Abstract
Background
Age-related brain changes are well-documented and influenced by genetics. Extensive research links apolipoprotein E (apoE) to brain function, with the E4 allele serving as a risk factor for brain disease, including Alzheimer's disease, and the E2 allele conferring protection. Recent evidence also supports protective effects of another gene, human leukocyte antigen (HLA) DRB1*13, on brain disease and age-related brain atrophy in cognitively healthy adults. Here we investigated the effects of apoE and HLA DRB1*13 on brain function by examining changes in neural network properties with age in healthy adults.
Methods
One hundred seventy-eight cognitively healthy women (28–99 y old) underwent a magnetoencephalography scan and provided a blood sample for genetic analysis. Age-related changes in neural network variability in genetic subgroups of DRB1*13 × apoE genotype combinations were assessed using linear regression of network variability against age.
Findings
For individuals lacking a DRB1*13 allele and/or carrying an apoE4 allele, network variability increased significantly with age. In contrast, no such increase was observed in the presence of DRB1*13 and/or apoE2.
Interpretation
These findings extend previous research documenting the protective effect of DRB1*13 on brain structure to include protection against age-related changes in brain function, and demonstrate similar protective effects on neural network variability for either DRB1*13 or apoE2. These protective effects could be due to reduction or elimination of factors known to disrupt brain function, including neuroinflammation and amyloid beta protein.
Funding
U.S. Department of Veterans Affairs, and University of Minnesota.
Keywords: Healthy brain aging, Human leukocyte antigen, DRB1*13, Apolipoprotein E, Neural network, Magnetoencephalography
Research in Context.
Genetic influence on age-related brain changes has been extensively studied, particularly with regard to apolipoprotein E (apoE). The effects of another gene - human leukocyte antigen (HLA) DRB1*13 - on age-related brain changes has just begun to be investigated with initial evidence pointing to neuroprotective effects. Here we investigated the joint effects of apoE and HLA DRB1*13 on age-related changes in neural network variability. We found that neural network variability increased significantly with age in individuals lacking a DRB1*13 allele and/or carrying an apoE4 allele. In contrast, no such increase was observed in the presence of DRB1*13 and/or apoE2. We hypothesize that the beneficial effects of DRB1*13 and apoE2 on network stability is due to reduction or elimination of factors known to disrupt brain function.
Alt-text: Unlabelled Box
1. Introduction
Aging-related brain changes are well-documented [1]; however, multiple factors, including physical and mental health conditions, lifestyle choices, and genetic influences contribute to individual variability in brain changes and cognitive functioning with age [2]. Among the most commonly cited genetic contributors to brain health is apolipoprotein E (apoE), which has three primary isoforms – E2, E3, and E4 – that vary in terms of structure, function, and disease susceptibility. For instance, the E2 allele is associated with preserved cognitive functioning and decreased risk of Alzheimer's disease [3], whereas the E4 allele has been associated with brain atrophy [4], functional network disturbances [5,6], susceptibility to Alzheimer's disease [7,8], and lower cognitive performance even among cognitively healthy individuals [9].
Increasing evidence also implicates human leukocyte antigen (HLA) genes in brain health. HLA genes are involved in immunity via elimination of external pathogens [10]. Recent research has linked several HLA gene variants to Alzheimer's disease as well as other neurological diseases [8,11,12]. Like apoE, however, some HLA genes appear to be protective in terms of cognitive function and brain health [[13], [14], [15]]. For example, the HLA DRB1 gene has been linked to enhanced cognitive performance among healthy adults [15], and studies of this gene at the protein level have shown that the DRB1*13:02 allele protects against age-related brain atrophy in cognitively healthy adults [14] and in Gulf War Illness [16]. Furthermore, the DRB1*13:02 allele has been shown to exert protective effects against several neuroimmune conditions [13,[17], [18], [19]]. The findings with regard to DRB1*13:01 (which differs from DRB1*13:02 by only a single amino acid residue) have been somewhat mixed. Specifically, while some protective effects have been observed for DRB1*13:01 [20], it has also been shown to be a risk factor for other primarily autoimmune conditions [19,21,22], many of which involve neurocognitive disruption. Despite some evidence relating these alleles to brain health and structure, the effects of these alleles on brain function are not yet well understood.
Healthy brain functioning depends on efficient and coordinated communication across a massive, interconnected network. Magnetoencephalography (MEG) captures integrated synaptic activity with the highest fidelity, providing a highly accurate measure of brain activity. Using MEG, our research group has demonstrated that patterns of neural network communication are virtually identical across healthy individuals and highly synchronous [23] to the extent that various brain diseases can be differentiated based on variation in synchronous neural interactions (SNI) [[24], [25], [26], [27]]. Furthermore, some of the variation in neural network properties can be attributed to genetic differences. For example, with regards to apoE, we have shown that the E2 allele is associated with the highest mean SNI and lowest variability of SNI whereas the E4 allele is associated with the lowest mean and highest variability in healthy adults [6]. Similarly increased variability induced by apoE4 has been demonstrated in in vitro brain cultures [28]. In terms of HLA, we have previously demonstrated protective effects on age-related brain atrophy in cognitively healthy individuals [14]; however, the effects on brain function in cognitively healthy individuals are unknown. Thus, here we extend the research on genetic effects on neural network properties and age-related brain changes by investigating the combined effects of apoE and HLA on changes in the strength of neural interactions and network variability with age in cognitively healthy adults.
2. Materials and methods
2.1. Participants
A total of 178 cognitively healthy women (60.3 ± 15.8 y, mean age ± SD; age range 28–99 y) participated in this study as paid volunteers after providing informed consent, in adherence to the Declaration of Helsinki. Their cognitive status was assessed using the Montreal Cognitive Assessment (MoCA [29] N = 166) or Modified Mini-Mental State exam (3MS [30]; N = 12). All participant's cognitive scores exceeded the suggested cut-offs indicative of healthy cognitive functioning. All study protocols were approved by the appropriate Institutional Review Boards.
