ABSTRACT
The RNA helicase Mtr4 is a versatile protein that is a crucial component of several distinct RNA surveillance complexes. Here we describe a novel complex that contains Mtr4, but has a role distinct from any of those previously described. We found that Mtr4 association with the human homolog of fission yeast Nrl1, NRDE-2, defines a novel function for Mtr4 in the DNA damage response pathway. We provide biochemical evidence that Mtr4 and NRDE-2 are part of the same complex and show that both proteins play a role in the DNA damage response by maintaining low DNA double-strand break levels. Importantly, the DNA damage response function of the Mtr4/NRDE-2 complex does not depend on the formation of R loops. We show however that NRDE-2 and Mtr4 can affect R-loop signals at a subset of distinct genes, possibly regulating their expression. Our work not only expands the wide range of Mtr4 functions, but also elucidates an important role of the less characterized human NRDE-2 protein.
KEYWORDS: NRDE-2, Mtr4, DNA damage, DDR, R loop, R-loop, double-strand breaks, DSBs, Nrl1, lncRNAs; NRDE2
1. Introduction
Mtr4 is an RNA helicase that is the centerpiece of several distinct complexes involved in turnover of specific RNAs [1]. It is part of exosome adaptor complexes such as TRAMP (Trf4-Air-Mtr4 polyadenylation) and NEXT (nuclear exosome targeting) [2]. We and others recently identified Mtr4 as a master player of another RNA surveillance complex, Mtr4/ZFC3H1 or PAXT (poly(A) tail exosome targeting), which plays a role in turnover of polyadenylated lncRNAs, such as ptRNAs (prematurely terminated RNAs), uaRNAs (upstream antisense RNAs) and eRNAs (enhancer RNAs) [3,4].
We and others previously identified NRDE-2 (nuclear RNAi defective-2; also known as C14ORF102) as an Mtr4-interacting protein in human cells [2,3]. While the function of human NRDE-2 is unknown, the S. pombe NRDE-2 homolog Nrl1 (NRDE-2 like 1) has been found to interact with the Mtr4-like protein Mtl1 and with splicing factors that help the RNAi-dependent assembly of heterochromatin at loci called HOODs (heterochromatin domains) on certain specific genes [5,6]. NRDE-2 was first identified in C. elegans as a factor required for RNAi in the nucleus [7]. Interestingly, its association with the RNAi machinery is necessary for both trimethylation of H3K9 at genomic loci targeted by siRNAs and also inhibition of transcription elongation downstream of the siRNA-targeted sequences, most likely achieved by inducing early transcription termination. Human NRDE-2 is a ~ 130 kDa protein that contains, like the C. elegans and S. pombe ortholgues, many Half-A-Tetratricopeptide (HAT) repeats (SMART accession #: SM00386) [8], usually found in RNA-binding proteins and often involved in protein-protein interactions [9].
Importantly, Nrl1 has been found to protect the S. pombe genome from instability by resolving R loops and promoting DNA repair through homologous recombination (HR) [8]. R loops are conserved co-transcriptional structures that arise from hybridization of a nascent RNA with the DNA template, and are thought to cover ~5% of mammalian genomes [10]. While R loops function in several important cellular processes, such as immunoglobulin class switch recombination in activated B cells [11], mitochondria replication [12,13], protection against epigenetic silencing at promoters [10] and transcription termination [10,14,15], their persistence or formation at inappropriate locations can lead to mutations, DNA double-strand breaks (DSBs) and chromosome rearrangements causing genome instability [16,17]. Indeed, R-loop accumulation has been linked to many diseases, from cancer to neurological disorders [18,19].
In this study, we have investigated the role of NRDE-2 and an Mtr4/NRDE-2 complex in human cells. We first conducted co-immunoprecipitation (co-IP) experiments of FLAG-tagged derivatives of both NRDE-2 and Mtr4 stably expressed in HEK293 cells. We identified common partners that confirm the existence of a specific Mtr4/NRDE-2 complex that interacts with splicing factors, similar to its yeast counterpart. We also identified new interacting proteins, including several involved in chromatin remodeling/DNA damage response (DDR), associated with the proteasome, or are cytoskeletal proteins. Importantly, we provide evidence that NRDE-2 plays a role in the DDR. However, in contrast with Nrl1, DSBs that accumulate after NRDE-2 or Mtr4 depletion are not dependent on R-loop accumulation. Finally, we found that NRDE-2 and Mtr4 siRNA-mediated knockdowns (KDs) can modestly but significantly affect R-loop profiles at specific and distinct loci, suggesting an independent connection of NRDE-2 and Mtr4 with transcriptional regulation through R-loop formation/resolution.
2. Results
2.1. Identification of human Mtr4/NRDE-2-associated proteins
With the goal of understanding NRDE-2 function, we first set out to identify NRDE-2-interacting proteins. NRDE-2 was previously identified as an Mtr4-interacting protein [2,3]; however, proteins associated with Mtr4/NRDE-2 remain undetermined. To identify NRDE-2-interacting proteins, we prepared extracts from HEK293 cells stably producing NRDE-2-3FLAG with expression equivalent to endogenous NRDE-2 (Fig. S1A). Similar to its S. pombe counterpart [6], NRDE-2-3FLAG localizes to the nucleus (Fig. S1B). Considering the possible association of NRDE-2 with insoluble chromatin-binding proteins, and to avoid obtaining possible indirect interactions mediated by DNA and/or RNA, we treated the extracts with Benzonase and RNase A (Fig. S1C). By treating with Benzonase, strong chromatin-binding proteins, such as histones, can be efficiently extracted even under physiological salt concentrations [20]. Nuclease-treated lysates were used for co-IP with FLAG antibody, and co-IPed proteins were eluted with 3×FLAG peptide (silver stain; Figure 1(a)) and subjected to mass spectrometry (MS) (Table S1). As expected, Mtr4 was detected with the highest peptide counts [2,3,6,8]. This interaction was further validated with another NRDE-2-3FLAG co-IP experiment performed under the same conditions as the one subjected to MS (Fig. S1D). Interestingly, despite the high abundance of Mtr4 in the MS analysis, no exosome subunits were identified. Additionally, no NEXT (RBM7 and ZCCHC8), TRAMP (PAPD5 and ZCCHC7) or Mtr4/ZFC3H1 (or PAXT) subunits were detected, confirming that the Mtr4/NRDE-2 complex is distinct from these complexes. Unlike in C. elegans [7], but similar to S. pombe [6,8,21], no RNAi factors were obtained.
Figure 1.
