ABSTRACT
Long noncoding RNAs (lncRNAs) often carry out their functions through associations with adaptor proteins. We recently identified heterogeneous ribonucleoprotein (hnRNP) A2/B1 as an adaptor of the human HOTAIR lncRNA. hnRNP A2 and B1 are splice isoforms of the same gene. The spliced version of HOTAIR preferentially associates with the B1 isoform, which we hypothesize contributes to RNA-RNA matching between HOTAIR and transcripts of target genes in breast cancer. Here we used enhanced cross-linking immunoprecipitation (eCLIP) to map the direct interactions between A2/B1 and RNA in breast cancer cells. Despite differing by only twelve amino acids, the A2 and B1 splice isoforms associate preferentially with distinct populations of RNA in vivo. Through cellular fractionation experiments we characterize the pattern of RNA association in chromatin, nucleoplasm, and cytoplasm. We find that a majority of interactions occur on chromatin, even those that do not contribute to co-transcriptional splicing. A2/B1 binding site locations on multiple RNAs hint at a contribution to the regulation and function of lncRNAs. Surprisingly, the strongest A2/B1 binding site occurs in a retained intron of HOTAIR, which interrupts an RNA-RNA interaction hotspot. In vitro eCLIP experiments highlight additional exonic B1 binding sites in HOTAIR which also surround the RNA-RNA interaction hotspot. Interestingly, a version of HOTAIR with the intron retained is still capable of making RNA-RNA interactions in vitro through the hotspot region. Our data further characterize the multiple functions of a repurposed splicing factor with isoform-biased interactions, and highlight that the majority of these functions occur on chromatin-associated RNA.
KEYWORDS: hnRNP A2/B1, long noncoding RNA, chromatin, HOTAIR, Xist, RNA-RNA interactions
1. Introduction
Long noncoding RNAs (lncRNAs) are defined as transcripts that are longer than 200 bases and have minimal evidence of being translated. These transcripts are often found in the nucleus, and have a number of functions incorporating a variety of mechanisms [1]. Many lncRNAs associate with adaptor proteins that mediate functional activity.
The hnRNP protein family is a complex and diverse set of proteins with numerous functions that bind RNA and associate primarily with nascent transcripts [2–4]. Our group recently identified hnRNP B1 as the most-enriched nuclear protein that bound preferentially to the heterochromatin-associated lncRNA HOTAIR[5]. HOTAIR targets histone methylation to silence the HoxD locus, potentially contributing to developmental patterning [6], though knockout of HOTAIR is not sufficient to disrupt overall mouse development [7]. HOTAIR is overexpressed in metastatic breast cancer, and knockdown of B1 can disrupt HOTAIR-dependent cancer phenotypes [5,8].
HnRNP B1 is one of two splice isoforms (the other being hnRNP A2) transcribed from a single genomic locus [9]. These isoforms each contain two RNA recognition motifs (RRMs) and a C-terminal glycine-rich domain, with the B1 isoform differing only by the inclusion of an additional twelve residues, encoded by exon 2, near the N-terminus (Figure 1(a)). HnRNP A2/B1 have been implicated in multiple aspects of RNA metabolism [10], including alternative splicing [11], mRNA stability [12], and mRNA degradation [13]. Mutations in the hnRNP A2/B1 glycine-rich domain can lead to diseases associated with multisystem proteinopathy such as ALS and frontotemporal dementia [14].
Figure 1.
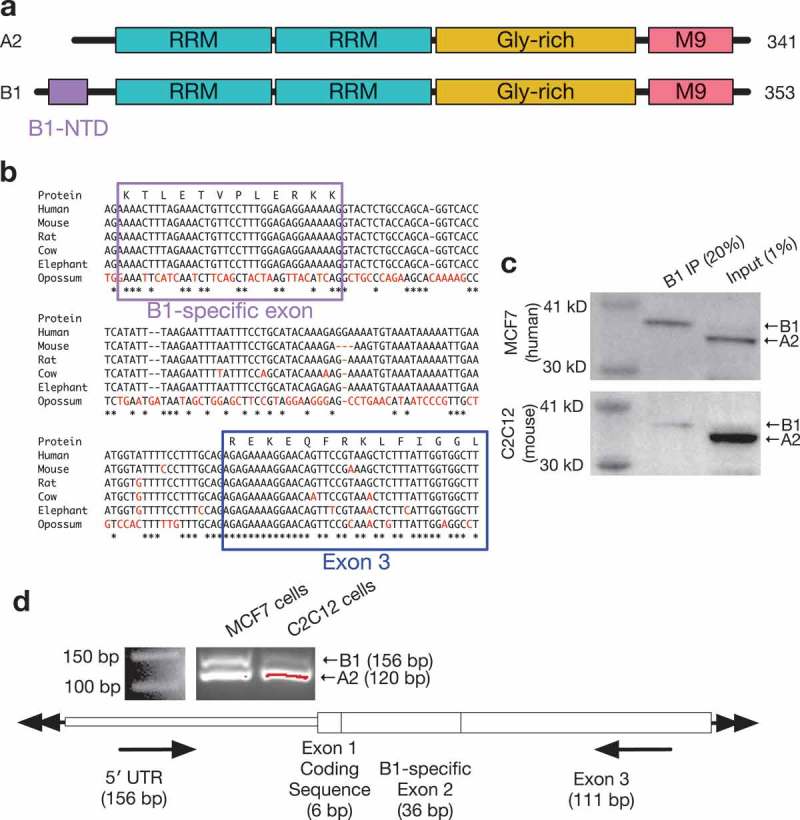
Conservation of the hnRNP B1-specific exon across species.
(a) Model of protein domains of hnRNP A2 and B1, with B1-specific domain highlighted. (b) Multiple sequence analysis of hnRNP B1 genomic sequence in human, mouse, rat, cow, elephant, and opossum reference sequences, with B1-specific exon and next downstream exon highlighted, demonstrating high degree of conservation of the B1-specific exon in all eutherian species. Mismatches from human sequence are colored in red. (c) Immunoprecipitation using antibody specific to hnRNP B1 specifically retrieves hnRNP B1 in both human and mouse samples. (d) RT-PCR primers surrounding B1-specific exon 2 demonstrate inclusion of B1 exon in total RNA from both human MCF7 and mouse C2C12 cells.
While the A2 isoform is the more common isoform in most cell types, the B1 isoform displays preferential binding to the spliced form of HOTAIR [5]. HnRNP B1 also associates preferentially with RNA transcripts of known HOTAIR target genes, and is also bound to target chromosomal loci. In addition, knockdown of A2/B1 specifically reduces HOTAIR-dependent histone methylation, catalyzed by the polycomb repressive complex 2 (PRC2), induced by over overexpression of a cDNA transgene in breast cancer cells. From these data, we have proposed a model in which hnRNP B1 can act as a matchmaker between HOTAIR and nascent transcripts at target gene loci. We determined that this matchmaking leads to direct RNA-RNA interactions between HOTAIR and target RNA, which may contribute to direct targeting of repressive histone methylation by PRC2.
In order to investigate the function of hnRNP B1 (as opposed to A2), we have characterized the RNAs to which both isoforms bind. We used the recently developed eCLIP method [15] to examine this in breast cancer cell lines, which have previously been used to study the effect of HOTAIR on cancer metastasis [8]. Our work indicates that hnRNP B1 and A2 bind many lncRNAs and mRNAs, with B1 binding preferentially to a subset of RNAs despite its lower abundance. A2/B1-RNA interactions occur primarily on chromatin, with the small fraction of interactions in the cytoplasm occurring primarily within the untranslated sequences of mRNAs. Interestingly, A2/B1 binding is highly enriched in a retained intronic sequence of HOTAIR that interrupts a ‘hotspot’ RNA-RNA interaction region. B1 also binds exons of HOTAIR, both in vivo and using purified protein and in vitro transcribed, spliced HOTAIR. In vitro RNA-RNA interactions are still allowed between HOTAIR with the retained intron and target gene RNA, suggesting that interruption of the primary sequence of the hotspot region does not disrupt the base-pairing activity. The multiple interaction sites between B1 and HOTAIR suggest a complex mechanism for how this matchmaker protein may contribute to HOTAIR function.
2. Results
2.1. The hnrnp B1 exon is well-conserved and expressed in mouse and human
The 36-nucleotide exon specific to hnRNP B1 is included in approximately 10% of A2/B1 transcripts in most human tissues. This B1-specific region has also been identified in the mouse and rat hnRNP A2/B1 transcript, along with transcripts corresponding to other minor pseudogenes processed from the same locus [16,17]. However, this exon is not annotated as being part of any hnRNP A2/B1 transcript in the mouse RefSeq or Ensembl databases. As the B1-specific exon has been shown to be included in mouse, rat, and cow A2/B1 [18], we decided to perform a more detailed analysis of the conservation of this exon.