2.2. Genetics
2.2.1. HLA genotyping
DNA isolation was carried out from 3 ml of whole blood drawn in EDTA tubes, using a commercially available kit (ArchivePure cat. 2300730) from 5Prime (distributed by Fisher Scientific or VWR) with an expected yield of 50-150 μg of DNA. The purified DNA samples were sent to Histogenetics (http://www.histogenetics.com/) for high-resolution HLA Sequence-based Typing (SBT; details are given in https://bioinformatics.bethematchclinical.org/HLA-Resources/HLA-Typing/High-Resolution-Typing-Procedures/ and https://bioinformatics.bethematchclinical.org/WorkArea/DownloadAsset.aspx?id=6482). Their sequencing DNA templates are produced by locus- and group-specific amplifications that include exon 2 and 3 for class I (A, B, C) and exon 2 for class II (DRB1, DRB3/4/5, DQB1, and DPB1) and reported as Antigen Recognition Site (ARS) alleles as per ASHI recommendation [31].
Given our previous findings of a protective role of DRB1*13:02 allele on gray matter volume reduction in healthy aging [14], we focused in this study on the DRB1*13 gene. In our sample of 178 healthy women, 47 carried the DRB1*13 gene. More specifically, 26 carried the DRB1*13:01 allele, 18 carried the DRB1*13:02 alleles, 2 carried the DRB1*13:03 allele, and 1 carried the DRB1*13:05 allele.
2.2.2. ApoE genotyping
DNA samples were genotyped using PCR amplification followed by restriction enzyme digestion [32]. Each amplification reaction contained PCR buffer with 15 mmol/L MgCl2 ng amounts of genomic DNA, 20 pmol apoE forward (5 N TAA GCT TGG CAC GGC TGT CCA AGG A 3 N) and reverse (5 N ATA AAT ATA AAA TAT AAA TAA CAG AAT TCG CCC CGG CCT GGT ACA C 3 N) primers, 1.25 mmol/L of each deoxynucleotide triphosphate, 10% dimethylsulfoxide, and 0.25 μL Amplitaq DNA polymerase. Reaction conditions in a thermocycler included an initial denaturing period of 3 min at 95 C, 1 min at 60 C, and 2 min at 72 C; followed by 32 cycles of 1 min at 95 C, 1 min at 60 C, and 2 min at 72 C; and a final extension of 1 min at 95 C, 1 min at 60 C, and 3 min at 72 C. PCR products were digested with HhaI and separated on a 4% Agarose gel which was stained with Ethidium Bromide. Known apoE isoform standards were included in the analysis.
2.2.3. Genetic groups
The frequencies of occurrence of DRB1*13 and the 6 apoE genotypes are given in Table 1. Based (a) on the presence or absence of DRB1*13, and (b) on the presence or absence of apoE alleles E2 and E4, seven genetic subgroups were distinguished (Table 2); the combination {DRB1*13 present, apoE E2/E4} did not occur in our sample.
Table 1.
Frequencies of DRB1*13 apoE genotypes.
| ApoE genotype | DRB1*13 |
Total | |
|---|---|---|---|
| Absent | Present | ||
| 2/2 | 2 | 0 | 2 |
| 2/3 | 13 | 6 | 19 |
| 2/4 | 5 | 0 | 5 |
| 3/3 | 78 | 31 | 109 |
| 3/4 | 30 | 8 | 38 |
| 4/4 | 3 | 2 | 5 |
| Total | 131 | 47 | 178 |
Table 2.
Genetic subgroups based on DRB1*13 and apoE genotypes. There was no participant with the combination {DRB1*13 present, apoE E2/E4}.
| Subgroup | DRB1*13 | ApoE | N |
|---|---|---|---|
| A | 0 | E2/E2, E2/E3 | 15 |
| B | 1 | E2/E2, E2/E3 | 5 |
| C | 0 | E4/E4, E3/E4 | 33 |
| D | 1 | E4/E4, E3/E4 | 10 |
| E | 0 | E2/E4 | 5 |
| F | 0 | E3/E3 | 79 |
| G | 1 | E3/E3 | 31 |
2.3. MEG data acquisition
All participants underwent a MEG scan. As described previously [25,26], subjects lay supine within the electromagnetically shielded chamber and fixated their eyes on a spot ~ 65 cm in front of them, for 45–60s. MEG data were acquired using a 248-channel axial gradiometer system (Magnes 3600WH, 4-D Neuroimaging, San Diego, CA), band-filtered between 0.1 and 400 Hz, and sampled at 1017.25 Hz. Data with artifacts (e.g. from non-removable metal or excessive subject motion) were eliminated from further analysis.
2.4. Data analysis
2.4.1. General
Standard statistical methods were used to analyze the data. The following packages were employed: IBM-SPSS statistical package, versions 23–25, and ad hoc FORTRAN computer programs employing the International Mathematics and Statistics Library (IMSL; Rogue Wave Software, Louisville, CO, USA) statistical and mathematical libraries. Prewhitening of the raw MEG series (see below) was performed using programs in Python [33].
2.4.2. Data preprocessing
Single trial MEG time series from all sensors (N = 60,000 time samples per series) underwent ‘prewhitening’ [34] using a [1, 3, 50] ARIMA model [33] to obtain practically white noise innovations (i.e. residuals). Given 248 MEG sensors, there were 30,628 pairs possible. All pairwise crosscorrelations, r, were computed for valid data (see above); of those, 90% came from valid data and were retained for further analysis.
2.4.3. Statistical analyses
2.4.3.1. General
Since we were interested in the strength of neural interactions irrespective of its sign, we took the absolute value of r and z-transformed [35] it:
| (1) |
For each participant, the standard deviation of z, SD(z), was computed as a measure of the variability of the neural network interactions. Finally, linear regression analysis was used to assess age-related effects on network variability. We assessed this effect on genetic subgroups of participants, based (a) on the presence or absence of HLA gene DRB1*13, and (b) on the presence or absence of apoE alleles E2 and E4.