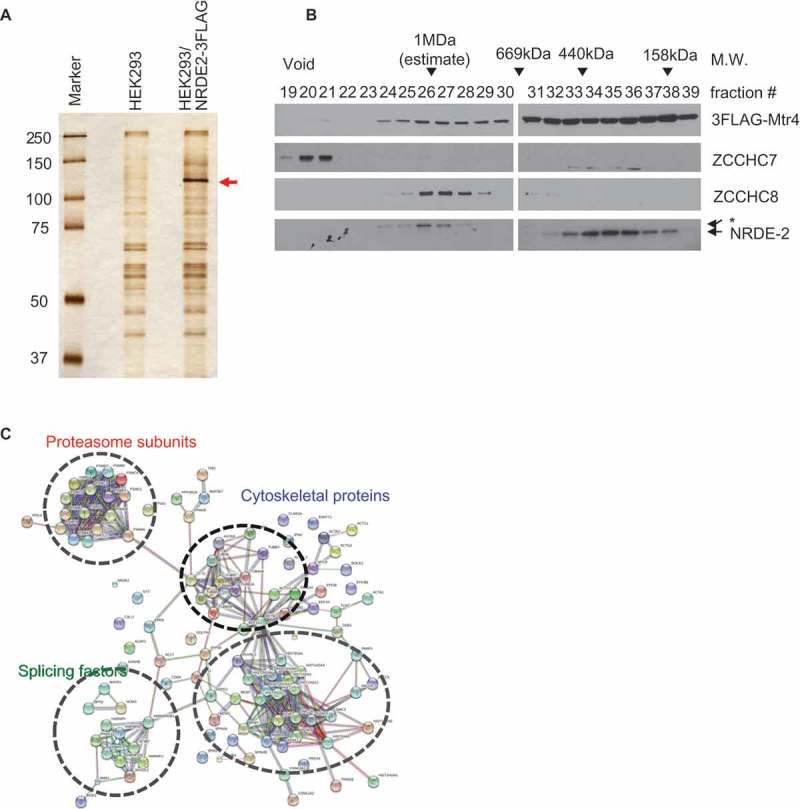
Identification of NRDE-2-interacting proteins by co-IP and mass spectrometry.
(a) Silver staining of FLAG co-IPs from control and NRDE-2-3FLAG stable HEK293 cell lines used for mass spectrometry. (b) Fractions from Superose 6 gel filtration of 3FLAG-Mtr4-expressing HEK293 cell lysates treated with Benzonase/RNase A were analyzed by WB, using antibodies against proteins shown on the right. Approximate molecular sizes are indicated at the top and fractions pooled at the bottom. The asterisk indicates a non-specific band. (c) The protein-protein interaction network among the proteins shared in NRDE-2-3FLAG and 3FLAG-Mtr4 co-IP that are absent in HEK293 control co-IP was constructed using STRING v10.5 (http://string-db.org) with the high confidence setting. Disconnected nodes are not shown in the network.
Next, we wished to determine which NRDE-2-associating proteins also co-purify with Mtr4. Since Mtr4 exists in multiple distinct protein complexes, we performed gel filtration before co-IP/MS analysis. Protein complexes in Benzonase/RNase A-treated extracts prepared from the stable HEK293 cell line expressing 3FLAG-Mtr4 [3] were fractionated by size using a Superose 6 column (Figure 1(b)), and the NRDE-2-containing fractions (158–669 kDa) were used for anti-FLAG co-IP followed by MS. We previously reported that 3FLAG-Mtr4 was broadly distributed between 158 kDa and void fractions in RNase A-treated extracts [3]. However, 3FLAG-Mtr4 in >1 MDa ~ void fractions was absent following Benzonase treatment (compare Figure 1(b) with Figure 2(b) in [3]). There were no significant changes in the distribution patterns of ZCCHC7, ZCCHC8 and NRDE-2 due to Benzonase (Figure 1(b), see Figure 2(b) in [3]). MS analysis successfully detected known Mtr4 partners, including NRDE-2 and exosome subunits (Table S1). Among the proteins that were not detected in a HEK293 control, 98 were detected in both NRDE-2 and Mtr4 interactomes. We note that 103 out of 252 (40.8%) of the proteins from the Benzonase/RNase A-treated 3FLAG-Mtr4 co-IP/MS overlap with the RNase A only-treated 3FLAG-Mtr4 co-IP/MS described in our previous work [3], despite the absence of most of the DNA-associated proteins (presence of benzonase) in our current analysis.
Figure 2.
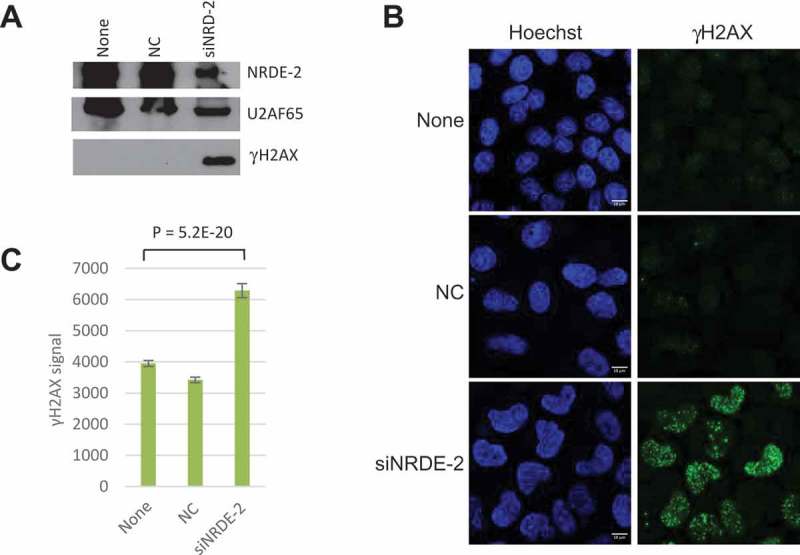
NRDE-2 KD leads to increased DSBs.
(a) γH2AX protein levels in non-transfected HeLa cells (None), cells transfected with an siRNA control (NC) or siNRDE-2 analyzed by WB. U2AF65 is used as a loading control. (b) γH2AX signal visualized by IFs in HeLa cells as in A. (c) Quantification of images as in B. n = 267 cells, SE is shown.
To gain better insight into the Mtr4/NRDE-2-interacting proteins, we uploaded the shared proteins to STRING v10.5 database [22] and created high confidence interaction networks (Figure 1(c)). The proteins were clustered largely into four groups: splicing factors, histone/chromatin/DDR-related proteins, cytoskeletal proteins and proteasome subunits. The interaction of Mtr4/NRDE-2 with splicing factors is reminiscent of the fission yeast Mtl1/Nrl1 complex, although an additional conserved protein, Ctr1, stably interacts with Mtl1/Nrl1 and splicing factors [6,8,21], but its human homolog CCDC174 was absent in both NRDE-2-3FLAG and 3FLAG-Mtr4 co-IP/MS analyses. Altogether these co-IP experiments highlight the existence of a human Mtr4/NRDE-2 complex distinct from any previously identified Mtr4 complexes, and which appears to share several features with the fission yeast complex.
