Abstract
Background
Gallbladder cancer (GBC) is a highly lethal disease and the most common biliary tract malignant tumor with poor prognosis. Accumulating evidence indicates that cluster of differentiation 44 (CD44) is overexpressed in several malignancies and has a crucial role in the development of cancer. However, its expression and function in GBC are unclear. The aim of this study was to explore CD44 expression and its role in GBC.
Materials and methods
The expression of CD44 was measured by immunohistochemistry. Tissue microarray analysis was used to confirm the relationship between CD44 expression and clinical outcomes of GBC patients. EDU assay, colony formation assay, cell migration and invasion assay were performed to detect the functions of CD44 in GBC-SD and NOZ transfected with si-RNA.
Results
CD44 was overexpressed and associated with poor outcomes in GBC patients. The univariate and multivariate analyses confirmed that elevated CD44 was an independent prognostic factor for the OS of GBC patients. Silencing CD44 could suppress the GBC cell proliferation, migration and invasion in vitro, as well as attenuated cancer stem cell functions.
Conclusion
CD44 markedly correlated with aggressive tumor behaviors and contributed to the progression of GBC, which could represent a novel prognostic marker and potential therapeutic target for GBC patients.
Keywords: CD44, gallbladder cancer, cancer stem cells, tumor progression
Introduction
Gallbladder cancer (GBC) is the most common biliary tract malignancy and highly lethal with a 5-year OS rate of less than 5%.1,2 The poor prognosis is due to, at least in part, aggressive biologic behaviors and delayed diagnosis for this disease. Although therapeutic options for advanced GBC (surgery combined with chemotherapy, radiation therapy, etc.) have been improved, the clinical outcome is still poor.3–5 Therefore, it is urgent to clarify novel and effective diagnostic biomarkers for GBC.
Several studies have indicated that cancer stem cells (CSCs), possessing the capabilities of self-renewal and differentiation, high tumorigenicity and resistance to current treatments, exist in numerous solid tumors.6–10 Additionally, genetic variants of CSCs have been showen to play significant biological roles in susceptibility and prognosis in GBC, suggesting a novel method for the treatment of GBC.11,12 Furthermore, many typical characteristic markers of CSCs have been observed at the cell membrane surface, and elevated expression of some specific cell surface proteins has been proven to facilitate tumor initiation, progression and poor clinical outcomes.13 Among these proteins, cluster of differentiation 44 (CD44), as the principal receptor, has been demonstrated to contribute to cell proliferation, invasion and metastasis of CSCs.14 Increasing evidence has indicated that CD44 is extensively overexpressed in various cancer types including prostate cancer,15 triple-negative breast cancer,16 lung cancer17 and pancreatic cancer,18 and correlated with aggressive biological behaviors and poor prognostic characteristics. Hence, it is unsurprising that CD44 has garnered increasing attention within the field of oncology in recent years. Nevertheless, the expression pattern and functional role of CD44 in GBC has not been elucidated clearly.
In this study, we found that CD44 mRNA was upregulated and related with prognosis in hepatobiliary tumors using The Cancer Genome Atlas (TCGA) data analysis. Moreover, we confirmed that CD44 was overexpressed and correlated with poor clinical outcomes in GBC patients via tissue microarray (TMA) analysis. We further explored the functional role of CD44 in the regulation of GBC cell proliferation, migration, invasive malignant behaviors, clonogenicity and CSC maintenance in vitro. In sum, these results revealed that increased expression of CD44 was related with aggressive tumor behavior and an unfavorable prognosis in GBC patients. Monitoring of the expression levels of CD44 protein may provide assistance in early diagnosis and predict prognosis of GBC.
Materials and methods
TCGA dataset analysis
In order to analyze the expression levels of CD44 mRNA in cancers, mRNA gene expression and clinical datasets were downloaded from TCGA (http://tcga-data.nci.nih.gov/) and analyzed.