2.4.3.2. Permutation tests
Finally, we examined more closely a specific comparison of the age effect on network variability between the subgroup that carried apoE4 but no DRB1*13 allele (subgroup C, Table 2) and the subgroup that carried both of the above (subgroup D, Table 2). This comparison is crucial for assessing the potential overriding protection conferred by DRB1*13 in the presence of apoE4. Since the sample size of subgroup D (N = 10) was less than that of subgroup C (N = 33), we performed a permutation test to account for the smaller sample size of subgroup D as follows. The number M of all possible subsamples of size N* = 10 out of 33 is very large:
| (2) |
Instead, we generated 1 million random subsamples (without replacement) from subgroup C of sample size N* = 10 each. We then performed a linear regression analysis of SD(z) against age for each one of these subsamples and retained the correlation coefficient. Finally, we calculated the proportion of correlations that were smaller than the correlation actually observed in subgroup D, rD. This is an estimate of the probability that a correlation equal to, or smaller than, rD occurs in subgroup C in subsamples matched for size N = 10 of subgroup C.
3. Results
3.1. Genetic groups
There was no statistically significant association between the occurrence of DRB1*13 and the 6 apoE genotypes (Table 1; χ[5]2 = 4.04, P = 0.543).
3.2. Age
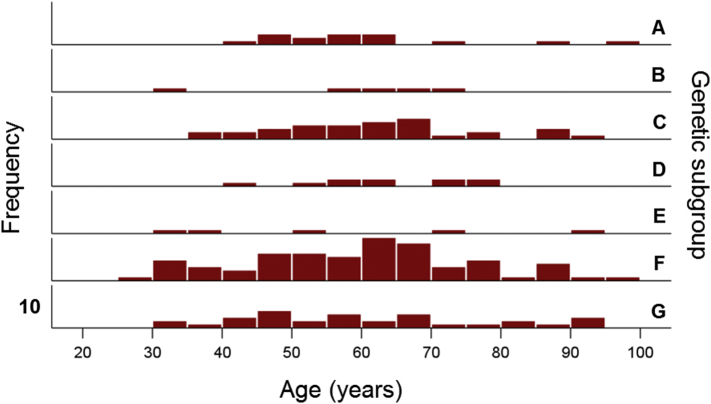
The frequency distribution of age in the whole sample of 178 women is shown in Fig. 1. The age distributions for the genetic subgroups (Table 2) are shown in Fig. 2; the mean ages did not differ significantly among the subgroups (P = 0.973, F-test, analysis of variance).
Fig. 1.
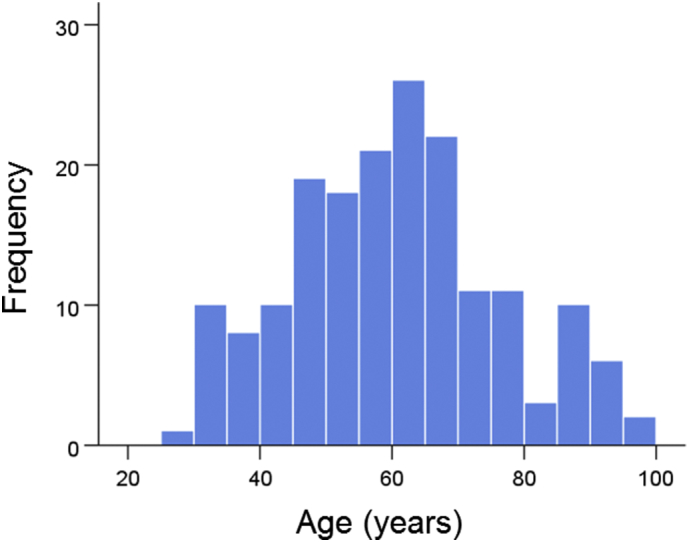
Frequency distributions of ages (N = 178).
Fig. 2.
Frequency distributions of ages in the genetic subgroups of Table 2. Vertical bar denotes a count of 10.
3.3. Effects of age on network variability
The results of the linear regression analysis of network variability against age for each genetic subgroup are shown in Table 3 and illustrated in Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8. We found the following. (a) In the presence of apoE2, network variability was not affected by age irrespective of whether DRB1*13 was absent (Fig. 3) or present (Fig. 4). (b) In the presence of apoE4, network variability increased significantly with age in the absence of DRB1*13 (Fig. 5) but not in its presence (Fig. 6). (c) In absence of both apoE2 and apoE4 (i.e. apoE3/3 genotype), network variability increased with age in the absence of DRB1*13 (Fig. 7) but not in its presence (Fig. 8). The lack of a significant relation to age held separately for DRB1*13:01 (P = 0.418, N = 26) and DRB1*13:02 (P = 0.582, N = 16). In summary, the presence of either DRB1*13 or apoE2 prevents the increase in network variability with age, whereas the presence of apoE4 increases network variability with age in the absence of DRB1*13.
Table 3.
Effect of age on network variability for DRB1*13/apoE genotype genetic subgroups. r, Pearson correlation coefficient.
| Subgroup | DRB1*13 | ApoE2 | ApoE4 | r | P-Value | Figure |
|---|---|---|---|---|---|---|
| A | Absent | Present | Absent | 0.101 | 0.721 | 3 |
| B | Present | Present | Absent | 0.378 | 0.530 | 4 |
| C | Absent | Absent | Present | 0.510 | 0.002 | 5 |
| D | Present | Absent | Present | −0.143 | 0.693 | 6 |
| E | Absent | Present | Present | 0.620 | 0.264 | No figure |
| F | Absent | Absent | Absent | 0.297 | 0.008 | 7 |
| G | Present | Absent | Absent | 0.141 | 0.448 | 8 |
Fig. 3.