2.2. NRDE-2 prevents DNA damage accumulation
For some time, splicing factors have been linked to genome stability maintenance through the prevention of R-loop accumulation [23]. Importantly, deletion of Nrl1 in fission yeast leads to an accumulation of R loops and DSBs [8]. Since we found NRDE-2 interacting with splicing factors, and to begin to test whether NRDE-2 is also involved in maintaining genome stability in human cells, we monitored, by western blot (WB) and immunofluorescence (IF), levels of γH2AX, a marker of DSBs, following siRNA-mediated KD of NRDE-2 in HeLa cells. Strikingly, the IFs, and quantification of γH2AX signal, showed a drastic increase (~ two-fold relative to control) of DNA damage in KDed cells compared to non-transfected cells or cells transfected with a control siRNA (NC) (Figure 2 + Fig. S2). Since NRDE-2 interacts with Mtr4, and Mtr4 KO in mouse B cells shows an increase in R loops at certain loci [24], we asked whether Mtr4 KD also leads to DSBs. Indeed, Mtr4 KD also resulted in a significant increase in DSBs as shown by the quantification of the γH2AX signal (Fig. S2). These data suggest that the Mtr4/NRDE-2 complex is involved in DNA damage prevention or repair.
2.3. DNA damage in NRDE-2 and Mtr4 depleted cells is R-loop independent
Since Nrl1 and mouse Mtr4 have both been linked to R-loop metabolism [8,24], we next asked whether the DSBs detected after NRDE-2 and Mtr4 KDs were caused by an increase in R-loop levels. To address this, we overexpressed GFP-RNase H1 in HeLa cells KDed for Mtr4 or NRDE-2 and quantified the γH2AX levels by WB analysis (Fig. S3). Confirming our IF data, Mtr4 and NRDE-2 KDs led to an accumulation of DSBs as shown by an increased γH2AX protein level compared to control cells. Interestingly, Mtr4 KD led to ~ 4 times more γH2AX than did NRDE-2 KD, a result that was not apparent by IF (Fig. S2), and which is discussed below. Importantly, RNase H1 overexpression did not significantly decrease γH2AX levels in NRDE-2 or Mtr4 KDed cells. To confirm these data, we performed IF to monitor γH2AX and GFP-RNase H1 signals simultaneously (Figure 3). The IF images show that γH2AX and high levels of RNase H1 expression can co-exist (see cells with white arrows) and that consequently there was no statistical difference in γH2AX staining after NRDE-2 KD between cells expressing RNase-H1 or not, as calculated by the quantification of the γH2AX signal in the RNase H1-expressing cells. However, RNase H1 overexpression had a significant and unexpected effect on DSB accumulation in Mtr4 KDed cells, as the γH2AX signal in these cells actually increased by 30% after RNase H1 overexpression. While we also detect a 10% increase of the γH2AX signal after RNase H1 overexpression in NRDE-2 KDed cells, this difference doesn’t appear significant (Figure 3, left histogram). These data confirm that DNA damage induced by NRDE-2 and/or Mtr4 KDs is not due to accumulation of R loops.
Figure 3.
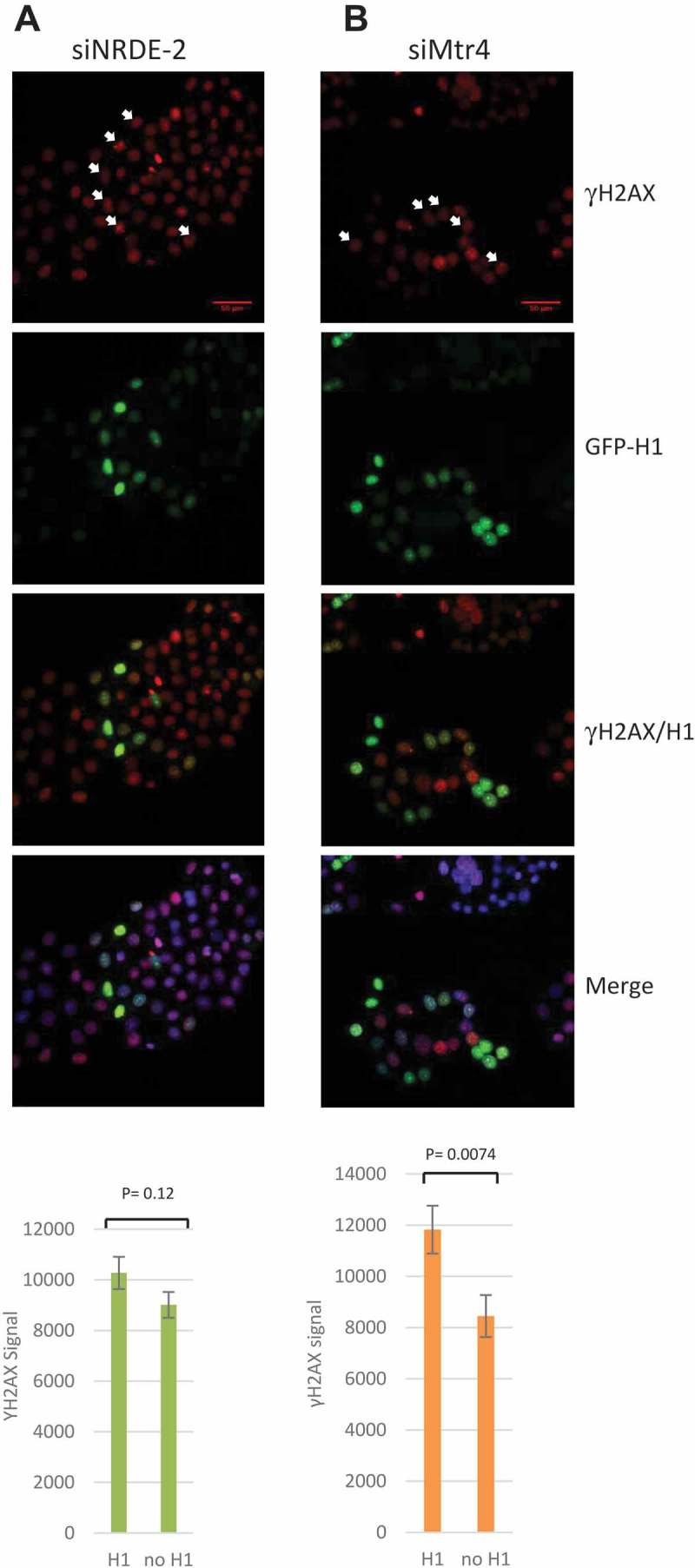
DSBs induced by NRDE-2 or Mtr4 KDs are R-loop independent.