Patients and specimens
As approved by the ethics committee of the First Affiliated Hospital, human GBC samples and the paired non-tumor samples were collected and immediately frozen in liquid nitrogen (–180°C) after surgery and then stored for use. Whether the selected tissues were tumor or normal was confirmed by the pathologist during the operation, and hematoxylineosin (H&E) staining was also performed to clarify GBC and normal tissues before scoring. The protocol for this special study was approved by the Ethical Committee of the First Affiliated Hospital of Zhengzhou University and we confirmed that patients whose tissue samples were used provided written informed consent.
Cell lines and culture
The cell lines used in this study are shown in Table S1. Cells were maintained in DMEM (Gibco, Carlsbad, CA, USA) with 10% FBS (Gibco) in a humidified atmosphere of 95% air and 5% CO2 at 37°C. All cell lines used in this study were passaged for less than 4 months in culture until the experiments were carried out.
TMA construction and immunohistochemistry analysis
IHC was performed as described previously.19 Briefly, TMA sections were deparaffinized, rehydrated and underwent antigen retrieval. They were then blocked for an hour at room temperature. After incubation with primary antibody CD44 (dilution 1:100, Proteintech Group, Wuhan, China) at 4°C overnight, the slide was probed with biotinylated goat anti-rabbit secondary antibody and then detected by SignalStain® DAB (CST, Danvers, MA, USA) and counterstained with Hematoxylin QS (Vector Laboratories, Burlingame, CA, USA). The images of IHC and H&E staining were obtained using the NanoZoomer 2.0-RS system (Hamamatsu Photonics Inc., Herrsching, Germany), and the digital slides were analyzed by the software of the NDP.view 2.5.14 version. Two pathologists (Xiaolong Chen and Yan Yu) scored the IHC simples in a blinded manner independently. Sections were semi-quantitatively scored for the CD44 staining patterns as follows: the staining extent in each core was scored as 1+ (<25% staining of tumor cells), 2+ (25%–50% staining of tumor cells), 3+ (50%–75% staining of tumor cells) or 4+ (>75% staining of tumor cells), who were blinded to the clinicopathological data, with a consensus reached in all cases. Scores of 3+ and 4+ were defined as exhibiting high expression and scores of 1+ and 2+ were deemed as exhibiting low expression.
Western blotting
Isolation of total protein from GBC cells was performed using RIPA buffer, and protein concentration was determined with bicinchoninic acid protocol. Equal protein was resolved by the SDS–polyacrylamide gel (Invitrogen, Carlsbad, CA, USA) and transferred to NC membrane (Millipore, Billerica, MA, USA). After blocking with blocking buffer, the membrane was incubated with primary antibody. The membranes were then washed with PBST three times; the membrane was then treated with secondary antibody. Signals were determined using the chemiluminescent reagents.
Primary and secondary antibodies used in this study are shown in Table S2.
siRNA transfection
The primer sequences used in this study are described in Table S3. CD44-siRNA was purchased from Sangon Biotech (Shanghai, China) to silence CD44. Cells were seeded in the six-well plates for 24 hours and then infected with siRNA using Lipofectamine 3000 transfection reagent (Invitrogen) according to the manufacturer’s instructions. The cells transfected 24–72 hours were then used for detecting the gene.
Cell proliferation assay
Initial cellular proliferation was evaluated using a CCK-8 Kit. NOZ or GBC-SD cells treated with CD44-siRNA were plated in a 96-well plate. Then proliferation was measured and determined at the established time point with 100 µL 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2, 4-disulfobenzene)-2H-tetrazolium monosodium salt (WST-8) (Beyotime, Shanghai, China) after incubating for 4 hours at 37°C and when absorbance was read at 560 nm with a spectrophotometer (Molecular Devices, San Jose, CA, USA).
Furthermore, an EDU kit (RiboBio, Guangzhou, China) was used to further assess the cell proliferation ability after CD44 knockdown. Images were taken with a microscope at 100× (Olympus, Tokyo, Japan) for further analysis. The ratio of EDU-stained cells (red fluorescence) to DAPI-stained cells (blue fluorescence) was used to evaluate the cell proliferation activity.
