ABSTRACT
The total number of protein-coding genes in the human genome is not significantly higher than those in much simpler eukaryotes, despite a general increase in genome size proportionate to the organismal complexity. The large non-coding transcriptome and extensive differential splicing, are increasingly being accepted as the factors contributing to the complex mammalian physiology and architecture. Recent studies reveal additional layers of functional complexity: some long non-coding RNAs have been re-defined as micropeptide or microprotein encoding transcripts, and in turn some protein-coding RNAs are bifunctional and display also non-coding functions. Moreover, several protein-coding genes express long non-coding RNA splice-forms and generate circular RNAs in addition to their canonical mRNA transcripts, revoking the strict definition of a gene as coding or non-coding. In this mini review, we discuss the current understanding of these hybrid genes and their possible roles and relevance.
KEYWORDS: cncRNAs, lncRNAs, translation, splicing, smORF, non-coding RNA, ceRNA, circRNA
Introduction
The earlier understanding on the evolution of complexity in higher eukaryotes necessitated a proportionate increase in the number of protein-coding genes in the human genome compared to the much simpler organisms like worms and flies. Hence, the original discovery that the human genome hardly contains 30,000 protein-coding genes, not much more than found in the genome of the worm – Caenorhabditis elegans was perplexing [1]. An optimistic view at the time suggested that the low numbers could mean fast realization of the objectives of functional genomics and easy unravelling of all the complexities within a few years [2]. Notwithstanding the optimism in the wake of the 21st century, we still have a long way to go before completely understanding the gene-protein-function correlation and associated complexity of the Homo sapiens. These miscalculations originally arose due to the devaluation of the significance of the non-coding parts of the genome, often referring to them as junk DNA. It is interesting to note that a typical bacteria harbors protein-coding genes in approximately 90% of its genome, while greater than 95% of the human genome does not code for proteins. By current estimates, the short and long non-coding RNA genes in the human genome out-number protein-coding genes. The importance of tRNAs and rRNAs as decoders of the genetic code was evident. However, the other spectrum of the non-coding transcriptome is slowly being characterized. Novel functions of these short regulatory RNAs and the more cryptic long non-coding RNAs (lncRNAs) are constantly being discovered (reviewed in [3,4]). These non-coding transcripts could be the missing link in explaining the complexity achieved by mammals, despite having similar number of protein-coding genes to that in invertebrates. Roles for lncRNAs in complex processes unique to higher eukaryotes like neuronal development and cancer metastasis support this notion [5–7]. Alternative splicing is another factor contributing to the functional diversity and complexity of the proteome as well as the non-coding transcriptome [8,9]. Differential splicing can give rise to multiple mRNAs from a single gene, coding for polypeptides displaying distinct sequences and functions. Non-coding RNA genes also undergo extensive splicing and posttranscriptional processing [10]. The importance of splicing in mammalian physiology and pathology is well documented and reviewed elsewhere [11,12].
A deeper understanding of the genomic ‘dark matter’ and of the role of alternative splicing has brought us to the question whether genes can be strictly classified as coding and non-coding. Classification of genes into coding or non-coding is often determined based on their protein coding potential indicated by the presence of long, conserved and translatable open reading frames (ORFs). Since majority of annotated eukaryotic proteins are larger than 100 amino acids, RNAs which do not harbor ORFs longer than 300 nt were classified as non-coding. However, recent evidences indicate that a large number of lncRNAs harbor small ORFs (smORFs) and encode for short micropeptides [3,13]. Another more reliable approach for coding/non-coding distinction relies on the eventual biological function of the transcripts and depends on whether they need to be translated to protein to perform their functions or whether they act as RNA-effectors. The credibility of this mode of transcript annotation is also questioned by the discovery of the so-called cncRNAs (coding and non-coding RNAs) or bifunctional RNAs, wherein a protein-coding RNA additionally performs non-coding functions (reviewed in [14]). For example, independent of the tumor suppressor function of the p53 protein, p53-encoding mRNA directly binds and suppresses the ubiquitin ligase activity of MDM2 [15]. Thus, the emergence of cncRNAs are blurring the boundaries between the coding and non-coding transcriptome [16], (Figure1). Here, we will focus our attention to the subset of protein-coding genes, which in addition to mRNAs also express non-coding transcript variants, often as a result of alternative splicing [17]. We will refer to these genes as bifunctional or hybrid genes and will summarize the current understanding on these transcript variants and discuss their possible functions.
Figure 1.
