ABSTRACT
In this article a novel mechanism of retrograde signaling by chloroplasts during stress is described. This mechanism involves the DNA/RNA binding protein WHIRLY1 as a regulator of microRNA levels. By virtue of its dual localization in chloroplasts and the nucleus of the same cell, WHIRLY1 was proposed as an excellent candidate coordinator of chloroplast function and nuclear gene expression. Comparison of wild-type and transgenic plants with an RNAi-mediated knockdown of WHIRLY1 showed, that the transgenic plants were unable to cope with continuous high light conditions. They were impaired in production of several microRNAs mediating post-transcriptional responses during stress. The results support a central role of WHIRLY1 in retrograde signaling and also underpin a so far underestimated role of microRNAs in this process.
KEYWORDS: Retrograde signaling, small RNA, stress response
WHIRLY1 belongs to a small plant specific family of DNA/RNA-binding proteins. By immunological methods, WHIRLY1 has been detected in chloroplasts and nucleus of the same cell [1]. Accordingly, functions of WHIRLY1 were reported for both compartments. In chloroplasts of barley, WHIRLY1 was shown to be the major compacting protein of nucleoids [2]. Moreover, WHIRLY1 has been found to bind to plastid RNAs [3,4]. In chloroplasts of Arabidopsis thaliana, WHIRLYs were reported to maintain plastid genome stability [5]. In the nucleus, WHIRLY1 was originally detected as a component of a transcriptional activator of the PR10a gene of potato [6]. Furthermore, it has been found to bind to telomeres [7].
Chloroplasts act as sensors of the environmental situation and produce diverse signals informing about the functionality of the photosynthetic apparatus [8,9]. These retrograde signals comprise redox changes and reactive oxygen species and regulate gene expression in the nucleus in particular during stress situations [10]. Although in recent years several compounds involved in chloroplast-to-nucleus communication have been identified, the full repertoire of molecular mechanisms adjusting nuclear gene expression to environmental cues remained obscure [11].
To investigate the impact of WHIRLY1 on stress resistance of barley plants, seedlings of three independent transgenic lines with an RNAi-mediated knockdown of WHIRLY1 (RNAi-W1–1, RNAi-W1-7 and RNAi-W1-9) were grown in continuous light at four different irradiances (50, 120, 200, 350 µmol photons m−2 s−1). Leaves had reduced levels of the WHIRLY1 protein ranging from undetectable traces (RNAi-W1-7) to 10% of the wild-type level (RNAi-W1-1, RNAi-W1-9) [2]. The reduction in length of the leaves was the same in both lines having 10% the WHILRLY1. Therefore, only the results obtained for lines W1-1 besides line W1-7 are presented in Figure 1. The reduction in leaf length occurred irrespective of the irradiance (Figure 1a) indicating that WHIRLY1 has a general positive effect on growth.
Figure 1.
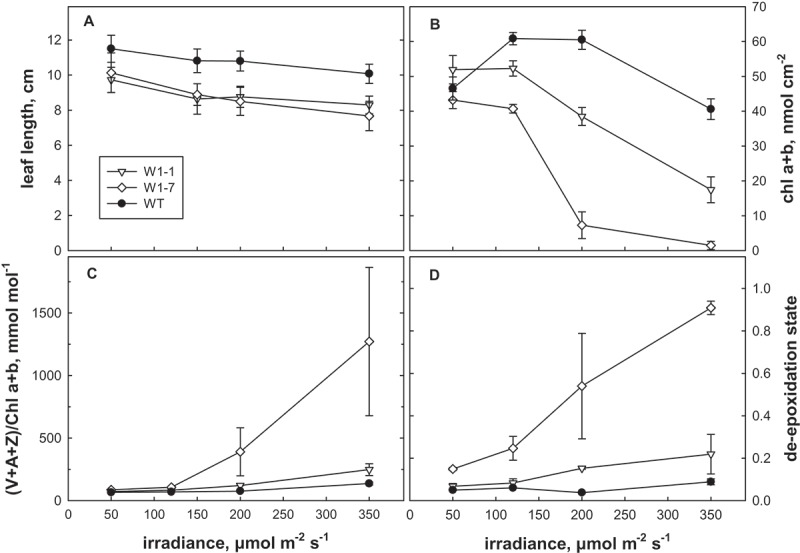
Characterization of WHIRLY1 knockdown lines at the seedling stage. Seedlings were exposed to continuous irradiation at 50, 120, 200 or 350 µmol photons m−2 s−1 for 7 days. Lengths of the primary leaves (cm) are indicated (a). Pigment extracts from the wild type (WT) and the RNAi-W1 lines (W1-1, W1-7, W1-9) were compared by HPLC for the content of chlorophylls/leaf area (b), the ratio of xanthophyll cycle pigments (VAZ) to chlorophyll (c) and the de-epoxidation state of VAZ (d). De-epoxidation state was calculated as (Z + 0.5A)/(V + A + Z). All data are means of 3 samples, error bars denote standard deviation. The results obtained for lines RNAi-W1-1 and RNAi-W1-9 are rather similar. Only the results of RNAi-W1-1 are therefore shown.