An analysis of the hnRNP A2/B1 genomic locus across several species indicates that the B1 exon is in fact highly conserved among placental mammals, with perfect nucleotide sequence identity to human in the mouse, rat, cow, and elephant genomes but no identifiable syntenic region in the opossum genome (Figure 1(b)). This is better conservation than is displayed by the next downstream exon or intervening intron, which are also well-conserved but only have ~ 90% sequence identity between these species. The B1 exon appears more strongly associated with placental mammals, suggesting that the sequence has been under strong stabilizing selection in that group of organisms. The intronic sequence surrounding this exon is also well-conserved, which may be a hint at a conserved mechanism of alternative splicing. Such isoform regulation is common among hnRNPs, with isoform diversity contributing to distinct functions[3].
In order to confirm whether or not the B1-specific exon is transcribed in non-human species, we first performed an immunoprecipitation, with an antibody designed to be specific to the human hnRNP B1 unique N-terminal region, on whole-cell protein lysate from mouse C2C12 cells or human MCF7 cells. This IP retrieved hnRNP B1 in both cell types, but a minimal amount of hnRNP A2 (Figure 1(c)). We also confirmed that the B1-specific exon is transcribed in C2C12 cells using RT-PCR primers specific to the 5′ UTR and the downstream exon, creating an amplicon spanning the exon (Figure 1(d)). B1 is therefore found in non-human cells, potentially through the placental mammalian lineage.
2.2. eCLIP-seq reveals the distribution of hnRNP A2/B1 RNA binding in breast cancer cells
Methods that examine direct protein-RNA interactions by UV crosslinking and immunoprecipitation (CLIP) are commonly used to identify transcripts bound to proteins of interest[19]. Recently the eCLIP method was developed, which has the advantages of individual nucleotide resolution, requiring less library amplification, normalization of the IP sample against a size-matched input (SMInput), and inclusion of a computational analysis pipeline [15]. The SMInput represents all abundant RNA-protein complexes that migrate in the same region of the gel/blot that is excised in the immunoprecipitation sample. Because hnRNPs primarily work in the nucleus, we included a nuclear isolation protocol previously used to isolate chromatin-associated CTCF-RNA complexes [20](Figure 2(a)). We performed eCLIP using antibodies to either hnRNP B1 specifically, or hnRNP A2/B1 in combination, using MCF7 breast adenocarcinoma cells. MCF7 cells were chosen since much of the prior characterization of HOTAIR has been performed in MCF7 cells [8,21], they express high levels of hnRNP A2/B1, and have been well-characterized by the ENCODE Project [22]. We also performed a parallel experiment using MCF10A cells as a non-tumorigenic counterpart to MCF7, derived from the same tissue [23].
Figure 2.
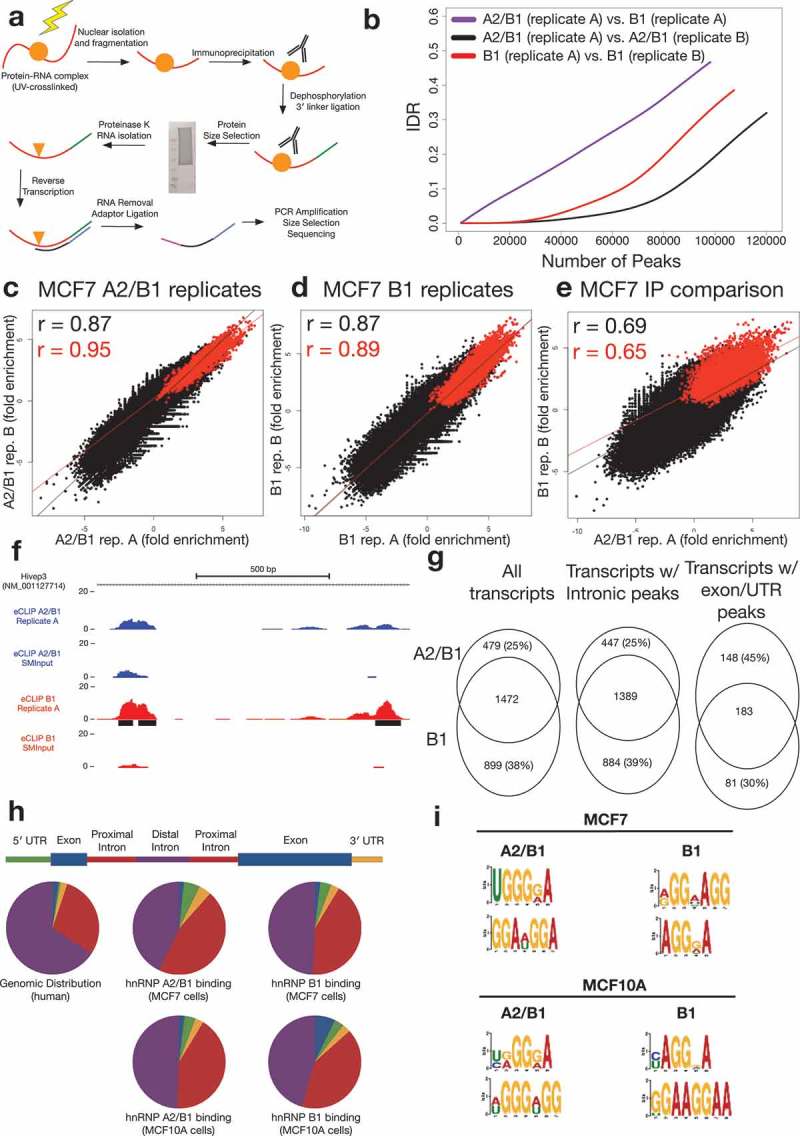
A2/B1 and B1 eCLIP results in MCF7 cells.
(a) Schematic of the eCLIP protocol(b) Irreproducible Discovery Rate (IDR) analysis comparing peak fold enrichment indicates increased reproducibility for replicates compared against one another (A2/B1, black; B1, red) than comparison of replicates from A2/B1 and B1 experiments (purple).(c) Correlation of A2/B1 eCLIP replicate fold enrichment at non-significant (black) or significant (red) peaks in replicate 1. R values are Pearson’s correlation coefficient.(d) Correlation of B1 eCLIP replicate fold enrichment at non-significant (black) or significant (red) peaks in replicate 1. R values are Pearson’s correlation coefficient.(e) Correlation of A2/B1 and B1 eCLIP fold enrichment at non-significant (black) or significant (red) peaks in A2/B1. R values are Pearson’s correlation coefficient.(f) Example hnRNP A2/B1 eCLIP data, demonstrating region of the transcription factor Hivep3 that contains binding sites (black) enriched specifically in hnRNP B1 (blue). Y-axis scale is normalized to reads per million. Region pictured is chr1:42,345,550–42,344,700.(g) Analysis of RefSeq transcripts containing input normalized peaks in both replicates of A2/B1 or B1 eCLIP-seq experiments. Also pictured are analyses of transcripts that contain peaks only in introns or exons/UTRs.(h) Distribution of peaks (conserved between both replicates) in different areas of transcripts in hnRNP A2/B1 and B1 eCLIP experiments, compared to experiments performed in non-tumorigenic MCF10A cells. ‘Proximal introns’ are defined as intronic regions within 2 kb of an exon.(i) Top two identified motifs in both replicates of hnRNP A2/B1 and B1 eCLIPs in MCF7 and MCF10A cells.
The eCLIP experiments produced transcriptome-wide maps of A2/B1 and B1 binding sites. We identified peaks of sequencing read buildup enriched over SMInput, with significant peaks being defined as those with low p-value (p < 10−5). In total, we identified 7626 A2/B1 peaks significant in both replicates, and, due to higher IP yield, 14,725 B1 peaks significant in both replicates. These significant peaks were highly correlated between replicates (A2/B1 r = 0.87; B1 r = 0.87), but less so between different antibodies (r = 0.69) (Figure 2(b-e)).
To further investigate the differences between A2/B1 and B1 RNA binding, we investigated the transcripts containing either A2/B1 or B1 input normalized peaks. Of the 54,064 transcripts annotated in the RefSeq database, 2,850 contained peaks enriched over input and conserved between replicates in either the A2/B1 or B1 experiment. A majority of the transcripts (1,472) identified in either eCLIP had peaks in both A2/B1 and B1 experiments; however, a minority of transcripts were unique to either A2/B1 (479) or B1 (899) experiment. Because A2 is the dominant isoform in MCF7 cells, the immunoprecipitation in the A2/B1 experiment captures more A2 interactions. Thus, the interactions that are unique to the A2/B1 eCLIP versus the B1 experiment are likely due to unique A2 preferential binding. More exonic and UTR peaks (148) were unique to the A2/B1 experiment compared to B1, suggesting that the A2 isoform may bind to more mature mRNAs than the B1 isoform, perhaps as a consequence of its relatively higher abundance (Figure 2(f-g)).
