffer (Roche, Basel, Switzerland) containing protease (1:100; Thermofisher) and phosphatase inhibitors (1:50; Thermofisher). After centrifugation at 15000 rpm for 15 min at 4 °C, the insoluble debris was removed, and the protein concentration was evaluated by using a BCA Protein Assay Kit (Thermo Fisher, USA). Total cellular protein (20 μg/well) and colorectal tissues protein (50 μg/well) were separated by SDS-PAGE and transferred onto polyvinylidene fluoride (PVDF) membranes. Then, the membranes were blocked with 5% bovine serum albumin (BSA) in Tris borate saline with 0.1% tween 20 (TBST) for 1.5 h at room temperature before being incubated with the corresponding primary antibodies (Supplementary Table 1) at 4 °C overnight. The protein bands were visualized by using a ChemiDoc XRS+ system (Bio-Rad, USA) and the band intensities were quantified using Image Lab software.
2.4. Animals
8-week-old Balb/c female mice (20–24 g) were purchased from the Experimental Animal Center of the Chinese Academy of Sciences (Shanghai, China) and housed in a specific pathogen-free (SPF) facility. All animal experiments were approved by the Institutional Animal Care and Use Committee of Xinhua hospital. The mice were allowed to acclimatized to the controlled environmental conditions (24 ± 1 °C temperature; 40–50% humidity; 12 h light/dark cycle) with free access to water and diet for more than one week prior to the experiments. Every mouse was weighed once a day and monitored for the appearance of bloody and loose stools. Mice were sacrificed after 7 days of LPS-induced acute inflammation or 4 weeks of LPS-induced chronic inflammation. The colorectal tissues were fixed in a 10% paraformaldehyde solution at 4 °C, or snap frozen in liquid nitrogen and stored in a − 80 °C refrigerator.
2.5. Gene silencing by CRISPR/Cas9 and small RNA interference (siRNA)
sgRNA sequence targeting mTOR was synthesized, annealed and subcloned into the lentiCRISPR v2 vector (Addgene, Cambridge, MA) to generate stable knockout cell lines in Caco-2 and HT29 cells, with an empty lentiCRISPR v2 vector used as a control. HEK293T cells were co-transfected with lentiCRISPR v2-control or lentiCRISPR v2-mTOR-KO and psPAX2 and pMD2G using Lipofectamine 2000 (Invitrogen) reagents. Caco-2 and HT29 cells were infected with lentiviruses and then selected by puromycin resistance to establish stable knockouts of mTOR for further experiments.
Caco-2 and HT-29 cells were seeded in 6-well culture dishes at a density of 1.5 × 105 cells/well respectively. After overnight incubation, cells were transfected with small interfering RNA targeting ATG5 (50 nM), TLR4 (50 nM), MyD88 (50 nM), NF-κB P65 (25 nM) or scrambled siRNA using INTERFERin transfection reagent (Polyplus-transfection Inc.) according to the manufacturer's instructions. Gene silencing efficiency was examined by RT-PCR and Western blot after 48 h of transfection. The sequences for siRNA used are shown in Supplementary Table 1.
2.6. ELISA and measurement of oxidative stress
Concentrations of IL-1β, IL-8 and TNF-α in Caco-2 and HT-29 cells were determined by using commercial ELISA quantitative kits according to the manufacturer's instructions. Intestinal and cellular T-AOC, SOD and MDA levels were determined by using commercial kits according to the manufacturer's instructions (Nanjing Jiancheng Bioengineering Institute, China). T-AOC was detected on the basis of the deoxidation ability of Fe3+ to Fe2+. SOD and MDA contents were assayed according to the xanthine oxidase (XO) and thiobarbituric acid methods, respectively.
2.7. Immunohistochemistry
Tissues slides were dehydrated in xylene and rehydrated with graded alcohols. After treatment with 3% peroxide-methanol at 37 °C in an incubator for 30 min, antigen retrieval was assayed by using 0.01 M citrate buffer (PH 6.0) at 95 °C for 20 min. Then, the tissue samples were incubated with diluted primary antibody (anti-LC3B at a 1:200 dilution, or anti-p-RSP6 at a 1:200 dilution) at 4 °C overnight, followed by incubations with biotinylated secondary antibody at 37 °C for 45 min and colouration with 3,3-diaminobenzidine (DAB) at room temperature in the dark. Finally, all sections were counterstained nuclei using hematoxylin. In addition, the expression of p-RPS6 and LC3B in respective groups of LPS-treated mice were scored according to the percentage of positive stained cells and the intensity of staining. The percentage scoring of immunoreactive cells was as follows: 0 (<10%), 1 (10–40%), 2 (40–70%) and 3 (>70%). The staining intensity was visually scored and stratified as follows: 0 (negative), 1 (yellowish), 2 (light brown), 3 (dark brown). The final immunoreactivity score was obtained by two independent researchers by multiplying the percentage and the score for each case was from 0 to 9.
2.8. Transmission Electron Microscopy
Caco-2, HT-29 cells and fresh human mucosal samples were isolated at a size of no >1 mm3 and fixed with 2.5% glutaraldehyde at 4 °C for 4 h. Post-fixation, the samples were dehydrated with a graded series of ethanol (30%, 50%, 70%, 80%, 90%, 95% and 100%) and propylene oxide and then were embedded in an epoxy resin. The embedded samples were then processed for transmission electron microscopy following standard procedures. /Finally, the ultrathin sections were examined with an HT7700 electron microscope (HITACHI, Tokyo, Japan).
2.9. Immunofluorescence staining
The paraffin sections were de-paraffinized, rehydrated, and treated according to a standard protocol. After incubation with the primary antibody (1:50) overnight, the slides were washed 3 times with PBS and incubated with the corresponding biotinylated secondary antibody for 40 min (BOSTER, USA). Then, the sections were washed again and incubated with fluorescein (1:100) for 1 h. Finally, all sections were counterstained with Prolong™ Diamond Antifade Mountant with DAPI and sealed with coverslips. Fluorescence images were captured by using an inverted fluorescence microscope (Leica, Wetzlar, Germany).
2.10. Tandem mRFP-GFP-LC3 fluorescence and confocal microscope
To assess the autophagic flux status in response to LPS stimulation, a tandem mRFP-GFP-LC3 adenovirus (HanBio Technology, Shanghai, China) was transfected into HT-29 cells for 48 h at an MOI of 50. The tandem mRFP-GFP-LC3 protein exhibited both red (mRFP) and green (GFP) fluorescence in neutral pH. The GFP signals could be quenched in the acidic environment of the lysosome, whereas the mRFP signal is more stable in an acidic environment. As a result, the yellow (red+green) and red-only puncta represent the formation of autophagosomes and autolysosomes, respectively. HT-29 cells were plated in a 12-well dish and permitted to reach 50%–70% confluence at the time of transfection. Adenovirus infection was performed in accordance with the manufacturer's guidance, and HT-29 cells were incubated in DMEM growth media with mRFP-GFP-LC3 adenovirus vectors for 2 h at 37 °C. The cells were then grown in normal growth media containing LPS or Bafilomycin A1 at different time points at 37 °C. Finally, LC3 puncta were examined with a Leica TCS SP5 Confocal microscope (Leica Microsystems, Wetzlar, Germany) fitted with a 40× objective.
2.11. RNA isolation and qRT-PCR
The total cellular and tissue RNA was isolated using the RNAiso Plus reagent (TaKaRa Biotechnology). Complementary DNA (cDNA) was reverse transcribed from 1000 ng of total RNA using the PrimeScript™ RT Master Mix (TaKaRa Biotechnology). Quantitative real-time PCR was performed using qPCR SYBR Green Master Mix (Cat No.11201, Yeasen, Shanghai, China) on a PikoReal 96 system (Thermo Scientific, USA) The relative expression levels were normalized to GAPDH (Sangon Biotech, Shanghai, China) and analyzed by the 2-ΔΔCt method [19,20]. All reactions were performed in triplicate. The sequences of primers used in this study are listed in Supplementary Table 1.
2.12. Statistical analysis
Data are presented as the mean ± standard error of mean (SEM). Differences between groups were performed using an unpaired Student's t-test, paired Student's t-test, and one-way analysis of variance (ANOVA). Clinical correlations were analyzed in the human samples using Pearson's correlation coefficient. Statistical analyses was performed using GraphPad Prism 7.0 software (La Jolla, CA, USA). *p < .05, **p < .01, and ***p < .001 were considered as statistical significance.
