ABSTRACT
DNA harbors the blueprint for life. However, the instructions stored in the DNA could be altered at the RNA level before they are executed. One of these processes is RNA editing, which was shown to modify RNA sequences in many organisms. The most abundant modification is the deamination of adenosine (A) into inosine (I). In turn, inosine can be identified as a guanosine (G) by the ribosome and other cellular machineries such as reverse transcriptase. In multicellular organisms, enzymes from the ADAR (adenosine deaminase acting on RNA) family mediate RNA editing in mRNA, whereas enzymes from the ADAT family mediate A-to-I editing on tRNAs. In bacteria however, until recently, only one editing site was described, in tRNAArg, but never in mRNA. The tRNA site was shown to be modified by tadA (tRNA specific adenosine deaminase) which is believed to be the ancestral enzyme for the RNA editing family of enzymes. In our recent work, we have shown for the first time, editing on multiple sites in bacterial mRNAs and identified tadA as the enzyme responsible for this editing activity. Focusing on one of the identified targets – the self-killing toxin hokB, we found that editing is physiologically regulated and that it increases protein activity. Here we discuss possible modes of regulation on hokB editing, potential roles of RNA editing in bacteria, possible implications, and future research directions.
KEYWORDS: RNA editing, bacteria, non-genetic variation, hokB, toxin-antitoxin, persistence, antibiotic tolerance, tadA, ADAT, ADAR
1. Introduction
RNA editing is a post-transcriptional process that results in RNA sequences that differ from the DNA sequence from which they are transcribed. Adenosine (A) to Inosine (I) RNA editing was observed in all domains of life [1]. Since inosine can be identified by the translation and genetic machineries (e.g., reverse transcriptase) as a guanosine (G) it has the potential to recode protein sequences [2,3]. In mammalians, millions of editing sites were identified, mainly in untranslated regions, while only a small fraction occurs in coding regions and results in protein recoding [4]. However, recently the full potential of A-to-I editing to massively recode the proteome was nicely demonstrated in Chepalopods such as squid, in which editing results in recoding of multiple proteins in their nervous system [5]. In metazoans, A-to-I editing on mRNAs was shown to be mediated by the ADAR family of enzymes [1,6]. Unlike eukaryotes, until recently editing on bacterial mRNAs was not known to occur. In fact, the only editing event known in bacterial RNA was an A-to-I editing on the wobble position of the ACG anticodon in the tRNAArg. This site is modified by the tRNA specific adenosine deaminase (tadA) enzyme, a homolog of the ADAT family which are responsible for A-to-I tRNA editing in metazoans. As the name of that enzyme indicates, tRNAArg was thought to be its sole target. The enzymes that mediate A-to-I RNA editing in metazoans’ mRNA belong to the ADAR (Adenosine deaminase acting on RNA) family. However, ADARs are not encoded in bacterial genomes. Thus, the accepted notion until recently was that there is no machinery in place allowing mRNA editing in bacteria.
High throughput sequencing has revolutionized the field of RNA editing research, enabling the discovery of RNA modifications and edited sites both in the nucleus [4,7–10] as well as in cellular organelles [11–14]. Therefore, it was surprising that until recently RNA editing in the entire domain of bacteria was reported to occur in only one nucleotide site in the entire bacterial transcriptome.
In our recent work, we have identified for the first time mRNA editing events in bacteria. We described 15 novel A-to-I events, 12 of which in coding regions [15]. In all 12 mRNAs sites, editing was predicted to recode a tyrosine codon (TAC) into a cysteine codon (TGC). Furthermore, by gene perturbation, we have demonstrated that tadA is the enzyme that introduces these editing events. Interestingly, 4 out of the 15 targets were toxins that belong to the Hok (Host-Killing) toxin – antitoxin family. We then focused on one family member, hokB, that displayed the highest editing levels (average of 76%, second only to tRNAArg). hokB encodes for a toxin that was shown to contribute to persistence and antibiotic tolerance by restricting growth of the small sub-population of bacterial cells that produce it [16,17]. Examining other toxins belonging to the hok family of toxins as well as hokB genes in diverse bacterial species revealed a reoccurring theme. Most genes contain one position encoding a cysteine codon and another position encoded by a particular tyrosine codon (TAC) that can be converted into cysteine codon upon editing. Thus, although the identity of the edited position in hokB can be altered in different bacteria or different hokB family members, the correlated pattern in which one cysteine is hardcoded in the DNA and the other can be generated via editing recurs in evolution. Furthermore, we also detected RNA editing in hokB in two additional pathogenic bacterial species.