Moreover the seedlings of the RNAi-W1 plants in contrast to the wildtype showed bleaching and a reduction of the chlorophyll content at 200 µmol photons m−2 s−1 (Figure 1b). The reduction was more prominent in case of the RNAi-W1-7 line, having the lowest level of WHIRLY1 protein [2], as compared to the two other lines.
Analyses of carotenoids showed that in leaves of the RNAi-W1 plants the ratio of VAZ (V = violaxanthin, A = antheraxanthin, Z = zeaxanthin) pool pigments to chlorophylls was enhanced at irradiances of 200 and 350 µmol photons m−2 s−1 (Figure 1c). The enhanced ratio of VAZ/chlorophyll in the RNAi-W1 plants coincided with a higher de-epoxidation state of the VAZ pool (Figure 1d) indicating synthesis of zeaxanthin from violaxanthin. In line RNAi-W1-7 with the most extreme knockdown of WHIRLY1, the alterations were more dramatic than in line RNAi-W1-1. At low light, no differences were detected between wild type and RNAi-W1 plants indicating that the alterations in the pigment composition are due to high light stress.
Zeaxanthin is known to have the highest antioxidative capacity of the xanthophylls and might protect thylakoid membrane lipids from oxidation [12]. Besides its direct effect as ROS scavenger, zeaxanthin plays an important role in non-photochemical quenching dissipating excess energy as heat and avoiding thereby the production of reactive oxygen species [13]. The enhanced de-epoxidation of the xanthophyll cycle pigments in the RNAi-W1 plants compared to the wild type therefore indicates that their photosynthetic apparatus absorbed more light than required for assimilation of carbon. ROS production by thylakoids from RNAi-W1 or from wild-type seedlings grown at 200 µmol photons m−2 s−1 was measured by electron paramagnetic spin resonance (EPR). Indirect spin trapping of superoxide/hydrogen peroxide using 4-POBN/ethanol/FeEDTA [14] showed that RNAi-W1 thylakoids generated in the light about two times larger signals as wild-type thylakoids (Figure 2a, b). To investigate whether also singlet oxygen production by thylakoids is enhanced in WHIRLY1 deficient chloroplasts, EPR measurements were performed with the specific spin probe TEMPD [15]. Using TEMPD as spin trap, no difference was observed between the wild type and the transgenic lines (Figure 2a).
Figure 2.
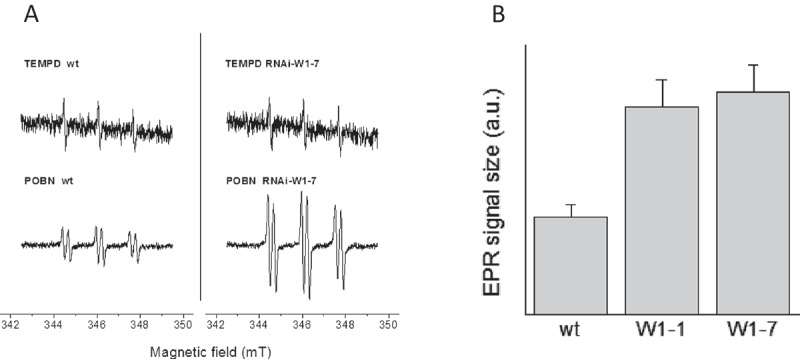
ROS production by thylakoids from wild type and RNAi-W1-1 and RNAi-W1-7 lines. Thylakoids were prepared from seedlings grown in continuous light of 200 µmol photons m−1s−1. Superoxide/hydrogen peroxide levels were measured by spin trapping EPR using 4-POBN/EtOH/FeEDTA as spintrap and singlet oxygen by the spin probe TEMPD-HCl (for experimental details, see [15]). Thylakoids were illuminated for two minutes with red light (500 µmol quanta m−2s−1) in the presence of the chemicals. Left: representative spectra, right: EPR signal sizes (4-POBN/EtOH/FeEDTA) were normalized to the signal obtained in wild-type thylakoids (mean ± SD, n = 6).