The distribution of input normalized peaks in both A2/B1 and B1 eCLIP experiments roughly mirrored the genomic distribution of exons, UTRs, and introns; however, we identified a shift in peak frequency towards regions of introns within 2 kilobases of splice junctions. Non-tumor MCF10A breast cells demonstrated a similar profile as MCF7 cells, with a slight overrepresentation of exonic peaks as compared to both the genomic distribution and the MCF7 eCLIP (Figure 2(h)).
Motif analysis revealed a preference for G-rich sequences in hnRNP A2/B1 binding sites. Previous analyses have suggested that hnRNP A2/B1 has a preference to bind UAGGG motifs in RNA, such as those found in telomeric RNA [24]; a similar UAGG motif has been identified using iCLIP in mouse spinal cord cells [25]. However, HITS-CLIP of A2/B1 in 293T cells did not identify a similar motif [11]. Our data indicates that, although hnRNP A2/B1 might have particular affinity for UAGGG sequences, it also binds a variety of AG-rich sequences. Motif analysis identified a slight difference in binding preference between A2/B1 and B1. B1 displays a strong enrichment for (AGG)n motifs in both MCF7 and MCF10A cells, while A2/B1 appears to have a weaker preference for AG-rich regions (Figure 2(i)). Motif analysis was not performed on the peak groups that were partitioned in Figure 2(h) due to the low number of peaks in exonic regions limiting the potential for statistically-significant motifs to be called. The higher abundance of A2 may lead to binding of additional RNAs with lower-affinity sites, out-competing B1 and leading to a difference in overall motif enrichment. Alternatively, the B1-specific N-terminal domain may impart a differential RNA sequence preference.
2.3. The hnrnp B1 binding profile differs in each cellular compartment
Because A2/B1 localizes to different nuclear compartments depending on cell state [26], we compared RNA-binding sites in the chromatin, nucleoplasm and cytoplasm. We performed B1 eCLIP on samples derived from a subcellular fractionation protocol [27] into cytoplasmic, nucleoplasmic, and chromatin-associated fractions (Figure 3(a), Supplementary Table S1). Our eCLIP experiment detected far more significant B1 binding peaks in the chromatin sample [27,539] than in the nucleoplasm (230) or cytoplasm (162), consistent with B1 protein abundance in each fraction. The signal detected in the chromatin sample included co-transcriptional binding, binding to lncRNAs, and binding to other chromatin-associated RNAs [28]. The majority of the nucleoplasmic and cytoplasmic peaks overlapped with chromatin peaks (Figure 3(b)). Very few of the remaining non-overlapping peaks were in protein-coding sequence, with the majority of binding sites found instead in proximal introns and UTRs (Figure 3(c, d)).
Figure 3.
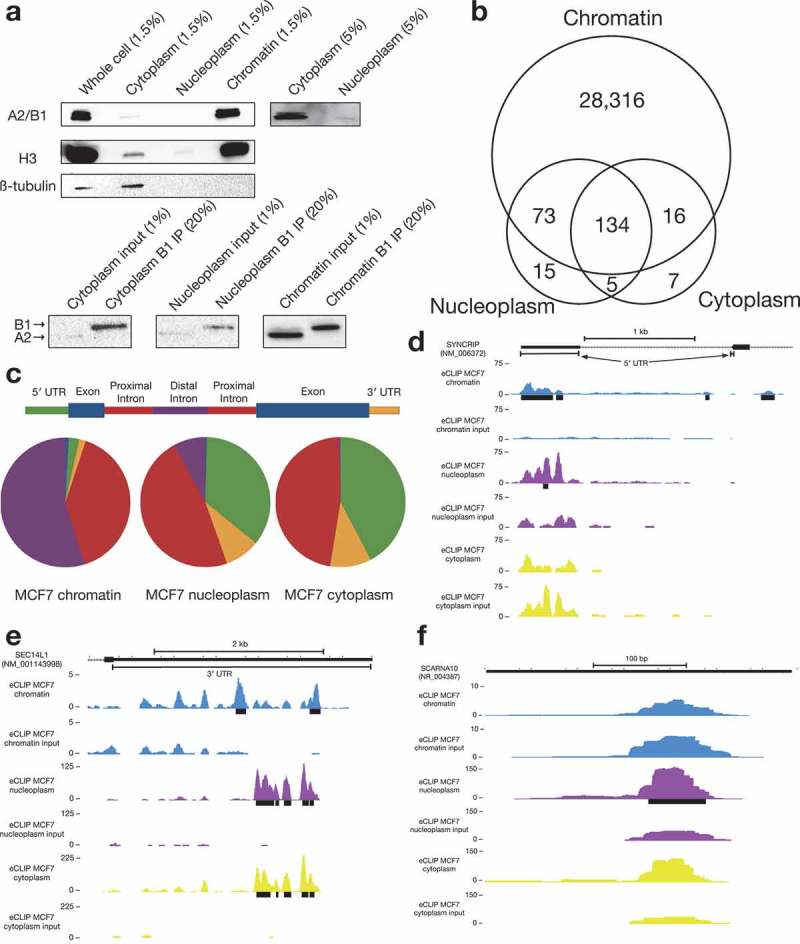
eCLIP of MCF7 chromatin, nucleoplasm, and cytoplasm.
(a)Western blot of fractionated MCF7 cells (top) demonstrates preferential localization of A2/B1 to chromatin in fractions (either 1.5% or 5%) of an eCLIP fractionation experiment. Western blot of eCLIP samples (bottom) indicates preferential localization of A2/B1 to chromatin, as well as specific immunoprecipitation of B1 isoform.(b)The vast majority of A2/B1 binding peaks were identified in the chromatin sample, with a small minority overlapping with a nucleoplasm or cytoplasm binding peak. A small fraction of binding peaks were unique to either nucleoplasm or cytoplasm.(c) Compared to the chromatin sample, the nucleoplasm and cytoplasm peaks were far more likely to identify binding to either 5′ UTR or proximal intronic sequence.(d) The 5′ UTR of SYNCRIP is bound by B1 in both chromatin and nucleoplasm fractions. However, nearby intronic binding peaks are found only in chromatin fraction.(e) In the 3′ UTR of SEC14L1, there are two distinct binding peaks for B1. However, in nucleoplasm (purple) and cytoplasm (yellow), B1 preferentially binds between the two chromatin peaks.(f) A nucleoplasm-specific B1 binding peak in the small Cajal body-associated RNA SCARNA10.
One example of a transcript with distinct B1-RNA interactions in the soluble, non-chromatin fraction is the SEC14L1 transcript. While B1 binds to the SEC14L1 3′ UTR in all fractions, the signal in the soluble fractions is both stronger and shifted as compared to the chromatin-specific binding peaks (Figure 3(e)), suggesting that B1 binding sites can change as a message matures, or that localization is correlated with a specific B1-RNA interaction. We also detected nucleoplasm/cytoplasm-enriched binding to a number of small Cajal body-specific RNAs (scaRNAs), which are a family of small transcripts that localize to Cajal bodies, nuclear organelles that are involved in the biogenesis of small nuclear ribonucleoproteins (snRNPs) (Figure 3(f)). In particular, scaRNAs are thought to act as guide RNAs in the modification of spliceosomal RNAs [29,30]. A splice isoform of A2/B1 missing exon 7–9, hnRNP A2*, has been shown to interact with telomerase at Cajal bodies [31].
B1 fractionation and eCLIP results provide further evidence for the distinct cellular localization of an RNA binding protein conferring specific function and association with RNA binding partners. A similar approach using the iCLIP method for the RNA binding protein TDP-43 has been performed in the nucleus versus cytoplasm of neuronal primary cells and neuroblastoma cells[32], which detected an enrichment for binding of the 3′ UTR of transcripts in the cytoplasmic fraction. Similarly, a recently-developed protocol, Fractionation iCLIP (Fr-iCLIP) has been used to examine the RNA binding partners of SR proteins in chromatin, nucleoplasmic, and cytoplasmic fractions [33], which detected SR protein binding to cytoplasmic RNAs with retained introns. Though B1 is primarily found associated with chromatin, the distinct patterns of binding certain RNAs off chromatin also suggest functional distinctions from chromatin binding, and hint at the possibility for dynamic binding of a single RBP molecule within a single transcript during RNA transport.
