ABSTRACT
All organisms contain RNA modifications in their ribosomal RNA (rRNA), but the importance, positions and exact function of these are still not fully elucidated. Various functions such as stabilizing structures, controlling ribosome assembly and facilitating interactions have been suggested and in some cases substantiated. Bacterial rRNA contains much fewer modifications than eukaryotic rRNA. The rRNA modification patterns in bacteria differ from each other, but too few organisms have been mapped to draw general conclusions. This study maps 23S ribosomal RNA modifications in Clostridium sporogenes that can be characterized as a non-toxin producing Clostridium botulinum. Clostridia are able to sporulate and thereby survive harsh conditions, and are in general considered to be resilient to antibiotics. Selected regions of the 23S rRNA were investigated by mass spectrometry and by primer extension analysis to pinpoint modified sites and the nature of the modifications. Apparently, C. sporogenes 23S rRNA contains few modifications compared to other investigated bacteria. No modifications were identified in domain II and III of 23S rRNA. Three modifications were identified in domain IV, all of which have also been found in other organisms. Two unusual modifications were identified in domain V, methylated dihydrouridine at position U2449 and dihydrouridine at position U2500 (Escherichia coli numbering), in addition to four previously known modified positions. The enzymes responsible for the modifications were searched for in the C. sporogenes genome using BLAST with characterized enzymes as query. The search identified genes potentially coding for RNA modifying enzymes responsible for most of the found modifications.
KEYWORDS: RNA modifications, RNA methylations, 23S RNA, oh5C, dihydrouridine, mass spectroscopy
1. Introduction
Protein synthesis is a highly regulated process and rRNA modifications are thought to play an important role as all organisms contain them, and many modified nucleotides are conserved among bacterial species. The majority of the modified nucleotides of the 50S ribosomal subunit observed so far are in domains II, IV and V of the 23S rRNA. Many are positioned in functional centers such as in the peptidyl transferase center [1]. Eukaryotic rRNAs contain many more modifications than bacteria, especially because of a high number of pseudouridines and ribose methylations. The whole 23S rRNA modification pattern has only been determined for a few bacteria and it is also difficult to conclude when a modification data set is complete. It is generally agreed that the total rRNA modification pattern has been elucidated in E. coli that contains 25 modifications in 23S rRNA and 11 in 16S rRNA, reviewed by e.g. [2,3]. E. coli 23S rRNA contains 14 methylations, nine pseudouridines, one methylpseudouridine, one dihydrouridine and one hydroxycytidine. In contrast Thermus thermophilus only seem to contain 11 modifications in 23 rRNA [4,5]. All E. coli genes involved in 23S rRNA modifications have been identified, except the one responsible for dihydrouridine formation [6,7]. In bacteria, the RNA modifying enzymes are often specific for one site and they differ in substrate preference such as bare rRNA, intermediates of assembly, or mature ribosomal subunits. Various steps of ribosome biogenesis may program certain modification enzyme actions, and most of the 23S rRNA modifications take place at early assembly steps [8]. This is in contrast to 16S sRNA modifications where many are applied on the assembled 30S subunit [8].
The full role of each of the RNA modifications is still not elucidated and they are singly dispensable but often not in combinations. For example, seven modified nucleotides of the peptidyl transferase center were found to be indispensable in the reconstruction of functionally active ribosomal particles in E. coli [9]. The modifications influence chemical properties of nucleotides, and can thus influence the structure, assembly and activity of the ribosome. The most common modifications in rRNA are methylations that increase nucleotide hydrophobicity and influence base stacking and in some cases affect hydrogen bonding, and pseudouridine formation that increases hydrogen bonding potential and also affects stacking interactions, reviewed in [2,10]. RNA nucleotide methylation occurs at a variety of atoms and the function of nucleotide methylations have been intensively studied in tRNA (reviewed in [11]). Especially for rRNA, the modifications may create ‘check-points’ for ribosomal subunits assembly [12]. Finally, rRNA methylation (or the lack of methylation) can cause resistance to antibiotics that bind to the ribosome by changing the antibiotic binding site, reviewed in [13,14].
Finding modifications in large RNAs is not a trivial task and there is no easy method to map the presence and position of all modifications in rRNA. The overall content of modifications can be determined by liquid chromatography–tandem mass spectrometry (LC-MS/MS) e.g. [15,16], but this does not tell where in the structure they are present. More extensive investigations rely on the rRNA being digested into defined products, which are then sequenced by tandem mass spectrometry [17]. In later years, this concept has been supplemented with the potential for quantitation based on LC-MS/MS analyses of isotopically labelled rRNA [18,19]. There are also new methods for profiling ribose methylations and pseudouridines based on next-generation nucleic acid sequencing (see e.g. [10,20]), which may help unravel the location and importance of RNA modifications in general.
In this study, we investigated the modification pattern in C. sporogenes 23S rRNA regions that are known to be hot spots for modifications in other bacteria. The purpose was to glean an impression of modification level and diversity in the 23S rRNA of a hitherto unexplored genus. The presence of ten modifications was determined by mass spectrometry and primer extension analysis of specific 23S rRNA regions. The ten modifications comprise nine different nucleotide positions and two of the ten differ from those seen in E. coli 23S rRNA. One modification appears at position U2500 where modifications have not previously been observed. Furthermore, BLAST and database searches were used to identify putative modifying enzymes coded for in the C. sporogenes genome.
2. Materials and methods
2.1. Growth conditions and purification of total RNA for primer extension analysis and mass spectrometry analysis
The strain Clostridium sporogenes ATCC 15579 was obtained from the American Type Culture Collection. It was grown anaerobically in ATCC® Medium: 2107 Modified Reinforced Clostridial Broth. 0.2 mL of an overnight culture of C. sporogenes was added to 100 mL aliquots of fresh broth and incubated at 37°C until significant visible growth was obtained. Cells were placed on ice for 10 min and then centrifuged at 4500 rpm for 10 min at 4°C. The pellets were resuspended in 10 mL of cold TMN buffer (100 mM NH4Cl, 10 mM MgCl2, 50 mM Tris-HCl pH 7.5). The cells were lysed by sonication, and cell debris was removed by centrifugation. RNA was extracted using phenol (equilibrated to pH 4.5 with Sodium acetate) and chloroform (3 x phenol, 1 x phenol/chloroform, 1x chloroform). The RNA was precipitated with 1/10 volume of 3M NaOAc, pH 4.5 and 2,5 volumes of 96% ethanol, incubated at −20ºC and centrifuged for 40 min. The pellet was washed with 70% cold ethanol, centrifuged and left to dry at room temperature before dissolving in water. E. coli RNA was used as control for the primer extension analysis. E. coli MRE600 was grown in LB medium at 37°C until middle exponential phase, harvested and then total RNA was obtained using GeneJET RNA purification kit (Thermo Scientific).
2.2. Mass spectrometric analysis of RNA
Selected fragments of the 23S rRNA sequence around potential modified sites were isolated for MALDI Time-of-Flight mass spectrometric analysis. Purified total RNA was hybridized with complementary oligodeoxynucleotides, followed by digestion with mung bean nuclease and RNase A as described in [21]. After sub-fragment purification on a denaturing polyacrylamide gel, the RNA was separately digested with RNase T1 (G-specific) and RNase A (U/C-specific) for mass spectrometric analysis as previously reported [22]. Briefly, 1–2 pmol rRNA sub-fragment were RNase digested to completion and analyzed directly using 3-hydroxypicolinic acid as matrix. Mass spectra were recorded in positive ion mode with a reflectron Time-of-Flight mass analyzer on a Bruker UltraFlex MALDI mass spectrometer and processed using the software ‘m/z’ (Proteometrics). The observed masses were compared to theoretical masses calculated using GPMAW software (Lighthouse Data, Denmark). Further analysis to confirm the exact position of the observed modifications was performed by MALDI tandem mass spectrometry using a Waters Q-TOF Premiere instrument (Waters, Manchester, UK) in positive ion mode as previously described [23].