3. Results
3.1. LPS activates mTOR and inhibits autophagy in acute and chronic experimental colitis in vivo
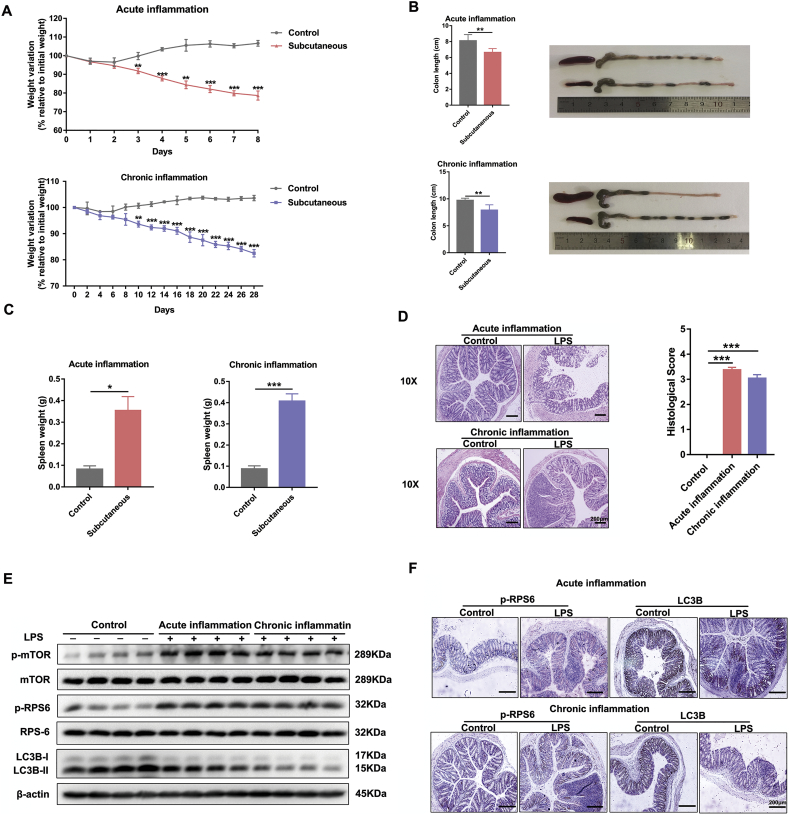
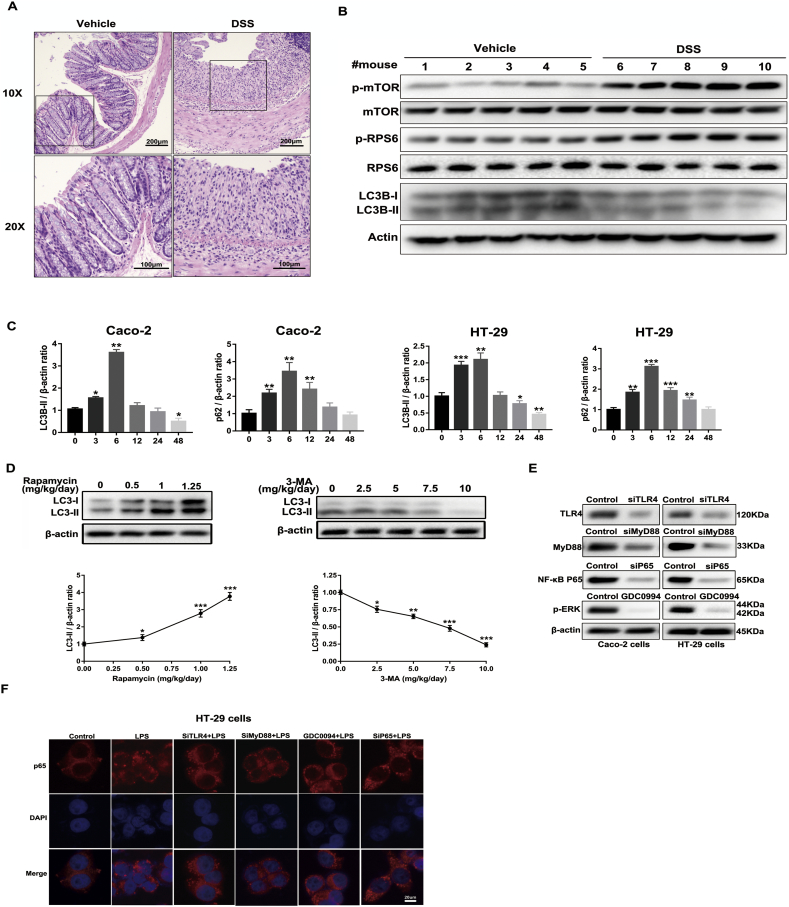
LPS was used to induce acute and chronic experimental colitis in Balb/c mice, and the results showed that administration of LPS markedly caused impairment of colon epithelium tissues, as evidenced by the body weight loss from initial weights (Fig. 1A), the diminution of colorectal length (Fig. 1B) and the obvious change in spleen weight (Fig. 1C) [21,22]. Furthermore, histological examination (H&E staining and histological score) of distal colon sections from the LPS-challenged mice exhibited remarkably higher mucosal inflammatory cell infiltration and more severe damage of the epithelial layer compared to that of the control group (Fig. 1D). To explore whether mTOR and autophagy are biologically modulated in LPS-induced intestinal inflammation, we examined the protein levels of the autophagy-related hallmark Microtubule associated protein 1 light chain 3 beta (LC3B), mTOR and related molecules. As shown in Fig. 1E, the expression of p-mTOR and its downstream indicator p-RPS6 was remarkably elevated in LPS-treated mice, which was in parallel with the diminished level of LC3B. These observations were reinforced by immunohistochemistry performed on colon tissues that revealed an enhanced p-RPS6 but reduced LC3B immunostaining in LPS-challenged mice compared with controls (Fig. 1F). These results were further verified in DSS-induced experimental colitis models (Supplementary Fig. 1A, B).
Fig. 1.
LPS activates mTOR and inhibits autophagy in acute and chronic experimental colitis in vivo. Mice were treated with subcutaneous administration of LPS (2.5 mg/kg/day for acute inflammation and 1.25 mg/kg/day for chronic inflammation) or vehicle and were killed 7 or 28 days after treatment (n = 6 for each group). (A) Loss of basal body weight, (B) colorectal length, (C) spleen weight change, and (D) representative H&E stained sections and histological score of the distal colon in differently treated mice as described above. Scale bar = 200 μm. (E) Proteins levels of p-mTOR, m-TOR, p-RPS6, RPS6, LC3B in colon tissues of vehicle- or LPS-treated mice. Data are the representative of three independent experiments with similar results. (F) Representative images showing p-RPS6 and LC3B immunostaining in the colon tissues injected with or without LPS. Scale bar = 200 μm. All data shown are representative of at least 2 independent experiments. In all cases, bars in the graphs represent mean ± s.e.m. Significant differences from the respective vehicle-treated mice are shown by *p < .05, ** p < .01, ***p < .001. Statistical analyses were performed by unpaired t-test.
Supplementary Fig. 1.
(A) Acute colitis was induced in 8-week-old C57BL/6 mice (20-24 g) with 3% DSS (MP Biomedicals, USA) dissolved in drinking water given ad libitum for 7 days. During the experimental period, clinical signs, body weight and stool consistency were monitored and recorded daily. On day 8, mice were sacrificed and the paraffin sections of colon tissues were stained with hematoxylin and eosin. (B) The protein levels of LC3B, p-mTOR and related molecules in DSS-induced model in C57BL/6 mice were determined by western blot. Data are the representative of three independent experiments with similar results. (C) Graphs shows a quantitative analysis of p62 and LC3B time-dependent expression in Caco-2 and HT-29 cells stimulated with LPS (1 μg/ml). Bars in the graphs represent mean ± s.e.m. Significant differences from vehicle-treated cells are shown by *p < .05, ** p < .01, ***p < .001. (D) In a preliminary dose-response experiment, mice were randomly assigned to four groups (n = 5) by different dose of rapamycin (0, 0.5, 1, 1.25 mg/kg/day) administrated for 10 days, then sacrificed for LC3B expression assessment by western blot. The results showed that 1.25 mg/kg/day rapamycin has the best inductive effects of autophagy activity. In another preliminary dose-response experiment, mice were randomly assigned to five groups (n = 5) by different dose of 3-MA (0, 2.5, 5, 7.5, 10 mg/kg/day) administrated for 10 days, then sacrificed for LC3B expression assessment by western blot. The results showed that 10 mg/kg/day 3-MA has the best inhibition effects of autophagy activity. Then the dose of 1.25 mg/kg/day rapamycin and 10 mg/kg/day 3-MA were chosen for subsequent experiment. Bars in the graphs represent mean ± s.e.m. Significant differences from vehicle-treated mice are shown by *p < .05, ** p < .01, ***p < .001. (E) The knockdown efficiency of TLR4, MyD88, NF-κB p65 and p-ERK inhibitor GDC-0994 in Caco-2 and HT-29 cells were analyzed by western blot. (F) HT-29 cells were pretreated with different pharmacological or genetic treatments before LPS stimulation for another 6 h. NF-κB p65 translocation was determined by immunofluorescence assay. Scale bar = 20 μm. The data shown were from three independent experiments. In all cases, bars in the graphs represent mean ± s.e.m. Statistical analyses were performed by unpaired t-test or one-way ANOVA. Significant differences from cells treated without LPS are shown by *p < .05, ** p < .01, ***p < .001.
3.2. LPS triggers mTOR phosphorylation and autophagic flux impairment in Caco-2 and HT-29 cells in vitro
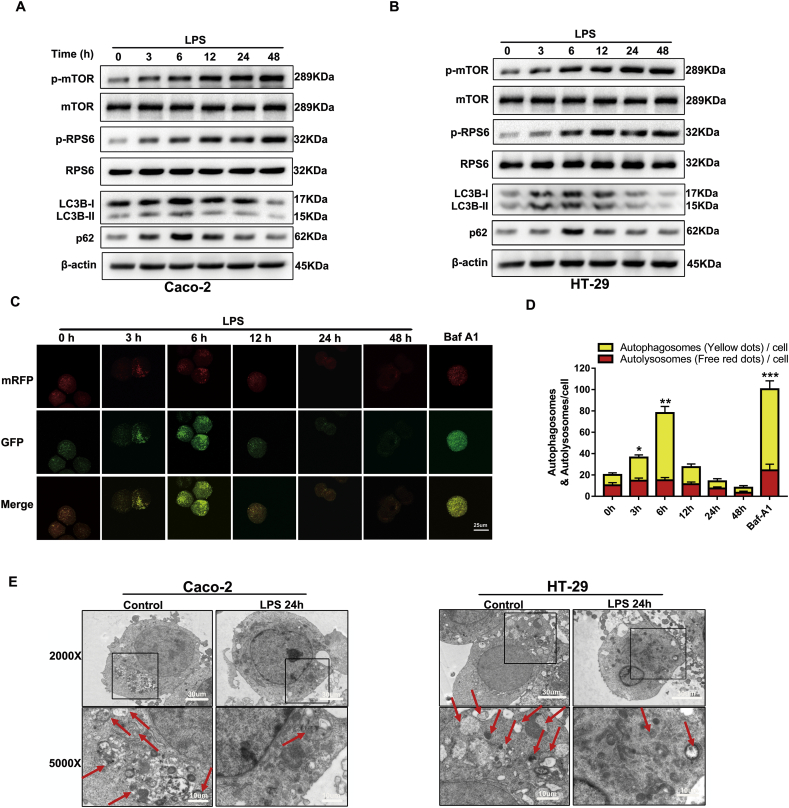
To determine the role of mTOR signalling and autophagy in human intestinal epithelial cells after LPS stimulation, we investigated the expression of p-mTOR, p-RPS6, LC3B and p62 by western blot. Notably, the protein levels of p-mTOR and p-RPS6 were gradually increased in a time-dependent manner upon LPS exposure, while the ratio of LC3B-II/β-actin reached a peak at 6 h and then declined gradually over the time-course (Fig. 2A, B and Supplementary Fig. 1C). The expression of p62 (a cargo protein degraded inside autolysosomes) also increased to its maximum at 6 h and then decreased with time prolonged, which indicated that the autophagic flux was blocked at autolysosome degradation (Fig. 2A, B and Supplementary Fig. 1C). To further confirm the autophagic flux status, we transfected cells with Ad-mRFP-GFP-LC3 adenovirus. The GFP signals can be quenched in the acidic environment of the lysosome, whereas the mRFP signal is more stable in an acidic environment. As a result, autolysosomes and autophagosomes are labelled with a red or yellow (red and green) colour, respectively. By detecting these two different fluorescent signals, the autophagic flux status after LPS stimulation can be monitored. Therefore, the autophagic flux is enhanced when both yellow and red puncta are increased in intestinal epithelial cells, while the autophagic flux is impeded when both yellow and red puncta are decreased in cells or when only yellow puncta are intensified without red puncta alteration. After transfection with mRFP-GFP-LC3 for 48 h and incubation with LPS at different time points, the cells were examined by confocal microscopy. As shown in Fig. 2C, treatment of the cells with bafilomycin A1, which is widely used to inhibit autophagosome-lysosome fusion, only increased the number of yellow dots. Furthermore, LPS treatment markedly enhanced the yellow dots and reached its maximum at 6 h, while it did not augment the number of red dots (Fig. 2C, D). This phenomenon indicated that the autophagic flux was retarded at the autolysosome stage after treatment with LPS. These results were consistent with transmission electron micrographs (Fig. 2E), suggesting that LPS activates mTOR and then blocks the autophagy process at the autolysosome stage in both in vivo and in vitro inflammatory models.