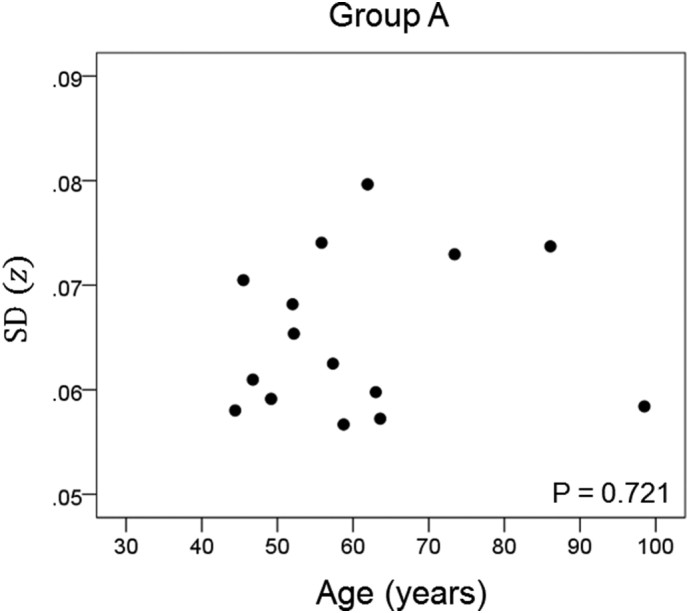
Network variability is plotted against age for subgroup A (Table 3), as indicated. (See text for details.)
Fig. 4.
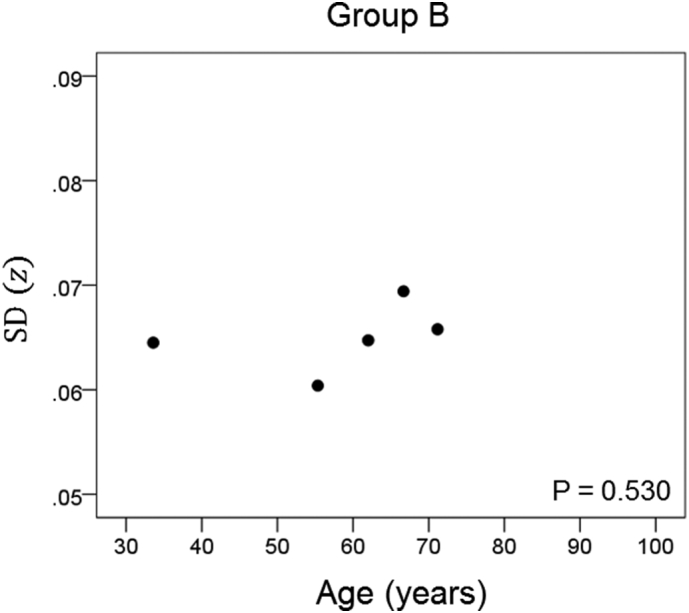
Network variability is plotted against age for subgroup B (Table 3), as indicated. (See text for details.)
Fig. 5.
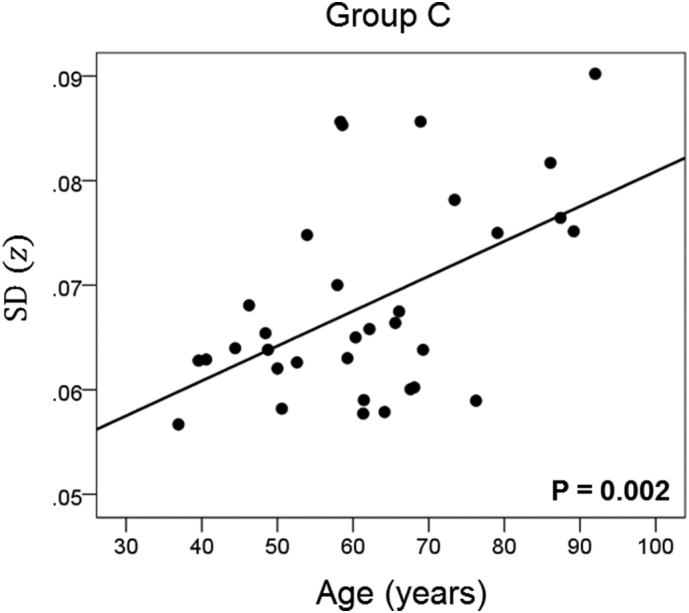
Network variability is plotted against age for subgroup C (Table 3), as indicated. (See text for details.)
Fig. 6.
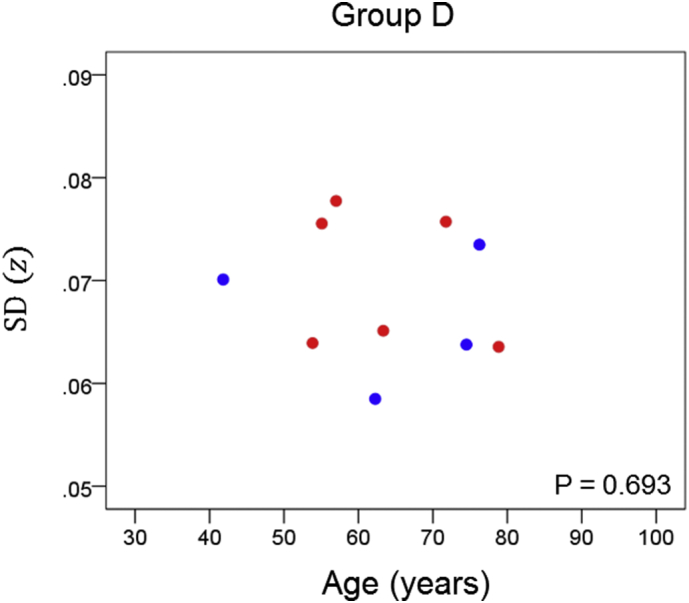
Network variability is plotted against age for subgroup D (Table 3), as indicated. (See text for details.) Blue, DRB1*13:01; red, DRB1*13:02; magenta, DRB1*13:05.
Fig. 7.