2.3. Primer extension analysis to verify the position of specific modifications
Selected modifications were studied by primer extension analysis [24] with AMV reverse transcriptase (Roche) and 5´-[32P] labelled oligodeoxynucleotide primers complementary to C. sporogenes 23S rRNA. To overcome presumed rRNA structure problems when using primer BV632 on C. sporogenes rRNA the extension temperature was raised by 2°C and the dideoxynucleotide concentration was doubled. The cDNA extension products were separated on 13% polyacrylamide sequencing gels. The positions of the stops were visualized by autoradiography on a Typhoon scanner and identified by reference to dideoxynucleotide sequencing reactions on 23S rRNA that were electrophoresed in parallel. The 23S rRNA sequence used was C. sporogenes ATCC 15579.
2.4. Potential genes for RNA modifying enzymes in C. sporogenes ATCC 15579
Potential genes for the modifying enzymes were sought based on information primarily from E. coli genes, but also from genes from other bacteria with known 23S rRNA modifying enzymes. BLAST searches using https://blast.ncbi.nlm.nih.gov/Blast.cgi and default parameters were performed with the query described in the text and in the non-redundant protein sequences from C. sporogenes ATCC 15579. The E values (Expect values) presented in the text are directly from the BLAST searches and express the number of BLAST hits you can expect to see by chance.
3. Results and discussion
3.1. Modifications in C. sporogenes 23S rRNA
This study focuses on mapping methylations and other mass-changing RNA modifications in C. sporogenes 23S rRNA. According to [1,2] and others, including the MODOMICS database [25], the modified nucleotides cluster in specific domains in 23S rRNA. As a detailed mapping is a major task, we have focused our investigations on the domains and areas where modifications can be expected to occur based on knowledge from other bacteria. The areas investigated are shown in green overlay in Figure 1. To ease comparison with other organisms all numbering in this paper is according to the E. coli 23S rRNA sequence. The C. sporogenes 23S rRNA is 2902 nucleotides in length while the E. coli 23S rRNA is 2904 with various insertions and deletions, so corrections are needed for exact C. sporogenes numbering.
Figure 1.
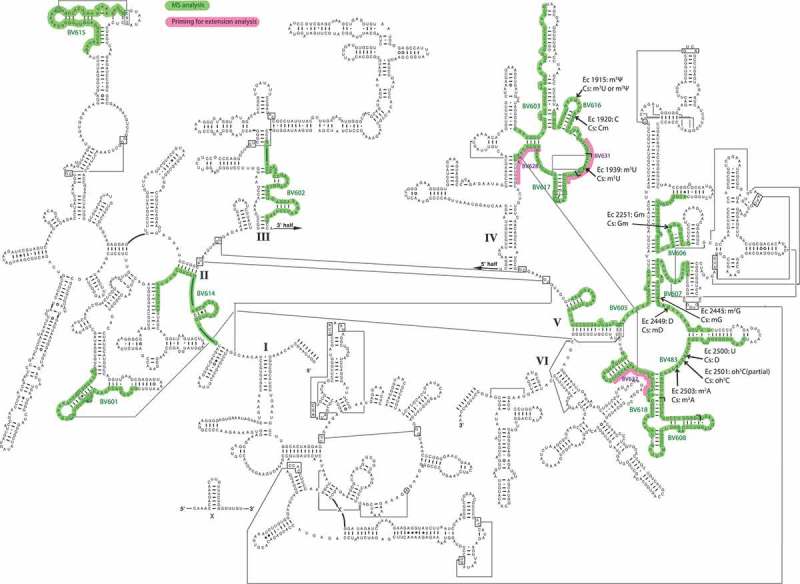
A secondary structure model of C. sporogenes 23S rRNA (http://www.rna.icmb.utexas.edu). Green overlay shows areas isolated for MS analysis with the names of the complementary oligodeoxynucleotides used for hybridization. The overlapse between BV616 and BV617 and between BV483 and BV608 are indicated by L-shaped symbols. Red overlay shows areas used for hybridizing DNA for primer extension analysis. Modified nucleotides are pointed to by arrows, using E. coli numbering though the 23S rRNA sequence is C. sporogenes. Ec is abbreviation for E. coli and Cs for C. sporogenes.
The amount of bacterial 23S rRNA modification data is still limited. In addition to the complete data from E. coli 23S rRNA modifications and the enzymes performing these modifications, there is a study of the 23S rRNA modifications from T. thermophilus [4], and some data on specific positions in Bacillus subtilis [26], Deinococcus radiodurans [27], Pseudomonas aeruginosa [6] and Mycobacterium tuberculosis [28]. Although using E. coli 23S rRNA as main information source for choosing areas of investigation, we took into account the data from the other organisms whenever relevant.
The modifications were pointed to, characterized and confirmed by analysis using mass spectrometry (MS), tandem mass spectrometry (MS-MS) and primer extension. Modifications might escape mass spectrometry analysis of RNase digestion products if the modifications end up in mononucleotides that are too low in mass for proper detection or if present in fragments that are difficult to detect. Also, not all modifications affect primer extension analysis, but even this information can be used to suggest the nature of modification, as will be illustrated later. The MS-MS analyses provide information on both the modified position as well as whether the modification is positioned on the base or the ribose moiety [29]. A summary of detected modifications is presented in Table 1 and the individual findings are described below.
Table 1.
Summary of the 23S rRNA modifications detected and a putative annotation of modifying enzymes. Ec = E. coli, Cspo = C. sporogenes, pos. = nucleotide position in E. coli 23S rRNA, mod = nucleotide modification, * The E value obtained by BLAST is presented after the accession code for the enzyme, see text for organism and % identity, ** = partial modified, *** indicate an alleged assignment.
| Ec pos. | Cspo mod. | Ec mod. | Enzyme E value* | Putative Cspo protein |
|---|---|---|---|---|
| 1915 | m3U | m3 Ψ | RlmH P0A8I8: 5e-25/RluD P3364: 4e-76 | EDU38179.1/EDU37299.1*** |
| 1920 | Cm | none | TlyA KBJ35054.1: 5e-61 TlyA WP_053414602.1: 6e-99 |
EDU36942.1 |
| 1939 | m5U | m5U | RlmD KIX81902.1: 3e-179 | EDU39155.1 |
| 2251 | Gm | Gm | RlmB P63177: 4e-53 | EDU39019.1 |
| 2445 | mG | m2G | RlmKL WP_001475205.1: 2e-62 Smu472 Q8DVL0: 3e-111 |
EDU37295.1 |
| 2449 | mD | D (mD**) | unknown | unknown |
| 2500 | D | none | unknown | unknown |
| 2501 | oh5C | oh5C ** | RlhA P76104: 8e-74 | EDU39416.1*** |
| 2503 | m2A | m2A | RlmN P36979.1: 8e-72 | EDU36326.1 |
3.2. RNA modifications in domain V of 23S rRNA
Domain V generally contains more posttranscriptional modifications than other 23S rRNA domains, and some of the modifications are also phylogenetically highly conserved. Hence, we started our investigations with this domain in C. sporogenes 23S rRNA, where we both re-found some of the generally modified nucleotides but also some new modified positions.