(a) IF of γH2AX signal in NRDE-2 KDed HeLa cells for 72h and transfected (H1) or not (no H1) with GFP-RNase H1 for 48h. Signal quantifications are shown at the bottom. n = 189 cells, SE shown. (b) Quantification of γH2AX signal in Mtr4 KDed cells expressing GFP-RNase H1 (H1) or not (no H1). n = 78 cells, SE shown. White arrows show cells expressing GFP-RNase H1 and high level of γH2AX signal.
We also examined a possible connection between R-loop formation and NRDE-2 and/or Mtr4 more directly. Specifically, we performed DRIP assays in HeLa cells after NRDE-2 and Mtr4 KDs, using the S9.6 antibody to IP RNA:DNA hybrids [25,26]. We examined R-loop formation at known positive loci, the RPL13A 3ʹ end and the BACT 5ʹ pause site, as well as an R-loop negative locus, EGR1 [10,14] (Figure 4(a–c)). While the DRIP assay detected the expected percentage of input at RPL13A (~10%), BACT (~2%) and EGR1 (<0.1%) loci in untransfected and siRNA control (NC) transfected cells [10], R-loop enrichment increased by 20% compared to control at RPL13A and BACT after NRDE-2 KD. In contrast, R-loop enrichment at the 5ʹ pause site of BACT did not change after Mtr4 KD, while ~20% less R-loop signal was detected at RPL13A. Although we only examined a very limited number of loci, the DRIP data suggests that while DSB accumulation after NRDE-2 or Mtr4 KDs is R-loop independent, it is very likely that NRDE-2 and Mtr4 play minor but significant and independent roles in R-loop resolution or formation at specific genes, possibly regulating their transcription.
Figure 4.
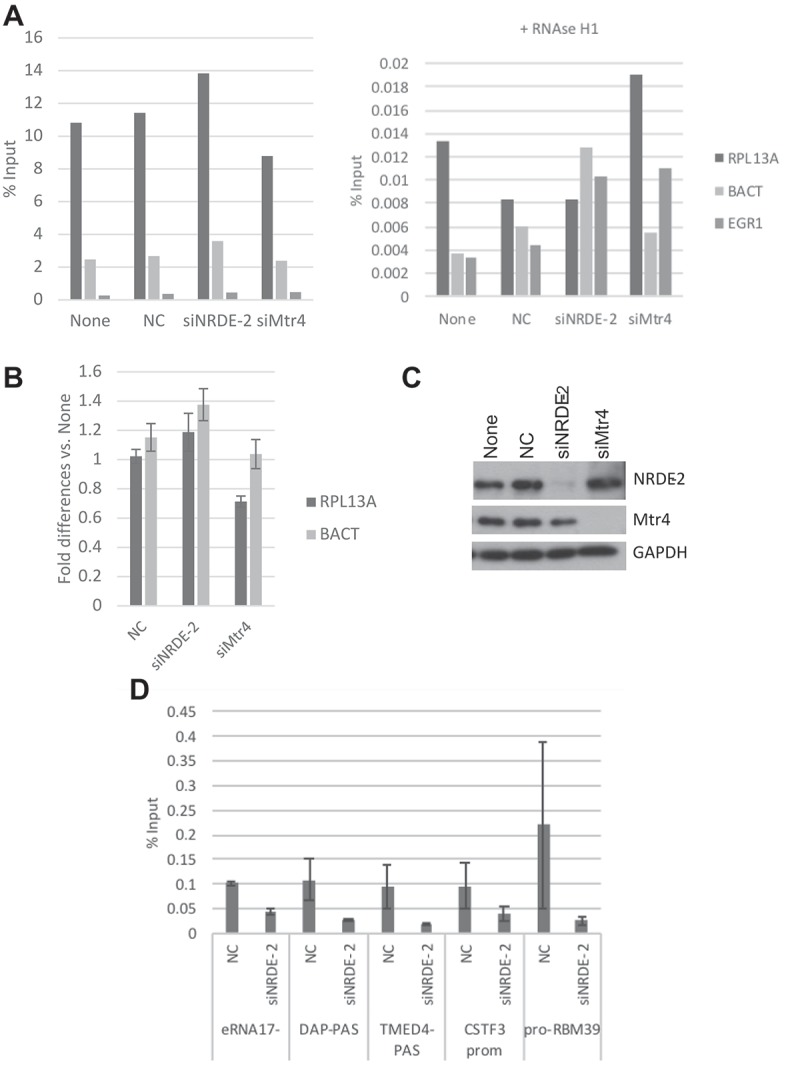
R-loop levels at RPL13A, BACT and EGR1 are not affected by NRDE-2 or Mtr4 KD.
(a) DRIP assays were performed in HeLa cell after 72 h of siRNAs transfection. DRIP signal was also measured after RNase H1 treatment (right). (b) DRIP signal was normalized to the signal in non-transfected cell. Error bars represent the average of two different experiments. SD shown. (c) NRDE-2 and Mtr4 KDs confirmation by WB. None = no transfection, NC: siRNA control transfection. (D) NRDE-2 ChIP at various loci. ChIP assays were performed in HeLa cell after 72 h of siRNAs transfection. n = 2
Since Mtr4 has recently been shown to regulate lncRNAs turnover as part of the Mtr4/ZFC3H1 (or PAXT) complex, we performed NRDE-2 ChIP assays to ask whether NRDE-2 could also play a role at genomic loci including those associated with lncRNAs expression. We indeed found that NRDE-2 binds various loci including eRNA (eRNA17−), ptRNAs intronic PAS (DAP-PAS and TMED4-PAS), PROMPT (pro-RBM39) as well as the promoter of CSTF3 (Figure 4(d); note that NRDE-2 and Mtr4 KDs did not significantly affect Mtr4 or NRDE-2 protein levels, respectively (Figure 4(c))). Importantly, NRDE-2 KD led to a weak but general increase in eRNA17−, ptTMED4, ptDAP, proRBM39 lncRNAs (Fig. S4). CSTF3 mRNA levels also increased significantly after NRDE-2 KD (three-fold relative to control), however CSTF3 ptRNA decreased (Fig. S4). While these data show some disparity, they indicate that NRDE-2 might function as a negative regulator of gene expression. While we could not detect R loops at eRNA17− loci, we were able to detect DRIP signals at ptRNAs intronic PAS (DAP and TMED4). These did not, however, show significant differences after NRDE-2 or Mtr4 KD compared to controls (data not shown), indicating that the potential NRDE-2 negative regulation of eRNA17−, ptDAP and ptTMED4 expression is R-loop independent. These data indicate that it is very likely that NRDE-2 plays a role in transcription regulation that in some cases might involve R-loop resolution or formation (Figure 5).