These observations motivated us to ask what is the functional importance of RNA editing in hokB. We found that RNA editing increases as function of culture density suggesting it is physiologically regulated. The conserved pattern described above, together with the increase in editing levels during cultural growth, suggest that this ‘soft’ coding of cysteine maybe important for the toxin activity. Such regulation could allow dynamic regulation of the toxin activity or stability perhaps via regulation on di-sulfide bond formation with another cysteine. Supporting this notion was the observation that converting the editable tyrosine codon of hokB to a hardcoded cysteine codon increases hokB’s toxicity. Our work was the first to describe mRNA editing in bacteria and it unveils tadA as an editing enzyme that acts both on tRNA and mRNA, similarly, to TRUB1 in eukaryotes that was recently shown to introduce pseudouridine both on mRNA and tRNA [18]. Below, we discuss how mRNA editing in bacteria can be regulated, what could be the functional implication of toxic editing in bacteria and what steps should be taken to unveil the full potential and importance of RNA editing in bacteria.
2. Regulation of RNA editing in bacteria
In our work, we observed that hokB editing levels increased as a function of cellular density. An intriguing question is how this increase in editing levels is achieved and what regulates mRNAs and in particular hokB editing levels. The most straightforward possibility is that dynamic in editing levels is governed by coordinated changes in the editing enzyme’s (i.e., tadA) expression or activity. According to this hypothesis, tadA’s expression increases as a function of cell density allowing free units of the enzyme to edit mRNA targets. Another intriguing possibility is that mRNA editing levels and in particular hokB editing is governed by the competition between tadA’s different substrates i.e., tRNAArg and mRNAs, on the availability of the editing enzyme tadA (Figure 1). During logarithmic growth tRNAArg expression levels are expected to be very high. hokB’s expression on the other hand, is governed by the levels of the stringent response alarmon ppGpp. During logarithmic phase only small portion of the cells in a population are expected to reach a high enough ppGpp levels to instruct hokB expression. Thus, hokB levels are predicted to be much lower during this phase (at least at its beginning). As density increases, so do the levels of ppGpp, and thus hokB mRNA levels are expected to rise [17,19]. Maybe even more important is the fact that overall, the translation machinery, and as a part of it tRNA levels, are known to decrease upon amino acid starvation [20], one of the characteristics of stationary phase. Thus, this overall decrease in tRNA level frees tadA molecules to edit other substrates (Figure 1). In particular the decline in the tRNA level, concomitant with the increase in hokB’s level shift the balance between tRNAArg/hokB-mRNA, thus enabling tadA to edit additional hokB mRNA copies. The effect of change in tRNA levels on mRNAs editing can be even more dramatic if one assumes different affinities between the different targets and the editing enzyme (Figure 1). This hypothesis is supported by the finding that tadA requires at least a 4-base motif (TACG) embedded within the anti-codon loop of tRNAArg [23,24], and was shown to directly interact with 7 base pairs within the anticodon loop of the tRNAArg. As we discuss in our original paper, differences in loop size or surrounding elements in the identified mRNA sites might affect tadA’s affinity. The editing sites in both hokB and tRNAArg are predicted to be embedded within an RNA loop and surrounded with the same 7bp sequence. However, itself differs between the two targets and this difference, together with additional sequence information in the vicinity of the sites, may lead to different affinities to the editing enzyme. Thus, it might be that the tRNAArg/hokB-mRNA ratio together with the different affinities will dictate the editing levels in hokB (and other mRNA targets) (Figure 1).
Figure 1.