Fig. 2.
LPS triggers mTOR phosphorylation and blocks autophagic flux in Caco-2 and HT-29 cells in vitro. Time-dependent expression of p-mTOR, mTOR, p-RPS6, RPS6, LC3B and p62 in epithelial Caco-2 (A) and HT-29 cells (B) stimulated with LPS (1 μg/ml). Data are the representative of three independent experiments with similar results. (C) Representative fluorescent images of HT29 cells transfected with ad-mRFP-GFP-LC3 adenovirus after incubation with or without LPS and Baf A1 at different time points. Scale bar = 25 μm. (D) Mean number of autophagosomes (dots with yellow colour in merged images) and autolysosomes (dots with only red colour in merged images) per cell. The number of red and yellow LC3 dots per cell were counted under confocal microscope (>30 cells/group).Adenovirus was transfected at 50 MOI. Data were shown as mean ± s.e.m. and replicated three times. (E) Representative TEM images of autophagosome/autolysosome (red sword) accumulation in Caco-2 and HT-29 cells treated with or without LPS. Scale bar = 30 μm/10 μm. The data shown were from three independent experiments. Significant differences from cells treated without LPS are shown by *p < .05, ** p < .01, ***p < .001. Statistical analyses were performed by one-way ANOVA.
3.3. Silencing mTOR remarkably reduces LPS-induced intestinal inflammation and oxidative stress in Caco-2 and HT-29 cells
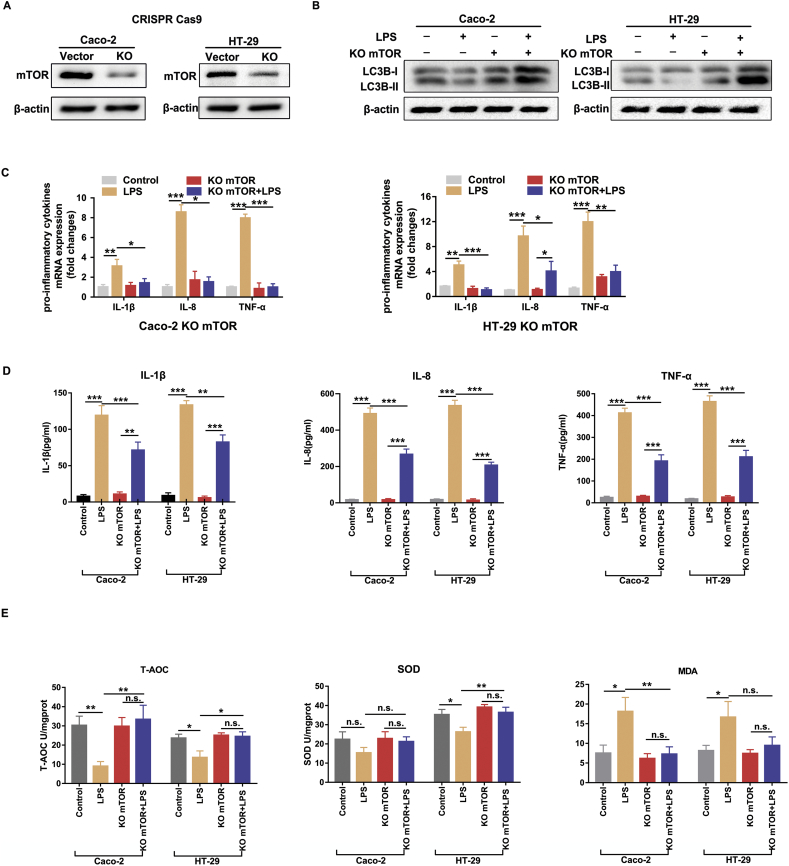
We next sought to investigate the effects of mTOR inhibition on LPS-induced intestinal inflammation and oxidative stress. We performed genetic knockout of mTOR using the CRISPR/Cas9 system, and immunoblotting confirmed that mTOR protein expression was efficiently down-regulated in Caco-2 and HT-29 cells (Fig. 3A). As expected, the silencing of mTOR slightly increased the basal level of autophagy and restored LPS-decreased levels of LC3B (Fig. 3B). Meanwhile, the real-time PCR and ELISA results demonstrated that the mRNA and protein levels of pro-inflammatory cytokines (IL-1β, IL-8 and TNF-α) were significantly down-regulated in mTOR silenced cells after LPS treatment (Fig. 3C, D). Malondialdehyde (MDA), total antioxidant capacity (T-AOC) and superoxide dismutase (SOD) have been widely served as oxidative stress markers. As shown in Fig. 3E, LPS-treated cells exhibited an obvious oxidative injury as indicated by the weakened T-AOC and SOD activities and elevated MDA levels. However, these detrimental effects of LPS stimulation were dramatically ameliorated by inhibiting mTOR in Caco-2 and HT-29 cells. These results indicated that the inhibition of mTOR protects against LPS-induced inflammation and oxidative injury, accompanied with an induction of autophagy.
Fig. 3.
Silencing mTOR significantly ameliorated LPS-induced intestinal inflammation and oxidant stress. Caco-2 and HT-29 cells were transfected with lenti-CRISPR vector and then incubated with LPS for subsequent quantitative real-time PCR, ELISA, western bolt and oxidative stress assays. (A) The silencing efficiency of mTOR in Caco-2 and HT-29 cells was determined by western blot. (B) The levels of LC3B were analyzed by western blot. Data are the representative of three independent experiments with similar results. (C) The mRNA expression of pro-inflammatory cytokines (IL-1β, IL-8, TNF-α) were determined by quantitative real-time PCR. (D) The protein levels of pro-inflammatory cytokines (IL-1β, IL-8, TNF-α) were determined by commercial ELISA kits according to the manufacturer's instructions. (E) Cellular oxidative stress markers (T-AOC, SOD and MDA levels) were tested by using commercially available kits according to the manufacturer's instructions. Bars in the graphs represent mean ± s.e.m. The data are representative of three independent experiments in triplicate. Statistical analyses were performed by unpaired t-test or one-way ANOVA. Significant differences are shown by *p < .05, ** p < .01, ***p < .001.
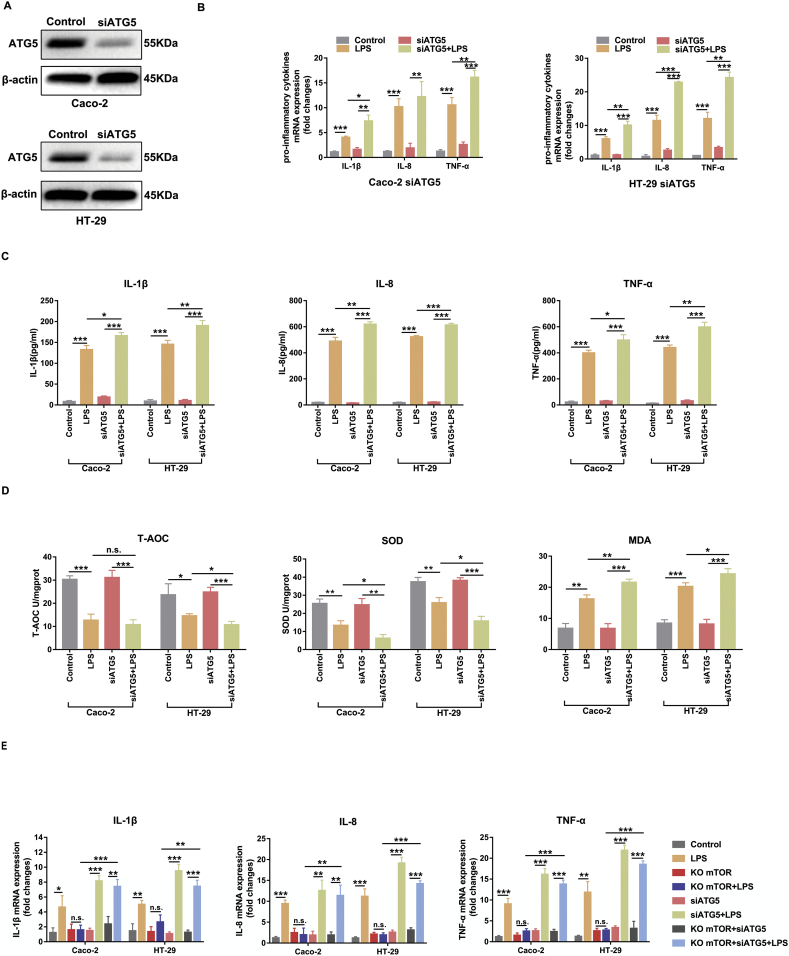
3.4. Inhibiting autophagy aggravated LPS-induced inflammation and oxidative injury in Caco-2 and HT-29 cells
Mounting evidence suggests that autophagy plays essential roles in inflammation, immunity and the maintenance of intestinal homeostasis. As indicated above, LPS triggered mTOR phosphorylation and inhibited autophagy, while silencing mTOR enhanced the autophagy level. Therefore, we next sought to explore the influence of autophagy on intestinal inflammation and oxidative stress. Considering that ATG5 is an important molecule involved in the autophagosome formation process, we applied siRNAs to block the expression of ATG5 and reduce autophagy activity. The effects of the knockdown of ATG5 in Caco-2 and HT-29 cells are shown in Fig. 4A. As expected, we found that siATG5 treatment induced the up-regulation of pro-inflammatory cytokines (IL-1β, IL-8 and TNF-α) and aggravated oxidative injury (MDA, T-AOC and SOD) in the presence of LPS stimulation in Caco-2 and HT-29 cells (Fig. 4B-D). We further examined whether knockdown of ATG5 in mTOR silenced cells could reverse the protective effects on inflammatory responses after LPS treatment. Notably, the mRNA levels of IL-1β, IL-8, and TNF-α were up regulated again (Fig. 4E). These data confirmed that LPS induced inflammation and oxidative injury via activating mTOR and suppressing autophagy in intestinal epithelial Caco-2 and HT-29 cells.
Fig. 4.