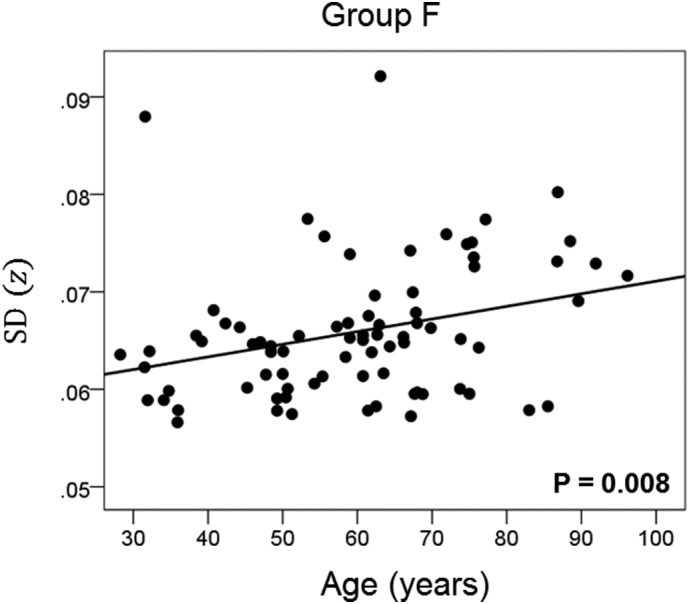
Network variability is plotted against age for subgroup F (Table 3), as indicated. (See text for details.)
Fig. 8.
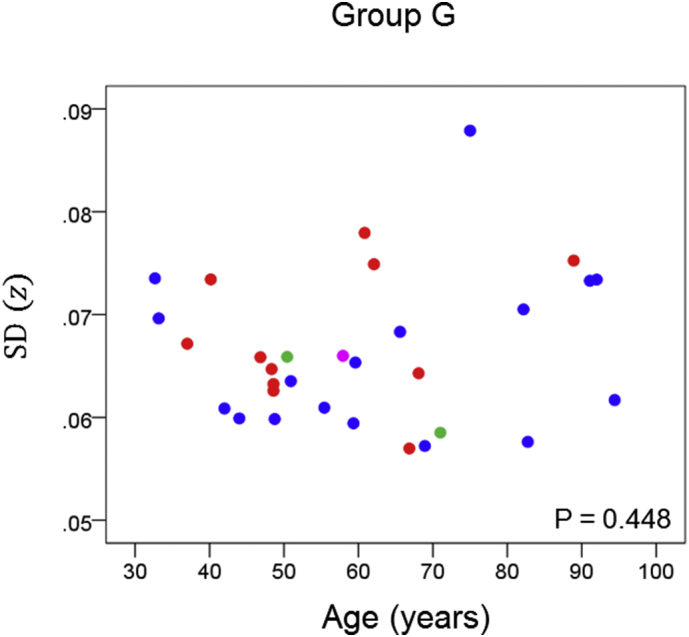
Network variability is plotted against age for subgroup G (Table 3), as indicated. (See text for details.) Blue, DRB1*13:01; red, DRB1*13:02; green, DRB1*13:03; magenta, DRB1*13:05.
Finally, we performed a permutations test to examine more closely the comparison of the age effect on network variability between the subgroup that carried apoE4 but no DRB1*13 allele (subgroup C) and the subgroup that carried both of the above (subgroup D). Since the sample size of subgroup D (N = 10) was less than that of subgroup C (N = 33), we performed a linear regression of SD(z) against age for each one of 1 million random subsamples of size N* = 10 (out of the 33 of subgroup C; see Methods). The positive correlation for subgroup C (rC = 0.510, N = 33) (Table 3) indicated a highly significant increase in network variability with age (P = 0.002); in contrast, that correlation in subgroup D was negative (rD = − 0.143, N = 10) but not statistically significant (P = 0.693). In the permutation test above, we found that in 98.4% of the 1 million regressions in subgroup C with N = 10, the correlations were >rD; in addition, 96.2% of the 1 million correlations were positive. These results add further support to the original finding that the presence of DRB1*13 eliminates the age-related increase in network variability associated with the presence of apoE4.
4. Discussion
In the present study we investigated the effects of HLA DRB1*13 and apoE on changes in neural network variability with age in cognitively healthy women. We found highly significant age-related changes in network variability that varied according to the presence of HLA DRB1*13 and apoE genotype. Specifically, apoE4 was associated with increased network variability with age, as was lack of HLA DRB1*13; in contrast, there were no changes in network variability with age in the presence of HLA DRB1*13 or apoE2. These findings add to the growing literature documenting protective effects of HLA DRB1*13 on the brain and extend the vast literature on apoE by shifting the focus from not only the deleterious effects of apoE4 but also to include the protective effects of apoE2, as proposed earlier [6].
Previous studies have demonstrated that healthy brain function can be distinguished from pathological conditions including Alzheimer's disease based on neural network characteristics [[24], [25], [26]] and that those network characteristics are genetically mediated [6,28], among other factors. Specifically, we have previously documented that apoE4, an allele that is a known risk factor for Alzheimer's disease, is associated with increased network variability both in vivo [6] and in vitro [28], whereas apoE2, an allele that is protective against Alzheimer's disease, is associated with decreased network variability [6]. Here we extend those findings and show additional genetic influence on network variability with HLA DRB1*13 alleles also conferring protection against variability and their absence associated with increased variability akin to that associated with apoE4. Notably, even in the presence of apoE4, protection against age-related changes in neural network properties is still conferred by HLA DRB1*13.
To our knowledge, this is the first study to evaluate the joint effects of apoE and HLA DRB1*13 on age-related brain changes in cognitively healthy individuals. Considerable research has focused on the deleterious effects of apoE4 on brain health including reductions in synaptic, vascular, and mitochondrial function, lipid glucose metabolism, neurogenesis, and amyloid-beta clearing, in addition to increases in brain atrophy, neuronal toxicity, amyloid-beta aggregation, and tangle formation [3,36]. In contrast, the mechanisms underlying the protection conferred by DRB1*13 and apoE2 are just beginning to be investigated; the current findings that both HLA DRB1*13 and apoE2 each prevent increased network variability with age suggest a common mechanism that promotes brain health.