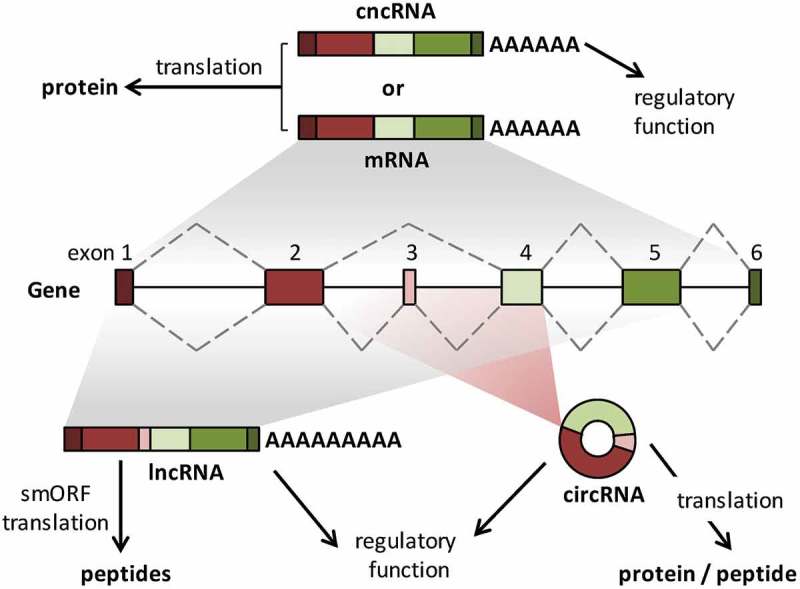
Can genes be strictly classified as coding and non-coding? Gene with multiple exons can be transcribed and spliced to generate an mRNA, which is translated to protein, which determines it’s cellular function. In some cases, the mRNAs can additionally have regulatory functions independent of translation. These protein-coding RNAs are classified as cncRNAs (coding/non-coding RNAs). The same gene can also generate long non-coding transcripts by alternative splicing. These lncRNAs can be genuine "non-coding RNAs" with regulatory functions or they can harbor smORFs (small ORFs) which encode micro-peptides. Back-splicing can also generate circRNAs (circular RNAs) from several genes. While circRNAs are usually stable non-coding RNAs with regulatory function, recent evidences suggest that at least some of them could be translated.
Prevalence of non-coding transcript variants of protein-coding genes
Most of the mammalian genes express more than one transcript, often generated as a result of alternative splicing [9]. This is in addition to the compact arrangement of the gene loci where multiple genes overlap with each other in the same as well as opposite strands of the chromosome. Moreover, several miRNAs and most of the small nucleolar RNAs (snoRNAs) are parts of the intronic regions of protein-coding and non-coding genes and are processed from the pre-mRNA, pre or post splicing [18]. Despite the presence of multiple splice variants per gene, most of the human genes express a single major transcript variant [19]. Interestingly, in case of approximately 17% of the protein-coding genes, the most prominent transcript variant is non-coding. Most of the major non-coding transcripts are predominantly nuclear and some of these detected transcripts are splicing intermediates carrying intron remnants [19]. Non-coding variants mainly arise by the skipping of the first or last exon, potentially resulting in the loss of start or stop codons, respectively. At least a few of these predicted candidates are possible evolutionary anomalies, since the direct consequence of alternative splicing in some cases is the generation of perfect non-sense mediated decay (NMD) substrates, which are degraded constitutively [20]. Nevertheless, several genes in the GenBank possess well annotated long non-coding transcript variants, which show moderate expression levels and could thus be stable enough to perform non-coding functions.
In addition to lncRNAs, another class of stable ncRNAs which are generated as a product of pre-mRNA splicing is the circular RNAs (circRNAs) [21,22]. They are usually generated by the back-splicing of one or more exons from a pre-mRNA, resulting in an end-joined circular RNA molecule [23,24]. CircRNAs are expressed from thousands of human genes and at least in some cases, show expression higher than the corresponding linear isoforms [21,25,26]. While a large number of circRNAs are regulatory non-coding RNAs, some of them have protein coding potential, even though they do not undergo capping and poly-adenylation [27–29]. Thus much like in the case of lncRNAs, direct experimental verification is the only means of assessing the translatability of circRNAs. Recent investigations have unraveled the functional relevance and interesting regulatory mechanisms of at least a few of these lncRNA/circRNA-mRNA hybrid genes (Table 1).
Table 1.