Taken together, analyses of pigments as well as ROS measurements revealed that the WHIRLY1 deficient plants experienced more photooxidative stress than the wild type when grown in continuous high light. This indicates that WHIRLY1, in addition to its positive effect on growth, also promotes stress resistance.
Since WHIRLY1 in chloroplasts was shown to bind to RNA as well as to DNA [3] it was obvious to investigate a putative role of WHIRLY1 in controlling the levels of microRNAs which play a central role in the control of plant development as well as in stress responses [16–18]. For the analysis of microRNAs, primary foliage leaves of wild-type plants and plants of the RNAi-W1-7 line, respectively, grown either at low light (100 µmol photons m−2 s−1) or at high light (350 µmol photons m−2 s1), were used. Eight conserved microRNAs reported to be stress responsive in Arabidopsis thaliana and in Triticeae [19,20] were selected and their levels were determined by Northern blot analyses as well as by RT-qPCR TaqMan MicroRNA assays.
In wild-type plants the levels of most of these microRNAs were enhanced at high light compared to low light conditions (Supplemental Fig. S1). These findings were confirmed by RT-qPCR TaqMan MicroRNA assays, although the changes were not statistically significant in each case (Figure 3a). While in Northern blot analyses at least several members of a microRNA family were detected (Supplemental Fig. S1), in RT-qPCR TaqMan MicroRNA assays only specific members of a family were measured. Therefore the results of both approaches are not always directly comparable, e.g. in case of miRNA159.
For all microRNAs tested in Northern blot hybridization the levels were reduced in leaves of the two RNAi-W1 lines (RNAi-W1-1 and RNAi-W1-7) (Supplemental Fig. S1). These results were confirmed by RT-qPCR TaqMan MicroRNA assays and were independent of the light conditions (Supplemental Fig. S2A, B).
mRNA targets for the tested microRNAs are known in several plant species including barley (Supplemental Table 1) [21–24]. All tested target mRNAs were additionally recognized using the psRNA-Target software [25,] (Supplemental Material S1). MicroRNAs hvu-miR159a and hvu-miR159b-3p target GAMyb mRNA, and microRNAs named hvu-miR164a, hvu-miR172b-3p, hvu-miR393h, and hvu-miR396b-5p target NAC, APETALA, TIR1 and GRF1 mRNAs, respectively. Barley HOX9 and AGO1 mRNAs have been shown to be targets of microRNA166a and microRNA168-5p, respectively.
The effects of the selected miRNAs on the levels of targeted mRNAs were tested by qRT-PCR. In primary foliage leaves of wild-type plants grown in high light, the upregulation of microRNAs coincided with a downregulation of targeted mRNAs (Figure 3b). In contrast, in the RNAi-W1 plants grown in high light most target gene mRNA levels were enhanced compared to the wild type (Figure 3c) whereas at low light the levels of target gene mRNAs were similar between the wild type and RNAi-W1-7 plants (Supplemental Fig. S3).
Figure 3.