Cell migration and invasion assay
A wound-healing assay was used to verify the GBC cell migration ability of CD44 downregulated cells. GBC-SD and NOZ cells were seeded in six-well plates and then infected with CD44-siRNA. When the cells reached about 90% confluency, cell wound was scratched by sterile plastic tips and then cultured for 48 hours. At the indicated time points, images were taken under the microscope.
Cell invasion assay was performed with Matrigel Invasion Chambers (BD Biosciences, San Jose, CA, USA). Here, 1×104 transfected cells were cultured on the upper chamber of the transwell insert in serum-free medium, and the bottom chamber is filled with DMEM with 10% FBS. After 24 hours, the invasive cells were stained with 0.5% crystal violet, imaged and counted.
Colony formation assay
Colony formation assay was further applied to identify the effect of CD44 silencing on GBC cell growth. Cell growth is considered to reflect the self-renewal ability and phenotype of CSCs. We diluted and plated the cells treated with siRNA in six-well plates with 2,000 cells per well. After 14 days of incubation in 37°C incubator, cells were fixed with 4% paraformaldehyde, stained with 2% crystal violet, imaged and counted.
Three-dimensional cell culture assay
To accurately mimic the in vivo environment, we performed a three-dimensional cell culture assay to assess the ability of GBC cell growth and proliferation after CD44 inhibition. In brief, after transfection with CD44-siRNA for 48 hours, cells were seeded into a Perfecta3D 96-well Hanging Drop plate (3D Biomatrix, Ann Arbor, MI, USA). After culture for 7 days in a 37°C incubator, cells were visualized and imaged.
qRT-PCR
Ten GBC tissues and paired normal tissues were selected for RT-PCR assay. Total RNA was extracted from tissue samples and with Trizol reagent (Invitrogen) according to the manufacturer instructions. The RNAs were reverse transcribed according to the protocol of the PrimeScript RT Master Mix Perfect Real Time (Takara, Shiga, Japan). PCR reaction was performed using an ABI7500 System (Applied Biosystems, Waltham, MA, USA) and SYBR Green PCR Master Mix (TaKaRa). The relative quantification of CD44 was calculated by using 2−DDCt method and β-actin was used as an internal control. The reactions were performed independently in triplicate. The primers sequences used in this study are displayed in Table S4, and the results are shown in Table S2.
Statistical analysis
All statistical analyses used the Student’s t-test as part of the SPSS 23.0 statistical software. The correlation between the expression of CD44 and outcomes of GBC patients was examined by Kaplan–Meier analysis with GraphPad Prism 7.0. In all cases, at least three independent experiments were performed. Results are expressed as mean ± SD and P<0.05 was assumed statistically significant.
Results
CD44 is overexpressed and correlated with poor prognosis in GBC
We performed TCGA data analysis to identify the expression level of CD44 mRNA in the most common cancers. As shown in Figure 1A, CD44 mRNA expression was frequently dys-regulated in cancers, and was especially upregulated in hepatobiliary tumors including liver hepatocellular carcinoma and cholangiocarcinoma. In order to detect the expression of CD44 in GBC, we used high-throughput TMA analysis and calculated the intensities of CD44 staining in GBC tissues and non-tumor tissues (Figures 1B and 2A). CD44 expression was increased in GBC tissues (N=132) compared to the paired non-tumor tissues (N=47) (P=0.0036, Figure 1C and D), which was consistent with PCR results (P=0.0001, Figure 1E). Additionally, we confirmed that CD44 expression was notably higher in GBC patients with an advanced TNM stage (III vs I, P=0.0210; IV vs I, P=0.0018, Figure 2A and B). Moreover, Kaplan–Meier analysis demonstrated that OS of patients with high CD44 expression was markedly shorter than those with low CD44 expression (P=0.0214, Figure 2C).
Figure 1.