In the lower right of the peptidyl transferase loop, the oligodeoxynucleotide BV483 (Figure 1) covers a modification ‘hotspot’. E. coli 23S rRNA has the ribose methylation Cm2498, a partial hydroxylation at C2501 (oh5C2501), m2A2503 and a pseudouridine at position 2504. In e.g. D. radiodurans, the methylation at C2498 is absent, but C2499 harbours a methyl group at C-5 [27]. The C. sporogenes 23S rRNA fragment from position 2480 to 2527 was isolated as described in ‘Materials and Methods’ and subsequently digested in parallel with RNase T1 and RNase A. The RNase products were directly analyzed by mass spectrometry. The RNase T1 digest (Figure 2(a)) revealed two products indicative of modifications. m/z 1013.1 suggests the presence of m2A2503, as this signal replaces the unmodified 2503AUG2505 signal at m/z 999.1. The RNase T1 product 2496CACCUCG2502 would appear at m/z 2219.3 in an unmodified version, but instead a signal at m/z 2237.3 appeared. As mentioned above, methylations at nucleotides 2498 and 2499, and hydroxyl group at 2501 have previously been reported, but combinations of these do not match the observed 18.0 Da. mass increase. The RNase A digestion (Figure 2(b)) showed an unmodified 2497AC2498 product at m/z 653.11 (ribose methylation of C2498 prevents RNase A cleavage, and would induce a mass increment of one nucleotide). In line with the RNase T1 digestion, m/z 1013.1 for 2502GAU2504 suggests m2A2503, and a base methylation of A2503 was verified by tandem mass spectrometry (data not shown). This was expected as m2A2503 seems to be a very well conserved modification in bacteria [30,31]. It is the only adenosine in E. coli rRNA, which is methylated at a carbon atom. The RlmN methyltransferase responsible for this modification belongs to the family of radical SAM enzymes that differs from the majority of other methyltransferases [31]. In the C. sporogenes genome we find a gene for a 348 aa enzyme (EDU36326.1) with a 36% identity (E-value 8e-72) to the E. coli 384 aa RlmN (P36979.1), and we consider this a likely candidate for performing the m2A2503 modification in C. sporogenes 23S rRNA. The modification is positioned in the peptidyl transferase center and it has been suggested to play a role in proofreading [31] and it might also be weakly related to antibiotic sensitivity.
Figure 2.
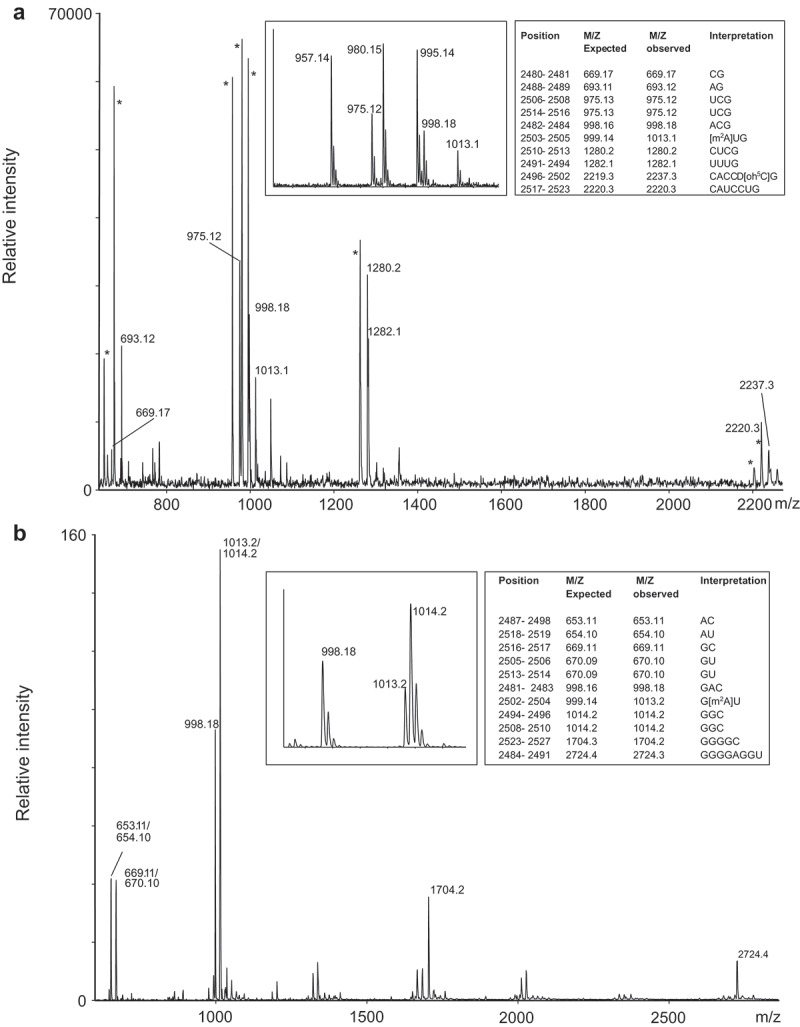
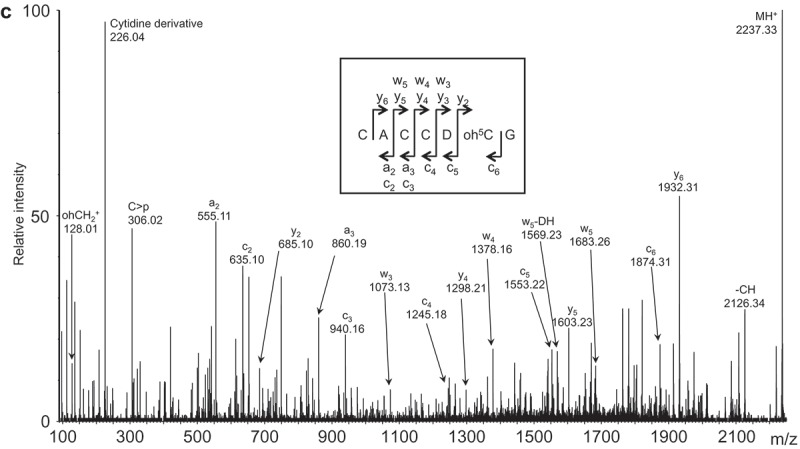
MS analyzes of the fragment isolated by the oligodeoxynucleotide BV483 covering position 2480 to 2527. (a) MALDI TOF mass spectrum of RNase T1 digested 23S rRNA sub-fragment. The labelled signals correspond to the 3´-phosphate products, but intense signals corresponding to 2´-3´-cyclic phosphate products (*) also appear approximately 18.0 Da. lower as exemplified in the insert. The table shows m/z values of unmodified RNA and compares to observed values and interpretation of the mass increase; the latter is based on modifications reported in literature, digestion with RNase A and tandem MS data as explained in the main text. (b) MALDI TOF mass spectrum of RNase A digested 23S rRNA sub-fragment. The table indicates m/z values of unmodified RNA and compares to observed values and interpretation of the mass increase; the latter is based on modifications reported in literature, digestion with RNase T1 and tandem MS data as explained in the main text. The insert illustrates m/z signals and isotope patterns leading to the assignment of m2A2503. Unlabelled signals have m/z values corresponding to gas phase-formed dimers of the RNase products around 660 and 1000 m/z. (c) Tandem mass spectrum of the m/z 2237.3 RNase T1 digestion product (position 2496–2502 of 23S rRNA) harbouring 18.0 Da. posttranscriptional modifications. Unambiguous backbone fragment ions, key N-glycosidic bond cleavage products and a few other prominent fragment signals are assigned. The insert indicates how backbone fragment ions contribute to the sequence determination. Fragment ion nomenclature is according to [50].