ApoE plays a critical role in the transport and metabolism of cholesterol and other lipids in addition to numerous central nervous system functions [37]. The three apoE isoforms – E2, E3, and E4 – are distinguished by single amino acid substitutions that result in substantial structural and functional differences [38]. For instance, critical brain processes including neurogenesis, synaptic functioning, lipoprotein receptor binding, and binding and clearance of amyloid-beta peptides have been found to differ according to apoE genotype with apoE2 evidencing relative superiority such that E2 ≥ E3 > E4 [3,37,39]. HLA involvement in brain health ultimately boils down to maintenance of the immune system via eradication of foreign antigens. Successful elimination of such antigens depends on a match between a specific antigen and one's HLA composition. A mismatch, on the other hand, prevents elimination of the antigen, and the “persistent antigen” [16] may lead to inflammation, cell damage, autoimmunity [40], and atrophy [14]. Evidence suggests that antigens related to several common illnesses including influenza A, hepatitis B and C, and herpes virus bind to HLA DRB1*13:02 and HLA DRB1*13:01 [41], thereby facilitating elimination of those antigens.
Consideration of the function of apoE and HLA genes in the context of the present findings lead us to speculate about a common mechanism that promotes network stability in both apoE2 and HLA DRB1*13 and, conversely, leads to network variability in apoE4. Specifically, we assert that the beneficial stable network effects observed for both apoE2 and HLA DRB1*13 are enabled by relative absence of neurotoxins compared to apoE4. That is, amyloid-beta plaques and/or other neurotoxic effects commonly associated with apoE4 are well-known to be reduced in E2 carriers and may be targeted for clearance in DRB1*13 carriers in the same manner as hepatitis B and other foreign antigens. At least 3 lines of research support the latter. First, mounting evidence supports the existence of antigen-presenting cells in the brain [42]. Second, it has been shown that autoantibodies against amyloid beta naturally occur in healthy individuals and facilitate degradation of amyloid beta [43], and are reduced in patients with Alzheimer's disease [44]. Consequently, clinical trials of intravenous immunoglobulin as immunotherapy for Alzheimer's disease and other neurodegenerative disorders are underway and have shown promising effects on reducing amyloid beta and improving cognition [45,46], particularly among apoE4 carriers [46]. And third, clearance of amyloid beta via activation of T cells has been shown to differ across different HLA alleles and DRB1*1301 is one of the alleles shown to stimulate amyloid beta T cells [47].
In summary, neuroprotection by DRB1*13 could be exerted by (a) eliminating persistent neurotoxic antigens [16], (b) limiting the presence of cathepsin S, a known endogenous harmful substance in brain aging [48], to which DRB1*13:02 binds with high, and DRB1*13:01 with lower, affinity [49], and (c) limiting the presence of amyloid A beta by facilitating the production of autoantibodies against it [47]. Concerning apoE, the detrimental effects of E4 and various mechanisms of action have been well established and investigated, as reviewed above. In contrast, the cell biological mechanism(s) by which apoE2 confers protection are not well understood but promising findings have been published recently [50].
All individuals in the current study were cognitively healthy; however, one might expect that those lacking the protective DRB1*13 or apoE2 alleles may ultimately be at greater risk for cognitive decline given evidence of age-related gray matter atrophy previously observed in women without DRB1*13:02 [14], evidence of relative protective effects of apoE2, and increased age-related variability in those lacking DRB1*13 and/or apoE2 documented in the present study. Longitudinal studies are currently underway in our lab to investigate genetic and other factors underlying healthy brain aging versus disease in addition to network characteristics associated with each.
5. Limitation of the study
The highly significant and novel protective effects of DRB1*13 and apoE2 on neural network variability observed in the present study must be considered within the context of the study limitations which include a relatively small sample size and limited allele diversity. Of the DRB1*13 alleles, DRB1*13:01 and DRB1*13:02 are the most common among US ethnic populations and thus our sample is representative of the US population at large [51]. Numerous other DRB1*13 alleles exist in the general population, yet representation of other DRB1*13 alleles was limited here precluding investigation into potential protective effects of those alleles. HLA, however, is highly polymorphic with some variants more strongly associated with disease and others with protection. For instance, HLA DRB1*15:01 has been associated with “pan-neuronal disease vulnerability” [11] given its association with Alzheimer's disease, multiple sclerosis, and Parkinson's disease. Thus, future studies may consider investigating the influence of other HLA alleles on age-related brain changes independently and in conjunction with other factors that may contribute to brain health and disease. Similarly, relatively limited sample size precluded us from investigating all possible HLA DRB1*13/apoE combinations or in some cases resulted in relatively small subsamples. Finally, although the results of this cross-sectional sample are compelling, longitudinal studies tracking changes in network properties with age and with regard to presence or absence of DRB1*13 alleles and apoE2 (vs apoE4) will permit more robust conclusions in terms of protective effects of DRB1*13 alleles and apoE2 as well as mitigating factors; this work is currently underway.
Author contributions
Contributed to data collection: ACL, SD. Contributed to study design: AG, APG. Contributed to data analysis: APG, LMJ, BEE, AG, ACL. Wrote the paper: LMJ, APG, AG. Contributed to editing the paper: All.
Declaration of interests
The authors have no conflicts of interest to declare.