Figure 5.
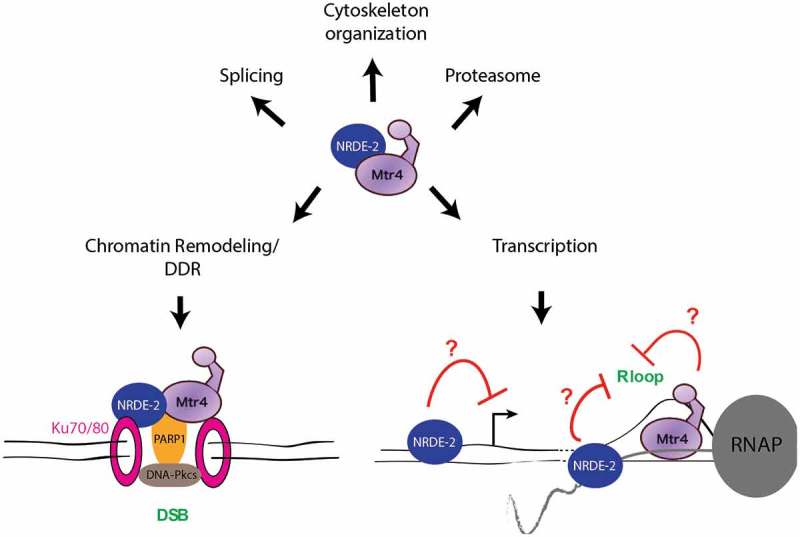
Possible functions of the Mtr4/NRDE-2 complex in various cellular processes.
The role of the Mtr4/NRDE-2 complex in the DDR might involve interaction with the Ku70/80 heterodimer, which recognizes DSBs. Its interaction with PARP1 may stimulate the activation of the DNA-dependent protein kinase catalytic subunit (DNA-Pkcs), a NHEJ factor that also interacts with the Ku complex. NRDE-2 and Mtr4 appear to have independent roles in transcription regulation. While NRDE-2 can have a negative effect on transcription, it seems to resolve R loops at the 3ʹ end of certain genes. According to [24], Mtr4 might also promote R-loop resolution, at least in certain circumstances.
3. Discussion
In this study, we investigated the functions and properties of the conserved human NRDE-2 protein. We identified several NRDE-2 associated proteins as well as proteins associated with one of its previously identified partners, the RNA helicase Mtr4. As strong evidence that NRDE-2 and Mtr4 are core components of a novel NRDE-2/Mtr4 complex, we found that the two proteins share many interacting factors that can be classified into four main categories. We identified proteins from the chromatin remodeling/DDR pathways, splicing factors, cytoskeletal proteins and proteasome subunits. The association of NRDE-2 with splicing factors has previously been observed in fission yeast, where nrl1 deletion is indeed associated with splicing defects of a subset of genes [6,8]. While it is therefore likely that human NRDE-2 also plays a role in pre-mRNA splicing we have not investigated this possibility.
Similar to the nrl1Δ strain, we found that NRDE-2 depleted cells accumulate DSBs. Because genome instability in nrl1Δ yeast has been attributed to an increase in R loops [8], we were in fact very surprised to find that the DSB increase after NRDE-2 or Mtr4 depletion was R-loop independent. Indeed, overexpression of RNase H1, which should eliminate R loops, did not reduce DNA damage after NRDE-2 or Mtr4 KD. On the contrary, RNase H1 overexpression appears to exacerbate DSBs, at least in the Mtr4 KDed cells. This data was however only observed by IF and its significance is not yet clear. While WBs would seem likely to be more quantitative than IF, we believe that the stress level (passage number, harvesting time, etc.) and/or cell cycle might have a significant impact on DSB accumulation. While we noticed that γH2AX levels can vary from one experiment to another, levels were always significantly higher after either NRDE-2 or Mtr4 KD. Nevertheless, our IF data showed that under certain circumstances, RNase H1 overexpression can trigger an increase in γH2AX signal in an Mtr4 KD background.
While we know that RNase H1 eliminates R loops, we also know that its overexpression or inhibition can have other effects [27], which include changes in gene expression and inhibition of HR-mediated DSB repair, at least in fission yeast [28,29]. It is thus possible that RNase H1 overexpression affects expression of genes involved in the DDR or interferes with repair in the Mtr4 KD background. The latter scenario implies that R loops might be required for repair of some of the DSBs triggered by Mtr4 KD. Another explanation would involve a role for Mtr4 in RNA-mediated HR repair [30]. While we cannot eliminate the last two possibilities, we find them unlikely because, as discussed below, Mtr4- and NRDE-2-interacting proteins favor a function for the complex in NHEJ rather than HR. It has however been shown in budding yeast that RNA can be used as a homologous template to repair DSBs [31]. Considering this scenario, RNase H1 overexpression could interfere with the repair process, which could explain the observed γH2AX signal increase in the Mtr4 KD background. However, despite the evidence that RNA oligos can function in repair of DSBs in humans [32], this kind of HR repair mechanism needs to be investigated more thoroughly.
How might Mtr4/NRDE-2 function in DNA repair? In S. pombe, it was proposed that the R loops formed in nrl1Δ strains sequester HR factors that are then no longer available to repair damage, leading to an increase in DSBs [8]. While Nrl1 has not been found to interact with HR factors, we found that NRDE-2 and Mtr4 both associated with several DDR factors, including PARP1, Ku70 and Ku80 as well as many histones and chromatin remodeling factors. The heterodimer Ku is part of the NHEJ DSB repair machinery, functioning by recognizing the breaks [33]. PARP1 is involved in numerous aspects of the DDR and functions in both the NHEJ and HR pathways [34]. Thus, it appears that while NRDE-2 and Nrl1 both function in the DDR, they likely do so by distinct mechanisms. We suggest that DSBs occurring after NRDE-2 and Mtr4 KDs result from a defect in the NHEJ repair process due to a lack of break recognition by the heterodimer Ku70/80 and/or PARP1 we found interacting with both proteins (Figure 5). The interaction with histones and chromatin remodeling factors make perfect sense in that scenario since their presence and particular organization at DNA damage sites provides the environment necessary for efficient and proper repair [35].