By combining the RNase digestion information (Figure 2(a,b)) the unaccounted 18.0 Da. modifications must reside in the sequence 2499CUC2501, which may be explained by a hydroxylation of C2501 and a dihydrouridine (D) at position 2500. We substantiated this by tandem mass spectrometry of the ‘2496CACCUCG2502 + 18.0 Da.’ RNAse T1 digestion product (Figure 2(c)). The data confirms an unmodified C2498, D2500 and oh5C2501. This is surprising as dihydrouridine in 23S rRNA has previously only been observed at position 2449 in E. coli. Either the observed D2500 is an exception to the common pattern of 23S rRNA modifications or its presence has escaped detection in other organisms. We observed full hydroxylation at C2501 as in D. radiodurans, while E. coli 23S rRNA only showed partial oh5C2501 [27]. A very recent study by Kimura et al. [7] has assigned the gene ydcP in E. coli to code for the enzyme responsible for oh5C2501, and the gene was renamed rlhA. Notably, they distinctly identified homologous of rlhA in Clostridia. The E. coli studies [7,27] observed a relation between modification level and growth phase and Kimura et al. showed that iron depletion decreased the modification level. It might be that in some organism this modification either reflects growth phase or response to changes in growth conditions. BLAST of the 653 aa RlhA E. coli protein sequence with protein sequences from C. sporogenes suggests the 788 aa long protein EDU39416.1/WP_003483554.1 (E-value 8e-74) with an extension in the C-terminal part relative to the E. coli protein and 33% identity in the region covered. This identification is tentative and needs verification.
Figure 2.
(Continued.)
Gm2251 is a highly conserved nucleotide modification that is found in all three kingdoms of life [2,5]. MS analysis of RNase digests of the RNA hybridized to oligodeoxynucleotide BV606 confirmed the presence of masses corresponding to a methylated RNase A product 2250GGGGC2254, and methylated GG from RNase T1 digestion in accordance with Gm2251 (ribose methylation prevents digestion with RNase T1). The exact position and the nature of the methylation were investigated by MS/MS of methylated 2250GGGGC2254 and confirmed methylation at the ribose of G2251 (data not shown). The methyltransferase RlmB is responsible for this 2ʹ-O methylated nucleotide [32]. A C. sporogenes protein (EDU39019.1/WP_003482898.1) with 36% identity to E. coli RlmB (E-value 4e-53) was found and is a putative candidate for the enzyme responsible for the G2251 ribose methylation. This nucleotide is located in the so-called P-loop of the 23S rRNA, interacting with the CCA-end of the P-site bound tRNA [33], which suggests an important function for the modification.
The region in domain V covered by oligodeoxynucleotide BV607 (Figure 1) has two modifications in E. coli 23S rRNA, namely m2G2445 and dihydrouridine (D) at 2449. Our MS analyses suggested an additional methyl group located at either A2448 or D2449 in C. sporogenes 23S rRNA: The RNase A digest showed a mass corresponding to dimethylated 2444GGGGAU2449 + 2.0 Da, the RNase T1 digest showed a mass corresponding to methylated 2448AUAACAG2454 + 2.0 Da and a methylated GG; there was absence of the corresponding unmodified fragments in all cases. Tandem MS analyses of modified 2444GGGGAU2449 and 2448AUAACAG2454 (Figure 3) showed methylation of G2445 and a methylated D2449 (or alternatively some other 16.0 Da modification of U2449). We favour a methylated D2449, because such a modification is sub-stoichiometrically present in E. coli 23S rRNA at position 2449 [34]. The enzyme responsible for the E. coli D2449 modification is not yet identified, but D is a common modification in the D-loop of tRNAs. Some dihydrouridine synthases (DUS) responsible for D modifications in tRNA have been characterized (reviewed by [35]). E. coli has three dus genes, and, so far, they have only been related to tRNA modifications; D2449 is thus the only 23S rRNA modification in E. coli where the modifying enzyme has not been identified. BLAST searches with the three E. coli Dus proteins stipulate two enzymes in C. sporogenes, EDU38226.1 with 34% identity to DusB and EDU38527.1 with 22% identity to DusA.
Figure 3.
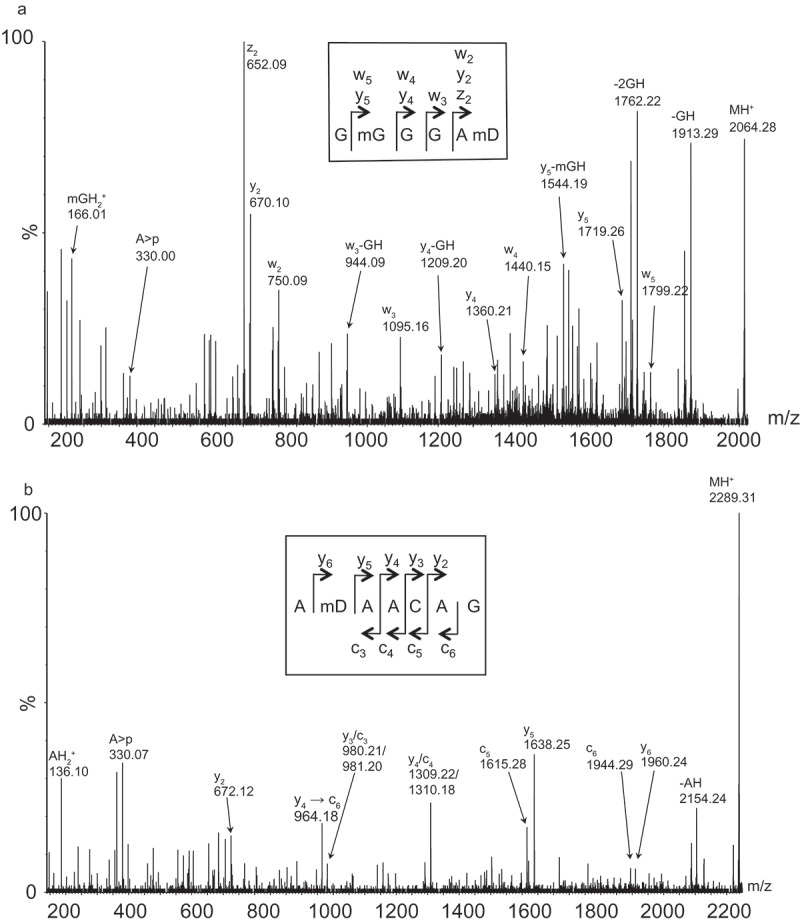
Tandem MS analyses of subfragments isolated by the oligodeoxynucleotide BV607 covering position 2418 to 2460. (a) Tandem mass spectrum of the m/z 2064.3 RNase A digestion product (position 2444–2449 of 23S rRNA) harbouring 30.0 Da. posttranscriptional modifications. Unambiguous backbone fragment ions, key N-glycosidic bond cleavage products and other prominent fragment signals are assigned. The insert indicates how fragment ions correlate to the sequence. (b) Tandem mass spectrum of the m/z 2289.3 RNase T1 digestion product (position 2448–2454 of 23S rRNA; 2´-3´cyclic phosphate) harbouring 16.0 Da. posttranscriptional modifications. Unambiguous backbone fragment and other prominent fragment signals are assigned. The insert indicates how backbone fragment ions correlate to the sequence.