A list of functionally characterized bifunctional genes expressing non-coding splice variants in addition to mRNA transcripts. Gene names and chromosomal locations of human genes are indicated. The lncRNA/circRNA and mRNA variants are mentioned with comments on their biological functions and related citations.
| Gene | Locus | Transcripts | Function | References |
|---|---|---|---|---|
| ASCC3 | 6q16.3 | ASCC3 ALE isoform | chromatin associated lncRNA | [32] |
| ASCC3 mRNA | encodes a DNA helicase involved in DNA repair | |||
| FOXO3 | 6q21 | circ-FOXO3 | scaffolds protein complexes and affects cell cycle progression and senescence | [75–77] |
| FOXO3 mRNA | encodes a forkhead family transcription factor with role in pro-apoptotic gene expression | |||
| HIPK3 | 11p13 | circHIPK3 | ceRNA for multiple miRNAs including miR-124 | [78] |
| HIPK3 mRNA | encodes homeodomain interacting protein kinase 3 | |||
| ITCH | 20q11.22 | circular RNA-ITCH | ceRNA for miR-7, miR-17 and miR-214, facilitating ITCH protein upregulation | [79,80] |
| ITCH mRNA | encodes a HECT-family E3-ubiquitin ligase | |||
| MBNL1 | 3q25.1 | circMBNL1 | directly binds MBNL1 protein and could regulate its own splicing | [48] |
| MBNL1 mRNA | encodes ‘muscleblind like splicing regulator-1ʹ, a C3H-type zinc finger protein involved in alternative splicing and generation of circMBNL1 | |||
| PABPN1 | 14q11.2 | circPABPN1 | sequestrates the RNA binding protein – ELAVL1/HuR, preventing PABPN1 mRNA translation | [59] |
| PABPN1 mRNA | encodes a nuclear poly(A)-binding protein | |||
| PPP1R10 | 6p21.33 | LncRNA-PNUTS | ceRNA for miR-205 | [41,42] |
| PNUTS mRNA | encodes regulatory subunit of protein phosphatase 1 | |||
| RASGEF1B | 4q21.21 | circRASGEF1B | is upregulated upon lipopolysaccharide stimulation and regulates ICAM-1 mRNA stability | [81] |
| RASGEF1B mRNA | guanine-nucleotide exchange factor for RAP2 GTPase | |||
| SRA1 | 5q31.3 | SRA1 ncRNA | transcriptional co-activation complexes | [34,37] |
| SRAP mRNA | encodes potential transcriptional repressor | |||
| UBAP2 | 9p13.3 | circUBAP2 | ceRNA for miR-143 | [82] |
| UBAP2 | encodes ubiquitin associated protein 2 |
Examples for functionally characterized hybrid genes transcribing mRNAs and lncRNAs/circRNA
Splicing is often coupled to transcriptional elongation and the speed of transcription affects the outcome of splicing, resulting in the inclusion of alternative exons or exon skipping [30]. Interestingly, UV irradiation induces alternative splicing by a mechanism which involves RNA Pol II hyperphosphorylation and subsequent slow-down of transcriptional elongation [31]. A recent study identified that the major splicing change induced by UV is the incorporation of alternative last exons (ALE) proximal to the transcriptional start site and the general shortening of the transcripts [32]. The ASCC3 gene encodes a helicase involved in DNA repair, which is expressed in the early stages of UV-induced DNA damage and participates in UV-induced transcriptional suppression [33]. UV induces the ALE switch of ASCC3 mRNA into a shorter lncRNA splice variant which participates in the transcription recovery in the later stages after UV-induced DNA damage [17,32]. This describes a unique case of conditional conversion of a protein-coding RNA into a non-coding RNA, perfectly synchronized with the temporal necessities of the DNA damage response.
One of the earliest and the best characterized human gene which expresses functional lncRNA splice variants in addition to protein-coding transcripts is SRA1 (steroid receptor RNA activator 1) [34]. Alternative splicing and the use of differential transcription start sites generates mRNAs and lncRNAs from this gene locus with variations at the 5` end [35]. While the lncRNA transcripts scaffold transcription co-activation complexes together with several nuclear receptors including the estrogen receptor and PPARγ, the mRNA-derived polypeptide (SRAP) is also a potential transcriptional regulator (reviewed in [36]). The preferential upregulation of the lncRNA isoform in invasive human breast cancer cell lines indicated a role for this splicing switch in malignancy, but later studies also identified high SRAP expression as a poor prognostic factor for survival in a subset of breast cancer patients [35,37]. Furthermore, the mouse homolog of SRA1 gene also displays similar gene architecture and expresses both coding and non-coding splice variants. Sra1−/- mice displayed defective adipogenesis, but it is not yet clear whether the loss of lncRNA or protein expression is crucial for this phenotype [38].