In C. elegans, NRDE-2 is essential for the nuclear RNAi pathway, which involves siRNA-directed H3K9me. NRDE-2 is directed to nascent transcripts through association with an siRNA-incorporated Argonaut (AGO) protein, NRDE-3, to introduce H3K9 methylation and block RNAP II elongation [7]. Although no RNAi factors were identified in the NRDE-2 (Nrl1)-proteomics analysis performed by us (human) and others (yeast), RNAi factor-directed introduction of H3K9me is observed in both organisms [15,36]. Notably, there are interconnections between R loops and the modification status of chromatin (reviewed by [37,38]). In human cells, R loops formed over RNAP II pause-site termination regions induce antisense transcription and consequent double-stranded RNA generation, which recruits RNAi factors such as DICER, AGOs, and the histone methyltransferase G9a to direct H3K9me at transcriptional termination sites [15]. In S. pombe, depletion of Nrl1 selectively abrogates RNAi-dependent heterochromatin assembly by decreasing H3K9me at HOODs [6]. The authors further suggested that RNAi-mediated heterochromatin assembly is defined by cryptic introns within HOODs and the spliceosome, since deletion of the cryptic introns or splicing factors resulted in reduced H3K9me3 levels [6]. Considering that splicing factor deficiency causes R-loop stabilization [23], it is possible that R-loop structures are enriched at specific loci such as HOODs in Nrl1-depleted cells and that H3K9 methylation is coupled with their resolution. It is then interesting to speculate that NRDE-2 is involved in R-loop resolution and H3K9 methylation at transcriptional termination sites, since NRDE-2 KD leads to an increase of R loops at the 3ʹend of RPL13A and BACT.
Several studies have suggested that changes in NRDE-2 expression could be involved in human health and disease. Array-based genome-wide copy number aberration analyses have suggested that NRDE-2 (14q32.11) haploinsufficiency can be associated with schizophrenia [39] and can be observed in circulating tumor cells that have been detected in the blood of patients with metastatic melanoma [40]. Moreover, an intronic single-nucleotide polymorphism (SNP) in NRDE-2 (refSNP Cluster Report ID: rs4904670) is associated with reduced lifespan, physiological aging changes, and major diseases like cancer and coronary heart disease [41]. Future analyses may uncover the possible link between the function of NRDE-2 and/or Mtr4/NRDE-2 and disease.
We identified here yet another complex containing Mtr4 that surprisingly has a function distinct from RNA surveillance. While our data reveal an R-loop independent role of the Mtr4/NRDE-2 complex in the DDR, each protein seems to individually (or part of two different complexes) affect R-loop levels at specific and distinct genes. It will be of great interest in the near future not only to dissect the molecular mechanisms that allow the Mtr4/NRDE-2 complex to protect the cell from DNA damage, but also to understand the independent role each protein plays at specific genes where R loops form. Our work shows that Mtr4 has an even broader role than expected from previous studies. In fact, it is very likely, according to our MS data, that the Mtr4/NRDE-2 complex participates in many other aspects of cell physiology through its interaction with subunits of the proteasome and cytoskeleton proteins (Figure 5).
4. Materials and methods
4.1. Gel filtration
Superose 6 gel filtration was performed as previously described [3] except that cell lysates were treated with >250 U/mL Benzonase (Sigma: #E1014) and 10 µg/mL RNase A (Sigma: #R5250) and that eluates were collected every 5 min (1 mL, flow rate = 0.2 mL/min).
4.2. Co-immunoprecipitation (co-IP) and mass spectrometry (MS)
co-IP using FLAG antibody and MS analysis were performed as previously described [3] except that HEK293 and HEK293/NRDE-2-3FLAG cells were lysed in 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40, 1x protease inhibitor cocktail (Biotools), 10μg/mL RNase A (Sigma: #R5250) and >250U/mL Benzonase (Sigma: #E1014).
4.3. Immunofluorescence (IF)
γH2AX IF were performed with HeLa cells after siRNAs transfection at 20 nM with RNAiMAX (Invitrogen: #13778) for 72h, methanol fixation for 10 mins and permeabilization with acetone for 1 min. FLAG IF in HEK293 cells were performed after fixation with 4% paraformaldehyde for 10 mins followed by fixation with 0.5% Triton X 100 for 10 mins. γH2AX antibody (Cell signaling Technology: #2577S) was used at 1:200 and anti-FLAG (Sigma: #F1804) at 1:250 for 2h. Secondary anti-rabbit Alexa 488 (Invitrogen: #A11008) and anti-mouse Alexa 568 (#A11031) at 1:500 for 1h at RT. Images were acquired using Zeiss LSM 700 confocal microscope and 20x/1.4 and 40x/1.4 oil objectives were used. Nuclear immunofluorescence signals were analyzed with ImageJ. 200 ng of GFP-RNase H1 and empty plasmid was transfected with Lipofectamine 2000 (Invitrogen: #11668) 24h after siRNA transfection for 48h.
4.4. siRNAs transfection and Western Blots
HeLa cells were not transfected (Ctrl) or transfected with an siRNA control (NC: TTCTCCGAACGTGTCACGT, GenePharma), siNRDE-2 (GCAAGCAGGUUGAACGCUA), siNRDE-2#2 (ID: S30064, Cat#: 4,427,037, Thermofisher), siMtr4 (CAAUUAAGGCUCUGAGUAA, GenePharma) for 72h at 20 nM with RNAiMAX (Invitrogen: #13778) prior extracts. The following antibodies were used: γH2AX (Cell signaling Technology: #2577S), U2AF65 (Sigma: #U4758), NRDE-2 (proteintech: #24968-I-AP), Mtr4 (Bethyl: #A300-614A), GFP (abm: #G095), GAPDH (Sigma: #G9545), FLAG (Sigma: #F1804). Western Blot quantifications were performed using ImageJ.
4.5. DRIP assay and qPCR
DRIP was performed as described in Ginno et al. [26]. Briefly, HeLa cells transfected with 20 nM siRNAs for 72h were lysed O/N in SDS and proteinase K. 50 μl of extracted gDNA was then digested O/N with a cocktail of restriction enzymes at 40 U each (HindIII/EcoRI/BsrGI/XbaI/SspI). After phenol/chloroform extraction and EtOH precipitation, 4.4 μg of digested DNA was treated O/N with 3 μl of Ribonuclease H (NEB, #M0297) as negative control. Treated and un-treated DNA were IPed O/N at 4°C with 10 μg of S9.6 antibody in binding buffer (10 mM NaPO4 pH 7.0, 140 mM NaCl, 0.5% Triton X-100). Next day, agarose A/G beads (Pierce #20421) were added for 2 hours. After washes, IPs were eluted at 55°C for 45 min in 250 μl elution buffer (50 mM Tris pH 8.0, 10 mM EDTA, 0.5% SDS) supplemented with 7 μl of Proteinase K at 20 mg/ml. DNA was analyzed by qPCR using the following primers: RPL13A (3ʹ of the gene): F: AGGTGCCTTGCTCACAGAGT, R: GGTTGCATTGCCCTCATTAC, βactin (5ʹ pause site): F: TTACCCAGAGTGCAGGTGTG, R: CCCCAATAAGCAGGAACAGA, EGR1 (intergenic region downstream of EGR1): F: GAACGTTCAGCCTCGTTCTC, R: GGAAGGTGGAAGGAAACACA.