Inhibition of ATG5 aggravated LPS-induced intestinal inflammation and oxidant stress. Caco-2 and HT-29 cells were transfected siRNA and then incubated with LPS for subsequent quantitative real-time PCR, ELISA, western bolt and oxidative stress assays. (A) The knockdown efficiency of ATG5 in Caco-2 and HT-29 cells was determined by western blot. Data are the representative of three independent experiments with similar results. (B) The mRNA expression of pro-inflammatory cytokines (IL-1β, IL-8, TNF-α) were determined by quantitative real-time PCR. (C) The protein levels of pro-inflammatory cytokines (IL-1β, IL-8, TNF-α) were determined by commercial ELISA kits according to the manufacturer's instructions. (D) Cellular oxidative stress markers (T-AOC, SOD and MDA levels) were analyzed by using commercially available kits according to the manufacturer's instructions. (E) To explore whether knocking down ATG5 in mTOR silencing cells could reverse its protective effects against LPS-induced inflammatory responses, quantitative real-time PCR was performed to analysis the mRNA levels of IL-1β, IL-8, and TNF-α. Bars in the graphs represent mean ± s.e.m. Statistical analyses were performed by unpaired t-test or one-way ANOVA. Significant differences are shown by *p < .05, ** p < .01, ***p < .001.
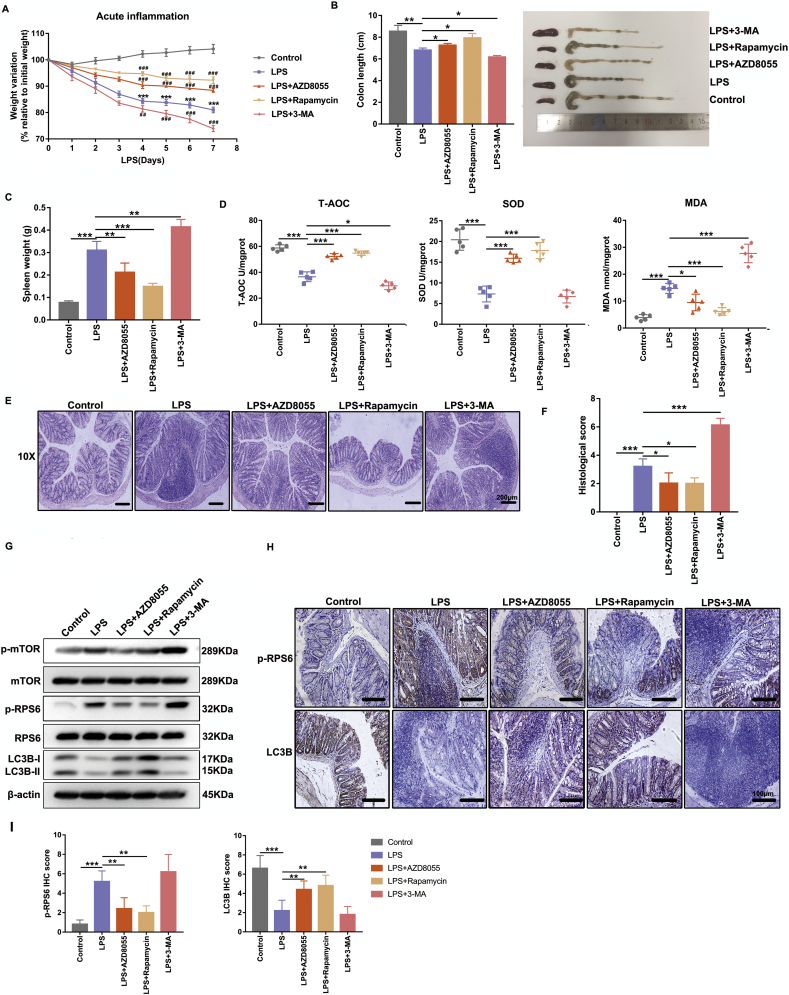
3.5. Pharmacological administration of mTOR inhibitors and autophagy stimulators markedly ameliorated LPS-induced experimental colitis and oxidative stress in vivo
To further elucidate the function of mTOR activation and autophagy suppression in intestinal epithelium in facilitating LPS-induced inflammatory responses and oxidative injury in vivo, we applied inhibitors or agonists to modulate mTOR and regulate the autophagy process. Interestingly, administration of AZD8055 (a selective ATP-competitive kinase inhibitor of mTOR) or rapamycin (an mTOR-dependent stimulator of autophagy) to LPS-treated mice significantly ameliorated intestinal inflammation and oxidant stress as judged by decreased body weight loss (Fig. 5A), colon length shortening (Fig. 5B), spleen weight augmentation (Fig. 5C), much lower MDA level, higher cellular T-AOC and SOD antioxidant activities (Fig. 5D), as well as less disruption of mucosal epithelium and inflammatory cell infiltration in colon tissues (Fig. 5E), and histological severity scores in comparison with those of LPS group (Fig. 5F). In contrast, inhibition of autophagy by 3-Methyladenine (3-MA) in LPS-treated mice remarkably aggravated oxidative injury and histological damage (Fig. 5A-F). In addition, the levels of p-mTOR and p-RPS6 were prominently slashed after AZD8055 or rapamycin administration, accompanied with the induction of autophagy in colon tissues as detected by Western blot (Fig. 5G). The levels of p-mTOR, p-RPS6 and LC3B stimulated by LPS were intensified by administering 3-MA to mice (Fig. 5H-I). Consistent with the above data, the immunostaining of p-RPS6 and LC3B further confirmed that inhibiting mTOR and triggering autophagy significantly attenuated LPS-induced intestinal inflammatory responses and oxidative stress in vivo (Fig. 5G-I).
Fig. 5.
Pharmacological administration of mTOR inhibitors and autophagy stimulators markedly ameliorated LPS-induced experimental colitis and oxidative stress in vivo. The 7- to 8-week Balb/c mice (20-24 g) were treated with subcutaneous injection of LPS (2.5 mg/kg/day) or vehicle and were killed 7 days after treatment (n = 5 for each group). Some mice received an additional administration of AZD8055 (10 mg/kg/day, i.p), rapamycin (1.25 mg/kg/day, i.p) or 3-Methyladenine (10 mg/kg/day, i.p) three days before LPS administration until sacrifice. Graphs show (A) body weight as a percentage of initial weight, measured daily after LPS treatment. *p < .05, ** p < .01, ***p < .001 versus control; #P < .05, ##P < .01 and ###p < .001 versus LPS group. Representative images of colon length shortening (B), spleen weight augment (C) from differently treated mice as described above. (D) The T-AOC activities, SOD activities and MDA abundance of colon tissues from differently treated mice were analyzed by using commercially oxidative stress kits. Representative images of H&E staining (E) and histological severity scores (F) of distal colon tissues. Scale bar = 200 μm. The sections from LPS-treated mice were scored on a scale of 0–4 based on % of colon involvement by inflammation, % of crypt loss, erosions, edema, and density of inflammatory cells. The scores display the sum of the parameters for a total severity score. (G) Effects of AZD8055, Rapamycin or 3-MA on p-mTOR, mTOR, p-RPS6, RPS6 and LC3B protein levels in the colon mucosa of differently treated mice were shown by representative western blot. Data are the representative of three independent experiments with similar results. Representative graphs showing p-RPS6 and LC3B immunostaining (H) and the final immunoreactivity scores (I) in the mucosa of mice receiving different treatments. Scale bar = 100 μm. All data shown are representative of at least 2 independent experiments. The bars in graphs represent mean ± s.e.m. Statistical analyses were performed by unpaired t-test or one-way ANOVA. Significant differences are shown by *p < .05, ** p < .01, ***p < .001.
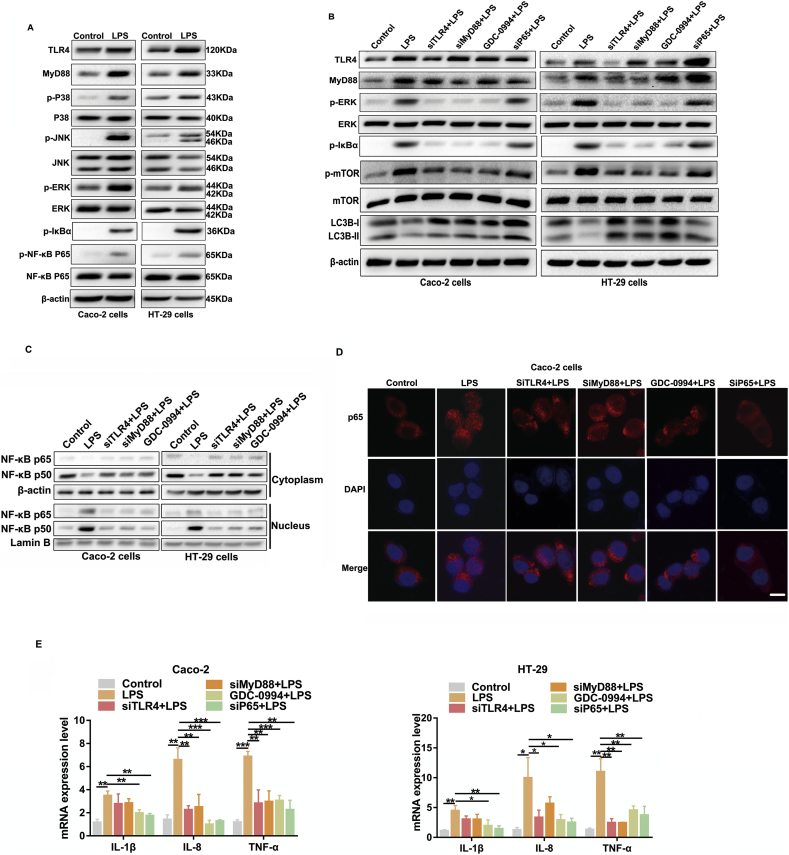
3.6. LPS activates mTOR via upstream TLR4-MyD88-MAPK signalling and inhibits autophagy through the NF-κB pathway in epithelial Caco-2 and HT-29 cells
To uncover the potential pathways that orchestrate LPS-induced mTOR activation and autophagy suppression in the intestinal epithelium, we focused on toll-like receptors (TLRs), which provoke a quick activation of innate immunity by inducing secretion of pro-inflammatory cytokines. TLR4, in particular, is a membrane receptor for LPS and abundantly expressed in various epithelial cells [23,24]. Notably, treatment with LPS in Caco-2 and HT-29 cells significantly enhanced the protein levels of TLR4, MyD88, p-p38 MAPK, p-JNK, p-ERK and p-NF-κB p65 (Fig. 6A). Moreover, we utilized an ERK inhibitor (GDC-0994) and three siRNAs to knockdown TLR4, MyD88 and NF-κB p65 to further validate the role of TLR4 signalling in modulating the mTOR-dependent autophagy (Fig. 6B and Supplementary Fig. 1E). As shown in Fig. 6B, knockdown of TLR4 and MyD88 significantly inhibited the phosphorylation level of ERK, IκBα, mTOR and promoted autophagy. The suppression of ERK phosphorylation by GDC-0994 also inhibited p-mTOR and activated autophagy. However, down-regulation of p65 remarkably strengthened the protein levels of autophagy while it had little effect on p-mTOR. As shown in Fig. 6C, D and Supplementary Fig. 1F, LPS significantly induced NF-κB p50 and p65 translocation into the nucleus, which was blocked by pretreating the cells with genetic or pharmacological inhibition agents in western blot and immunofluorescence staining assays. Furthermore, genetic or pharmacological knockdown of TLR4, MyD88, p-ERK and NF-κB p65 effectively decreased the mRNA levels of pro-inflammatory cytokines induced by LPS (Fig. 6E), which was in parallel with high levels of autophagy and low levels of mTOR phosphorylation. Taken together, these results present convincing evidence to support our emerging view that LPS activates mTOR via upstream TLR4-MyD88-MAPK signalling and inhibits autophagy through the NF-κB pathway in intestinal epithelial cells.