CD44 is dysregulated in cancers and upregulated in hepatobiliary tumors.
Notes: (A) CD44 mRNA expression level in TCGA data analysis. CD44 mRNA was upregulated in tumor tissues of LIHC and CHOL. (B) Panoramic scanning for GBC TMA. (C) Representative CD44 staining in GBC tissues and normal samples. (D) Histological scoring of CD44 in GBC tissues and normal samples; CD44 was overexpressed in GBC tissues compared with the non-tumor tissues (P=0.0036). (E) Related CD44 mRNA expression in GBC tissues compared with paired non-tumor tissues (P<0.0001). *P<0.05.
Abbreviations: CD44, cluster of differentiation 44; TCGA, The Cancer Genome Atlas; LIHC, liver hepatocellular carcinoma; CHOL, cholangiocarcinoma; GBC, gallbladder cancer; TMA, tissue microarray; BLCA, bladder urothelial carcinoma; BRCA, breast cancer; COAD, colon cancer; ESCA, esophagus cancer; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIRC, renal cancer; KIRP, kidney renal papillary cell carcinoma; LUAD, lung cancer; LUSC, lung squamous cell carcinoma; PRAD, prostate adenocarcinoma; READ, rectal cancer; STAD, stomach cancer; THCA, thyroid cancer; UCEC, uterine corpus endometrial carcinoma; ACC, adrenocortical carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; LAML, acute myeloid leukemia; LGG, brain lower drade glioma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and paraganglioma; SARC, sarcoma; SKCM, skin cutaneous melanoma; TGCT, testicular germ cell tumors; THYM, thymoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma.
Figure 2.

CD44 is overexpressed in GBC tissues and negatively correlated with OS.
Notes: (A) Representative CD44 and immunohistochemical staining patterns with different staining scores in GBC tissues. (B) Histological scoring of CD44 according to TNM classification. CD44 expression was notably high in GBC patients with advanced TNM stage (*P<0.05, **P<0.01). (C) Kaplan–Meier survival analysis between expression of CD44 (red, high CD44 expression; green, low CD44 expression). GBC patients with high CD44 expression had markedly shorter OS than those with low CD44 expression (P=0.0214).
Abbreviations: CD44, cluster of differentiation 44; GBC, gallbladder cancer; H&E, hematoxylin-eosin.
To further evaluate the relationship between CD44 expression and clinicopathological characteristics in GBC, we found that upregulated CD44 was significantly correlated to GBC TNM stage (III and IV vs I and II, P=0.012), distant metastasis (present vs absence, P=0.044) and differentiation grade (III–IV vs I–II, P=0.001). While there was no obvious difference in patients’ age, tumor size and gender between high CD44 expression and low CD44 expression patients by Pearson’s chi-squared test analysis (all P>0.05) (Table 1). These data revealed that CD44 was overexpressed in GBC tissues, and high CD44 expression exhibited a worse prognosis than those with low CD44 expression, indicating the important role for oncogene CD44 in GBC.
Table 1.
The relationship between CD44 expression and clinicopathological features of gallbladder cancer
| Clinicopathological features | No. of cases (%) | CD44
|
P-value | ||
|---|---|---|---|---|---|
| Low | high | ||||
| Age (years) | ≤Median | 33 | 18 | 15 | 0.403 |
| >Median | 33 | 16 | 17 | ||
| Gender | Male | 19 | 9 | 10 | 0.437 |
| Female | 47 | 25 | 22 | ||
| TNM stage | Stage I and II | 31 | 21 | 10 | 0.012 |
| Stage III and IV | 35 | 13 | 22 | ||
| Metastasis | Absent | 37 | 23 | 14 | 0.044 |
| Present | 29 | 11 | 18 | ||
| Neoplasm histologicgrade | I and II | 29 | 21 | 7 | 0.001 |
| III and IV | 38 | 13 | 25 | ||
| Tumor size | ≤4 cm | 31 | 17 | 14 | 0.397 |
| >4 cm | 35 | 17 | 18 | ||
Abbreviation: CD44, cluster of differentiation 44.