2.4. Binding of hnRNP A2/B1 to noncoding RNAs
Based on the fact that A2/B1 has previously been shown to interact with lncRNAs [5,34], we identified which lncRNAs are associated with A2/B1 in the MCF7 eCLIP. A2/B1 displays strong binding to a small fraction of lncRNAs from both a commonly used lncRNA database [35] and a recently published database of lncRNAs expressed in MCF7 cells [36] (Figure 4(a)). For example, there are a series of A2/B1 and B1 binding sites in Xist, the lncRNA that contributes to dosage compensation through inactivation of one X chromosome in mammals [37]. Xist spreads in cis along with PRC2 across one X chromosome, leading to repression of nearly all genes on that chromosome [38]. The 5′ end of the mouse Xist transcript contains a 1.6-kilobase region known as RepA, which has previously been shown to recruit proteins including PRC2[39], and folds into three independent structural modules [40]. The downstream portion of this region, which is structurally conserved between mouse and human [41], contains four strong, reproducible A2/B1 binding sites in our MCF7 eCLIP dataset that are also identified in A2/B1 eCLIP experiments using mouse C2C12 cells (Supplementary Figure 2). These binding sites lie immediately downstream of iCLIP-derived binding sites for other RNA-binding proteins, RBM15 and RBM15b (Figure 4(b)). RBM15 and RBM15b have been shown to mediate the formation of m6A methylation on Xist [42]. HnRNP A2/B1has been proposed to be a reader of m6A marks [43], though whether there is a direct physical association between the protein and modified base is not clear.
Figure 4.
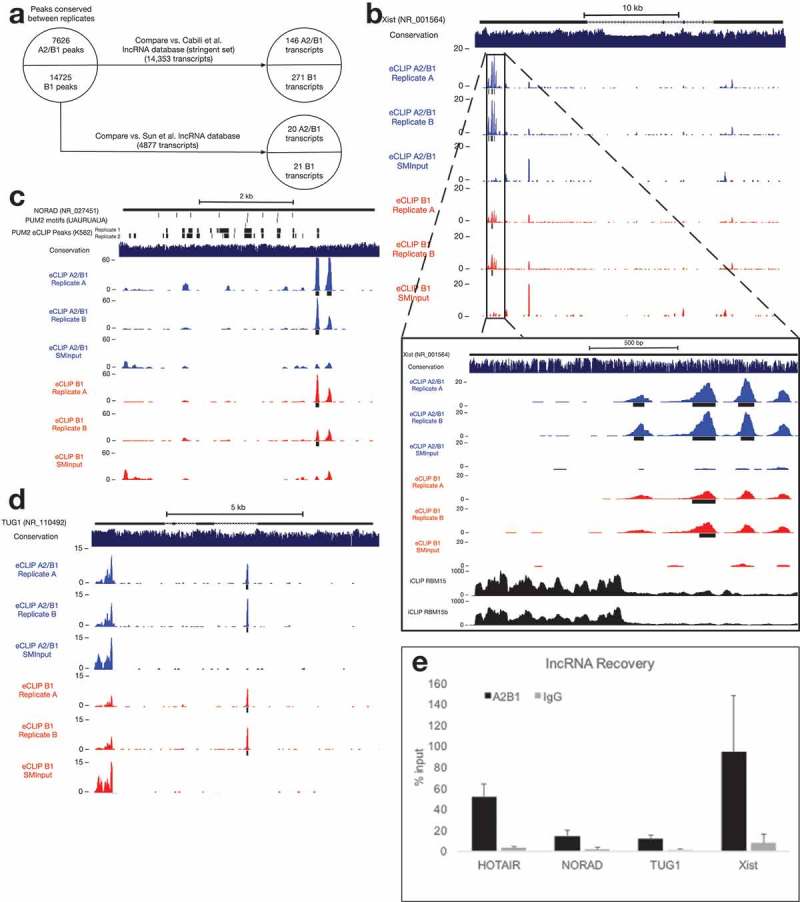
Binding of lncRNAs by hnRNP A2/B1.
(a) hnRNP A2/B1 interacts with a small number of previously-identified lncRNAs.(b) hnRNP A2/B1 binding sites within the RepA region of Xist. RBM15 and RBM15b iCLIP tracks published in [42].(c) An hnRNP A2/B1 binding site within the lncRNA NORAD. PUM2 eCLIP replicate peaks retrieved from ENCODE Project experiment accession ENCSR661ICQ[15].(d) An hnRNP A2/B1 binding site within an intron of the lncRNA TUG1.(e) Native RNA immunoprecipitation demonstrating association of A2/B1 and selected lncRNAs.
We also identified hnRNP A2/B1 and B1 peaks in the lncRNA NORAD (Figure 4(c), which has been shown to regulate genomic stability through sequestration of PUM1 and PUM2 proteins [44,45]. The A2/B1 binding site we identified is near the 3′ end of NORAD, overlapping a potential PUM2 binding site identified by eCLIP in K562 cells [15], but not overlapping any UGURUAUA PUM2 consensus sequences. NORAD contains no introns, highlighting the fact that A2/B1 makes interactions with RNA that do not involve splicing.
Another lncRNA containing A2/B1 and B1 peaks is TUG1, which is a Notch pathway-regulated lncRNA that functions in the maintenance of stemness in glioma stem cells through recruitment of PRC2 to neuronal differentiation genes [46,47]. TUG1 has also been investigated as a potential biomarker in a variety of different cancers [48]. The hnRNP A2/B1 binding site appears in the intron immediately following the proposed PRC2-interacting region of TUG1 (Figure 4(d)).
The A2/B1 lncRNA eCLIP peaks are located in regions with higher average phastCons scores than a random sampling of all lncRNA sequences, suggesting a potential for conservation of these binding events (Supplementary Figure 1). Native RNA immunoprecipitation was used to validate the binding of A2/B1 to each of these lncRNAs (Figure 4(e)). Each lncRNA was significantly enriched in an A2/B1 immunoprecipitation, compared to IgG. Interestingly, these A2/B1-lncRNA interactions were not identified in prior hnRNP A2/B1 eCLIP in neuronally differentiated iPSCs [25], hinting that they might be promoted in cells, such as MCF7 cells, that have a malignant phenotype.
2.5. Binding of the lncRNA HOTAIR by hnRNP A2/B1
The eCLIP profiles of HOTAIR identified one strong hnRNP A2/B1 binding site (p < 10−5 cutoff for peak significance). This peak lies downstream from the previously identified PRC2 binding site [49], which is likely responsible for the input signal identified in the first three exons (Figure 5(a)). This was surprising because A2/B1 serve as an adaptor of a cDNA HOTAIR transgene in breast cancer cells and the protein can directly bind to this version of HOTAIR, which is missing intron 3 [5]. To begin to validate the eCLIP result, we designed PCR primers spanning either intron 3 or intron 2. By RT-PCR of MCF7 total RNA, we found that intron 3 is retained in a fraction of HOTAIR transcripts, while intron 2 is undetectable at the same degree of amplification (Figure 5(b)). This raises the question as to why this particular intron is being specifically retained, and whether or not that is related to its being bound by A2/B1. We found no detectable change in intron 3 when A2/B1 is knocked down in these cells (Supplementary Figure 3), suggesting that this binding event may serve a different purpose than regulating the retention of the intron.
Figure 5.
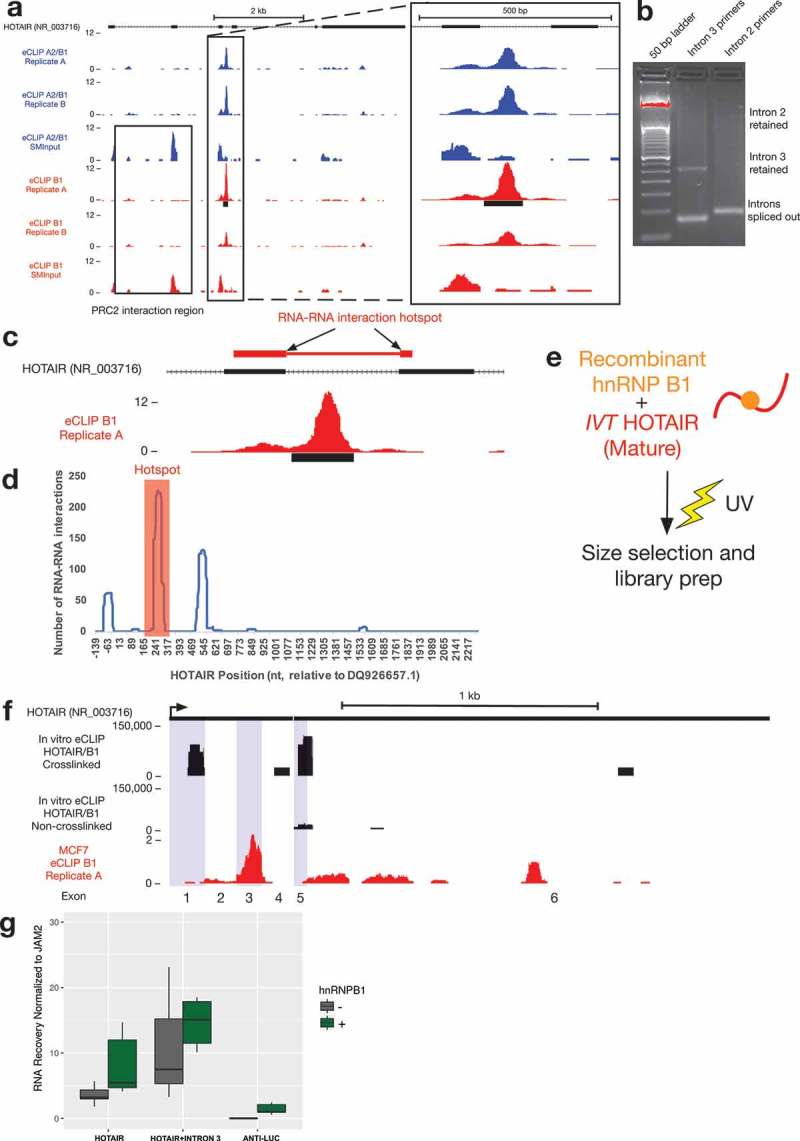
Investigation of a novel hnRNP A2/B1 binding site in the lncRNA HOTAIR.