In E. coli, the large bifunctional enzyme, RlmKL modifies both m7G2069 and m2G2445 [34,36,37]. We analyzed the region around G2069 in C. sporogenes 23S rRNA (covered by oligodeoxynucleotide BV605, Figure 1) by MS, but observed lack of G2069 methylation (or other modifications in this area). This is in accordance with unmodified G2069 in the Gram-positive rRNAs investigated (including B. subtilis, Mycobacterium smegmatis and Streptomyces coelicolor) [34]. The Gram-positive Streptococcus mutans harbours two proteins Smu472 and Smu776 similar to the N- and C- terminal part of RlmKL, respectively, where Smu472 add the m2G2445 modification [34]. As we do not see m7G2069, we only expect a gene for an RlmL homologue in the C. sporogenes genome. BLAST of the Smu472 found only one C. sporogenes protein: a 380 aa EDU37295.1 (WP_003486512.1) with a 45% identity (E-value 3e-111). BLAST with E. coli RlmKL (WP_001475205.1) gives the same enzyme with 32% identity (E-value 2e-62) and with only homology in the N-terminal end. We thus expect EDU37295.1 (hypothetical protein CLOSPO_03464) to be an RlmL analogue and responsible for the m2G2445 modification in C. sporogenes. m2G2445 is in a helix structure close to the peptidyl transferase loop but might be important for interaction with other nucleotides (reviewed in [2]) .
We also investigated the loop region covered by oligodeoxynucleotide BV608 (Figure 1) where E. coli 23S rRNA has Um2552. The nucleotide is conserved, and the modification seems to be widespread although at least some Bacillus species seem to have methylation at the neighbouring G2553 [38]. Our MS analyses did not show any signals that could be assigned to a U2552 modification. We therefore performed a primer extension analysis with varying dNTP concentration to see if that could indicate a modification [39]. The results are shown in Figure 4(a) and compared to E. coli 23S rRNA. It shows a clear dNTP dependent stop at U2552 E. coli 23S rRNA, while the data for C. sporogenes are less clear. The experiment has been repeated several times with the adjustments mentioned in ‘Materials and Methods’. At very low dNTP concentration there are four strong stops in the area. The stops at 2552 and 2553 persist at higher concentration but are eventually partly overcome at the highest concentration, but the pattern is not the one typically seen with probing of ribose methylations. We believe these stops are caused by strong secondary structures, which inhibit progression of the reverse transcriptase. A strong RNA helix structure is present neighbouring the loop with the 2552 modification that might cause the dNTP dependent stop at positions 2558 and 2559. When succeeding in opening the helix, the possibility of forming two new helix structures appears by parring 2520–2524 with 2533–2536 and 2538–2542 with 2546–2550 and we suggest this to be the cause of the double band seen at positions 2552–2553.
U2552 in E. coli is modified by the methyltransferase RlmE and is located in the so-called A-loop that interacts with aminoacyl tRNA [40] and the methylation has been suggested to be a negative modulator of translational accuracy [41]. A BLAST search with WP_000145975.1 (E. coli RlmE) does not find C. sporogenes proteins with significant similarity. Based on the MS data, the primer extension data and the absence of an RlmE orthologue we suggest that U2552 is unmodified in C. sporogenes 23S rRNA.
3.3. RNA modifications in domain IV of 23S rRNA
As E. coli has an m2G modification at position 1835, we analyzed the region covered by oligodeoxynucleotide BV603 (Figure 1) by MS and observed lack of methylation at the position corresponding to G1835. In contrast, the investigation of the next region of domain IV revealed three methylations at U1915, C1920 and U1939. The methylation at C1920 is indicated by the presence of a mass signifying a specific inhibition of RNase A digestion at C1920 (data not shown). Furthermore, the fragment 1911UAACUAUAAC1920 contains an additional methylation in the mass spectrum of the RNase T1 products. Its position could be pinpointed to be at U1915 as the primer extension analysis with primer BV631 (Figure 1) shows a very strong stop at U1915 (Figure 4(b)). m3U will prevent primer extension and thus give a very strong stop [42], while m5U will not prevent primer extension [26], and ribose methylation will give weaker stops that are dependent of the dNTP concentration used for primer extension [39]. Based on this, we propose an m3U at position U1915, and it is likely an m3Ψ (m3pseudouridine), as this modification appears to be universal in bacteria. The same primer extension points to a dNTP dependent stop just below C1920 consistent with a ribose methylation at this position. E. coli 23S rRNA is not modified at this position but other bacteria are, as discussed below.
Figure 4.
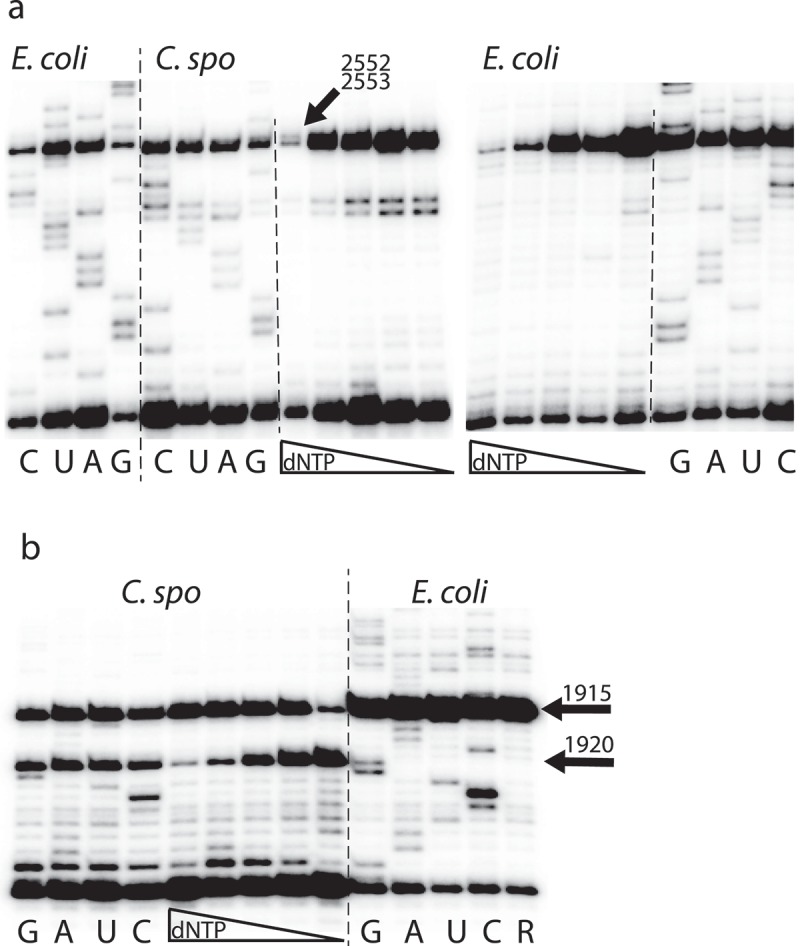
Primer extension analysis to pinpoint nucleotides with modifications. The long vertical lines separate E. coli and C. sporogenes samples and the shorter lines separate sequencing and dNTP variation. G, A, U, and C indicate sequence lanes with dideoxynucleotide-sequencing. (a) Extension from primer BV632 (Figure 1). The first four lanes are sequencing of 23S rRNA from E. coli, the next four lanes are sequencing of 23S rRNA from C. sporogenes, then follows five lanes with decreasing dNTP concentrations (0.64, 0.16, 0.08, 0.02, and 0.01mM), see text for discussion. The next five lanes are with decreasing dNTP concentrations on E. coli 23S rRNA (0.5, 0.1, 0.04, 0.02, and 0.004mM) and show a dNTP-dependent stop at U2552, indicating a ribose methylation. The rightmost four lanes show sequencing. (b) Extension from primer BV631 (Figure 1). The first four lanes are sequencing lanes with 23S rRNA from C. sporogenes. There is not clear sequencing close to the primer as is often seen, but there is a complete stop at position 1915 and a partial stop at 1920. The next five lanes show extension on the same RNA with decreasing dNTP concentrations (0.5, 0.1, 0.04, 0.02, and 0.004mM). This shows a dNTP-dependent stop at 1920, indicating a ribose methylation. The last five lanes show sequencing of 23S rRNA from E. coli plus a control (R) without ddNTPs. The absence of stop at C1920 and the clear stop at U1915 are indicated by arrows.