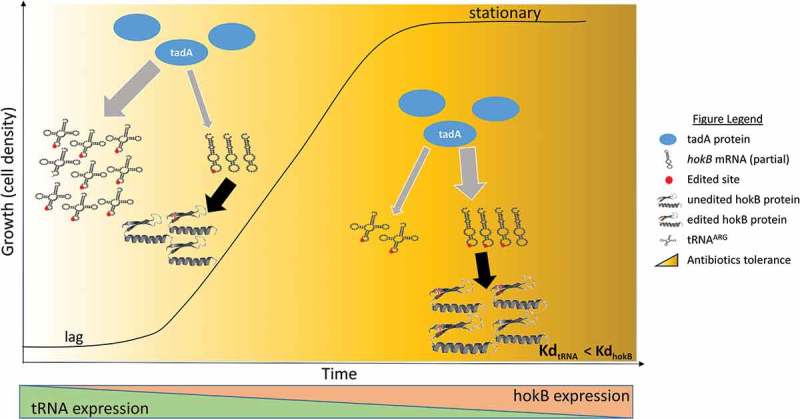
Hypothesized RNA editing regulation in bacteria. In lag and early log phase tRNAArg is abundant and therefore limits the available tadA units for hokB mRNA editing. As cell density increase (i.e., approaching to stationary phase), tRNAs are found in lower numbers, freeing tadA units to edit additional copies of hokB mRNA (as well as other mRNAs), making the toxin more effective (by recoding a tyrosine codon into a cysteine, presumably allowing the generation of an S-S bond), thus contributing to hokB mediated antibiotic tolerance. This scenario can be accompanied with different affinities of tadA to its tRNAArg/mRNA substrates (lower Kd = higher affinity). In parallel hokB expression is also elevated upon approaching stationary phase further increasing hokB activity and antibiotic tolerance. The tRNA and hokB predicted structures are as shown in our original paper [15] and were predicted using the Vienna RNA Websuite [21] and RaptorX [22].
Whatever are the principles that governs RNA editing in bacteria, the question is, how will it affect the cell, and maybe more intriguing, will it affect all cells in the same way?
3. RNA-mediated variation between and within individual cells
When we analyzed all the edited RNAs of E. coli, none of the sites displayed 100% editing levels, in fact we observed a wide variability in editing levels ranging from 1% to 87% (at mid log phase) [15]. Thus, looking at the population level, not all copies of the edited transcripts are in fact edited (at least at the time points examined). What can account for partial editing in a certain transcript observed in our experiment? It could be that during sequencing, we also captured the pre-edited version of the transcript(s) that would eventually be edited. Alternatively, it is possible that only a subset of the transcript’s copies within each cell will be edited during their lifetime. According to this possibility RNA editing diversifies the transcriptome within cells, as was previously reported in cephalopods [5]. Another interesting possibility is that the extent of RNA editing differs between cells, and it could thus contribute to non-genetic variation within isogenic populations (Figure 2). Consider a hypothetical measurement of 50% editing level of a certain transcript as measured in a population of cells. This observation could result from a whole range of possibilities. In one extreme case, in each of the cells, 50% of that transcript are edited. On the other extreme, in 50% of the cells all the copies of a transcript are edited, and in the other half of the cell there is no editing at all. Thus, editing can contribute both to within cells and between cell variability in the transcriptome (Figure 2). One way to test for the contribution of RNA editing to inter – or intra- individual variation is by single cell RNA sequencing. Unfortunately, to our knowledge, this method is not yet established for bacteria. In hokB’s case we have shown that editing enhances its toxicity. What could be the importance of inter-individual variation, if it exists, in the case of hokB editing?
Figure 2.