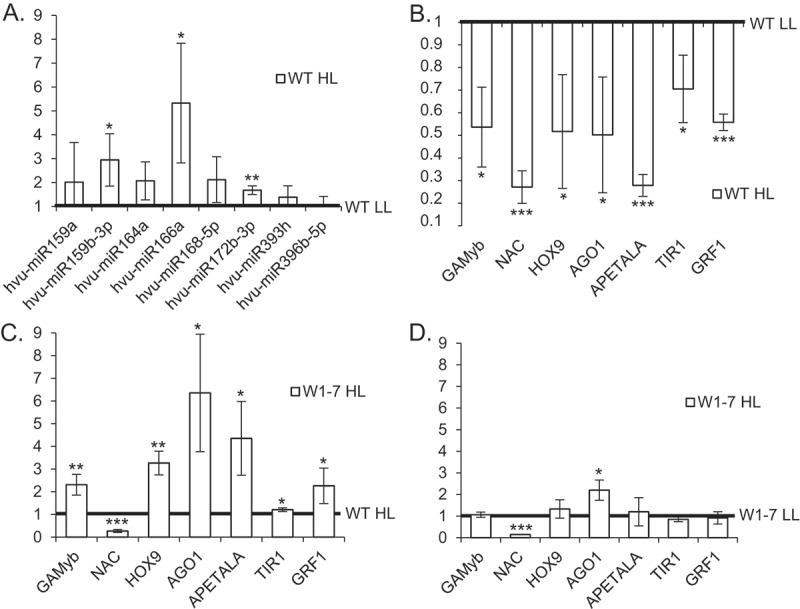
RT-qPCR analysis of microRNAs and expression of target genes in wild type (WT) and transgenic RNAi -W1-7 plants exposed to either low (LL) or high light (HL). (a) In WT plants exposed to HL the levels of microRNAs were enhanced. Results are presented as fold changes and results for WT plants grown in LL are treated as 1. (b) In the wild type plants HL lead to a downregulation of the levels of target mRNAs. (c) Levels of most target mRNAs were enhanced in HL treated W1-7 plants when compared to the WT. (d) Target mRNAs expression stayed mostly unchanged when RNAi-W1-7 plants exposed to LL and HL are compared. Error bars indicate SD (n = 3), and the asterisk indicates a significant difference between the sample and control (t test, *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001).
MicroRNAs analyzed in the study respond to multiple stresses [reviewed in 20]. The expression of a particular microRNA can vary between organs or within time after stress treatment, as in case of microRNA166a being upregulated in the barley leaf and downregulated in roots during desiccation [26]. The results indicate that high light induced signals from chloroplasts stimulate a WHIRLY1 dependent downregulation of the level of mRNAs targeted by the tested microRNAs being upregulated in the wild type. In contrast to the wild type, plants of the RNAi-W1-7 line did neither show a light-induced increase in microRNAs nor a decrease in the mRNA levels of their target genes. This indicates that the WHIRLY1 deficient plants can’t respond to stress and thereby suffer from a higher ROS production. In many cases abiotic stress treatment leads to the expression fluctuation of microRNAs, negatively correlated with target mRNA levels. WHIRLY1 deficiency abolished microRNA based target mRNAs expression regulation.
In the transgenic plants grown either in low light or in high light, the levels of targeted mRNAs did not show essential differences (Figure 3d). The only exception is the mRNA encoding a NAC transcription factor (GenBank: AK356223.1) that has a lower level in W1-7 plants grown either in high light or low light despite the low level of its potential cognate microRNA164a. The reason for this result remains unclear. NAC transcription factors comprise one of the largest gene families and are involved in the regulation of plant development, senescence and responses to various stresses. Their activities can be regulated at different levels (transcription efficiency, alternative splicing, and posttranslational regulation) that possibly might affect the final level of NAC mRNAs [27].
WHIRLY1 has been proposed to move from the chloroplast to the nucleus in response to environmental cues such as high light intensity [28]. In this study it has been demonstrated that the repertoire of the plants’ responses towards high light involves a WHIRLY1 dependent increase in the levels of diverse nuclear microRNAs. As WHIRLY1 can bind to RNA it might be a general factor influencing the biogenesis and/or stability of microRNAs. The observed phenomenon might be caused either by direct binding of WHIRLY1 to the nuclear microRNAs and/or its architectural impact on nuclear chromatin as observed in chloroplasts [2]. To elucidate the specific role of WHIRLY1 in the regulation of the levels of microRNAs and targeted mRNAs during retrograde signaling further detailed studies are required.
1. Material and methods
1.1. Growth of barley plants
Barley seedlings were sown in multipots on soil (Einheitserde ED73, Einheitswerk Werner Tantau, Ütersen, Germany). After three days in darkness and low temperature (6°C) the seedlings were transferred in a chamber with continuous light. Continuous light was adjusted to 50, 120, 200 or 350 photons µmol s−1 m−2. Temperature was 21–26°C. Ten days after sowing, i.e. after 7 days in continuous light, the primary foliage leaves of the seedlings were analyzed.