High expression of CD44 is an adverse prognostic factor in GBC patients
Subsequently, we analyzed prognostic factors for GBC patients, and found that TNM stage, differentiation grade, distant metastasis and CD44 expression were potential prognostic factors by univariate analyses (Table 2). Notably, in multivariate analyses, TNM stage (III–IV), distant metastasis and CD44 expression (high) were demonstrated to be independent prognostic factors for GBC patients (Table 2). All of these findings implied that upregulated CD44 was closely associated with poor survival in GBC patients, and so elevated CD44 expression was an independent prognostic factor of GBC.
Table 2.
Univariate and multivariate analyses of OS of gallbladder carcinoma
| Clinicopathological features | Univariate analyses
|
Multivariate analyses
|
|||||
|---|---|---|---|---|---|---|---|
| HR | 95% (CI) | P-value | HR | 95% (CI) | P-value | ||
| Age (years) | <Median | 1.000 | 0.607–1.998 | 0.750 | |||
| >Median | 1.102 | ||||||
| Gender | Male | 1.000 | 0.399–1.508 | 0.454 | |||
| Female | 1.289 | ||||||
| Tumor size | ≤4 cm | 1.000 | 0.664–2.168 | 0.547 | |||
| >4 cm | 1.199 | ||||||
| TNM stage | Stage I–II | 1.000 | 2.296–8.919 | <0.001 | 1.000 | 1.301–7.226 | 0.010 |
| Stage III–IV | 4.525 | 3.066 | |||||
| Histologic grade | Stage I–II | 1.000 | 1.616–5.666 | 0.001 | 1.000 | 0.640–4.383 | 0.293 |
| Stage III–IV | 3.026 | 1.675 | |||||
| Distant metastasis | Absent | 1.000 | 1.448–5.783 | 0.003 | 1.000 | 0.275–0.995 | 0.048 |
| Present | 2.894 | 0.523 | |||||
| CD44 expression | Low | 1.000 | 1.483–2.683 | <0.001 | 1.000 | 1.247–2.557 | 0.002 |
| High | 1.995 | 1.786 | |||||
Abbreviation: CD44, cluster of differentiation 44.
CD44 silencing inhibits GBC cell proliferation, migration and invasion
CD44 silencing was accomplished by RNA interference (RNAi) to further elucidate the functional role of CD44 on GBC cells. The efficiency of CD44-siRNA was confirmed in GBC cells by western blotting, with results showing that CD44 expression was reduced in a dose-related manner in NOZ and GBC-SD cells (Figure 3A). A CCK-8 assay was used to determine that RNAi-mediated CD44 downregulation could inhibit growth of GBC cells in both NOZ and GBC-SD cells (Figure 3B). Moreover, the proliferation ability of GBC cells could be suppressed after CD44 knockdown was further affirmed with EDU assay (Figure 3C). In addition, decreased migration after transfection with CD44-siRNA was verified by wound-healing assay (Figure 3D). Using a Matrigel invasion assay, we observed that downregulation of CD44 resulted in reduced invasive potential in both NOZ and GBC-SD cells when compared with the control group (Figure 3E). Our data collectively demonstrated that the activity of GBC cell growth, invasive and migratory was remarkably suppressed after CD44 knockdown.
Figure 3.
RNAi-mediated CD44 silencing inhibits in vitro GBC cells’ proliferation, migration and invasion.
Notes: (A) Dose-dependent CD44-siRNA downregulated the expression of CD44. (B) CCK-8 assay showed that CD44 silencing attenuated proliferation of GBC-SD and NOZ cells. (C) EDU assay confirmed that CD44 knockdown suppresses proliferation of GBC-SD and NOZ cells. (D) CD44 silencing caused a remarkable suppression of cell migration in GBC-SD and NOZ cells using wound-healing assay. (E) The invasiveness of GBC-SD and NOZ cells infected with CD44-siRNA was significantly suppressed according to cell invasion assay. **P<0.01.