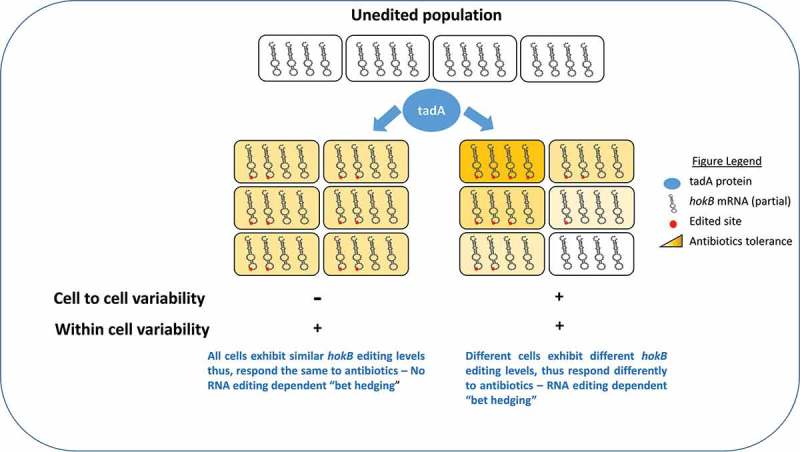
RNA editing in bacteria can diversify the transcriptome and contribute to ‘non-genetic’ cell to cell variability. RNA editing (marked here as a red dot) can be found in similar levels in bacterial cells (affecting some or all hokB transcripts within each cell term here ‘within cell variability’). Alternatively, it could differ from cell to cell (term here ‘cell to cell variability’), thus contributing a second level of variability (the first being hokB transcription). Such a scenario can produce different sub-populations of cells expressing hokB with different editing levels that will respond differently to antibiotics (i.e, RNA editing dependent ‘bet hedging’). Because RNA is extracted from populations of bacteria, both scenarios shown here, will display editing level of 50%.
4. Persistence and antibiotic tolerance regulation
One of the major problems in treating infectious bacterial diseases is the phenomenon of persistence that is characterized by the existence of sub-populations of bacteria that are tolerant to an antibiotic treatment [25]. Persistence-mediated antibiotic tolerance is sometimes thought of as a ‘bet-hedging’ strategy. Since many antibiotic drugs kill only dividing cells, a reduced growth could be a tolerance mechanism. Yet non-growing cells clearly reduce the fitness of the population at the absence of the drug. Thus, if a certain proportion of the cells in an isogenic population stochastically adopts a non-growing state they might survive an antibiotics attack when it arrives, thus trading off growth and survival. This strategy was shown to be mediated in some cases by the stochastic activity of many toxin-antitoxin systems [25]. hokB which is a part of a type I hokB/sokB toxin-antitoxin system is a potent toxin that can kill the cells expressing it, however it was shown that under certain scenarios i.e., exposure to antibiotics it can be beneficial. Intermediates levels of hokB expression were shown to result in membrane depolarization, which leads to sharp reduction in membrane potential and growth arrest but not to cell death [17]. Such growth arrest contributes to antibiotic tolerance through a mechanism of persistence. In a logarithmic population, expression of hokB could be governed by stochastic fluctuation of the second messenger alarmon ppGpp and only a tiny minority of the cells (~ 0.01%) express hokB [19]. Thus, hokB’s expression triggers growth arrest in a fraction of the cells in the culture, which in turn, are not sensitive to antibiotics agents if encountered and can resume growth and rescue the population upon antibiotic exposure. Can RNA editing further increase the cell-to-cell variability in hokB activity and add another level of stochasticity that can fine tune persistence and antibiotic tolerance? When we compared growth of bacterial cultures that express either unedited or edited versions of hokB we observed that editing enhances hokB’s toxicity. Thus, RNA editing regulates hokB’s toxicity levels post-transcriptionally. In addition, as discussed earlier, RNA editing could in theory promote non-genetic variation between cells, some with low and some with high hokB editing levels. Thus, in addition to the stochastic event that leads to random expression of hokB in a portion of the cells, cell-to-cell variability can also operate at the level of hokB’s editing in those cells that are expressing hokB (Figure 2). The combination of both random expression and variable editing level can further increase the heterogeneity in hokB’s activity in the population and further shape the persisters landscape. In other words, different cells will demonstrate growth arrest for different periods, allowing them to respond differently to the removal of the damaging agent (i.e., antibiotics). Therefore, RNA editing in hokB can fine-tune the balance between, growth arrest and antibiotic sensitivity and could thus contribute to the formation of a hokB RNA editing-dependent persistence spectrum (Figures 1 and 2).