Protein Phosphatase 1 Nuclear Targeting Subunit (PNUTS or PPP1R10) was originally described as a protein-coding gene encoding a regulatory subunit of protein phosphatase-1 (PP1). PNUTS protein suppresses PP1 activity and participates in cell cycle regulation and DNA damage response [39–41]. PNUTS gene locus is highly conserved between human and mouse and expresses both coding and non-coding transcripts. In the mouse genome, lncRNA-PNUTS is generated by alternative splice site integration at the 5ʹ end of exon 12 and consequent generation of a premature stop codon [42]. Interestingly, siRNA mediated depletion of the RNA binding protein hnRNP E1 leads to a splicing switch resulting in the preferential expression of the lncRNA [42]. Moreover, actinomycin-D and cycloheximide, pharmacological modulators of transcription and translation, respectively, also alters the mRNA-lncRNA switch at this gene locus, indicating that this process is highly dynamic. LncRNA-PNUTS seems to regulate epithelial-to-mesenchymal transition (EMT) and cell migration by acting as a competing endogenous RNA (ceRNA) for miR-205, a master regulator of EMT-related transcription factors.
CircRNAs are resistant to exonuclease digestion, which has enabled the enrichment and sequencing analysis of this class of RNAs in several physiological and pathological settings [43]. However, there are only a handful of examples for functionally characterized mRNA-circRNA hybrid genes (Table 1).
General themes for the regulatory role of the non-coding transcript variants
The coding-to-non-coding switch in the case of ASCC3 and PNUTS certainly means that there could be many more inducible non-coding RNA expressing genes and the currently approximated number of lncRNAs could be well underestimated. SRA1 and ASCC3 genes represent interesting cases of regulatory complexity wherein the non-coding transcript variants of protein-coding genes function as genuine lncRNAs with scaffolding function attributable to their specific folding and structure. While there could be many such candidate lncRNA-mRNA hybrid genes and interesting regulatory mechanisms waiting to be discovered, some general themes can be applied to explain the functional relevance of this category of alternative splicing products. The major functions attributable to the non-coding transcripts arising from hybrid genes are (i) the regulation of splicing and mRNA processing (ii) miRNA sponge activity (iii) competitive RBP-binding/sequestration and (iv) micropeptide/microprotein expression (Figure 2).
Figure 2.
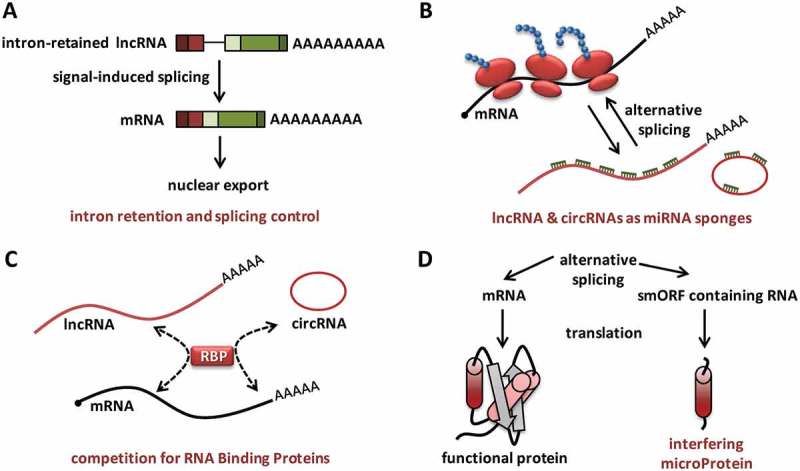
Roles of ``non-coding‘‘ variants arising from protein-coding genes. (a). Intron retention could lead to the generation of nuclear lncRNA intermediates which can undergo signal-induced posttranscriptional splicing to translatable mRNAs, which are then exported from the nucleus. (b). Some lncRNA transcript variants and circRNAs function as miRNA sponges, facilitating efficient translation of mRNAs. (c) lncRNAs and circRNAs compete with mRNAs for binding to regulatory RNA-binding proteins (RBPs), sequester RBPs and alter the translation and stability of mRNAs. (d). mRNAs and smORF-containing transcript variants could encode full-length proteins and truncated micropeptides respectively. Some micropeptides could function as miPs (microProteins) and interfere with full length protein function, owing to their common origins and sequence identity.
LncRNAs are active regulators of splicing and many antisense lncRNA transcripts participate in alternative splicing of their overlapping protein-coding RNAs [44]. The alternatively spliced non-coding variants could also be involved in the specific recruitment or sequestration of spliceosome factors and RNA binding proteins (RBPs). Major non-coding splice variants arise from the retention of introns [19]. Intron retention has been accepted as a mechanism of gene regulation in lower eukaryotes and plants, while its role is less understood in mammals [45,46]. One major consequence of intron retention is the nuclear accumulation of the resulting transcripts, which may undergo signal-induced posttranscriptional splicing [19,47], (Figure 2(a)). In addition, co-transcriptional back-splicing of circRNAs seems to modulate gene expression by competing with linear-splicing [48]. Interestingly, depletion of general splicing or transcription termination factors seems to enhance circRNA biogenesis from hybrid gene loci [49].