Role of the funding source
Partial funding for this study was provided by the University of Minnesota (the Kunin Professorship for Women's Healthy Brain Aging, the Brain and Genomics Fund, the McKnight Presidential Chair of Cognitive Neuroscience, and the American Legion Brain Sciences Chair). The sponsors had no role in the current study design, analysis or interpretation, or in the writing of this paper. The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
References
- 1.Cabeza R., Nyberg L., Park D.C., editors. Cognitive neuroscience of aging: Linking cognitive and cerebral aging. Oxford University Press; 2016. [Google Scholar]
- 2.Enzinger C., Fazekas F., Matthews P.M. Risk factors for progression of brain atrophy in aging: six-year follow-up of normal subjects. Neurology. 2005;64 doi: 10.1212/01.WNL.0000161871.83614.BB. [DOI] [PubMed] [Google Scholar]
- 3.Liu C.C., Kanekiyo T., Xu H., Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen K., Reiman E.M., Alexander G.E. Correlations between apolipoprotein E4 gene dose and whole brain atrophy rates. Am J Psychiatry. 2007;164:916–921. doi: 10.1176/ajp.2007.164.6.916. [DOI] [PubMed] [Google Scholar]
- 5.Canuet L., Tellado I., Couceiro V. Resting-state network disruption and APOE genotype in Alzheimer's disease: a lagged functional connectivity study. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leuthold A.C., Mahan M.Y., Stanwyck J.J., Georgopoulos A., Georgopoulos A.P. The number of cysteine residues per mole in apolipoprotein E affects systematically synchronous neural interactions in women's healthy brains. Exp Brain Res. 2013;226:525–536. doi: 10.1007/s00221-013-3464-x. [DOI] [PubMed] [Google Scholar]
- 7.Corder E.J., Saunders A.M., Strittmetter W.J. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 8.Lambert J.C., Ibrahim-Verbaas C.A., Harold D. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wisdom N.M., Callahan J.L., Hawkins K.A. The effects of apolipoprotein E on non-impaired cognitive functioning: a meta-analysis. Neurobiol Aging. 2011;32:63–74. doi: 10.1016/j.neurobiolaging.2009.02.003. [doi:10/1016/j.neurobiolaging.2009.02.003] [DOI] [PubMed] [Google Scholar]
- 10.Meuer S.C., Hussey R.E., Hodgdon J.C. Surface structures involved in target recognition by human cytotoxic T lymphocytes. Science. 1982;218:471–473. doi: 10.1126/science.6981845. [DOI] [PubMed] [Google Scholar]
- 11.Steele N.Z., Carr J.S., Bonham L.W. Fine-mapping of the human leukocyte antigen locus as a risk factor for Alzheimer disease: a case–control study. PLOS. 2017 doi: 10.1371/journal.pmed.1002272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Z.X., Wan Y., Tan L. Genetic association of HLA gene variants with MRI brain structure in Alzheimer's disease. Mol Neurobiol. 2017 doi: 10.1007/s12035-016-9889-z. [DOI] [PubMed] [Google Scholar]
- 13.Georgopoulos A.P., James L.M., Mahan M.Y. Reduced Human Leukocyte Antigen (HLA) protection in Gulf War Illness (GWI) EBioMedicine. 2016;3:79–85. doi: 10.1016/j.ebiom.2015.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.James L.M., Christova P., Lewis S.M., Engdahl B.E., Georgopoulos A., Georgopoulos A.P. Protective effect of Human Leukocyte Antigen (HLA) allele DRB1* 13: 02 on age-related brain gray natter volume reduction in healthy women. EBioMedicine. 2018;29:31–37. doi: 10.1016/j.ebiom.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Payton A., van den Boogerd E., Davidson Y. Influence and interactions of cathepsin D, HLA-DRB1 and APOE on cognitive abilities in an older non-demented population. Genes Brain Behav. 2006 doi: 10.1111/j.1601-183X.2006.00191.x. [DOI] [PubMed] [Google Scholar]
- 16.James L.M., Christova P., Engdahl B.E., Lewis S.M., Carpenter A.F., Georgopoulos A.P. Human leukocyte antigen (HLA) and Gulf War Illness (GWI): HLA-DRB1*13:02 spares subcortical atrophy in Gulf War veterans. EBioMedicine. 2017;26:126–131. doi: 10.1016/j.ebiom.2017.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bettencourt A., Carvalho C., Leal B. The protective role of HLA-DRB1*13 in autoimmune diseases. J Immunol Res. 2015;15 doi: 10.1155/2015/948723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furukawa H., Oka S., Tsuchiya N. The role of common protective alleles HLA-DRB1*13 among systemic autoimmune diseases. Genes Immun. 2017;18:1–7. doi: 10.1038/gene.2016.40. [DOI] [PubMed] [Google Scholar]
- 19.Hov J.R., Kosmoliaptsis V., Traherne J.A. Electrostatic modifications of the HLA-DR P9 peptide-binding pocket and susceptibility to primary sclerosing cholangitis. Hepatology. 2011;53:1967–1976. doi: 10.1002/hep.24299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van der Woude D., Lie B.A., Lundström E. Protection against anti-citrullinated protein antibody-positive rheumatoid arthritis is predominantly associated with HLA-DRB1*1301: a meta-analysis of HLA-DRB1 associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in four European populations. Arthritis Rheum. 2010;62:1236–1245. doi: 10.1002/art.27366. [DOI] [PubMed] [Google Scholar]
- 21.Fainboim L., Canero V.M.C. Protracted, but not acute, hepatitis a virus infection is strongly associated with HLA-DRB*1301, a marker for pediatric autoimmune hepatitis. Hepatology. 2001;33:1512–1517. doi: 10.1053/jhep.2001.24562. [DOI] [PubMed] [Google Scholar]
- 22.Pando M., Larriba J., Fernandez G.C. Pediatric and adult forms of type I autoimmune hepatitis in Argentina: evidence for differential genetic predisposition. Hepatology. 1999;20:1374–1380. doi: 10.1002/hep.510300611. [DOI] [PubMed] [Google Scholar]