A base methylation of U1939 was determined by combinations of RNase digestion, MS and tandem MS analyses (data not shown). A primer extension analysis covering the relevant region with primer BV604 (Figure 1) was performed but no stops were seen, and as the modification does not affect primer extension, we assume it is m5U1939. This modification has also been observed at the same position in 23S rRNA from e.g. E. coli [2,3], T. thermophilus [4], and P. aeruginosa [6].
E. coli 23S rRNA has m3Ψ1915 and the methylating enzyme is RlmH (previously named YbeA) [42]. It has been reported that the methyltransferase RlmH prefers to work on pseudouridine, and requires associated 70S ribosome as a substrate [42,43]. A recent structural study elucidates that this small enzyme works as a functional asymmetric dimer [44]. The C. sporogenes genome contains a gene annotated to code for an RlmH (EDU38179.1) but with only 31% identity (E-value 5e-25) to the E. coli RlmH sequence. The m3Ψ1915 modification is also present in Pseudomonas (putative modifying gene PA4004) [6]. T. thermophilus also have mU at this position where Mengel-Jørgensen et al. [4] in addition to mass spectrometry data see a strong primer extension stop, thus pointing to a m3U while Polikanov et al. [5] report m5U from X-ray modelling. U1915 is positioned in the loop of helix 69 in a region where the two ribosomal subunits interact and close to helix 44 of the 30S subunit [5].
This study does not look for pseudouridines, which cannot be detected directly by MS methods, but as the U1915 methylation likely implies a pseudouridine, the C. sporogenes genome was also searched for pseudouridine synthase genes. A BLAST with E. coli RluD pseudouridine synthase (WP_000079112.1) that is responsible for the Ψ1915 shows three C. sporogenes protein sequences belonging to the RluA family of pseudouridine synthase with 43% (EDU37299.1, E-value 4e-76), 35% (EDU35865.1) and 24% (EDU37345.1) identity to E. coli RluD, respectively. EDU37299.1 is thus a candidate for a U1915 pseudouridine synthase in C. sporogenes, but a confirmation requires experimental investigation.
The enzyme TlyA responsible for Cm1920 ribose methylation was first identified in M. tuberculosis and later found to exist in two variants [45]. Absence of this methylation causes resistance to capreomycin and viomycin [28,46]. Searching the C. sporogenes genes for possible TlyA orthologues suggests a 273 aa enzyme (EDU36942.1/WP_003485867.1) with 36% identity to the 268 aa M. tuberculosis TlyII variant (KBJ35054.1) (E-value 5e-61); 37% identity to the 269 aa M. smegmatis MC2 155 TlyII variant (WP_011729305.1) (E-value 4e-61); 40% identity to the 235 aa T. thermophilus TlyI variant (WP_011228016.1) (E-value 1e-59); 42% identity to the 240 aa Brachyspira hyodysenteriae TlyI variant (AIS71962.1) (E-value 2e-57) and 52% identity to the 282 aa Geobacillus stearothermophilus TlyII variant (WP_053414602.1) (E-value 6e-99). From this we infer that C. sporogenes contains a TlyA orthologue responsible for the Cm1920. This nucleotide is positioned in helix 69 right at a bridge where the two ribosomal subunits interact.
The methyltransferase RlmD (previously named YgcA and RumA) in E. coli is responsible for the m5U1939 modification [47,48] and uses free 23S rRNA as substrate [47]. The search for a similar enzyme in C. sporogenes (using WP_000046812.1, 433 aa as query) resulted in the proteins EDU35986.1/WP_003484135.1 (E-value 2e-53) and EDU39155.1/WP_003483097.1 (E-value 1e-52) with only 28% and 27% identity, respectively. The Bacillus subtilis RlmCD has been shown to methylate both m5U747 and m5U1939 [26]. A search for similarity with this 459 aa RlmCD (KIX81902.1) in C. sporogenes resulted in the protein EDU35986.1/WP_003484135.1 (E-value 2e-82) with 33% identity and EDU39155.1/WP_003483097.1 (E-value 3e-179) with 52% identity. EDU39155.1 is thus the most likely candidate for the m5U1939 modifying enzyme in C. sporogenes. The m5U1939 modification has also been found in P. aeruginosa (putative modifying gene PA0933) [6] and in T. thermophilus [5] and in Mycoplasma capricolum. As the modifying enzyme in M. capricolum (RlmFO) uses a different, folate dependent mechanism [49], we also checked if a gene for a similar enzyme is present in C. sporogenes, but this is not the case. U1939 is positioned near the the CCA end of ribosome bound tRNAs and the modification may thus play a role in this interaction.
3.4. Domain II and III of 23S rRNA from C. sporogenes
We finally investigated the domain II and domain III regions shown in Figure 1 with MS to search for specific modifications seen in other bacteria. No mass changing modifications were observed in the RNase digestion products. As pointed out above we cannot rule out any possibility of modified nucleotides in these regions with our method, but the analyses determine lack of methylations at the following positions modified in other bacteria; U571, G745, U747, A1067 and A1618, as only unmodified RNase digestion products appeared in the mass spectra.
4. Concluding remarks
It is well known from other studies that rRNA modifications tend to cluster in specific areas in bacterial rRNA and this is coherent with our study of 23S rRNA modifications in C. sporogenes ATCC 15579. We have detected nine mass-changing modifications, all of which cluster in domain IV and VI of 23S rRNA. Eight of these modifications are also found in other bacteria at the corresponding positions while one is at a position not previously found to be modified, namely D2500. The presumed mD2449 is also a particular case, as this modification is found at sub-stoichiometrical levels in E. coli [34], but is fully modified in C. sporogenes 23S rRNA. This resembles the situation for the oh5C2501, where the modification level depends on E. coli’s environmental conditions [7]. Although we find it unlikely, we cannot exclude the presence of modifications in the areas not investigated. Also, a modification might escape MS detection if it ends up in fragments too small to be detected in either of the two RNase digests. In general, the modification pattern in 23S rRNA from C. sporogenes is more similar to the modification pattern seen in T. thermophilus than the pattern in E. coli. This includes the apparent absence of methylations in domain II and III and the lower number of methylations overall, seven and eight [4,5], respectively, compared to the 14 methylations in E. coli 23S rRNA. All detected modifications occur in regions either engaged in subunit interaction or tRNA binding, which is consistent with the suggestion from Mengel-Jørgensen et al [7] that the role is to modulate inter-molecular RNA-RNA contacts.