Fig. 6.
LPS activates mTOR via upstream TLR4-MyD88-MAPK signalling and inhibits autophagy through NF-κB pathway in epithelial Caco-2 and HT-29 cells. (A) The protein levels of TLR4, MyD88, p-p38 MAPK, p38 MAPK, p-JNK, JNK, p-ERK, ERK, p-NF-κB p65 and NF-κB p65 in epithelial Caco-2 and HT-29 cells stimulated with LPS were analyzed by western blot. (B) Representative western blots showing TLR4 signalling associated molecules, p-mTOR and LC3B protein levels in vehicle or LPS-treated cells receiving different pharmacological or genetic interference. (C) Representative western blot images are shown for NF–κB p65 and p50 nuclear translocation and cytoplasmic levels in LPS-treated Caco-2 and HT-29 cells with genetic or pharmacological administration. NF-κB subunit signals were normalized to the nuclear marker lamin B1. Data are the representative of three independent experiments with similar results. (D) Caco-2 cells were pretreated with different pharmacological or genetic treatments before LPS stimulation for another 6 h. NF-κB p65 translocation was determined by immunofluorescence assay. Scale bar = 20 μm. (E) Graphs show the mRNA levels of pro-inflammatory cytokines (IL-1β, IL-8, TNF-α) in vehicle or LPS-treated cells receiving different pharmacological or genetic treatments. The data are representative of three independent experiments in triplicate. Bars in the graphs represent mean ± s.e.m. Statistical analyses were performed by unpaired t-test or one-way ANOVA. Significant differences are shown by *p < .05, ** p < .01, ***p < .001.
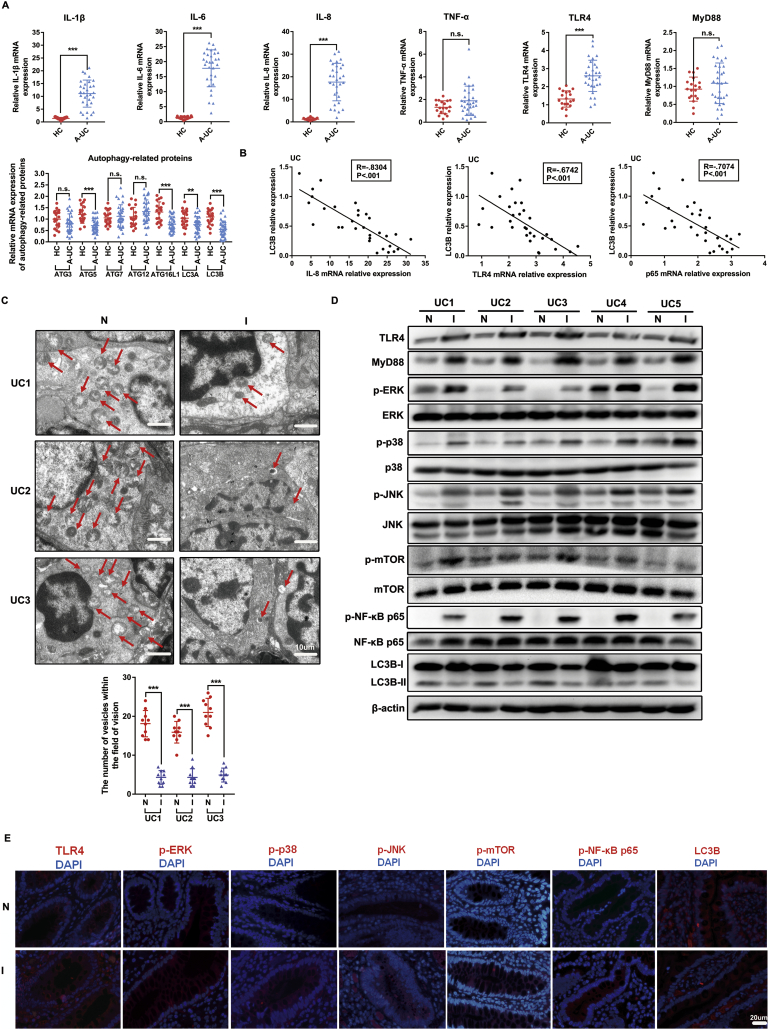
3.7. TLR4-MyD88-MAPK signalling and the NF-κB pathway are involved in the dysregulation of p-mTOR and autophagy in colon tissues of UC patients
We further assessed whether there was a dysregulation of mTOR phosphorylation and autophagy-related proteins in human patients with A-UC and the possible participation of TLR4-MyD88-MAPK signalling and the NF-κB pathway in such regulation. As shown in Fig. 7A, the qRT-PCR results showed increased IL-1β, IL-6, IL-8, TLR4 and NF-κB p65 expression and decreased autophagy-related molecules (such as ATG5, ATG16L1, LC3A and LC3B) mRNA expression in the inflamed mucosa of patients with A-UC (n = 31), compared with that in healthy controls (n = 19). However, there was no notable difference in the expression of TNF-α, MyD88, ATG3, ATG7 and ATG12 between the inflamed mucosa of A-UC and healthy controls. We found that the expression of the autophagy hallmark LC3B in A-UC was negatively correlated with the levels of IL-8 (Spearman's rank correlation coefficient in UC, R = −0.8304, p < .001), TLR4 (R = −0.6742, p < .001) and NF-κB p65 (R = −0.7074, p < .001) mRNA (Fig. 7B), indicating TLR4 and the NF-κB pathway probably inversely regulated the expression of autophagy-related proteins. As shown in Fig. 7C, we examined the number of autophagic vesicles in mucosa tissues of 3 UC patients from colonoscopy using TEM. Autophagosomes, also known as initial autophagic vacuoles (AVi), were generally defined as vacuolar structures containing cellular contents similar to intracellular cytosol and organelles, as presented in Fig. 7C. The number of autophagic vesicles was counted in at least 10 fields of view in each section at the same magnification (Fig. 7C). A lower number of autophagic vesicles was observed in inflamed tissues compared to adjacent normal tissues, confirming that the inflamed intestine mucosa exhibited more impaired autophagy activity than adjacent normal mucosa. We also noticed increased protein levels of p-mTOR, TLR4, MyD88, p-P38 MAPK, p-JNK, p-ERK and p-NF-κB p65 and decreased LC3B protein levels in 5 pairs of inflamed colon biopsy tissues compared with their corresponding adjacent normal colon tissues from the same A-UC patients (Fig. 7D). Corroborating the above data, the immunofluorescence staining of p-mTOR, LC3B and other molecules in the proposed pathway further confirmed that the activated p-mTOR and impaired autophagy in inflamed colon tissues of UC patients were regulated by TLR4-MyD88-MAPK signalling and the NF-κB pathway (Fig. 7E).
Fig. 7.
TLR4-MyD88-MAPK signalling and NF-κB pathway are involved in the regulation of p-mTOR and autophagy in colon tissues of UC patients. (A) Expression of IL-1β, IL-6, IL-8, TNF-α, TLR4, MyD88 and autophagy-related proteins mRNA in human biopsies were quantified by qRT-PCR. The data are representative of three independent experiments. Bars in the graphs represent mean ± s.e.m. *p < .05, ** p < .01, ***p < .001 versus healthy controls. (B) Correlation analysis were performed between the relative levels of LC3B mRNA and IL-8 expression (Spearman's rank correlation coefficient, R = −0.8304, p < .001), LC3B mRNA and TLR4 expression (Spearman's rank correlation coefficient, R = −0.6742, p < .001) and LC3B mRNA and p65 expression (Spearman's rank correlation coefficient, R = −0.7074, p < .001) in inflamed mucosa from 31 A-UC patients. (C) Representative TEM images of autophagosome/autolysosome (red sword) accumulation in inflamed mucosa tissues (I) and adjacent normal mucosa tissues (N). Scale bar = 10 μm. The quantification of the number of autophagic vesicles in 10 visions in each inflamed mucosa tissues and adjacent normal mucosa tissues. Bars in the graphs represent mean ± s.e.m. Significant differences are shown by *p < .05, ** p < .01, ***p < .001. (D) The protein levels of TLR4, MyD88, p-P38 MAPK, p-JNK, p-ERK, p-mTOR, p-NF-κB p65 and LC3B in 5 paired of inflamed colon biopsy tissues (I) and adjacent normal colon tissues (N) were determined by western blot. Data are the representative of three independent experiments with similar results. (E) Representative immunofluorescent images for detection of TLR4, p-ERK1/2, p-P38 MAPK, p-JNK, p-mTOR, p-NF-κB p65, NF-κB p65 and LC3B (red) and DAPI (blue). Scale bar = 20 μm. The data shown are from three independent experiments. Statistical analyses were performed by either unpaired t-test or paired t-test.
4. Discussion
The major findings of our study can be summarized as follows. (1) Activated mTOR and impaired autophagy play pivotal roles in intestinal inflammatory responses and oxidative stress injury. (2) Mechanistically, mTOR-dependent autophagy regulates gut inflammatory responses via its upstream TLR4-MyD88-MAPK signalling and the downstream NF-κB pathway. (3) The pharmacological inhibition of mTOR and activation of autophagy remarkably ameliorated intestinal colitis through inhibiting inflammation and oxidative stress.
The gut microbiome, containing at least 1014 bacteria, resides in the mucosal surface of the human intestine. The direct or indirect homeostasis of gut microbiota influences the physiology of the gastrointestinal tract and links it to the pathogenesis of IBD. LPS, as an important constituent of the cell wall of gram-negative bacteria, trigger lots of inflammatory signals through the LPS component of gram-negative bacteria. Therefore, in this study, LPS was used as an inflammatory agent to identify a novel role for mTOR and autophagy in intestinal inflammation and oxidative stress. Inflammation represents a crucial line of defence against microorganisms and other pathogens. Increasing evidence suggests that autophagy has important effects on the induction and modulation of the inflammatory reaction, however, the role of mTOR and autophagy in intestinal inflammation, the functions and the underlying mechanisms by which mTOR and autophagy interrelate in IBD pathogenesis, especially in the intestinal epithelial cells, have yet to be fully understood. Therefore, in this study, we modulate mTOR and autophagy to explore the balance between these two processes with the aim of seeking the possibilities for further clinical therapeutic targeting.