One of the most investigated roles of lncRNAs and circRNAs is their ability to function as ceRNAs or miRNA sponges [50–52]. The over-representation of ceRNA activity among functionally characterized lncRNAs/circRNAs is mainly because of the availability of sequence analysis algorithms which help in their prediction and relative ease with which one can prove such a function [53,54]. However, these ceRNAs are important for the complex fine-tuning of the proteome by modulating miRNA-mediated gene silencing [55]. Like the lncRNA-PNUTS, many of the lncRNA transcript variants could be potential ceRNAs (Figure 2(b)). One intriguing question regarding lncRNA-PNUTS left unanswered is the fact that its cognate mRNA does not function as a ceRNA, despite harboring the miR-205 binding sites. It could well be argued that the miRNA binding sites present within the coding region cannot efficiently perform the sponge function due to altered RNA secondary structure or ribosome occupancy. If this is true, the non-coding splice variants of coding transcripts could act as professional and specific miRNA scavengers. It is known that certain pseudogenes/lncRNAs impart miRNA resistance upon their functional protein-coding counterparts by acting as specific ceRNAs [56,57]. Due to their long half-lives, circRNAs are very efficient ceRNAs and there are many mRNA/circRNA expressing bifunctional genes, where the protein expression is fine-tuned by the cognate circRNA [52]. In a similar manner, the lncRNA splice variants may also act as suppressors of miRNA activity on their cognate mRNAs. Alternative splicing is known to modulate miRNA-dependent gene expression by generating transcripts with variable 3ʹ ends with and without miRNA binding sites [58]. The generation of splice forms which lack protein-coding potential could be an additional level of complexity.
The non-coding variants of hybrid genes may act as competitive interactors of RBPs, thus sequestering them from their target mRNAs (Figure 2(c)). Such regulation is relevant in the context of pre-mRNA splicing as well as mRNA translation and decay. For instance, binding of circPABN1 to ELAVL1/HuR prevents its binding to the linear PABN1 mRNA, resulting in the suppression of PABN1 translation [59]. Competitive RBP-binding by non-coding variants could also modulate the miRNA-RBP cross-talk [60]. In addition to circRNAs generated by simple back-splicing of exons, sequencing data also confirm the existence of exon-intron circular RNAs (EIciRNAs) and circular intronic RNAs (ciRNA) [61,62]. These intron-retained circRNAs promote transcription of their host genes in cis by associating with RBPs and RNA-pol II [58,59].
It is increasingly being accepted that a large proportion of transcripts annotated as lncRNAs harbor smORFs and express micropeptides which may be stable enough to be functional [63–65]. Majority of the lncRNA products from the bifunctional or hybrid genes could be of this category and may express micropeptides which are functionally related to their canonical mRNA encoded proteins. Translatable exonic-circRNAs also very often generate polypeptides which resemble truncated versions of the proteins translated from their cognate mRNA pair [66]. Analogous to microRNAs, microproteins (miPs) are defined as small (< 120 residues), single domain polypeptides, which exert a dominant negative function by heterodimerizing their homologous target proteins into non-functional protein complexes [67,68]. Many of the non-coding splice-forms originally predicted to be NMD targets may be actively translated, providing miPs which interfere with the biological functions of the full length proteins encoded by their mRNA splice variants (Figure 2(d)). The short ORF database (http://www.sorfs.org) lists close to 2 million smORFs in the human transcriptome [69]. Interestingly, the database generated by extensive filtering of ribosome profiling data, assigns half of these smORFs to genes which are already annotated as ‘’protein-coding’’. If this is true, this would mean that there are more than 1 million 10–100 residue long peptides in the human cells, which are generated by the alternatively spliced transcript variants. More importantly, around 200,000 of them show no overlap with other known ORFs, suggesting that they are unique polypeptides generated by the use of alternative exons and/or frames of translation.
Outlook
The general skepticism in accepting concepts which deviate from the central dogma of molecular biology was the main reason for ignoring the importance of the non-coding regions of the genome as ‘’junk DNA’’. We have come a long way forward and the non-coding transcripts are now receiving the attention which they deserve. However, the lack of conserved sequences/domains and limitations in the predictability of non-coding RNA functions are causes for concern. The predictability of function is the main reason why antisense transcripts and ceRNAs dominate the list of functional lncRNAs/circRNAs in the literature. This will be the same for the specific set of non-coding transcripts discussed here. Novel hybrid genes and inducible ncRNA-mRNA switches could be revealed by studies which couple functional screens with splice-form specific expression analysis. However, transcript variants with ceRNAs/miRNA sponge function will be easier to identify and investigate. In this context, it should be noted that there is general skepticism regarding the ceRNA hypothesis [70]. The ceRNA hypothesis states that pseudogenes, lncRNAs and circRNAs compete with mRNA targets for binding to miRNAs, thus suppressing miRNA function [71]. Many of the studies describing miRNA sponges in the past have relied on over-expression approaches and whether those lncRNAs can function as ceRNAs at physiologically relevant copy numbers is debatable. However functional endogenous miRNA sponges have been reported and large-scale studies have provided support to the existence of extensive ceRNA regulatory networks [52,55,72].