(a) Location of an hnRNP A2/B1 eCLIP peak in an intron of HOTAIR, downstream of the proposed PRC2 binding site (visible in the input library).(b) The intron between exons 3 and 4 of HOTAIR retained in a small fraction of HOTAIR MCF7 total RNA.(c) Intron 3 disrupts an RNA-RNA interaction site between HOTAIR and multiple gene target transcripts.(d) Graph depicting the total number of interactions (y-axis) between each nucleotide of the HOTAIR transcript and the top 1% of transcripts (440 RNAs) predicted to interact with HOTAIR[50]. The nucleotide position within the HOTAIR transcript (x-axis) has been shifted upstream (−139nt) so that it accurately corresponds to the TSS of the transcript variant GenBank accession DQ926657.1. The highest number of RNA-RNA interaction events is found between 226 to 297 nt (red box).(e) Schematic of in vitro eCLIP experiment using recombinant B1 protein and in vitro transcribed HOTAIR.(f) In vitro eCLIP of recombinant B1 and in vitro transcribed mature form of HOTAIR identified binding sites in exon 1 and in exons 5–6 of HOTAIR (exons alternately shaded blue), with minimal binding identified in non-crosslinked control. Comparison is made to exonic in vivo B1 eCLIP signal (red).(g) In vitro RNA-RNA interaction assays with RAT-tagged JAM2 and either full-length HOTAIR, HOTAIR with intron 3 or Anti-Luc RNA in the presence or absence of hnRNP B1. The association of HOTAIR and Anti-Luc RNA with JAM2 was quantified by RT-qPCR (n = 6).
We have previously proposed a model where hnRNP B1 can facilitate RNA-RNA interaction sites between HOTAIR and transcripts of its target genes [5,8]. Interestingly, intron 3 disrupts an RNA-RNA interaction site that we have previously shown allows HOTAIR to directly interact with the transcript of one of its target genes [5] (Figure 5(c)). Multiple predicted RNA-RNA interaction sites between HOTAIR and HOXD transcripts can also be found in a ‘hotspot’ in the exons surrounding intron 3. Analysis of a publically-available dataset of RNA-RNA interaction predictions between HOTAIR and the entire human transcriptome [50] demonstrates that this ‘hotspot’ region makes thermodynamically-favorable interactions to other transcripts more than any other region of HOTAIR (Figure 5(d), Supplementary Figure 4). The existence of a strong A2/B1 binding site so close to these predicted RNA-RNA interaction sites provides support for the model of A2/B1 as a ‘matchmaker’ protein, but also raises the question of whether this role might be regulated by splicing of intron 3 of HOTAIR.
In order to test how A2/B1 might bind HOTAIR in the absence of intron 3, we performed eCLIP on an in vitro spliced form of the HOTAIR transcript cross-linked with purified recombinant hnRNP B1 (Figure 5(e)). This in vitro eCLIP experiment revealed two strong binding regions within HOTAIR (Figure 5(f)), but little binding in an experiment with a control long noncoding RNA (data not shown). The first binding site is located in exon 1 of HOTAIR. According to the proposed secondary structure of HOTAIR [51], this region basepairs with the RNA-RNA interaction hotspot of HOTAIR (Supplementary Figure 5). In vivo eCLIP read pileup occurs within the RNA-RNA interaction hotspot region, and may reflect a similar region of binding as the in vitro event. The other in vitro eCLIP-identified binding site resides in exons 5 and 6, and overlaps reads from both the in vivo eCLIP data and previous HITS-CLIP in 293T cells [11].
To test the potential impact of retention of intron 3 on HOTAIR activity, we used our previously-described in vitro RNA-RNA interaction assay [5]. As we observed previously, the mature version of HOTAIR with no introns can interact with the JAM2 RNA and addition of recombinant B1 stimulates this interaction (Figure 5(g)). Somewhat surprisingly, use of intron 3-containing HOTAIR did not prevent the direct intermolecular RNA-RNA interactions with the JAM2 target RNA (Figure 5(g)). In fact, an increase in interaction was observed. Addition of B1 into these assays further increased the interactions of HOTAIR+ intron 3 and JAM2, suggesting that binding of B1 in intron 3 may contribute additional potential for these interactions. RNA folding prediction of otherwise mature HOTAIR containing this intron suggests that the intron may fold independently and minimize the disruption to RNA structure in the RNA-RNA interaction hotspot region (Supplementary Figure 5). These results demonstrate that A2/B1 binds multiple regions of HOTAIR and that each of these binding events may contribute to a molecular mechanism of RNA matchmaking with transcripts of target genes.
3. Discussion
Our profiling of A2/B1 binding sites transcriptome-wide has led to a number of new findings. We find that both A2 and B1 isoforms have some level of preferential association with many transcripts, potentially stemming either from recognition of distinct sequence motifs or simply based on abundance of each isoform. We describe the strong preferential association of A2/B1 with chromatin-associated RNAs rather than those in the soluble fraction of the nucleus, suggesting additional roles on chromatin beyond co-transcriptional splicing. We identified novel lncRNA interactions with A2/B1, the positions of which hint at ways that A2/B1 may contribute to the mechanisms of these RNAs. The surprising manner in which A2/B1 encounters the endogenous version of the lncRNA partner HOTAIR, before HOTAIR is fully processed, suggests additional regulatory potential in the model of RNA-RNA matchmaking that we have previously proposed [5].
3.1. Conservation of the hnRNP B1 isoform
To date, functional studies of the hnRNP A2 and B1 isoforms have, with rare exception [17,18], treated the two isoforms as a single protein. However, we have shown that the B1 isoform preferentially interacts with certain RNAs, likely due to contributions of direct interactions between the B1-specific N-terminal region and the target RNA. The proximity of the B1-specific exon to the RRMs suggests that the B1-specific exon may be able to regulate the activity of the RRMs when bound to RNA.
The strong conservation of the sequence of the B1-specific exon implies that it has functional significance, leading to its being subjected to stabilizing selection across a number of species. Interestingly, the strong conservation of the B1-specific protein sequence extends to conservation of the RNA and DNA sequence of the B1-specific exon, which also has identical sequence across eutherians. While this portion of the B1 transcript does not appear to be heavily protein-bound according to the eCLIP input libraries, it is possible that it is transiently, or context-specifically, bound by splicing factors that require a particular sequence in order to generate the ideal proportions of A2 and B1.
3.2. Interactions between A2/B1 and additional noncoding RNAs
The preference in hnRNP A2/B1 binding towards proximal introns, as compared to the overall transcriptome, is indicative of the role that A2/B1 is known to play in the regulation of splicing [10]. Our work also expands the knowledge of A2/B1 RNA interactions, with possible implications for regulation and function outside of splicing. We hypothesize that A2/B1 bind to both nascent RNAs co-transcriptionally as well as mature lncRNAs such as HOTAIR that interact with chromatin in trans, and in certain circumstances facilitating RNA-RNA interactions between the two types of RNA (Figure 6(b)).
Figure 6.