1.2. Analysis of leaf pigment content
A segment of a length of 1.4 cm (area: 1-1.3 cm2) was excised from the mid part of a primary foliage leaf and was immediately frozen in liquid nitrogen. Until analysis by HPLC the samples were stored in a freezer at −80°C. For extraction, the leaf discs along with five glass beads were ground in the frozen state in a Geno Grinder (Type 2000, SPEX, CertiPrep, Munich, Germany) with 0.5 ml 80% (v/v) acetone buffered with 20 mM Tris, pH 7.8. After centrifugation, the pellet was extracted twice with 200 µl 100% acetone. From the unified extracts, 50 µl were used for HPLC analysis on an Agilent 1100 system (Agilent, Waldbronn, Germany) with DAD detection. The protocol was the same as published before [29]. De-epoxidation state was calculated as (Z + 0.5A)/(V + A + Z), with V, violaxanthin, A, antheraxanthin and Z, zeaxanthin.
1.3. Room-temperature electron paramagnetic spin resonance measurements
Spin-trapping assays with 4-pyridyl-1-oxide-N-tert-butylnitrone (4-POBN) (Sigma-Aldrich, Darmstadt, Germany) were carried out to detect the formation of hydroxyl radicals. The spin trapping assay contained 100 mM TEMPD-HCl for the detection of 1O2 and 50 mM 4-POBN, 4% ethanol, 50 µM Fe-EDTA for the detection of hydroxyl radicals. Fe-EDTA is used as a Fenton reagent catalyzing the reduction of H2O2 into −OH and •OH, the latter being detected in the spin trapping assay. The samples contained thylakoid membranes at a concentration of 10 µg Chl/ml and were illuminated for 2 min with red light (RG 630, Calflex C) of an intensity of 500 µmol quanta m−2s−1. EPR spectra were recorded at room temperature in a standard quartz flat cell using an ESP-300 X-band (9.73 GHz) spectrometer (Bruker, Rheinstetten, Germany). The following parameters were used: microwave frequency 9.73 GHz, modulation frequency 100 kHz, modulation amplitude: 1G, microwave power: 6.3 milliwatt receiver gain: 2x104, time constant: 40.96 ms; number of scans: 4.
1.4. RNA isolation
Total RNA was extracted from 100 mg of powdered tissue with 38% (w/v) phenol solution. The method allowed for efficient isolation of small RNAs and is described in detail by [30]. RNA integrity was evaluated on agarose gels. DNA traces were removed with Ambion TURBO DNase (ThermoFisher Scientific, Waltham, MA, USA).
1.5. Northern blot microRNA analyses
MicroRNAs were analysed using Northern blot hybridization for enhanced detection of small RNAs [31]. Fifteen µg of RNA was separated on Urea PAGE (15%), transferred onto Amersham Hybond-NX nitrocellulose (GE Healthcare, Little Chalfont, Buckinghamshire, UK) and fixed with EDC chemical crosslinking. MicroRNAs were detected with γ32P ATP-labeled (6000 Ci/mmol; HARTMANN ANALYTIC GmbH, Braunschweig, Germany) DNA oligo probes (Sigma-Aldrich, St. Louis, MO, USA). The U6 hybridization signal was taken as a loading control. Blots were exposed to phosphorimaging screen (Fujifilm), scanned with Fujifilm FLA5100 reader (Fujifilm Co., Ltd., Tokyo, Japan) and quantified with Multi Gauge V2.2 software. Probe sequences are listed in Supplemental Table 2.
1.6. Quantitative real-time PCR (RT-qPCR)
mRNA levels were quantified using cDNA achieved with 3 µg of DNA-depleted RNA, Invitrogen SuperScript III Reverse Transcriptase (ThermoFisher Scientific, Waltham, MA, USA) and 0.5 µg Oligo(dT)18 Primer (ThermoFisher Scientific, Waltham, MA, USA). cDNA templates were 4-fold diluted. MicroRNA expression was detected and quantified with TaqMan™ MicroRNA Assays (ThermoFisher Scientific, Waltham, MA, USA) following the instructions of the manufacturer. 10 ng of DNA-depleted RNA was reversely transcribed with TaqMan MicroRNA Reverse Transcription Kit (ThermoFisher Scientific, Waltham, MA, USA). Reactions were proceeded with the Power SYBR Green PCR Master MIX (Applied Biosystems, Warrington, UK), two specific primers (final concentration of 200 nM each) for mRNA RT-qPCR. TaqMan Universal PCR Master Mix II with UNG (ThermoFisher Scientific, Waltham, MA, USA) was used for microRNA RT-qPCR. Each RT-qPCR reaction was run for three biological replicates with the 7900HT Fast Real-Time PCR System (Applied Biosystems, Waltham, MA, USA) in 384-well plates. mRNA results were normalized to the mRNA level of the barley ADP-ribosylation factor 1-like [GenBank: AJ508228.2] [32]. MicroRNA levels were normalized to the U6 snRNA expression. Expression levels were calculated with the relative quantification method (2-ΔΔCt) and presented as a fold-change value. The R2 values of the analyzed data (≥ 0.997) were calculated with LinRegPCR software [33,34]. The significance of the fold-change values was tested with a T-test. TaqMan Small RNA Assays, together with microRNA sequences are listed in Supplemental Table 2. Primers are listed in Supplemental Table 3.