The distinction between the coding and non-coding transcriptome is slowly being blurred by the emergence of micropeptide encoding smORFs. The huge potential for the hybrid gene-derived lncRNAs in generating diverse micropeptides is underlined by the sORF database entries. Do these riboseq-derived smORFs really encode micropeptides? How many of them give rise to physiologically relevant micropeptides or miPs? Technical advancements in the specific and quantitative analysis of micropeptides will be key to filtering out the noise in these datasets. Another question is whether the transcripts predicted to be NMD substrates are genuine targets of NMD? Some RNAs which are indeed NMD targets could well have cancer-specific implications as NMD is known to be inhibited in cancer [73]. Translatability of the circRNAs is also a variable factor in the analysis and functional characterization of bifunctional genes. Methylation, presence of ORFs and additional structural elements like IRES (internal ribosome-entry sites) seems to regulate circRNA translation and algorithms have already been established to evaluate their translation potential [27–29,66,74]. In addition to the native mRNA-lncRNA and mRNA-circRNA hybrid genes, splice-site mutations in cancer can also potentially generate pro-oncogenic non-coding transcript variants. Identifying genuine hybrid genes and categorizing their potential non-coding transcripts into micropeptide encoders, potential ceRNAs and other classes will be the way forward.
Biography
Sonam Dhamija Both authors equally contributed to the drafting and revision of the manuscript and approved it for publication.
Acknowledgments
We thank Dr. Sven Diederichs (DKFZ, Heidelberg) and Dr. Matthias Gaestel (MHH, Hannover) for critical reading of the manuscript. Sonam Dhamija thanks the CellNetworks- Cluster of Excellence post-doctoral fellowship program, Heidelberg for support.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].Venter JC, Adams MD, Myers EW, et al. The sequence of the human genome. Science. 2001;291:1304–1351. [DOI] [PubMed] [Google Scholar]
- [2].Claverie JM. Gene number. What if there are only 30,000 human genes? Science. 2001;291:1255–1257. [DOI] [PubMed] [Google Scholar]
- [3].Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Biol. 2017;19:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155. [DOI] [PubMed] [Google Scholar]
- [5].Ramos AD, Andersen RE, Liu SJ, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell. 2015;16:439–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Dhamija S, Diederichs S. From junk to master regulators of invasion: lncRNA functions in migration, EMT and metastasis. Int J Cancer. 2016;139:269–280. [DOI] [PubMed] [Google Scholar]
- [7].Gutschner T, Hammerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liu Y, Gonzalez-Porta M, Santos S, et al. Impact of alternative splicing on the human proteome. Cell Rep. 2017;20:1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Kim E, Magen A, Ast G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 2007;35:125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Deveson IW, Brunck ME, Blackburn J, et al. Universal alternative splicing of noncoding exons. Cell Syst. 2018;6:245–55 e5. [DOI] [PubMed] [Google Scholar]
- [11].Baralle FE, Giudice J. Alternative splicing as a regulator of development and tissue identity. Nat Rev Mol Cell Biol. 2017;18:437–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Park E, Pan Z, Zhang Z, et al. The expanding landscape of alternative splicing variation in human populations. Am J Hum Genet. 2018;102:11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kondo T, Plaza S, Zanet J, et al. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. [DOI] [PubMed] [Google Scholar]
- [14].Sampath K, Ephrussi A. CncRNAs: rNAs with both coding and non-coding roles in development. Development. 2016;143:1234–1241. [DOI] [PubMed] [Google Scholar]
- [15].Candeias MM, Malbert-Colas L, Powell DJ, et al. P53 mRNA controls p53 activity by managing Mdm2 functions. Nat Cell Biol. 2008;10:1098–1105. [DOI] [PubMed] [Google Scholar]
- [16].Nam JW, Choi SW, You BH. Incredible RNA: dual functions of coding and noncoding. Mol Cells. 2016;39:367–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Szempruch A, Guttman M. Linking protein and RNA function within the same gene. Cell. 2017;168:753–755. [DOI] [PubMed] [Google Scholar]
- [18].Yang L. Splicing noncoding RNAs from the inside out. Wiley Interdiscip Rev RNA. 2015;6:651–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gonzalez-Porta M, Frankish A, Rung J, et al. Transcriptome analysis of human tissues and cell lines reveals one dominant transcript per gene. Genome Biol. 2013;14:R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100:189–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333. [DOI] [PubMed] [Google Scholar]