Funding Statement
This work was supported by the Natur og Univers, Det Frie Forskningsråd [0602-02289B].
Acknowledgments
Financial support from the Danish Council for Independent Research—Natural
Sciences (grant 0602-02289B) is gratefully acknowledged.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Decatur WA, Fournier MJ.. rRNA modifications and ribosome function. Trends Biochem Sci. 2002. July;27(7):344–351. PubMed PMID: 12114023. [DOI] [PubMed] [Google Scholar]
- 2.Sergeeva OV, Bogdanov AA, Sergiev PV. What do we know about ribosomal RNA methylation in Escherichia coli? Biochimie. 2015. October;117:110–118. PubMed PMID: 25511423. [DOI] [PubMed] [Google Scholar]
- 3.Fischer N, Neumann P, Konevega AL, et al. Structure of the E. coli ribosome-EF-Tu complex at <3 A resolution by Cs-corrected cryo-EM. Nature. 2015. April 23;520(7548):567–570. PubMed PMID: 25707802. [DOI] [PubMed] [Google Scholar]
- 4.Mengel-Jorgensen J, Jensen SS, Rasmussen A, et al. Modifications in Thermus thermophilus 23 S ribosomal RNA are centered in regions of RNA-RNA contact. J Biol Chem. 2006. August 4;281(31):22108–22117. PubMed PMID: 16731530. [DOI] [PubMed] [Google Scholar]
- 5.Polikanov YS, Melnikov SV, Soll D, et al. Structural insights into the role of rRNA modifications in protein synthesis and ribosome assembly. Nat Struct Mol Biol. 2015. April;22(4):342–U93. PubMed PMID: WOS:000352317000014; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutierrez B, Douthwaite S, Gonzalez-Zorn B. Indigenous and acquired modifications in the aminoglycoside binding sites of Pseudomonas aeruginosa rRNAs. RNA Biol. 2013. August;10(8):1324–1332. PubMed PMID: 23948732; PubMed Central PMCID: PMCPMC3817154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kimura S, Sakai Y, Ishiguro K, et al. Biogenesis and iron-dependency of ribosomal RNA hydroxylation. Nucleic Acids Res. 2017. December 15;45(22):12974–12986. PubMed PMID: 29069499; PubMed Central PMCID: PMCPMC5727448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siibak T, Remme J. Subribosomal particle analysis reveals the stages of bacterial ribosome assembly at which rRNA nucleotides are modified. RNA. 2010. October;16(10):2023–2032. PubMed PMID: 20719918; PubMed Central PMCID: PMCPMC2941110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Green R, Noller HF. In vitro complementation analysis localizes 23S rRNA posttranscriptional modifications that are required for Escherichia coli 50S ribosomal subunit assembly and function. RNA. 1996. October;2(10):1011–1021. PubMed PMID: 8849777; PubMed Central PMCID: PMCPMC1369433. [PMC free article] [PubMed] [Google Scholar]
- 10.Zaringhalam M, Papavasiliou FN. Pseudouridylation meets next-generation sequencing. Methods. 2016. September 1;107:63–72. PubMed PMID: WOS:000383733500009; English. [DOI] [PubMed] [Google Scholar]
- 11.Motorin Y, Helm M. RNA nucleotide methylation. Wiley Interdiscip Rev RNA. 2011. Sep-Oct;2(5):611–631. PubMed PMID: 21823225. [DOI] [PubMed] [Google Scholar]
- 12.Woodson SA. RNA folding and ribosome assembly. Curr Opin Chem Biol. 2008. December;12(6):667–673. PubMed PMID: 18935976; PubMed Central PMCID: PMCPMC2651837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conn GL, Savic M, Macmaster R. Antibiotic resistance in bacteria through modification of nucleosides in16S RibosomalRNA In: Grosjean H, editor. DNA and RNA modification enzymes: structure, mechanism, function and evolution: landes bioscience. ISBN: 978-1-58706-329-9 2009. p. 524–536. [Google Scholar]
- 14.Long KS, Vester B. Antibiotics resistance in bacteria caused by modified nucleosides in 23S ribosomal RNA In: Grosjean H, editor. DNA and RNA modification enzymes: structure, mechanism, function and evolution: landes bioscience. 2009. p. 537–549. [Google Scholar]
- 15.Noon KR, Bruenger E, McCloskey JA. Posttranscriptional modifications in 16S and 23S rRNAs of the archaeal hyperthermophile Sulfolobus solfataricus. J Bacteriol. 1998. June;180(11):2883–2888. PubMed PMID: 9603876; PubMed Central PMCID: PMCPMC107253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thuring K, Schmid K, Keller P, et al. Analysis of RNA modifications by liquid chromatography-tandem mass spectrometry. Methods. 2016. September 1;107:48–56. PubMed PMID: WOS:000383733500007; English. [DOI] [PubMed] [Google Scholar]
- 17.Guymon R, Pomerantz SC, Crain PF, et al. Influence of phylogeny on posttranscriptional modification of rRNA in thermophilic prokaryotes: the complete modification map of 16S rRNA of Thermus thermophilus. Biochemistry. 2006. April 18;45(15):4888–4899. PubMed PMID: 16605256. [DOI] [PubMed] [Google Scholar]
- 18.Popova AM, Williamson JR. Quantitative analysis of rRNA modifications using stable isotope labeling and mass spectrometry. J Am Chem Soc. 2014. February 5;136(5):2058–2069. PubMed PMID: WOS:000331493700058; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Taoka M, Nobe Y, Hori M, et al. A mass spectrometry-based method for comprehensive quantitative determination of post-transcriptional RNA modifications: the complete chemical structure of Schizosaccharomyces pombe ribosomal RNAs. Nucleic Acids Res. 2015. October 15;43(18). DOI: 10.1093/nar/gkv560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birkedal U, Christensen-Dalsgaard M, Krogh N, et al. Profiling of ribose methylations in RNA by high-throughput sequencing. Angew Chem Int Edit. 2015. January 7;54(2):451–455. PubMed PMID: WOS:000347238800008; English. [DOI] [PubMed] [Google Scholar]
- 21.Andersen TE, Porse BT, Kirpekar F. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004. June;10(6):907–913. PubMed PMID: 15146074; PubMed Central PMCID: PMCPMC1370582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Douthwaite S, Kirpekar F. Identifying modifications in RNA by MALDI mass spectrometry. Methods Enzymol. 2007;425:3–20. PubMed PMID: 17673077 [DOI] [PubMed] [Google Scholar]
- 23.Mengel-Jorgensen J, Kirpekar F. Detection of pseudouridine and other modifications in tRNA by cyanoethylation and MALDI mass spectrometry. Nucleic Acids Res. 2002. December 1;30(23):e135 PubMed PMID: 12466567; PubMed Central PMCID: PMCPMC137990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stern S, Moazed D, Noller HF. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. PubMed PMID: 2468070. [DOI] [PubMed] [Google Scholar]