Abbreviations: RNAi, RNA interference; CD44, cluster of differentiation 44; GBC, gallbladder cancer; NC, nonspecific control siRNA.
Downregulation of CD44 suppresses colony formation ability of CSCs
Reports have shown that CD44+ GBC cells had higher spheroid colony forming ability in vitro, and this growth of spherical colonies was considered to reflect the self-renewal ability and phenotype of CSCs. The colony formation assay showed that GBC-SD cells treated with CD44-siRNA generated less colonies compared with the untreated control group (Figure 4A). We further performed a three-dimensional cell culture assay to confirm the abovementioned results in a mimic in vivo environment. In agreement with the results obtained from the two-dimensional colony formation assay, the cancer spheroids formed from GBC-SD cells under CD44 silencing were significantly smaller than the cells of the control group (Figure 4B). Hence, CD44 downregulation decreased the colony formation ability in GBC, which further revealed that CD44 might serve as a crucial factor in the progression and development of GBC, in part, through the suppression of CSCs.
Figure 4.
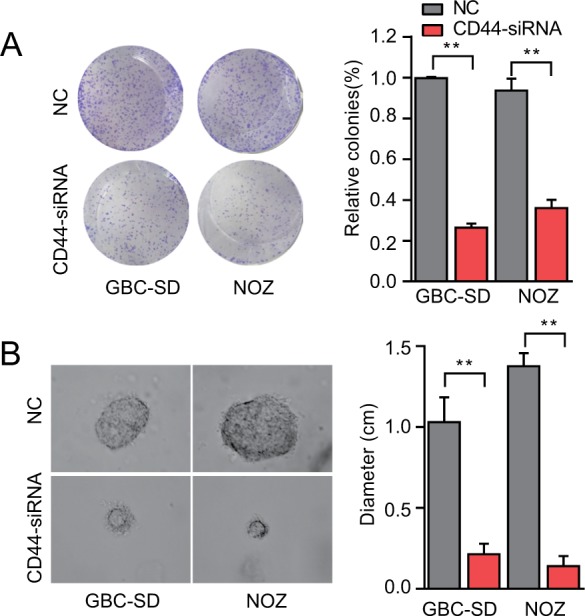
CD44 downregulation suppresses colony formation ability of CSCs.
Notes: (A) The colony formation ability of CSCs treated with CD44-siRNA was decreased. (B) Formation of spheres from GBC cells transfected with CD44-siRNA accessed by three-dimensional cell culture. The diameter of cancer spheroids formed from GBC-SD cells with CD44 silencing was significantly smaller than the control group. All the results were reproducible in three independent experiments. **P<0.01.
Abbreviations: CD44, cluster of differentiation 44; CSCs, cancer stem cells; GBC, gallbladder cancer; NC, nonspecific control siRNA.
Discussion
Despite advances in identifying risk factors for GBC, its mortality still remains high due to its delayed diagnosis and a lack of effective treatment strategies.20 Among GBC patients, five-year OS is less than 5% and mean survival time is less than 6 months. In this regard, it is critical to determine reliable tumor markers for the early diagnosis and a novel therapeutic strategy for patients with advanced GBC.
In the present study, we demonstrated that CD44 mRNA was clearly upregulated in hepatobiliary tumors and CD44 protein was overexpressed in GBC tissues in comparison with paired non-tumor tissues. Additionally, Kaplan–Meier survival analysis demonstrated that OS of patients with CD44 high-expression was significantly poorer than that of patients with CD44 low-expression. Meanwhile, we confirmed that elevated CD44 was an independent prognostic factor in GBC patients along with other well-established factors including metastasis and TNM stage by univariate and multivariate analyses. Consistent with our results, Kumar et al reported that CD44 was overexpressed and contributed to poor 5-year OS rate in pancreatic cancer patients.21 Furthermore, CD44 expression was notably high and associated with dismal clinical outcomes in cancers including non-small-cell lung cancer,22 breast cancer,16 oral squamous cell carcinoma,23 ovarian cancer24 and gastric cancer.25 All these findings suggested that CD44 could be a biomarker for early diagnosis or prognosis prediction of GBC.