This dynamics is especially interesting since hokB is also encoded in pathogenic species of bacteria. Indeed, we have shown that editing can be detected in hokB of Klebsiella pneumoniae and Yersinia enterocolitica. Therefore, RNA editing in bacteria could potentially have medical implications as a potential contributor to drug persistence. Since so far we only shown editing in three bacterial species, and in focusing on a one gene, future efforts should be made towards better cataloging RNA editing events in other species and identifying their functional implications. Having said that, what lesson can we learn now from RNA editing in bacteria in regard to our own editome? Could it be that tadA is also active in our cells in editing mRNAs?
5. tadA (ADAT) mRNA editing in multicellular organisms
Although the main focus of this paper is RNA editing in bacteria, it is noteworthy that all eukaryotic genomes encode a tadA homolog, called hetADAT. In humans, this heterodimeric enzyme is encoded by ADAT2 and ADAT3, the homologs of TAD2 and TAD3 in yeast [26,27]. In contrast to ADARs that recognize dsRNAs as their substrates, hetADAT and their tadA homologs in bacteria can identify only single stranded RNAs that reside in a loop structure. In metazoans hetADAT were shown to edit the adenosine at position 34 of the anticodon of eight tRNAs [26,27]. From evolutionary perspective, it is believed that the ADAR family has evolved from an ADAT ancestor, via the addition of a double-stranded RNA binding domain [6]. Currently the number of targets edited by ADAR in any transcriptome explored is much larger than those identified from ADAT. It is tempting to speculate that the acquisition of a double-stranded binding domain that change the RNA secondary structure preference of the enzyme also resolves the competition between editing tRNAs and mRNA, in a way that can free the ADAR enzyme to identify many new mRNA targets. In prokaryotes however, the same enzyme i.e., tadA, is responsible for both tRNA and mRNA editing. Given the high expression levels of tRNAs this may limit the enzyme activity on other targets. Nonetheless, the human transcriptome is much larger than its bacterial counterpart and many sites should correspond to the tRNA sequence and structure edited by hetADAT. It would be therefore exciting to test if hetADAT is also responsible for mRNA editing (or editing of additional types of RNAs beside tRNAs). The first step in such an analysis will be to search for editing sites in mRNAs harboring the edited tRNAs recognition sequence, as well as predicted secondary sequence. Such an analysis should be followed by complementary gene perturbation assays (e.g., RNAi and overexpression of hetADAT) to validate the authenticity of these sites. The alternative is that hetADAT do not edit mRNAs in metazoan and that this function is entirely reserved for the ADARs enzymes, and by doing so breaking the link between tRNA and mRNA editing.
6. Conclusion
Our work described A-to-I mRNA editing in bacteria for the first time and discovered that tadA edits both tRNAs and mRNAs. Future research should focus on unveiling other sites in more bacterial species. Additionally, we need to better understand how RNA editing in bacteria is regulated and what is its functional importance. This is especially true for RNA editing in hokB which could have biomedical implications due to potential effect on bacterial drug tolerance. Finally, the possibility that humans’/metazoans’ transcriptomes could be modified by hetADAT, like its bacterial counterpart, should be investigated. In conclusion, sequence variation at the RNA level in bacteria, and perhaps human as well, is more frequent and complex than previously thought.
Funding Statement
This work was supported by the Israel Science Foundation (ISF-1332/14); the Gruss Lipper Post Doctoral Fellowship; Minerva Center (AZ 5746940763).