Funding Statement
This work was supported by the Deutsche Forschungsgemeinschaft [Kr1350/7]; Deutsche Forschungsgemeinschaft [Kr1350/9]; Narodowe Centrum Nauki [UMO-2016/23/B/NZ9/00862]; Ministerstwo Nauki i Szkolnictwa Wyższego, KNOW RNAResearch Centre in Poznan [01/ KNOW2/2014]; A. K. L. benefits from the support of the LabEx Saclay Plant Sciences-SPS (ANR-10-LABX-0040-SPS) and the French Infrastructure for Integrated Structural Biology (FRISBI) ANR-10-INSB-05.
Acknowledgments
We would like to thank Artur Jarmolowski (AMU Poznan, Poland) for fruitful discussions. Jens Hermann (CAU Kiel, Germany) is thanked for technical assistance.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary Material
Supplemental data for this article can be accessed here.
References
- 1.Grabowski E, Miao Y, Mulisch M, et al. Single-stranded DNA binding protein Whirly1 in barley leaves is located in plastids and the nucleus of the same cell. Plant Physiol. 2008;147:1800–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krupinska K, Oetke S, Desel C, et al. WHIRLY1 is a major organizer of chloroplast nucleoids. Front Plant Sci. 2014;5:432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Melonek J, Mulisch M, Schmitz-Linneweber C, et al. Whirly1 in chloroplasts associates with intron containing RNAs and rarely co-localizes with nucleoids. Planta. 2010;232:471–481. [DOI] [PubMed] [Google Scholar]
- 4.Prikryl J, Watkins KP, Friso G, et al. A member of the Whirly family is a multifunctional RNA- and DNA-binding protein that is essential for chloroplast biogenesis. Nucleic Acids Res. 2008;36:5152–5165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marèchal A, Parent J-S, Vèronneau Lafortune, et al. Whirly proteins maintain genome stability Arabidopsis. Proc Natl Ac Sci USA. 2009;106:14693–14698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desveaux D, Despres C, Joyeux A, et al. PBF-2 is a novel single-stranded DNA binding factor implicated in PR-10a gene activation in potato. Plant Cell. 2000;12:1477–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoo HH, Kwon C, Lee MM, et al. Single-stranded DNA binding factor AtWHY1 modulates telomere length homeostasis in Arabidopsis. Plant J. 2007;49:442–451. [DOI] [PubMed] [Google Scholar]
- 8.Pfalz J, Liebers M, Hirth M, et al. Environmental control of nuclear gene expression by chloroplast redox signals. Front Plant Sci. 2012;3:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleine T, Leister D.. Retrograde signaling: organelles go networking. Biochim Biophys Acta. 2016;1857:1313–1325. [DOI] [PubMed] [Google Scholar]
- 10.Dietz K-J. Efficient high light acclimation involves rapid processes at multiple mechanistic levels. J Exp Bot. 2015;66:2401–2414. [DOI] [PubMed] [Google Scholar]
- 11.Chan KX, Phua SY, Crisp P, et al. Learning the languages of the chloroplast: retrograde signaling and beyond. Ann Rev Plant Biol. 2016;67:25–53. [DOI] [PubMed] [Google Scholar]
- 12.Havaux M, Dall’Osto L, Bassi R. Zeaxanthin has enhanced antioxidant capacity with respect to all other xanthophylls in Arabidopsis leaves and functions independent of binding to PSII antennae. Plant Physiol. 2007;145:1506–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Wakao S, Fischer BB, et al. Sensing and responding to excess light. Ann Rev Plant Biol. 2009;60:239–260. [DOI] [PubMed] [Google Scholar]
- 14.Mubarakshina MM, Ivanov BN, Naydov IA, et al. Production and diffusion of chloroplastic H2O2 and its implication to signaling. J Exp Bot. 2010;61:3577–3587. [DOI] [PubMed] [Google Scholar]
- 15.Krieger-Liszkay A, Trösch M, Krupinska K. Generation of reactive oxygen species in thylakoids from senescing flag leaves of the barley varieties Lomerit and Carina. Planta. 2015;241:1497–1508. [DOI] [PubMed] [Google Scholar]