In recent years, the crosstalk between autophagy and inflammation has been reported. Emerging literature has explored the function of autophagy in diverse cellular models of inflammation, showing that autophagy plays an essential role in the monitoring of inflammation and restoring intestinal homeostasis [10]. In the intestinal, autophagy has been reported to mediate critical functions in innate and adaptive immunity such as antigen presentation by dendritic cells, cytokine secretion by macrophages and antimicrobial peptide secretion by paneth cells [25]. Following treatment with LPS, ATG16L1- or Atg7-deficient macrophages produce large amounts of pro-inflammatory cytokines such as IL-1β and IL-18. Mice lacking ATG16L1 in haematopoietic cells are more susceptible to dextran sulphate sodium (DSS)-induced experimental acute colitis, which could be alleviated by administration with anti-IL-1β antibodies, demonstrating the significance of the autophagy-related protein ATG16L1 in the repression of gut inflammation [26]. Subsequent studies demonstrated that anything that blocks the autophagy process, be it Atg gene deletion or pharmacological intervention, leads to enhanced caspase-1 activation and expanded production of pro-inflammatory cytokines such as IL-1β [27]. Furthermore, IL-10−/− mice show an impaired autophagic flux in the intestinal mucosa, together with the increased expression of pro-inflammatory genes and the activation of the NF-κB pathway [28]. Ortiz-Masia et al. reported that HIF-1-dependent induction of Wnt1 in hypoxic macrophages undermined the autophagy process in epithelial cells, indicating the participation of Wnt signalling in the impaired autophagy in the mucosa of IBD patients [28]. Moreover, a growing body of literatures has demonstrated the close link between autophagy and antioxidant defence in the gastrointestinal tract [29]. On the one hand, autophagy modulates the inflammatory response through several mechanisms, including the selective degradation of both pro-inflammatory complexes, such as NEMO or BCL10 or inflammasome stimuli like mitochondrial ROS or mitochondrial DNA [30]. On the other hand, high levels of cytokines production and ROS, in response to damage have been reported to disrupt the signalling needed to control autophagy process [31]. Therefore, the protein levels of autophagy hallmark LC3B in Caco-2 and HT-29 cells increased from 0 to 6 h under the stimulation of LPS probably due to it's a highly sensitive process in response to stressful condition in order to maintain cellular homeostasis. However, the high levels of cytokines production and ROS may reversely influence and disrupt autophagy process and lead to the impaired autophagy. Moreover, p62, a cargo protein degraded inside autolysosomes, also increased to its maximum at 6 h and then decreased with time prolonged, which indicated that the autophagic flux was blocked in the combination with the change of LC3B during the autolysosome degradation process. And we will explore the underlying mechanisms of the changing LC3B, p62 with time prolonged in depth in further studies. In addition, activation of autophagy has been identified as cytoprotective in some settings, whereas in other cases, sustained or excessive activation of autophagy has been associated with inflammatory pathologies [32,33]. Recent investigations have shown that the phagocytic uptake of cells dying through autophagy induction in human macrophages evokes a pro-inflammatory response as characterized by the induction and secretion of cytokines such as TNF-α, IL-6, IL-8 and IL-10 [34,35]. Consistent with the former viewpoint, in the current study, we discovered that the impeded autophagic flux associated with the phosphorylation of mTOR in LPS-induced Caco-2 and HT-29 cells, murine colitis and A-UC patients, extending the previous knowledge of defective autophagy in the regulation of intestinal inflammation.
mTOR governs the programmed cell death pathways of apoptosis and autophagy that can determine cell fate. The pathways of PI3K, Akt, and AMPK play a significant role in mTOR signalling. In regards to AMPK, this protein can inhibit mTORC1 activity through activation of tuberous sclerosis 1/tuberous sclerosis 2 (TSC1/TSC2) complex. AMPK suppress and block mTORC1 activity via controlling the activity of TSC1/TSC2 [36]. Therefore, coupled to the cellular biology of mTOR are a number of considerations for the development of novel treatments involving the fine control of mTOR signalling and autophagy in the intestinal inflammation. In the present study, the complexity of mTOR signalling has been taken into considerations in pharmacological administration the mice with inhibitors or agonists to modulate mTOR and autophagy. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mTOR kinase inhibitor with excellent selectivity (approximately 1000-fold) against all class I phosphatidylinositol 3-kinase (PI3K) isoforms and other members of the PI3K-like kinase family [37]. AZD8055 inhibits the phosphorylation of mTORC1 substrates p70S6K and 4E-BP1 as well as phosphorylation of the mTORC2 substrate AKT and downstream proteins [38]. Moreover, 3-methyladenine (3-MA) inhibits class III PI3K and is widely used as an autophagy inhibitor in various studies [39,40]. Interestingly, our in vivo data showed that systemic administration of a mTOR inhibitor (AZD8055) or a mTOR-dependent autophagy stimulator (rapamycin) to LPS-treated mice significantly ameliorated gut inflammation and oxidant stress as shown by decreased body weight loss and histological damage, where the inhibition of autophagy by 3-MA in LPS-treated mice remarkably aggravated oxidative injury and histological damage. Indeed, the gut-specific Atg5 or ATG16 knockout mouse models could mimic human IBD patients in which autophagy is genetically impaired. Whereas, our results proposed that mTOR blockade with autophagy stimulation could be of interest in the clinical management of human IBD in those patients in which autophagy is not genetically impaired. So far, several potential mechanisms, such as the activation of the inflammasome in myeloid cells or the functional deficiency in intestinal mucus secretion has been put forward to link this with the increased susceptibility to colitis in murine models with impaired autophagy [41,42]. However, little is known about the underlying mechanisms that mediate the alleviation of gut inflammation and oxidative stress via triggering autophagy. Saitoh et al. have reported that activating autophagy not only directly abolished exaggerated activation of the NLRP3 inflammasome pathway, but it also indirectly inhibited the production of mitochondrial ROS, which were closely associated with inflammatory mediators' maturation [43]. Some additional studies have supported the concept that the inflammasome structures, destructive mitochondria, the macrophage polarization, and the selective degradation of inflammatory complexes probably explain how autophagy prevent inflammation progression [5,44].
One clinical trial by Lai et al. on the safety, tolerance, and efficacy of sirolimus in patients with clinically active SLE showed that sirolimus could significantly expand the CD4 + CD25 + FoxP3+ regulatory T cell and CD8+ memory T-cell populations and could inhibit IL-4 and IL-17 production after 12 months of treatment. These in vivo data show that blockade of mTOR by rapamycin inhibits mitochondrial oxidative stress and remits disease activity via correcting the pro-inflammatory T-cell lineage specification in patients with active SLE [45]. As recognized by several case reports in recent years, some immunosuppressive agents (sirolimus and everolimus) that are routinely utilized in transplantation have also been applied as a “rescue” therapy for IBD who were refractory to conventional anti-TNF-α therapies and showed excellent clinical efficacy in inducing and maintaining clinical remission [17,18,46]. Therefore, our present study could be the first step of a further evaluation of mTOR inhibitors and autophagy activators such as sirolimus, rapamycin and its derivative everolimus in IBD. This could represent a new therapeutic alternative, with huge clinical application values.
To explore the potential signalling pathways that are involved in LPS-induced mTOR activation and autophagy suppression in the intestinal epithelium, we focused on TLRs, which are responsible for detecting microbial pathogens and generating innate immune responses. Pathogen recognition by TLRs stirs up rapid activation of host innate immunity by inducing the production of pro-inflammatory cytokines and upregulation of co-stimulatory molecules [47]. TLR4, in particular, is a membrane receptor for LPS and is abundantly expressed in various epithelial cells. The regulative relationship between TLR4 and mTOR has been explored in few studies. A recent study has suggested that supplemental dietary L-arginine attenuates intestinal mucosal disruption through inhibiting the TLR4 and mTOR pathways [48]. mTOR activation plays an essential role in TLR4-triggered neutrophil and macrophage activation [49]. In addition, it is well established that MAPK activation (including p38, ERK and JNK) is involved in the production of various inflammatory mediators present in autoimmune and inflammation associated diseases such as COPD [50]. Intriguingly, our in vitro data revealed that upon knockdown of TLR4, MyD88 and ERK phosphorylation, the reduction of mTOR accompanied an upregulation of LC3B. The functions of mTOR and those dependent on LC3B and ATG5 were exactly opposite in regulation of LPS-induced inflammation in Caco-2 and HT-29 cells. Thus, these results together suggested that LPS-induced autophagy impairment in intestinal epithelial cells was most likely regulated by mTOR and its upstream TLR4-MyD88-MAPK signalling, and the fine control of mTOR signalling and autophagy should be considered in further drug development and application for disorders of gut inflammation in order to generate rewarding clinical outcomes.
Therefore, our results suggested that mTOR activation and autophagy impairment were involved in the process of intestinal inflammation, while the precise pathway through which mTOR and autophagy regulate inflammatory responses remains ambiguous. Previous evidence has shown that NF-κB p65, as well as other NF-κB related molecules, can act as the target for degradation by autophagy, simultaneously regulating the autophagy process [51]. For example, Lorne et al. has reported that stimulating autophagy by rapamycin significantly inhibits NF-κB p65 phosphorylation in LPS-stimulated neutrophils [52]. Moreover, a recent murine study has shown that impaired mucosal autophagy level is closely associated with BCL10 accumulation, the improvement of IκB phosphorylation and nuclear translocation of NF-κB, with following increase in pro-inflammatory cytokines and M1 markers, while pharmacological induction of autophagy can inhibit the activation of NF-κB signalling and reverse intestinal inflammatory responses [53]. In line with the above studies, our current results show that knockdown of TLR4 and MyD88 significantly inhibited the phosphorylation level of ERK, mTOR and p65 and promoted autophagy. The inhibition of ERK phosphorylation by GDC-0994 also inhibited p-mTOR and activated autophagy. However, silencing of NF-κB p65 remarkably strengthened the protein levels of autophagy while it had little effect on p-mTOR. Furthermore, genetic or pharmacological knockdown of TLR4, MyD88, p-ERK and NF-κB p65 effectively diminished the mRNA levels of pro-inflammatory cytokines induced by LPS, which was in parallel with high levels of autophagy and low levels of mTOR phosphorylation. In summary, our current data suggested that LPS activates mTOR via upstream TLR4-MyD88-MAPK signalling and inhibits autophagy through the NF-κB pathway to regulate intestinal inflammation (Fig. 8). Consistent with the above in vitro and vivo findings, our clinical data further demonstrated the proposed TLR4-MyD88-MAPK signalling and NF-κB pathway were probably implicated in the dysregulation of mTOR-dependent autophagy in the pathogenesis of UC.