The primers used to detect ptRNAs, full-length mRNA and proRBM39 after NRDE-2 KD are described in [3]. Primers for eRNA17- are the same as the ones used for ChIP.
4.6. ChIP assay
A confluent 10 cm dish of HeLa cells were cross-linked in 1% formaldehyde for 10 minutes at room temperature (RT). Cross-links were quenched in 125 mM glycine for 5 mins, cells were rinsed in PBS and then collected by centrifugation (500xg for 5 mins). Cells were collected in 400 μl RIPA buffer (150 mM NaCl, 1% NP40, 0.5% DOC, 0.1% SDS, 50 mM Tris.Cl pH8, 5 mM EDTA pH 8) supplemented with PMSF and a protease inhibitor cocktail (0.2 mM Pepstatin A, 72 μM Leupeptin, 26 μM Aprotinin) and sonicated (30s x 10 times using a bioruptor sonicator). Extracts were clarified by centrifugation at 13000xg for 15 mins. NRDE-2 ChIP was performed O/N at 4°C with 2 μg of antibody (proteintech: #24968-I-AP) and 20 μl of protein A/G sepharose (Invitrogen). No antibody control was performed in parallel. Beads were washed 3x in RIPA buffer without SDS. Immune complexes were eluted in 0.1M NaHCO3/1%SDS (15 mins rotation at RT). Cross-links were reversed for 5 hours at 65°C (250mM NaCl and 1μg RNase A). DNA was purified using QIAquick PCR purification kit (#28106) from Qiagen. Generally, 1/50th was used for each q-PCR reaction. The primers used for DAP int PAS, TMED4 int PAS and proRBM39 detection are described in [3]. eRNA17- and CSTF3 primers are (F: GAGCCATGGATGGGTGATAA, R: AACCCATCTTGTCAGGCAGA) and (F: ACTGATTTGGGGGTGGTTTT, R: GGCCTCAGCTGATTACAACG), respectively.
Funding Statement
This work was supported by the Muscular Dystrophy Association [MDA 377780];National Institutes of Health [P41 GM 103533];National Institutes of Health [R35 GM 118136];
Acknowledgments
We thank Robert Crouch for the GFP-RNase H1 plasmid, Frédéric Chédin and Lionel Sanz for the S9.6 antibody and for their insightful advices and comments to perform DRIP assay. We also thank Juan Irizarry-Cole for his help with the IF quantifications.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- 1.Ogami K, Chen Y, Manley JL.. RNA surveillance by the nuclear RNA exosome: mechanisms and significance. Non-Coding RNA. 2018. March 11;4(1):8 PubMed Central PMCID: PMC5886371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lubas M, Christensen MS, Kristiansen MS, et al. Interaction profiling identifies the human nuclear exosome targeting complex. Mol Cell. 2011. August 19;43(4):624–637. PubMed PMID: 21855801. [DOI] [PubMed] [Google Scholar]
- 3.Ogami K, Richard P, Chen Y, et al. An Mtr4/ZFC3H1 complex facilitates turnover of unstable nuclear RNAs to prevent their cytoplasmic transport and global translational repression. Genes Dev. 2017. July 21 PubMed PMID: 28733371; PubMed Central PMCID: PMC5558927 DOI: 10.1101/gad.302604.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meola N, Domanski M, Karadoulama E, et al. Identification of a nuclear exosome decay pathway for processed transcripts. Mol Cell. 2016. November 3;64(3):520–533. PubMed PMID: 27871484. [DOI] [PubMed] [Google Scholar]
- 5.Yamanaka S, Mehta S, Reyes-Turcu FE, et al. RNAi triggered by specialized machinery silences developmental genes and retrotransposons. Nature. 2013. January 24;493(7433):557–560. PubMed PMID: 23151475; PubMed Central PMCID: PMC3554839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee NN, Chalamcharla VR, Reyes-Turcu F, et al. Mtr4-like protein coordinates nuclear RNA processing for heterochromatin assembly and for telomere maintenance. Cell. 2013. November 21;155(5):1061–1074. PubMed PMID: 24210919; PubMed Central PMCID: PMC3974623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guang S, Bochner AF, Burkhart KB, et al. Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature. 2010. June 24;465(7301):1097–1101. PubMed PMID: 20543824; PubMed Central PMCID: PMC2892551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aronica L, Kasparek T, Ruchman D, et al. The spliceosome-associated protein Nrl1 suppresses homologous recombination-dependent R-loop formation in fission yeast. Nucleic Acids Res. 2016. February 29;44(4):1703–1717. PubMed PMID: 26682798; PubMed Central PMCID: PMC4770224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Preker PJ, Keller W. The HAT helix, a repetitive motif implicated in RNA processing. Trends Biochem Sci. 1998. January;23(1):15–16. PubMed PMID: 9478129. [DOI] [PubMed] [Google Scholar]
- 10.Sanz LA, Hartono SR, Lim YW, et al. Prevalent, dynamic, and conserved R-loop structures associate with specific epigenomic signatures in mammals. Mol Cell. 2016. June 29 PubMed PMID: 27373332 DOI: 10.1016/j.molcel.2016.05.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu K, Chedin F, Hsieh CL, et al. R-loops at immunoglobulin class switch regions in the chromosomes of stimulated B cells. Nat Immunol. 2003. May;4(5):442–451. PubMed PMID: 12679812. [DOI] [PubMed] [Google Scholar]
- 12.Xu B, Clayton DA. RNA-DNA hybrid formation at the human mitochondrial heavy-strand origin ceases at replication start sites: an implication for RNA-DNA hybrids serving as primers. The EMBO Journal. 1996. June 17;15(12):3135–3143. PubMed PMID: 8670814; PubMed Central PMCID: PMC450256. [PMC free article] [PubMed] [Google Scholar]
- 13.Pohjoismaki JL, Holmes JB, Wood SR, et al. Mammalian mitochondrial DNA replication intermediates are essentially duplex but contain extensive tracts of RNA/DNA hybrid. J Mol Biol. 2010. April 16;397(5):1144–1155. PubMed PMID: 20184890; PubMed Central PMCID: PMC2857715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Skourti-Stathaki K, Proudfoot NJ, Gromak N. Human senataxin resolves RNA/DNA hybrids formed at transcriptional pause sites to promote Xrn2-dependent termination. Mol Cell. 2011. June 24;42(6):794–805. PubMed PMID: 21700224; PubMed Central PMCID: PMC3145960. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Skourti-Stathaki K, Kamieniarz-Gdula K, Proudfoot NJ. R-loops induce repressive chromatin marks over mammalian gene terminators. Nature. 2014. October 5 PubMed PMID: 25296254 DOI: 10.1038/nature13787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguilera A, Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol Cell. 2012. April 27;46(2):115–124. PubMed PMID: 22541554; eng. [DOI] [PubMed] [Google Scholar]