- [22].Enuka Y, Lauriola M, Feldman ME, et al. Circular RNAs are long-lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 2016;44:1370–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Schindewolf C, Braun S, Domdey H. In vitro generation of a circular exon from a linear pre-mRNA transcript. Nucleic Acids Res. 1996;24:1260–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep. 2015;10:103–111. [DOI] [PubMed] [Google Scholar]
- [25].Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19:141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Salzman J, Chen RE, Olsen MN, et al. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Yang Y, Fan X, Mao M, et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell Res. 2017;27:626–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Legnini I, Di Timoteo G, Rossi F, et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell. 2017;66:22–37 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pamudurti NR, Bartok O, Jens M, et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Dujardin G, Lafaille C, de la Mata M, et al. How slow RNA polymerase II elongation favors alternative exon skipping. Mol Cell. 2014;54:683–690. [DOI] [PubMed] [Google Scholar]
- [31].Munoz MJ, Perez Santangelo MS, Paronetto MP, et al. DNA damage regulates alternative splicing through inhibition of RNA polymerase II elongation. Cell. 2009;137:708–720. [DOI] [PubMed] [Google Scholar]
- [32].Williamson L, Saponaro M, Boeing S, et al. UV irradiation induces a non-coding RNA that functionally opposes the protein encoded by the same gene. Cell. 2017;168:843–55 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Boeing S, Williamson L, Encheva V, et al. Multiomic analysis of the UV-induced DNA damage response. Cell Rep. 2016;15:1597–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Kawashima H, Takano H, Sugita S, et al. A novel steroid receptor co-activator protein (SRAP) as an alternative form of steroid receptor RNA-activator gene: expression in prostate cancer cells and enhancement of androgen receptor activity. Biochem J. 2003;369:163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hube F, Guo J, Chooniedass-Kothari S, et al. Alternative splicing of the first intron of the steroid receptor RNA activator (SRA) participates in the generation of coding and noncoding RNA isoforms in breast cancer cell lines. DNA Cell Biol. 2006;25:418–428. [DOI] [PubMed] [Google Scholar]
- [36].Leygue E. Steroid receptor RNA activator (SRA1): unusual bifaceted gene products with suspected relevance to breast cancer. Nucl Recept Signal. 2007;5:e006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Yan Y, Skliris GP, Penner C, et al. Steroid Receptor RNA Activator Protein (SRAP): a potential new prognostic marker for estrogen receptor-positive/node-negative/younger breast cancer patients. Breast Cancer Res. 2009;11:R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu S, Sheng L, Miao H, et al. SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J Biol Chem. 2014;289:13000–13009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Allen PB, Kwon YG, Nairn AC, et al. Isolation and characterization of PNUTS, a putative protein phosphatase 1 nuclear targeting subunit. J Biol Chem. 1998;273:4089–4095. [DOI] [PubMed] [Google Scholar]
- [40].Landsverk HB, Kirkhus M, Bollen M, et al. PNUTS enhances in vitro chromosome decondensation in a PP1-dependent manner. Biochem J. 2005;390:709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Landsverk HB, Mora-Bermudez F, Landsverk OJ, et al. The protein phosphatase 1 regulator PNUTS is a new component of the DNA damage response. EMBO Rep. 2010;11:868–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Grelet S, Link LA, Howley B, et al. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat Cell Biol. 2017;19:1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Romero-Barrios N, Legascue MF, Benhamed M, et al. Splicing regulation by long noncoding RNAs. Nucleic Acids Res. 2018;46:2169–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Pleiss JA, Whitworth GB, Bergkessel M, et al. Rapid, transcript-specific changes in splicing in response to environmental stress. Mol Cell. 2007;27:928–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Jacob AG, Smith CWJ. Intron retention as a component of regulated gene expression programs. Hum Genet. 2017;136:1043–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mauger O, Lemoine F, Scheiffele P. Targeted intron retention and excision for rapid gene regulation in response to neuronal activity. Neuron. 2016;92:1266–1278. [DOI] [PubMed] [Google Scholar]
- [48].Ashwal-Fluss R, Meyer M, Pamudurti NR, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. [DOI] [PubMed] [Google Scholar]
- [49].Liang D, Tatomer DC, Luo Z, et al. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol Cell. 2017;68:940–54 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Du Z, Sun T, Hacisuleyman E, et al. Integrative analyses reveal a long noncoding RNA-mediated sponge regulatory network in prostate cancer. Nat Commun. 2016;7:10982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Paci P, Colombo T, Farina L. Computational analysis identifies a sponge interaction network between long non-coding RNAs and messenger RNAs in human breast cancer. BMC Syst Biol. 2014;8:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384. [DOI] [PubMed] [Google Scholar]