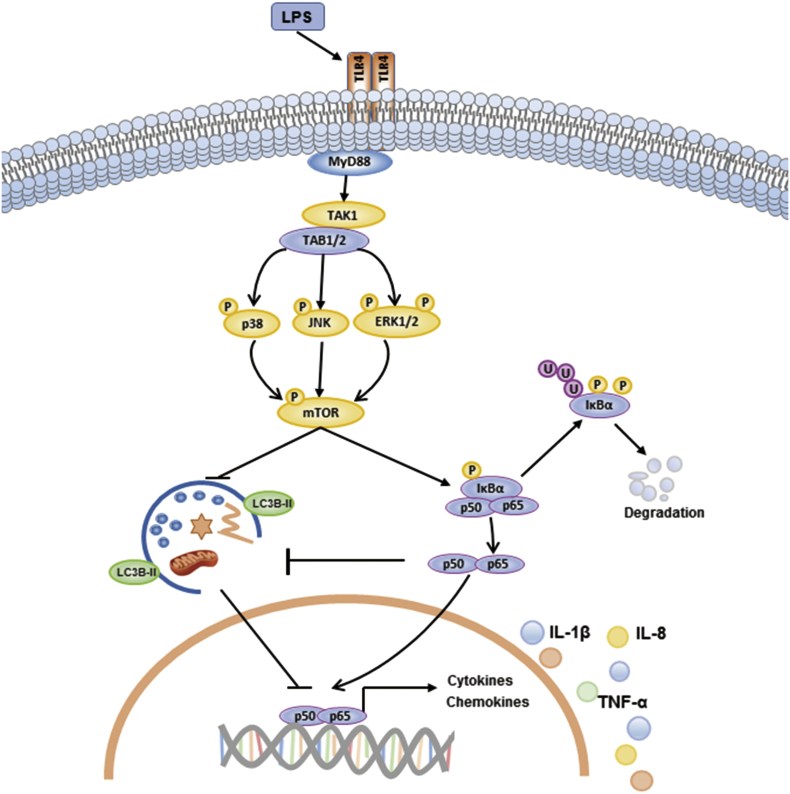
Fig. 8.
Schematic representation of the proposed pathways activated by LPS to modulate mTOR-dependent autophagy and the production of pro-inflammatory cytokines in Caco-2 and HT-29 cells. Mammalian TLR4 (Toll like receptor 4) is essential for LPS recognition and subsequent immune responses against gram-negative bacteria. Formation of LPS-TLR4 complex causes the recruitment of intracellular adaptor protein MyD88 (myeloid differentiation primary response gene 88) and leads to the formation of TAK1/TAB1/TAB2 complex. Then the activation of MAPKs (p38 kinase, JNK and ERK) induces the phosphorylation of mTOR and the activation of canonical NF-κB pathway. The IKK complex (Fig. not shown) phosphorylates IκBα, which leads to its ubiquitylaiton and subsequent degradation. The mTOR-dependent inhibition of autophagy was further suppressed by activated NF-κB signalling. This allows p50 and p65 to translocated to the nucleus and induce the expression of pro-inflammatory cytokines.
Despite the increasing researches about the role of autophagy in modulating inflammation, this field is still in its infancy. Taken together, our data elucidated that the activation of mTOR and subsequent inhibition of autophagy are indispensable for intestinal inflammation and oxidative injury in vitro and in vivo. Mechanistically, classic TLR4-MyD88-MAPK signalling and the NF-κB pathway serve as the critical upstream and downstream regulators, respectively, in modulating the functions of mTOR-dependent autophagy in the pathogenesis of intestinal inflammation. Therefore, our findings, for the first time, reveal a mechanism concerning the regulation of mTOR-dependent autophagy and provide credible evidence for further clinical trials of mTOR inhibitors and autophagy stimulators (e.g., rapamycin) as a promising therapy for intestinal inflammatory diseases such as refractory IBD. And intestinal epithelial cell specific mTOR knockout mice will help us to further verify the function of mTOR in intestinal epithelium in the regulation of LPS-induced inflammation and oxidative stress.
The following are the supplementary data related to this article.
DSS-induced experimental colitis in vivo and quantitative analysis of autophagy-related proteins.
Acknowledgments
Acknowledgements
The authors wish to thank all the patients enrolled in this study.
Funding
This work was supported by National Natural Science Foundation of China (Grant no. 81370485).
Conflicts of interest
The authors declare no conflicts of interest.
Author contributions
MZ designed research plan, conducted the experiments, analyzed all data, and wrote the manuscript. WX and JW helped to designed research plan and in mice experimentation and writing of the manuscript. JY, CZ and JW assisted with some experiments. WG and YS collaborated to collect endoscopy biopsies. PD and YC supervised the experimental work and data analyses. All authors participated in revising the manuscript and agreed to the final version.
Contributor Information
Mingxia Zhou, Email: mingxiazhou@sjtu.edu.cn.
Weimin Xu, Email: xuweimin@sjtu.edu.cn.
Jiazheng Wang, Email: wangjiazheng1991@sjtu.edu.cn.
Junkai Yan, Email: yanjunkai2015@sjtu.edu.cn.
Yingying Shi, Email: syy_0802@sjtu.edu.cn.
Cong Zhang, Email: zhang-cong@alumni.sjtu.edu.cn.
Wensong Ge, Email: gewensong@xinhuamed.com.cn.
Jin Wu, Email: wujin51@yahoo.com.
Peng Du, Email: dupeng@xinhuamed.com.cn.
Yingwei Chen, Email: chenyingwei@xinhuamed.com.cn.
References
- 1.Wadwa M., Klopfleisch R., Adamczyk A., Frede A., Pastille E., Mahnke K. IL-10 downregulates CXCR3 expression on Th1 cells and interferes with their migration to intestinal inflammatory sites. Mucosal Immunol. 2016;9(5):1263–1277. doi: 10.1038/mi.2015.132. [DOI] [PubMed] [Google Scholar]
- 2.Das S., Maras J.S., Hussain M.S., Sharma S., David P., Sukriti S. Hyperoxidized albumin modulates neutrophils to induce oxidative stress and inflammation in severe alcoholic hepatitis. Hepatology. 2017;65(2):631–646. doi: 10.1002/hep.28897. [DOI] [PubMed] [Google Scholar]
- 3.Zhou M., He J., Shen Y., Zhang C., Wang J., Chen Y. New frontiers in genetics, gut microbiota, and immunity: A rosetta stone for the pathogenesis of inflammatory bowel disease. Biomed Res Int. 2017;2017:8201672. doi: 10.1155/2017/8201672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen G., Ran X., Li B., Li Y., He D., Huang B. Sodium Butyrate Inhibits Inflammation and Maintains Epithelium Barrier Integrity in a TNBS-induced Inflammatory Bowel Disease Mice Model. EBioMedicine. 2018;30:317–325. doi: 10.1016/j.ebiom.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Netea-Maier R.T., Plantinga T.S., van de Veerdonk F.L., Smit J.W., Netea M.G. Modulation of inflammation by autophagy: Consequences for human disease. Autophagy. 2016;12(2):245–260. doi: 10.1080/15548627.2015.1071759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun M., He C., Cong Y., Liu Z. Regulatory immune cells in regulation of intestinal inflammatory response to microbiota. Mucosal Immunol. 2015;8(5):969–978. doi: 10.1038/mi.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scharl M., Rogler G. Inflammatory bowel disease: dysfunction of autophagy? Dig Dis. 2012;30(Suppl. 3):12–19. doi: 10.1159/000342588. [DOI] [PubMed] [Google Scholar]
- 8.Strisciuglio C., Duijvestein M., Verhaar A.P., Vos A.C., van den Brink G.R., Hommes D.W. Impaired autophagy leads to abnormal dendritic cell-epithelial cell interactions. J Crohns Colitis. 2013;7(7):534–541. doi: 10.1016/j.crohns.2012.08.009. [DOI] [PubMed] [Google Scholar]
- 9.Kuenzig M.E., Yim J., Coward S., Eksteen B., Seow C.H., Barnabe C. The NOD2-smoking Interaction in Crohn's disease is likely specific to the 1007fs mutation and may be explained by age at diagnosis: A meta-analysis and case-only study. EBioMedicine. 2017;21:188–196. doi: 10.1016/j.ebiom.2017.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwerd T., Pandey S., Yang H.T., Bagola K., Jameson E., Jung J. Impaired antibacterial autophagy links granulomatous intestinal inflammation in Niemann-Pick disease type C1 and XIAP deficiency with NOD2 variants in Crohn's disease. Gut. 2017;66(6):1060–1073. doi: 10.1136/gutjnl-2015-310382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosin-Roger J., Simmen S., Melhem H., Atrott K., Frey-Wagner I., Hausmann M. Hypoxia ameliorates intestinal inflammation through NLRP3/mTOR downregulation and autophagy activation. Nat Commun. 2017;8(1):98. doi: 10.1038/s41467-017-00213-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newton P.T., Vuppalapati K.K., Bouderlique T., Chagin A.S. Pharmacological inhibition of lysosomes activates the MTORC1 signaling pathway in chondrocytes in an autophagy-independent manner. Autophagy. 2015;11(9):1594–1607. doi: 10.1080/15548627.2015.1068489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oaks Z., Winans T., Caza T., Fernandez D., Liu Y., Landas S.K. Mitochondrial Dysfunction in the Liver and Antiphospholipid Antibody Production Precede Disease Onset and Respond to Rapamycin in Lupus-Prone Mice. Arthritis Rheumatol. 2016;68(11):2728–2739. doi: 10.1002/art.39791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y., Wang X., Wu H., Chen S., Zhu H., Zhang J. Glycine enhances muscle protein mass associated with maintaining Akt-mTOR-FOXO1 signaling and suppressing TLR4 and NOD2 signaling in piglets challenged with LPS. Am J Physiol Regul Integr Comp Physiol. 2016;311(2):R365–R373. doi: 10.1152/ajpregu.00043.2016. [DOI] [PubMed] [Google Scholar]
- 15.Caza T.N., Fernandez D.R., Talaber G., Oaks Z., Haas M., Madaio M.P. HRES-1/Rab4-mediated depletion of Drp1 impairs mitochondrial homeostasis and represents a target for treatment in SLE. Ann Rheum Dis. 2014;73(10):1888–1897. doi: 10.1136/annrheumdis-2013-203794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez D., Bonilla E., Mirza N., Niland B., Perl A. Rapamycin reduces disease activity and normalizes T cell activation-induced calcium fluxing in patients with systemic lupus erythematosus. Arthritis Rheum. 2006;54(9):2983–2988. doi: 10.1002/art.22085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massey D.C., Bredin F., Parkes M. Use of sirolimus (rapamycin) to treat refractory Crohn's disease. Gut. 2008;57(9):1294–1296. doi: 10.1136/gut.2008.157297. [DOI] [PubMed] [Google Scholar]
- 18.Mutalib M., Borrelli O., Blackstock S., Kiparissi F., Elawad M., Shah N. The use of sirolimus (rapamycin) in the management of refractory inflammatory bowel disease in children. J Crohns Colitis. 2014;8(12):1730–1734. doi: 10.1016/j.crohns.2014.08.014. [DOI] [PubMed] [Google Scholar]
- 19.Lv P., Zhou M., He J., Meng W., Ma X., Dong S. Circulating miR-208b and miR-34a are associated with left ventricular remodeling after acute myocardial infarction. Int J Mol Sci. 2014;15(4):5774–5788. doi: 10.3390/ijms15045774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He F., Lv P., Zhao X., Wang X., Ma X., Meng W. Predictive value of circulating miR-328 and miR-134 for acute myocardial infarction. Mol Cell Biochem. 2014;394(1–2):137–144. doi: 10.1007/s11010-014-2089-0. [DOI] [PubMed] [Google Scholar]