- 23.Langheim F.P.J., Leuthold A.C., Georgopoulos A.P. Synchronous dynamic brain networks revealed by magnetoencephalography. Proc Natl Acad Sci U S A. 2006;103:455–459. doi: 10.1073/pnas.0509623102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georgopoulos A.P., James L.M., Carpenter A.F., Engdahl B.E., Leuthold A.C., Lewis S.M. Gulf War illness (GWI) as a neuroimmune disease. Exp Brain Res. 2017 doi: 10.1007/s00221-017-5050-0. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 25.Georgopoulos A.P., Karageorgiou E., Leuthold A. Synchronous neural interactions assessed by magnetoencephalography: a functional biomarker for brain disorders. J Neural Eng. 2007;4:349–355. doi: 10.1088/1741-2560/4/4/001. [DOI] [PubMed] [Google Scholar]
- 26.Georgopoulos A.P., Tan H.M., Lewis S.M. The synchronous neural interactions test as a functional neuromarker for post-traumatic stress disorder (PTSD): a robust classification method based on the bootstrap. J Neural Eng. 2010;7 doi: 10.1088/1741-2560/7/1/016011. [DOI] [PubMed] [Google Scholar]
- 27.Engdahl B., Leuthold A.C., Tan H.R. Post-traumatic stress disorder: a right temporal lobe syndrome? J Neural Eng. 2010;7 doi: 10.1088/1741-2560/7/6/066005. [DOI] [PubMed] [Google Scholar]
- 28.Christopoulos V., Georgopoulos A., Georgopoulos A.P. The effect of apolipoprotein E4 on synchronous neural interactions in brain cultures. Exp Brain Res. 2015;233:1977–1982. doi: 10.1007/s00221-015-4270-4. [DOI] [PubMed] [Google Scholar]
- 29.Nasreddine Z.S., Phillips N.A., Bédirian V. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc. 2005;53:695–699. doi: 10.1111/j.1532-5415.2005.53221.x. [DOI] [PubMed] [Google Scholar]
- 30.Teng E.L., Chui H.C. The modified mini-mental state examination (3MS) Can J Psychiatry. 1987;41:114–121. [PubMed] [Google Scholar]
- 31.Cano P., Klitz W., Mack S.J. Common and well-documented HLA alleles: report of the Ad-Hoc committee of the American society for histocompatiblity and immunogenetics. Hum Immunol. 2007;68:392–417. doi: 10.1016/j.humimm.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 32.Reymer W.A., Groenemeyer B.E., Van de Burg R. Apolipoprotein E genotyping on agarose gels. Clin Chem. 1995;41:1046–1047. [PubMed] [Google Scholar]
- 33.Mahan M.Y., Chorn C.R., Georgopoulos A.P. Proc 14th Python In Science Conference. 2015. White Noise Test: detecting autocorrelation and nonstationarities in long time series after ARIMA modeling. [Scipy Austin, TX] [Google Scholar]
- 34.Box G.E.P., Jenkins G.M. Holden-Day; San Francisco: 1976. Time series analysis: Forecasting and control. [Google Scholar]
- 35.Fisher R.A. 13th edition. Oliver and Boyd; Edinburgh: 1958. Statistical methods for research workers. [Google Scholar]
- 36.Mahley R.W., Weisgraber K.H., Huang Y. Apolipoprotein E: structure determines function, from atherosclerosis to Alzheimer's disease to AIDS. J Lipid Res. 2009;50:183–188. doi: 10.1194/jlr.R800069-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahley R.W., Rall S.C. Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 38.Weisgraber K.H., Rall S.C., Mahley R.W. Human E apoprotein heterogeneity. Cysteine-arginine interchanges in the amino acid sequence of the apo-E isoforms. J Biol Chem. 1981;256:9077–9083. [PubMed] [Google Scholar]
- 39.Hatters D.M., Peters-Libeu C.A., Weisgraber K.H. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 40.Institute of medicine. Adverse effects of vaccines: Evidence and causality. National Academies Press; Washington, DC: 2012. [PubMed] [Google Scholar]
- 41.Vita R., Overton J.A., Greenbaum J.A. The immune epitope database (IEDB) 3.0. Nucleic Acids Res. 2015;43(Database issue) doi: 10.1093/nar/gku938. D405–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D'Agostino P.M.D., Gottfried-Blacvkmore A., Anandasabapathy N., Bullock K. Brain dendritic cells: biology and pathology. Acta Neuropathol. 2012 doi: 10.1007/s00401-012-1018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dodel R., Balakrishnan K., Keyvani K. Naturally occurring autoantibodies against β-amyloid: investigating their role in transgenic animal and in vitro models of Alzheimers' disease. J Neurosci. 2011;31:5847–5854. doi: 10.1523/JNEUROSCI.4401-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du Y., Dodel R., Hampel H. Reduced levels of amyloid beta-peptide antibody in Alzheimer disease. Neurology. 2001;57:801–805. doi: 10.1212/wnl.57.5.801. [DOI] [PubMed] [Google Scholar]
- 45.Dodel R., Neff F., Noeker C. Intravenous immunoglobulins as a treatment for Alzheimer's disease: rationale and current evidence. Drugs. 2010;70:513–528. doi: 10.2165/11533070-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 46.Relkin N. Clinical trials of intravenous immunoglobulin for Alzheimer's disease. J Clin Immunol. 2014;34:74–79. doi: 10.1007/s10875-014-0041-4. [DOI] [PubMed] [Google Scholar]
- 47.Zota V., Nemeriovsky A., Baron R. HLA-DR alleles in amyloid β-peptide autoimmunity: a highly immunogenic role for the DRB1*1501 allele. J Immunol. 2009;183:3522–3530. doi: 10.4049/jimmunol.0900620. [DOI] [PubMed] [Google Scholar]
- 48.Wendt W., Lübbert H., Stichel C.C. Upregulation of cathepsin S in the aging and pathological nervous system of mice. Brain Res. 2008;26(1232):7–20. doi: 10.1016/j.brainres.2008.07.067. [DOI] [PubMed] [Google Scholar]
- 49.Davenport M.P., Quinn C.L., Chicz R.M. Naturally processed peptides from two disease-resistance-associated HLA-DR13 alleles show related sequence motifs and the effects of the dimorphism at position 86 of the HLA-DR beta chain. Proc Natl Acad Sci U S A. 1995;92:6567–6571. doi: 10.1073/pnas.92.14.6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conejero-Goldberg C., Gomar J.J., Bobes-Bascaran T. APOE2 enhances neuroprotection against Alzheimer's disease through multiple molecular mechanisms. Mol Psychiatry. 2014;9:1243–1250. doi: 10.1038/mp.2013.194. [DOI] [PubMed] [Google Scholar]
- 51.Sintasath D.M., Tang T., Slack R. Relative HLA-DRB1∗ 13 allele frequencies and DRB3 associations of unrelated individuals from five US populations. Hum Immunol. 1999;60:1001–1010. doi: 10.1016/s0198-8859(99)00085-3. [DOI] [PubMed] [Google Scholar]