- [53].Furio-Tari P, Tarazona S, Gabaldon T, et al. spongeScan: A web for detecting microRNA binding elements in lncRNA sequences. Nucleic Acids Res. 2016;44:W176–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Militello G, Weirick T, John D, et al. Screening and validation of lncRNAs and circRNAs as miRNA sponges. Brief Bioinform. 2017;18:780–788. [DOI] [PubMed] [Google Scholar]
- [55].Chiu HS, Martinez MR, Bansal M, et al. High-throughput validation of ceRNA regulatory networks. BMC Genomics. 2017;18:418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Poliseno L, Salmena L, Zhang J, et al. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature. 2010;465:1033–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Wang J, Liu X, Wu H, et al. CREB up-regulates long non-coding RNA, HULC expression through interaction with microRNA-372 in liver cancer. Nucleic Acids Res. 2010;38:5366–5383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Han S, Kim D, Shivakumar M, et al. The effects of alternative splicing on miRNA binding sites in bladder cancer. PLoS One. 2018;13:e0190708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Abdelmohsen K, Panda AC, Munk R, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14:361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Suzuki HI, Spengler RM, Grigelioniene G, et al. Deconvolution of seed and RNA-binding protein crosstalk in RNAi-based functional genomics. Nat Genet. 2018;50:657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhang Y, Zhang XO, Chen T, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51:792–806. [DOI] [PubMed] [Google Scholar]
- [62].Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. [DOI] [PubMed] [Google Scholar]
- [63].Mackowiak SD, Zauber H, Bielow C, et al. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol. 2015;16:179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Couso JP, Patraquim P. Classification and function of small open reading frames. Nat Rev Mol Cell Biol. 2017;18:575–589. [DOI] [PubMed] [Google Scholar]
- [65].Polycarpou-Schwarz M, Gross M, Mestdagh P, et al. The cancer-associated microprotein CASIMO1 controls cell proliferation and interacts with squalene epoxidase modulating lipid droplet formation. Oncogene. Forthcoming 2018;37:4750–4768. [DOI] [PubMed] [Google Scholar]
- [66].Chen X, Han P, Zhou T, et al. circRNADb: A comprehensive database for human circular RNAs with protein-coding annotations. Sci Rep. 2016;6:34985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Straub D, Wenkel S. Cross-species genome-wide identification of evolutionary conserved MicroProteins. Genome Biol Evol. 2017;9:777–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Eguen T, Straub D, Graeff M, et al. MicroProteins: small size-big impact. Trends Plant Sci. 2015;20:477–482. [DOI] [PubMed] [Google Scholar]
- [69].Olexiouk V, Van Criekinge W, Menschaert G. An update on sORFs.org: a repository of small ORFs identified by ribosome profiling. Nucleic Acids Res. 2018;46:D497–D502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283. [DOI] [PubMed] [Google Scholar]
- [71].Salmena L, Poliseno L, Tay Y, et al. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Gilot D, Migault M, Bachelot L, et al. A non-coding function of TYRP1 mRNA promotes melanoma growth. Nat Cell Biol. 2017;19:1348–1357. [DOI] [PubMed] [Google Scholar]
- [73].Wang D, Zavadil J, Martin L, et al. Inhibition of nonsense-mediated RNA decay by the tumor microenvironment promotes tumorigenesis. Mol Cell Biol. 2011;31:3670–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Li X, Liu CX, Xue W, et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol Cell. 2017;67:214–27 e7. [DOI] [PubMed] [Google Scholar]
- [75].Du WW, Yang W, Liu E, et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yang W, Du WW, Li X, et al. Foxo3 activity promoted by non-coding effects of circular RNA and Foxo3 pseudogene in the inhibition of tumor growth and angiogenesis. Oncogene. 2016;35:3919–3931. [DOI] [PubMed] [Google Scholar]
- [77].Du WW, Yang W, Chen Y, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:1402–1412. [DOI] [PubMed] [Google Scholar]
- [78].Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget. 2015;6:6001–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Huang G, Zhu H, Shi Y, et al. cir-ITCH plays an inhibitory role in colorectal cancer by regulating the Wnt/beta-catenin pathway. PLoS One. 2015;10:e0131225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Ng WL, Marinov GK, Liau ES, et al. Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway. RNA Biol. 2016;13:861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Zhang H, Wang G, Ding C, et al. Increased circular RNA UBAP2 acts as a sponge of miR-143 to promote osteosarcoma progression. Oncotarget. 2017;8:61687–61697. [DOI] [PMC free article] [PubMed] [Google Scholar]