- 16.Kruszka K, Pieczynski M, Windels D, et al. Role of microRNAs and other sRNAs of plants in their changing environments. J Plant Physiol. 2012;169:1664–1672. [DOI] [PubMed] [Google Scholar]
- 17.Budak H, Kantar M, Bulut R, et al. Stress responsive miRNAs and isomiRs in cereals. Plant Sci. 2015;235:1–13. [DOI] [PubMed] [Google Scholar]
- 18.Li S, Castillo-González C, Yu B, et al. The functions of plant small RNAs in development and in stress responses. Plant J. 2016;90:654–670. [DOI] [PubMed] [Google Scholar]
- 19.Barciszewska-Pacak M, Milanowska K, Knop K, et al. Arabidopsis microRNA expression regulation in a wide range of abiotic stress responses. Front Plant Sci. 2015;6:410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alptekin B, Langridge P, Budak H. Abiotic stress miRNomes in the Triticeae. Funct Integr Genomics. 2017;17:145–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ozhuner E, Eldem V, Ipek A, et al. Boron stress responsive microRNAs and their targets in barley. PLoS One. 2013;8:e59543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kruszka K, Pacak A, Swida-Barteczka A, et al. Transcriptionally and post-transcriptionally regulated microRNAs in heat stress response in barley. J Exp Bot. 2014;65:6123–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pacak A, Kruszka K, Swida-Bateczka A, et al. Developmental changes in microRNA expression profiles coupled with miRNA target analysis. Acta Biochim Pol. 2016;63:799–809. [DOI] [PubMed] [Google Scholar]
- 24.Deng P, Wang L, Cui L, et al. Global Identification of microRNAs and their targets in barley under salinity stress. PLoS One. 2015;10:e0137990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dai X, Zhao PX. psRNATarget: a plant small RNA target analysis server. Nucl Ac Res. 2011;Web Server issue:W155–9. http://plantgrn.noble.org/psRNATarget/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kantar M, Unver T, Budak H. Regulation of barley miRNAs upon dehydration stress correlated with target gene expression. Funct Integr Genomics. 2010;10:493–507. [DOI] [PubMed] [Google Scholar]
- 27.Shao H, Wang H, Tang X. NAC transcription factors in plant multiple abiotic stress responses: progress and prospects. Front Plant Sci. 2015;6:902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foyer CH, Karpinska B, Krupinska K. The functions of WHIRLY1 and REDOX-RESPONSIVE TRANSCRIPTION FACTOR1 in cross tolerance responses in plants: a hypothesis. Phil Trans R Soc B. 2014;369:20130226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niinemets Ü, Bilger W, Kull O, et al. Acclimation to high irradiance in temperate deciduous trees in the field: changes in xanthophyll cycle pool size and in photosynthetic capacity along a canopy light gradient. Plant Cell Environ. 1998;21:1205–1218. [Google Scholar]
- 30.Kruszka K, Pacak A, Swida-Barteczka A, et al. Developmentally regulated expression and complex processing of barley pri-microRNAs. BMC Genomics. 2013;14:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pall GS, Hamilton AJ. Improved northern blot method for enhanced detection of small RNA. Nat. 2008;3:1077–1084. [DOI] [PubMed] [Google Scholar]
- 32.Rapacz M, Stepien A, Skorupa K. Internal standards for quantitative RT-PCR studies of gene expression under drought treatment in barley (Hordeum vulgare L.): the effects of developmental stage and leaf age. Acta Physiologiae Plantarum. 2012;34:1723. [Google Scholar]
- 33.Ramakers C, Ruijter JM, Deprez RH, et al. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett. 2003;339:62–66. [DOI] [PubMed] [Google Scholar]
- 34. http://www.hartfaalcentrum.nl/index.php?main=files&sub=LinRegPCR
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