- 21.Song Z., Tong G., Xiao K., Jiao Le F., Ke Y., Hu C. L-cysteine protects intestinal integrity, attenuates intestinal inflammation and oxidant stress, and modulates NF-kappaB and Nrf2 pathways in weaned piglets after LPS challenge. Innate Immun. 2016;22(3):152–161. doi: 10.1177/1753425916632303. [DOI] [PubMed] [Google Scholar]
- 22.Zong X., Hu W., Song D., Li Z., Du H., Lu Z. Porcine lactoferrin-derived peptide LFP-20 protects intestinal barrier by maintaining tight junction complex and modulating inflammatory response. Biochem Pharmacol. 2016;104:74–82. doi: 10.1016/j.bcp.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 23.Okamoto N., Mizote K., Honda H., Saeki A., Watanabe Y., Yamaguchi-Miyamoto T. Funiculosin variants and phosphorylated derivatives promote innate immune responses via the Toll-like receptor 4/myeloid differentiation factor-2 complex. J Biol Chem. 2017;292(37):15378–15394. doi: 10.1074/jbc.M117.791780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ryu J.K., Kim S.J., Rah S.H., Kang J.I., Jung H.E., Lee D. Reconstruction of LPS transfer cascade reveals structural determinants within LBP, CD14, and TLR4-MD2 for efficient LPS recognition and transfer. Immunity. 2017;46(1):38–50. doi: 10.1016/j.immuni.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 25.Baxt L.A., Xavier R.J. Role of autophagy in the maintenance of intestinal homeostasis. Gastroenterology. 2015;149(3):553–562. doi: 10.1053/j.gastro.2015.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saitoh T., Fujita N., Jang M.H., Uematsu S., Yang B.G., Satoh T. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456(7219):264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
- 27.Hooper K.M., Barlow P.G., Stevens C., Henderson P. Inflammatory bowel disease drugs: A Focus on Autophagy. J Crohns Colitis. 2017;11(1):118–127. doi: 10.1093/ecco-jcc/jjw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ortiz-Masia D., Cosin-Roger J., Calatayud S., Hernandez C., Alos R., Hinojosa J. Hypoxic macrophages impair autophagy in epithelial cells through Wnt1: relevance in IBD. Mucosal Immunol. 2014;7(4):929–938. doi: 10.1038/mi.2013.108. [DOI] [PubMed] [Google Scholar]
- 29.Lee J., Giordano S., Zhang J. Autophagy, mitochondria and oxidative stress: cross-talk and redox signalling. Biochem J. 2012;441(2):523–540. doi: 10.1042/BJ20111451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bhattacharyya S., Xue L., Devkota S., Chang E., Morris S., Tobacman J.K. Carrageenan-induced colonic inflammation is reduced in Bcl10 null mice and increased in IL-10-deficient mice. Mediators Inflamm. 2013;2013:397642. doi: 10.1155/2013/397642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dodson M., Darley-Usmar V., Zhang J. Cellular metabolic and autophagic pathways: traffic control by redox signaling. Free Radic Biol Med. 2013;63:207–221. doi: 10.1016/j.freeradbiomed.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thorburn A. Autophagy and its effects: making sense of double-edged swords. PLoS Biol. 2014;12(10) doi: 10.1371/journal.pbio.1001967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lassen K.G., Xavier R.J. Genetic control of autophagy underlies pathogenesis of inflammatory bowel disease. Mucosal Immunol. 2017;10(3):589–597. doi: 10.1038/mi.2017.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrovski G., Zahuczky G., Majai G., Fesus L. Phagocytosis of cells dying through autophagy evokes a pro-inflammatory response in macrophages. Autophagy. 2007;3(5):509–511. doi: 10.4161/auto.4731. [DOI] [PubMed] [Google Scholar]
- 35.Petrovski G., Ayna G., Majai G., Hodrea J., Benko S., Madi A. Phagocytosis of cells dying through autophagy induces inflammasome activation and IL-1beta release in human macrophages. Autophagy. 2011;7(3):321–330. doi: 10.4161/auto.7.3.14583. [DOI] [PubMed] [Google Scholar]
- 36.Yu X., Long Y.C., Shen H.M. Differential regulatory functions of three classes of phosphatidylinositol and phosphoinositide 3-kinases in autophagy. Autophagy. 2015;11(10):1711–1728. doi: 10.1080/15548627.2015.1043076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chresta C.M., Davies B.R., Hickson I., Harding T., Cosulich S., Critchlow S.E. AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 2010;70(1):288–298. doi: 10.1158/0008-5472.CAN-09-1751. [DOI] [PubMed] [Google Scholar]
- 38.Renshaw J., Taylor K.R., Bishop R., Valenti M., De Haven Brandon A., Gowan S. Dual blockade of the PI3K/AKT/mTOR (AZD8055) and RAS/MEK/ERK (AZD6244) pathways synergistically inhibits rhabdomyosarcoma cell growth in vitro and in vivo. Clin Cancer Res. 2013;19(21):5940–5951. doi: 10.1158/1078-0432.CCR-13-0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gierisch M.E., Pfistner F., Lopez-Garcia L.A., Harder L., Schafer B.W., Niggli F.K. Proteasomal Degradation of the EWS-FLI1 Fusion Protein is Regulated by a Single Lysine Residue. J Biol Chem. 2016;291(52):26922–26933. doi: 10.1074/jbc.M116.752063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xue H., Yuan G., Guo X., Liu Q., Zhang J., Gao X. A novel tumor-promoting mechanism of IL6 and the therapeutic efficacy of tocilizumab: Hypoxia-induced IL6 is a potent autophagy initiator in glioblastoma via the p-STAT3-MIR155-3p-CREBRF pathway. Autophagy. 2016;12(7):1129–1152. doi: 10.1080/15548627.2016.1178446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsuboi K., Nishitani M., Takakura A., Imai Y., Komatsu M., Kawashima H. Autophagy Protects against Colitis by the Maintenance of Normal Gut Microflora and Secretion of Mucus. J Biol Chem. 2015;290(33):20511–20526. doi: 10.1074/jbc.M114.632257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee H.Y., Kim J., Quan W., Lee J.C., Kim M.S., Kim S.H. Autophagy deficiency in myeloid cells increases susceptibility to obesity-induced diabetes and experimental colitis. Autophagy. 2016;12(8):1390–1403. doi: 10.1080/15548627.2016.1184799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nakahira K., Haspel J.A., Rathinam V.A., Lee S.J., Dolinay T., Lam H.C. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lapaquette P., Guzzo J., Bretillon L., Bringer M.A. Cellular and molecular connections between autophagy and inflammation. Mediators Inflamm. 2015;2015:398483. doi: 10.1155/2015/398483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lai Z.-W., Kelly R., Winans T., Marchena I., Shadakshari A., Yu J. Sirolimus in patients with clinically active systemic lupus erythematosus resistant to, or intolerant of, conventional medications: a single-arm, open-label, phase 1/2 trial. Lancet. 2018;391(10126):1186–1196. doi: 10.1016/S0140-6736(18)30485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dumortier J., Lapalus M.G., Guillaud O., Poncet G., Gagnieu M.C., Partensky C. Everolimus for refractory Crohn's disease: a case report. Inflamm Bowel Dis. 2008;14(6):874–877. doi: 10.1002/ibd.20395. [DOI] [PubMed] [Google Scholar]
- 47.Chen J.Q., Szodoray P., Zeher M. Toll-like Receptor Pathways in Autoimmune Diseases. Clin Rev Allergy Immunol. 2016;50(1):1–17. doi: 10.1007/s12016-015-8473-z. [DOI] [PubMed] [Google Scholar]
- 48.Tan J., Applegate T.J., Liu S., Guo Y., Eicher S.D. Supplemental dietary L-arginine attenuates intestinal mucosal disruption during a coccidial vaccine challenge in broiler chickens. Br J Nutr. 2014;112(7):1098–1109. doi: 10.1017/S0007114514001846. [DOI] [PubMed] [Google Scholar]
- 49.Yu M., Kang X., Xue H., Yin H. Toll-like receptor 4 is up-regulated by mTOR activation during THP-1 macrophage foam cells formation. Acta Biochim Biophys Sin. 2011;43(12):940–947. doi: 10.1093/abbs/gmr093. [DOI] [PubMed] [Google Scholar]
- 50.Kyriakis J.M., Avruch J. Mammalian MAPK signal transduction pathways activated by stress and inflammation: a 10-year update. Physiol Rev. 2012;92(2):689–737. doi: 10.1152/physrev.00028.2011. [DOI] [PubMed] [Google Scholar]
- 51.Nivon M., Fort L., Muller P., Richet E., Simon S., Guey B. NFkappaB is a central regulator of protein quality control in response to protein aggregation stresses via autophagy modulation. Mol Biol Cell. 2016;27(11):1712–1727. doi: 10.1091/mbc.E15-12-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lorne E., Zhao X., Zmijewski J.W., Liu G., Park Y.J., Tsuruta Y. Participation of mammalian target of rapamycin complex 1 in Toll-like receptor 2- and 4-induced neutrophil activation and acute lung injury. Am J Respir Cell Mol Biol. 2009;41(2):237–245. doi: 10.1165/rcmb.2008-0290OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao J., Sun Y., Shi P., Dong J.N., Zuo L.G., Wang H.G. Celastrol ameliorates experimental colitis in IL-10 deficient mice via the up-regulation of autophagy. Int Immunopharmacol. 2015;26(1):221–228. doi: 10.1016/j.intimp.2015.03.033. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DSS-induced experimental colitis in vivo and quantitative analysis of autophagy-related proteins.