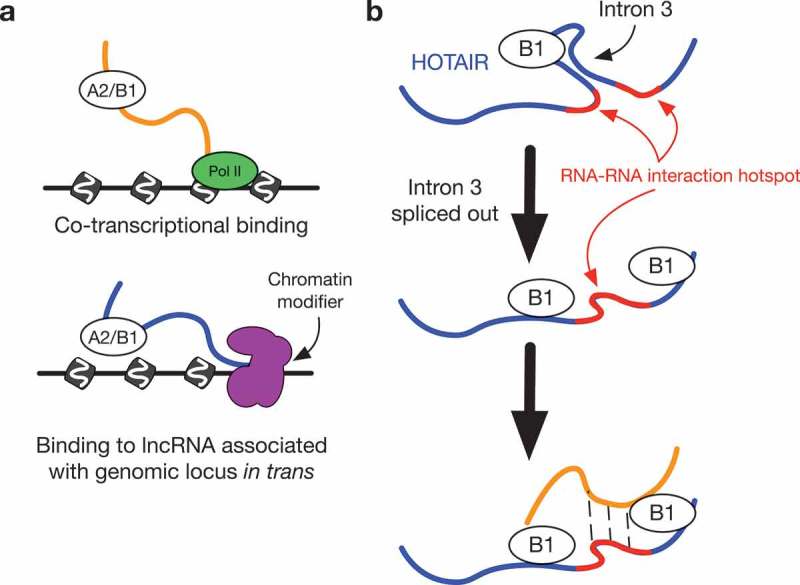
Model of hnRNP B1 interaction with HOTAIR.
(a) hnRNP B1 on chromatin can bind to transcripts co-transcriptionally, or bind to lncRNAs associating with other loci in trans.(b) On the lncRNA HOTAIR, hnRNP B1 preferentially binds to intron 3, which bisects an RNA-RNA interaction hotspot. Retention of intron 3 may not disrupt the ability of HOTAIR to pair with other RNAs in the hotspot region. Following splicing of intron 3, B1 remains bound to nearby regions of HOTAIR, continuing to promote RNA-RNA interactions from the hotspot region.
The binding of A2/B1 to NORAD, TUG1, and Xist (Figure 4) hints that A2/B1 may play a role in their mechanisms of action. The A2/B1 binding site on NORAD overlaps one of the PUM2 binding sites that titrate Pumilio proteins away from mRNAs [44,45]. On TUG1 and Xist, A2/B1 also binds downstream of binding sites for other proteins that contribute to lncRNA function. We suspect that, as with HOTAIR in the RNA matchmaker model, A2/B1 often acts in concert with other proteins on RNA to carry out a variety of lncRNA molecular mechanisms.
Though most A2/B1-RNA interactions are on chromatin, we also identified B1-RNA interactions that are enriched in the nucleoplasm and cytoplasm. These include the nucleoplasm-enriched interaction between B1 and certain scaRNAs that suggests localization of B1 to Cajal bodies. We suspect that B1 promotes RNA-RNA interactions between scaRNAs and their snRNA targets [29]. We also identified B1 binding sites that are shifted depending on the cellular compartment, such as on the 3′ UTR of SEC14L1 (Figure 3(e)), which we hypothesize to be a reflection of how B1 binding activity may be dynamic and may redistribute during RNA maturation based on local cellular environment.
3.3. Potential roles of hnRNP A2/B1 interaction with HOTAIR
The multiple binding sites of B1 on HOTAIR that we have uncovered through in vivo and in vitro eCLIP suggest a complex mode of engagement. Each binding site places B1 either in proximity to or overlapping the RNA-RNA interaction hotspot on HOTAIR (Figure 5 and Supplementary Figure 5), suggesting potential for regulation of these RNA interactions, either positively or negatively. The strong binding site for A2/B1 that we have identified in intron 3 of HOTAIR, though not indicative of direct splicing repression by A2/B1, may still indicate a role for repression of splicing in the HOTAIR mechanism. Intron 3 binding is not required for A2/B1 to contribute to the HOTAIR mechanism, since cDNA transgenes are sufficient. However, in the context of the endogenous gene, this interaction may still be important in ways that are not yet fully clear. The results of the RNA-RNA interaction assays testing HOTAIR-JAM2 matching (Figure 5(g)) suggest that intron 3 may actually be a positive regulator of the HOTAIR-B1 mechanism, both in an hnRNP B1-dependent and -independent manner. This may be due in part to the ability of intron 3 to fold independently and potentially ‘present’ the RNA-RNA interaction hotspot, though further work is needed to understand this better.
3.4. Conclusion
Our transcriptome-wide study of A2/B1 RNA binding partners provides additional examples further emphasizing that A2/B1 has more roles than simply acting as a splicing regulator. We identify a number of cases in which A2/B1 may bind RNA in concert with other proteins, demonstrate how the splicing function of A2/B1 is potentially re-purposed for additional molecular mechanisms with lncRNAs, and identify a number of A2/B1 binding sites in distal introns far removed from splice sites. These results solidify the view that A2/B1 is a multifunctional RNA-binding protein with a diverse set of roles across the transcriptome.
4. Materials and methods
4.1. Cell culture
MCF7 cells were maintained in RPMI (Gibco) with 10% FBS and 1x Pen-Strep (Life Technologies). MCF10A cells were maintained in DMEM/F12 media (Sigma) supplemented with 5% horse serum (Gibco), 20 ng/mL EGF (Sigma), 0.5 mg/mL hydrocortisone (Sigma), 100 ng/mL cholera toxin (Sigma), 10 µg/mL insulin (Life Technologies), and Pen-Strep (Life Technologies). C2C12 cells were grown in DMEM (Gibco) with 20% FBS and 1x Pen-Strep (Life Technologies).
4.2. eCLIP-seq
eCLIP-seq was performed as previously described [15] with minor modifications. Briefly, we UV-crosslinked confluent cells at 150 mJ and 254 nm wavelength in 13 mL of PBS in 15 cm plates; these cells were then spun down, flash frozen in liquid nitrogen, and stored at – 80ºC. Nuclear isolation was performed as previously described [20], followed by further digestion by Bioruptor, RNase I, and Turbo DNase as specified in the original eCLIP protocol. Overnight immunoprecipitation was performed using 2 mg Protein G Dynabeads per 10 µg antibody, and appropriate antibodies for hnRNP A2/B1 (Abcam ab31645, 9 µg/IP), hnRNP B1 (IBL 18,941, 2.5 µg/IP), or IgG (Novus NB810-56,910, 5 µg/IP). Following end repair and 3′ adaptor ligation (using X1A and X1B adaptors as previously described, see Table S2 in Supplementary Material), size selection was carried out using Nupage 4–12% Bis-Tris protein gels followed by overnight transfer at 30V to nitrocellulose membranes. Protein-RNA complexes on nitrocellulose were then digested with Proteinase K (Roche) and reverse transcribed using SuperScript IV (Thermo Fisher). These cDNAs were then prepared for sequencing as described using Illumina TruSeq HT dual indexed primers, and sequenced on an Illumina NextSeq 500 for high-output 2 × 75 bp run.
4.3. Cellular fractionation of MCF7 cells and eCLIP
Cellular fractionation of cross-linked cells into cytoplasm, nucleoplasm, and chromatin subcompartments was performed as previously described [27] and tested using antibodies specific to each subcompartment (Figure 3(a)). Each fraction was raised to 1 mL final volume using Buffer D (20 mM HEPES pH 7.5, 210 mM NaCl, 7.5% glycerol, 0.75 mM MgCl2, 0.25 mM PMSF, and 1x proteinase inhibitor) then incubated with TURBO DNase (40U for chromatin sample, 10U for soluble samples) for 30 minutes at 37ºC, then quenched with 10 mM EDTA. Fractions were then further digested with Bioruptor and RNase (but no additional DNase) as specified in the original eCLIP protocol, then immunoprecipitated overnight with antibody to hnRNP B1(IBL 18,941, 2.5 µg/IP). The rest of the eCLIP protocol was performed identically to other samples. Validation of fractionation was performed with Western analysis for A2/B1, histone H3 (HRP-conjugated 1º antibody, Abcam ab21054-200), and beta-tubulin (Invitrogen MA5-16,308).
4.4. Computational analysis for eCLIP
eCLIP-seq libraries were analyzed using the previously published computational pipeline [15], with minor changes provided to us by the Yeo group (G. Pratt, personal communication). These changes included a modified version of the clipper peak calling program [52] modified to perform more accurately when followed by input normalization of peak signal.
Peaks were then normalized against the size matched input sample by calculating fold enrichment of number of reads in IP versus number of reads in the input sample. Peaks were called significant if IP read count was higher than input and the peaks had a Bonferroni-corrected Fisher exact p-value of less than 10–5.
Motifs were determined using the DREME program [53] with the options -rna -m 4. IDR analysis was performed on input-normalized peaks by ranking peaks by enrichment P-value and performing the ENCODE pipelines described at https://sites.google.com/site/anshulkundaje/projects/idr/deprecated.