- 17.Chan YA, Hieter P, Stirling PC. Mechanisms of genome instability induced by RNA-processing defects. Trends Genet. 2014. June;30(6):245–253. PubMed PMID: 24794811; PubMed Central PMCID: PMC4039741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richard P, Manley JL. R loops and links to human disease. J Mol Biol. 2016. September 4 PubMed PMID: 27600412; PubMed Central PMCID: PMC5478472 DOI: 10.1016/j.jmb.2016.08.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groh M, Gromak N. Out of balance: R-loops in human disease. PLoS Genet. 2014. September;10(9):e1004630 PubMed PMID: 25233079; PubMed Central PMCID: PMC4169248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anindya R, Aygun O, Svejstrup JQ. Damage-induced ubiquitylation of human RNA polymerase II by the ubiquitin ligase Nedd4, but not Cockayne syndrome proteins or BRCA1. Mol Cell. 2007. November 9;28(3):386–397. PubMed PMID: 17996703. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y, Zhu J, Schermann G, et al. The fission yeast MTREC complex targets CUTs and unspliced pre-mRNAs to the nuclear exosome. Nat Commun. 2015. May 20;6:7050 PubMed PMID: 25989903; PubMed Central PMCID: PMC4455066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015. January;43(Database issue):D447–D452. PubMed PMID: 25352553; PubMed Central PMCID: PMC4383874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Manley JL. Inactivation of the SR protein splicing factor ASF/SF2 results in genomic instability. Cell. 2005. August 12;122(3):365–378. PubMed PMID: 16096057; eng. [DOI] [PubMed] [Google Scholar]
- 24.Lim J, Giri PK, Kazadi D, et al. Nuclear proximity of Mtr4 to RNA exosome restricts DNA mutational asymmetry. Cell. 2017. April 20;169(3):523–537 e15. PubMed PMID: 28431250; PubMed Central PMCID: PMC5515252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boguslawski SJ, Smith DE, Michalak MA, et al. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J Immunol Methods. 1986. May 1;89(1):123–130. PubMed PMID: 2422282. [DOI] [PubMed] [Google Scholar]
- 26.Ginno PA, Lott PL, Christensen HC, et al. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012. March 30;45(6):814–825. PubMed PMID: 22387027; PubMed Central PMCID: PMC3319272. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vanoosthuyse V. Strengths and weaknesses of the current strategies to map and characterize R-loops. Non-Coding RNA. 2018;4(2):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hartono SR, Malapert A, Legros P, et al. The affinity of the S9.6 antibody for double-stranded RNAs impacts the accurate mapping of R-loops in fission yeast. J Mol Biol. 2017. December 28 PubMed PMID: 29289567 DOI: 10.1016/j.jmb.2017.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohle C, Tesorero R, Schermann G, et al. Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell. 2016. November 3;167(4):1001–1013 e7. PubMed PMID: 27881299. [DOI] [PubMed] [Google Scholar]
- 30.Meers C, Keskin H, Storici F. DNA repair by RNA: templated, or not templated, that is the question. DNA Repair. 2016. August;44:17–21. PubMed PMID: 27237587; PubMed Central PMCID: PMC4958532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keskin H, Shen Y, Huang F, et al. Transcript-RNA-templated DNA recombination and repair. Nature. 2014. November 20;515(7527):436–439. PubMed PMID: 25186730; PubMed Central PMCID: PMC4899968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shen Y, Nandi P, Taylor MB, et al. RNA-driven genetic changes in bacteria and in human cells. Mutat Res. 2011. December 1;717(1–2):91–98. PubMed PMID: 21515292. [DOI] [PubMed] [Google Scholar]
- 33.Mahaney BL, Meek K, Lees-Miller SP. Repair of ionizing radiation-induced DNA double-strand breaks by non-homologous end-joining. Biochem J. 2009. February 1;417:639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ray Chaudhuri A, Nussenzweig A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat Rev Mol Cell Biol. 2017. October;18(10):610–621. PubMed PMID: 28676700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Escargueil AE, Soares DG, Salvador M, et al. What histone code for DNA repair? Mutat Res. 2008. Mar-Apr;658(3):259–270. PubMed PMID: 18296106. [DOI] [PubMed] [Google Scholar]
- 36.Holoch D, Moazed D. RNAi in fission yeast finds new targets and new ways of targeting at the nuclear periphery. Genes Dev. 2012. April 15;26(8):741–745. PubMed PMID: 22508721; PubMed Central PMCID: PMC3337448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Santos-Pereira JM, Aguilera A. R loops: new modulators of genome dynamics and function. Nat Rev Genet. 2015. October;16(10):583–597. PubMed PMID: 26370899. [DOI] [PubMed] [Google Scholar]
- 38.Chedin F. Nascent connections: R-loops and chromatin patterning. Trends Genet. 2016. December;32(12):828–838. PubMed PMID: 27793359; PubMed Central PMCID: PMC5123964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maiti S, Kumar KH, Castellani CA, et al. Ontogenetic de novo copy number variations (CNVs) as a source of genetic individuality: studies on two families with MZD twins for schizophrenia. PLoS One. 2011. March 2;6(3):e17125 PubMed PMID: 21399695; PubMed Central PMCID: PMC3047561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chiu CG, Nakamura Y, Chong KK, et al. Genome-wide characterization of circulating tumor cells identifies novel prognostic genomic alterations in systemic melanoma metastasis. Clin Chem. 2014. June;60(6):873–885. PubMed PMID: 24718909. [DOI] [PubMed] [Google Scholar]
- 41.Yashin AI, Wu D, Arbeeva LS, et al. Genetics of aging, health, and survival: dynamic regulation of human longevity related traits. Front Genet. 2015;6:122 PubMed PMID: 25918517; PubMed Central PMCID: PMC4394697. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