Acknowledgments
We thank the EGL fellowship for supporting Dan Bar-Yaacov, the Minerva Foundation and the Israel Science Foundation for grant support.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- 1.Knoop V. When you can’t trust the DNA: RNA editing changes transcript sequences. Cell Mol Life Sci. 2011;68:567–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sommer B, Köhler M, Sprengel R, et al. RNA editing in brain controls a determinant of ion flow in glutamate-gated channels. Cell. 1991;67:11–19. [DOI] [PubMed] [Google Scholar]
- 4.Bazak L, Haviv A, Barak M, et al. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 2014;24:365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liscovitch-Brauer N, Alon S, Porath HT, et al. Trade-off between transcriptome plasticity and genome evolution in cephalopods. Cell. 2017;169(191–202):e111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grice LF, Degnan BM. The origin of the ADAR gene family and animal RNA editing. BMC Evol Biol. 2015;15:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramaswami G, Li JB. RADAR: a rigorously annotated database of A-to-I RNA editing. Nucleic Acids Res. 2013;42:996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu H, Wang Q, He Y, et al. Genome-wide A-to-I RNA editing in fungi independent of ADAR enzymes. Genome Res. 2016;26:499–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang IX, Grunseich C, Chung YG, et al. RNA–DNA sequence differences in Saccharomyces cerevisiae. Genome Res. 2016;26:1544–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz S, Bernstein DA, Mumbach MR, et al. Transcriptome-wide mapping reveals widespread dynamic-regulated pseudouridylation of ncRNA and mRNA. Cell. 2014;159:148–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bentolila S, Oh J, Hanson MR, et al. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 2013;9:e1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bar-Yaacov D, Avital G, Levin L, et al. RNA–DNA differences in human mitochondria restore ancestral form of 16S ribosomal RNA. Genome Res. 2013;23:1789–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bar-Yaacov D, Frumkin I, Yashiro Y, et al. Mitochondrial 16S rRNA Is methylated by tRNA methyltransferase TRMT61B in all vertebrates. PLoS Biol. 2016;14:e1002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oldenkott B, Yamaguchi K, Tsuji-Tsukinoki S, et al. Chloroplast RNA editing going extreme: more than 3400 events of C-to-U editing in the chloroplast transcriptome of the lycophyte Selaginella uncinata. Rna. 2014;20:1499–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bar-Yaacov D, Mordret E, Towers R, et al. RNA editing in bacteria recodes multiple proteins and regulates an evolutionarily conserved toxin-antitoxin system. Genome Res. 2017;27:1696–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pedersen K, Gerdes K. Multiple hok genes on the chromosome of Escherichia coli. Mol Microbiol. 1999;32:1090–1102. [DOI] [PubMed] [Google Scholar]
- 17.Verstraeten N, Knapen WJ, Kint CI, et al. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Mol Cell. 2015;59:9–21. [DOI] [PubMed] [Google Scholar]
- 18.Safra M, Nir R, Farouq D, et al. TRUB1 is the predominant pseudouridine synthase acting on mammalian mRNA via a predictable and conserved code. Genome Res. 2017;27:393–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdes K, Maisonneuve E. Remarkable functional convergence: alarmone ppGpp mediates persistence by activating type I and II toxin-antitoxins. Mol Cell. 2015;59:1–3. [DOI] [PubMed] [Google Scholar]
- 20.Svenningsen SL, Kongstad M, Stenum TS, et al. Transfer RNA is highly unstable during early amino acid starvation in Escherichia coli. Nucleic Acids Res. 2016;45:793–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruber AR, Lorenz R, Bernhart SH, et al. The vienna RNA websuite. Nucleic Acids Res. 2008;36:W70–W74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Källberg M, Wang H, Wang S, et al. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7:1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losey HC, Ruthenburg AJ, Verdine GL. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat Struct Mol Biol. 2006;13:153–159. [DOI] [PubMed] [Google Scholar]
- 24.Wolf J, Gerber AP, Keller W. tadA, an essential tRNA‐specific adenosine deaminase from Escherichia coli. EMBO J. 2002;21:3841–3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harms A, Maisonneuve E, Gerdes K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science. 2016;354. [DOI] [PubMed] [Google Scholar]
- 26.Torres AG, Piñeyro D, Rodríguez-Escribà M, et al. Inosine modifications in human tRNAs are incorporated at the precursor tRNA level. Nucleic Acids Res. 2015;43:5145–5157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gerber AP, Keller W. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science. 1999;286:1146–1149. [DOI] [PubMed] [Google Scholar]