4.5. Native RNA immunoprecipitations
MCF-7 cells were washed in PBS and lysed in 10 mM Hepes 7.4, 150 mM KCl, 3 mM MgCl2, 2 mM DTT, 0.5% NP-40, 1 mM PMSF, 10% glycerol with protease and RNase inhibitors (Roche). Immunoprecipitations were performed overnight at 4°C with 1 mg of lysate and 5 μg of the following antibodies: Mouse Total IgG (EMD Millipore) and hnRNP A2/B1 (Novus NB120-6102). For each IP, 25 µL of Protein A/G magnetic beads (Thermo-Fisher) were added to each sample and incubated an additional 2 h. Beads were washed 5X with the wash buffer: 20 mM Tris at pH 7.4, 200 mM NaCl, 2 mM MgCl2, 1 mM DTT, and split for RNA and protein analysis.
4.6. RNA isolation and PCR
RNA was isolated with 500 μL TRIzol (Life Technologies) followed by purification by RNeasy kit (QIAGEN). Samples were DNase treated using the TURBO DNase kit (Ambion). Two µg of each RNA sample was reverse transcribed using a cDNA High Capacity Kit (Life Technologies). cDNA was PCR amplified using Phusion polymerase and 1 minute extension times to accurately amplify reads over 1 kb in length.
4.7. In vitro eCLIP-seq
Cloning and purification of recombinant hnRNP B1 was performed as previously described [5]. HOTAIR and antisense luciferase control RNA (see [5] for sequence details) were in vitro transcribed using the MEGAscript T7 Transcription Kit, treated with TURBO DNase, and purified with RNeasy Qiagen Kit. In a 1:10 RNA:protein molar ratio, 1.2 ug of control or HOTAIR RNA was incubated with recombinant B1 in 100 µL buffer (20 mM HEPES-KOH pH 7.9, 100 mM KCl, 0.2 EDTA pH 8.0. 20% Glycerol, 0.5 mM PMSF, 0.5 DTT) for 20 minutes at room temperature. The mixture was diluted to 250 µL in refolding buffer and UV-crosslinked twice in one well of a 24-well plate at 250 mJ and 254 nm wavelength, with mixing by pipette in between. B1-RNA crosslinked and non-crosslinked samples were treated with 0.1 ng RNase A for 3 minutes at 37ºC and 1200 rpm, then stopped with 200 U Murine RNase Inhibitor (NEB). Following this, the in vitro samples were subjected to end repair, adaptor ligation, SDS-PAGE and transfer to nitrocellulose, and the remainder of the eCLIP-seq protocol, then sequenced multiplexed with other eCLIP-seq libraries. Compared to the control sample, HOTAIR-B1 libraries produced ~ 6-fold higher concentration of final product after library preparation and size selection, using the same number of PCR amplification cycles for each sample.
4.8. RNA-RNA interaction analysis
Using a previously published database of computationally predicted interactions between human lncRNAs and the entire transcriptome [50], a list of all 44,006 annotated mature transcripts and computational predictions for RNA-RNA interaction with HOTAIR were analyzed. This list was sorted by the interaction energy (i.e. the minimum predicted free energy found among the interactions contained within each pair of RNA sequences) and the top and bottom 1% by rank (440 transcripts) were used as input for a Java program that plots the total number of interactions for each nucleotide of the 2,422nt HOTAIR transcript.
4.9. RNA-RNA interaction assays
Performed as previously described [5] using in vitro-transcribed versions of HOTAIR, HOTAIR + intron 3, and the RAT-tagged JAM2 target as well as recombinant hnRNP B1 purified from E. coli.
Funding Statement
This work was supported by the National Institute of General Medical Sciences [T32GM008497]; National Institute of General Medical Sciences [R35GM119575]; Victor and Earleen Bolie Scholarship; University of Colorado School of Medicine RNA Bioscience Initiative; Paul O’Hara II Seed Grant from the University of Colorado Cancer Center ACS-IRG.
Acknowledgments
We would like to thank G. Pratt and E. van Nostrand for invaluable assistance with the eCLIP protocol, as well as R. Parker and J. Wheeler for experimental interpretation advice. Thanks to O. Rissland and N. Mukherjee for comments on the manuscript. We thank the University of Colorado Cancer Center Genomics Core (supported by NIH grant P30-CA46934) for technical support. C2C12 cells were a gift from the B. Olwin lab at the University of Colorado Boulder. This work was supported by NIH grant R35GM119575 (A.M.J), the University of Colorado-Denver MD/PhD Program NIH Training Grant T32GM008497, a Paul O’Hara II Seed Grant from the University of Colorado Cancer Center ACS-IRG Grant Program (A.M.J.), the Victor and Earleen Bolie Scholarship Fund (M.M.B), and the University of Colorado School of Medicine RNA Bioscience Initiative (M.M.B.).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here.
References
- 1.Wang KC, Chang HY.. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43(6):904–914. Epub 2011/ 09/20. S1097-2765(11)00636-8 [pii] PubMed PMID: 21925379; PubMed Central PMCID: PMC3199020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tang YH, Han SP, Kassahn KS, et al. Complex evolutionary relationships among four classes of modular RNA-binding splicing regulators in eukaryotes: the hnRNP, SR, ELAV-like and CELF proteins. J Mol Evol. 2012;75(5–6):214–228. PubMed PMID: 23179353. [DOI] [PubMed] [Google Scholar]
- 3.Gueroussov S, Weatheritt RJ, O’Hanlon D, et al. Regulatory expansion in mammals of multivalent hnrnp assemblies that globally control alternative splicing. Cell. 2017;170(2):324–39 e23. .PubMed PMID: 28709000. [DOI] [PubMed] [Google Scholar]
- 4.Han SP, Friend LR, Carson JH, et al. Differential subcellular distributions and trafficking functions of hnRNP A2/B1 spliceoforms. Traffic. 2010;11(7):886–898. PubMed PMID: 20406423; PubMed Central PMCID: PMC2906249 Epub 2010/ 04/22. TRA1072 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meredith EK, Balas MM, Sindy K, et al. An RNA matchmaker protein regulates the activity of the long noncoding RNA HOTAIR. RNA. 2016;22(7):995–1010. . PubMed PMID: 27146324; PubMed Central PMCID: PMCPMC4911922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. PubMed PMID: 17604720; PubMed Central PMCID: PMC2084369 Epub 2007/ 07/03. S0092-8674(07)00659-9 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amandio AR, Necsulea A, Joye E, et al. Hotair Is dispensible for mouse development. PLoS Genet. 2016;12(12):e1006232 . PubMed PMID: 27977683; PubMed Central PMCID: PMCPMC5157951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464(7291):1071–1076. PubMed PMID: 20393566; PubMed Central PMCID: PMC3049919 Epub 2010/ 04/16.nature08975 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burd CG, Swanson MS, Gorlach M, et al. Primary structures of the heterogeneous nuclear ribonucleoprotein A2, B1, and C2 proteins: a diversity of RNA binding proteins is generated by small peptide inserts. Proc Natl Acad Sci U S A. 1989;86(24):9788–9792. PubMed PMID: 2557628; PubMed Central PMCID: PMCPMC298587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Contreras R, Cloutier P, Shkreta L, et al. hnRNP proteins and splicing control. Adv Exp Med Biol. 2007;623:123–147. Epub 2008/ 04/03.PubMed PMID: 18380344. [DOI] [PubMed] [Google Scholar]
- 11.Huelga SC, Vu AQ, Arnold JD, et al. Integrative genome-wide analysis reveals cooperative regulation of alternative splicing by hnRNP proteins. Cell Rep. 2012;1(2):167–178. PubMed PMID: 22574288; PubMed Central PMCID: PMC3345519 Epub 2012/05/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goodarzi H, Najafabadi HS, Oikonomou P, et al. Systematic discovery of structural elements governing stability of mammalian messenger RNAs. Nature. 2012;485(7397):264–268. PubMed PMID: 22495308; PubMed Central PMCID: PMC3350620 Epub 2012/ 04/13. nature11013 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geissler R, Simkin A, Floss D, et al. A widespread sequence-specific mRNA decay pathway mediated by hnRNPs A1 and A2/B1. Genes Dev. 2016;30(9):1070–1085. .PubMed PMID: 27151978; PubMed Central PMCID: PMCPMC4863738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim HJ, Kim NC, Wang YD, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495(7442):467–473. .PubMed PMID: 23455423; PubMed Central PMCID: PMCPMC3756911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Nostrand EL, Pratt GA, Shishkin AA, et al. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods. 2016;13(6):508–514. .PubMed PMID: 27018577; PubMed Central PMCID: PMCPMC4887338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatfield JT, Rothnagel JA, Smith R. Characterization of the mouse hnRNP A2/B1/B0 gene and identification of processed pseudogenes. Gene. 2002;295(1):33–42. Epub 2002/09/21.S0378111902008004 [pii]. PubMed PMID: 12242009. [DOI] [PubMed] [Google Scholar]
- 17.Kamma H, Horiguchi H, Wan L, et al. Molecular characterization of the hnRNP A2/B1 proteins: tissue-specific expression and novel isoforms. Exp Cell Res. 1999;246(2):399–411. PubMed PMID: 9925756 Epub 1999/ 02/02. S0014-4827(98)94323-3 [pii]. [DOI] [PubMed] [Google Scholar]