- 25.Boccaletto P, Machnicka MA, Purta E, et al. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res. 2018. January 4;46(D1):D303–D307. PubMed PMID: 29106616; PubMed Central PMCID: PMCPMC5753262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Desmolaize B, Fabret C, Bregeon D, et al. A single methyltransferase YefA (RlmCD) catalyses both m5U747 and m5U1939 modifications in Bacillus subtilis 23S rRNA. Nucleic Acids Res. 2011. November;39(21):9368–9375. PubMed PMID: 21824914; PubMed Central PMCID: PMCPMC3241648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Havelund JF, Giessing AM, Hansen T, et al. Identification of 5-hydroxycytidine at position 2501 concludes characterization of modified nucleotides in E. coli 23S rRNA. J Mol Biol. 2011. August 19;411(3):529–536. PubMed PMID: 21723290. [DOI] [PubMed] [Google Scholar]
- 28.Johansen SK, Maus CE, Plikaytis BB, et al. Capreomycin binds across the ribosomal subunit interface using tlyA-encoded 2ʹ-O-methylations in 16S and 23S rRNAs. Mol Cell. 2006. July 21;23(2):173–182. PubMed PMID: 16857584. [DOI] [PubMed] [Google Scholar]
- 29.Andersen TE, Kirpekar F, Haselmann KF. RNA fragmentation in MALDI mass spectrometry studied by H/D-exchange: mechanisms of general applicability to nucleic acids. J Am Soc Mass Spectrom. 2006. October;17(10):1353–1368. PubMed PMID: 16875834. [DOI] [PubMed] [Google Scholar]
- 30.Atkinson GC, Hansen LH, Tenson T, et al. Distinction between the Cfr methyltransferase conferring antibiotic resistance and the housekeeping RlmN methyltransferase. Antimicrob Agents Chemother. 2013. August;57(8):4019–4026. PubMed PMID: 23752511; PubMed Central PMCID: PMCPMC3719738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benitez-Paez A, Villarroya M, Armengod ME. The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA. 2012. October;18(10):1783–1795. PubMed PMID: 22891362; PubMed Central PMCID: PMCPMC3446703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovgren JM, Wikstrom PM. The rlmB gene is essential for formation of Gm2251 in 23S rRNA but not for ribosome maturation in Escherichia coli. J Bacteriol. 2001. December;183(23):6957–6960. PubMed PMID: WOS:000172158600030; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moazed D, Noller HF. Sites of interaction of the CCA end of peptidyl-tRNA with 23S rRNA. Proc Natl Acad Sci U S A. 1991. May 1;88(9):3725–3728. PubMed PMID: 2023922; PubMed Central PMCID: PMCPMC51525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang KT, Desmolaize B, Nan J, et al. Structure of the bifunctional methyltransferase YcbY (RlmKL) that adds the m7G2069 and m2G2445 modifications in Escherichia coli 23S rRNA. Nucleic Acids Res. 2012. June;40(11):5138–5148. PubMed PMID: 22362734; PubMed Central PMCID: PMCPMC3367195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kasprzak JM, Czerwoniec A, Bujnicki JM. Molecular evolution of dihydrouridine synthases. Bmc Bioinformatics. 2012. June 28;13:153 PubMed PMID: 22741570; PubMed Central PMCID: PMCPMC3674756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lesnyak DV, Sergiev PV, Bogdanov AA, et al. Identification of Escherichia coli m2G methyltransferases: I. the ycbY gene encodes a methyltransferase specific for G2445 of the 23 S rRNA. J Mol Biol. 2006. November 17;364(1):20–25. PubMed PMID: 17010378. [DOI] [PubMed] [Google Scholar]
- 37.Kimura S, Ikeuchi Y, Kitahara K, et al. Base methylations in the double-stranded RNA by a fused methyltransferase bearing unwinding activity. Nucleic Acids Res. 2012. May;40(9):4071–4085. PubMed PMID: 22210896; PubMed Central PMCID: PMCPMC3351187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hansen MA, Kirpekar F, Ritterbusch W, et al. Posttranscriptional modifications in the A-loop of 23S rRNAs from selected archaea and eubacteria. Rna-A Publ Rna Soc. 2002. February;8(2):202–213. PubMed PMID: WOS:000173794200008; English [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maden BEH, Corbett ME, Heeney PA, et al. Classical and novel approaches to the detection and localization of the numerous modified nucleotides in eukaryotic Ribosomal-Rna. Biochimie. 1995;77(1–2):22–29. PubMed PMID: WOS:A1995QP54800004; English. [DOI] [PubMed] [Google Scholar]
- 40.Caldas T, Binet E, Bouloc P, et al. The FtsJ/RrmJ heat shock protein of Escherichia coli is a 23 S ribosomal RNA methyltransferase. J Biol Chem. 2000. June 2;275(22):16414–16419. PubMed PMID: 10748051. [DOI] [PubMed] [Google Scholar]
- 41.Widerak M, Kern R, Malki A, et al. U2552 methylation at the ribosomal A-site is a negative modulator of translational accuracy. Gene. 2005. February 28;347(1):109–114. PubMed PMID: 15715963. [DOI] [PubMed] [Google Scholar]
- 42.Purta E, Kaminska KH, Kasprzak JM, et al. YbeA is the m3Psi methyltransferase RlmH that targets nucleotide 1915 in 23S rRNA. RNA. 2008. October;14(10):2234–2244. PubMed PMID: 18755835; PubMed Central PMCID: PMCPMC2553730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ero R, Peil L, Liiv A, et al. Identification of pseudouridine methyltransferase in Escherichia coli. Rna-A Publ Rna Soc. 2008. October;14(10):2223–2233. PubMed PMID: WOS:000259568700023; English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koh CS, Madireddy R, Beane TJ, et al. Small methyltransferase RlmH assembles a composite active site to methylate a ribosomal pseudouridine. Sci Rep. 2017. April 20;7(1):969 PubMed PMID: 28428565; PubMed Central PMCID: PMCPMC5430550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monshupanee T, Johansen SK, Dahlberg AE, et al. Capreomycin susceptibility is increased by TlyA-directed 2ʹ-O-methylation on both ribosomal subunits. Mol Microbiol. 2012. September;85(6):1194–1203. PubMed PMID: 22779429; PubMed Central PMCID: PMCPMC3438285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maus CE, Plikaytis BB, Shinnick TM. Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2005. February;49(2):571–577. PubMed PMID: 15673735; PubMed Central PMCID: PMCPMC547314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Agarwalla S, Kealey JT, Santi DV, et al. Characterization of the 23 S ribosomal RNA m5U1939 methyltransferase from Escherichia coli. J Biol Chem. 2002. March 15;277(11):8835–8840. PubMed PMID: 11779873. [DOI] [PubMed] [Google Scholar]
- 48.Madsen CT, Mengel-Jorgensen J, Kirpekar F, et al. Identifying the methyltransferases for m(5)U747 and m(5)U1939 in 23S rRNA using MALDI mass spectrometry. Nucleic Acids Res. 2003. August 15;31(16):4738–4746. PubMed PMID: 12907714; PubMed Central PMCID: PMCPMC169892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lartigue C, Lebaudy A, Blanchard A, et al. The flavoprotein Mcap0476 (RlmFO) catalyzes m5U1939 modification in Mycoplasma capricolum 23S rRNA. Nucleic Acids Res. 2014. July;42(12):8073–8082. PubMed PMID: 24939895; PubMed Central PMCID: PMCPMC4081110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mcluckey SA, Vanberkel GJ, Glish GL. Tandem mass-spectrometry of small, multiply charged oligonucleotides. J Am Soc Mass Spectr. 1992. January;3(1):60–70. PubMed PMID: WOS:A1992GZ12100008; English. [DOI] [PubMed] [Google Scholar]