- 18.Han SP, Kassahn KS, Skarshewski A, et al. Functional implications of the emergence of alternative splicing in hnRNP A/B transcripts. RNA. 2010;16(9):1760–1768. . PubMed PMID: 20651029; PubMed Central PMCID: PMCPMC2924535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wheeler EC, Van Nostrand EL, Yeo GW. Advances and challenges in the detection of transcriptome-wide protein-RNA interactions. Wiley Interdiscip Rev RNA. 2018;9(1). DOI: 10.1002/wrna.1436 PubMed PMID: 28853213; PubMed Central PMCID: PMCPMC5739989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kung JT, Kesner B, An JY, et al. Locus-Specific Targeting to the X chromosome revealed by the RNA Interactome of CTCF. Mol Cell. 2015;57(2):361–375. PubMed PMID: 25578877 Epub 2015/ 01/13. S1097-2765(14)00955-1 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y, Lv F, Liang D, et al. HOTAIR may regulate proliferation, apoptosis, migration and invasion of MCF-7 cells through regulating the P53/Akt/JNK signaling pathway. Biomed Pharmacother. 2017;90:555–561. .PubMed PMID: 28407576 [DOI] [PubMed] [Google Scholar]
- 22.Consortium EP An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. PubMed PMID: 22955616; PubMed Central PMCID: PMCPMC3439153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soule HD, Maloney TM, Wolman SR, et al. Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 1990;50(18):6075–6086. PubMed PMID: 1975513. [PubMed] [Google Scholar]
- 24.McKay SJ, Cooke H. hnRNP A2/B1 binds specifically to single stranded vertebrate telomeric repeat TTAGGGn. Nucleic Acids Res. 1992;20(24):6461–6464. Epub 1992/12/25 PubMed PMID: 1282701; PubMed Central PMCID: PMC334558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez FJ, Pratt GA, Van Nostrand EL, et al. Protein-RNA networks regulated by normal and als-associated mutant hnrnpa2b1 in the nervous system. Neuron. 2016;92(4):780–795. .PubMed PMID: 27773581; PubMed Central PMCID: PMCPMC5123850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friend LR, Han SP, Rothnagel JA, et al. Differential subnuclear localisation of hnRNPs A/B is dependent on transcription and cell cycle stage. Biochim Biophys Acta. 2008;1783(10):1972–1980. . PubMed PMID: 18588922. [DOI] [PubMed] [Google Scholar]
- 27.Wysocka J, Reilly PT, Herr W. Loss of HCF-1-chromatin association precedes temperature-induced growth arrest of tsBN67 cells. Mol Cell Biol. 2001;21(11):3820–3829. . PubMed PMID: 11340173; PubMed Central PMCID: PMCPMC87041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemieux B, Blanchette M, Monette A, et al. A Function for the hnRNP A1/A2 proteins in transcription elongation. PLoS One. 2015;10(5):e0126654 PubMed PMID: 26011126; PubMed Central PMCID: PMCPMC4444011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meier UT. RNA modification in Cajal bodies. RNA Biol. 2017;14(6):693–700. PubMed PMID: 27775477; PubMed Central PMCID: PMCPMC5519239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darzacq X, Jady BE, Verheggen C, et al. Cajal body-specific small nuclear RNAs: a novel class of 2ʹ-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21(11):2746–2756. PubMed PMID: 12032087; PubMed Central PMCID: PMCPMC126017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang F, Tang ML, Zeng ZX, et al. Telomere- and telomerase-interacting protein that unfolds telomere G-quadruplex and promotes telomere extension in mammalian cells. Proc Natl Acad Sci U S A. 2012;109(50):20413–20418. .PubMed PMID: 23184978; PubMed Central PMCID: PMCPMC3528488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tollervey JR, Curk T, Rogelj B, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nat Neurosci. 2011;14(4):452–458. .PubMed PMID: 21358640; PubMed Central PMCID: PMCPMC3108889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brugiolo M, Botti V, Liu N, et al. Fractionation iCLIP detects persistent SR protein binding to conserved, retained introns in chromatin, nucleoplasm and cytoplasm. Nucleic Acids Res. 2017;45(18):10452–10465. . PubMed PMID: 28977534; PubMed Central PMCID: PMCPMC5737842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carpenter S, Aiello D, Atianand MK, et al. A long noncoding RNA mediates both activation and repression of immune response genes. Science. 2013;341(6147):789–792. PubMed PMID: 23907535 Epub 2013/ 08/03. doi: science.1240925 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–1927. .PubMed PMID: 21890647; PubMed Central PMCID: PMCPMC3185964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun M, Gadad SS, Kim DS, et al. Discovery, annotation, and functional analysis of long noncoding rnas controlling cell-cycle gene expression and proliferation in breast cancer cells. Mol Cell. 2015;59(4):698–711. . PubMed PMID: 26236012; PubMed Central PMCID: PMCPMC4546522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Augui S, Nora EP, Heard E. Regulation of X-chromosome inactivation by the X-inactivation centre. Nat Rev Genet. 2011;12(6):429–442. . PubMed PMID: 21587299. [DOI] [PubMed] [Google Scholar]
- 38.Simon MD, Pinter SF, Fang R, et al. High-resolution Xist binding maps reveal two-step spreading during X-chromosome inactivation. Nature. 2013;504(7480):465–469. PubMed PMID: 24162848; PubMed Central PMCID: PMC3904790 Epub 2013/ 10/29 nature12719 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao J, Ohsumi TK, Kung JT, et al. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol Cell. 2010;40(6):939–953. .PubMed PMID: 21172659; PubMed Central PMCID: PMCPMC3021903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu F, Somarowthu S, Pyle AM. Visualizing the secondary and tertiary architectural domains of lncRNA RepA. Nat Chem Biol. 2017;13(3):282–289. . PubMed PMID: 28068310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yen ZC, Meyer IM, Karalic S, et al. A cross-species comparison of X-chromosome inactivation in Eutheria. Genomics. 2007;90(4):453–463. . PubMed PMID: 17728098. [DOI] [PubMed] [Google Scholar]
- 42.Patil DP, Chen CK, Pickering BF, et al. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537(7620):369–373. .PubMed PMID: 27602518; PubMed Central PMCID: PMCPMC5509218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Alarcon CR, Goodarzi H, Lee H, et al. HNRNPA2B1 Is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162(6):1299–1308. . PubMed PMID: 26321680; PubMed Central PMCID: PMCPMC4673968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee S, Kopp F, Chang TC, et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164(1–2):69–80. .PubMed PMID: 26724866; PubMed Central PMCID: PMCPMC4715682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tichon A, Gil N, Lubelsky Y, et al. A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Commun. 2016;7:12209 .PubMed PMID: 27406171; PubMed Central PMCID: PMCPMC4947167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khalil AM, Guttman M, Huarte M, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106(28):11667–11672. .PubMed PMID: 19571010; PubMed Central PMCID: PMCPMC2704857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katsushima K, Natsume A, Ohka F, et al. Targeting the Notch-regulated non-coding RNA TUG1 for glioma treatment. Nat Commun. 2016;7:13616 .PubMed PMID: 27922002; PubMed Central PMCID: PMCPMC5150648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z, Shen J, Chan MT, et al. TUG1: a pivotal oncogenic long non-coding RNA of human cancers. Cell Prolif. 2016;49(4):471–475. . PubMed PMID: 27339553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tsai MC, Manor O, Wan Y, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329(5992):689–693. PubMed PMID: 20616235; PubMed Central PMCID: PMC2967777 Epub 2010/ 07/10. science.1192002 [pii]. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Terai G, Iwakiri J, Kameda T, et al. Comprehensive prediction of lncRNA-RNA interactions in human transcriptome. BMC Genomics. 2016;17(Suppl 1):12 . PubMed PMID: 26818453; PubMed Central PMCID: PMCPMC4895283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Somarowthu S, Legiewicz M, Chillon I, et al. HOTAIR forms an intricate and modular secondary structure. Mol Cell. 2015;58(2):353–361. Epub 2015/ 04/14. S1097-2765(15)00171-9 [pii] PubMed PMID: 25866246; PubMed Central PMCID: PMC4406478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lovci MT, Ghanem D, Marr H, et al. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol. 2013;20(12):1434–1442. .PubMed PMID: 24213538; PubMed Central PMCID: PMCPMC3918504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27(12):1653–1659. . PubMed PMID: 21543442; PubMed Central PMCID: PMCPMC3106199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