In order to detect the biological function of CD44 on GBC progression, we performed RNAi to disrupt CD44 expression in GBC cells in vitro. Our data showed that downregulation of CD44 led to significant suppression of cell proliferation. Meanwhile, the migration and invasion abilities of GBC cells were obviously decreased after treatment with CD44-siRNA, implying that cD44 may function as an oncogene in GBC progression. In agreement with our results, Hu et al reported that knockdown of CD44 expression inhibited cell proliferation and colony formation in non-small cell lung carcinoma.26 Additionally, CD44 silencing could suppress triple-negative breast cancer cells’ growth and migration.27 Similar observations have also been reported in bladder cancer, malignant pleural mesothelioma as well as head and neck squamous cell carcinoma,28–30 strongly suggesting new research directions based on the potential of CD44 as a prognostic biomarker and therapeutic target in GBC.
CSCs, a small part of cells, are present in heterogeneous tumors, and play a critical role in the initiation and progression of cancer and exhibition of strong chemoresistance to conventional chemotherapeutic drug treatment.31–35 For these reasons, understanding the mechanism by which cancer cells enhance stemness may further facilitate cancer therapies. Yadav et al investigated germ line variants in CSC gene variants, and confirmed the essential role of CSCs in GBC susceptibility, prognosis and survival outcomes.12 Another study demonstrated that a CD44-positive/CD133-positive subpopulation revealed CSC-like characteristics and might represent the true CSC/tumor-initiating cell population in GBC.36 Given CD44 is the main surface marker typical for CSCs, we performed colony formation assay to explore the effect of CD44 downregulation on the growth of CSCs. Results showed that the colony forming ability of GBC was signally inhibited after being infected with CD44-siRNA. Moreover, a three-dimensional cell culture was used to mimic the in vivo environment to exclude the possibility of artifact induced by in vitro culture.
Conclusion
In summary, our findings provide significant insights into the progression of GBC and confirm that CD44 is overex-pressed in GBC and associated with prognosis and disease progression. This evidence indicates that CD44 can act as a CSC marker and a critical regulator of GBC progression. Additionally, CD44 silencing suppresses cell proliferation, migration, invasion and colony forming in GBC cells. Thus, CD44 is a potential novel inhibition target in the treatment of GBC.
Supplementary materials
Table S1.
Cell lines used in this study
| Cell lines | Cell type | Source | Country |
|---|---|---|---|
| GBC-SD | Gallbladder carcinoma cell | Cell Bank of the Chinese Academy of Science | Shanghai, China |
| NOZ | Gallbladder carcinoma cell | Cell Bank of the Chinese Academy of Science | Shanghai, China |
Abbreviation: GBC, gallbladder cancer.
Table S2.
Information on antibodies used in this study
| Antibody | WB | IHC | Specificity | Company |
|---|---|---|---|---|
| β-Actin | 1:5,000 | / | Mouse monoclonal | Proteintech Group, Wuhan, China |
| CD44 | 1:2000 | 1:100 | Rabbit monoclonal | Proteintech Group, Wuhan, China |
Abbreviations: WB, western blotting; IHC, imunohistochemistry
Table S3.
siRNA sequence used in this study
| siRNA | Sense sequence | Anti-sense sequence |
|---|---|---|
| CD44 | 5′-UGCCUUUGAUGGACCAAUU-3′ | 5′-AUUGGUCCAUCAAAGGCAU-3′ |
| NC | 5′-UUCUCCGAACGUGUCACGU-3′ | 5′-CGUGACACGUUCGGAGAAU-3′ |
Abbreviation: NC, nonspecifc control siRNA.
Table S4.
qRT-PCR primer sequence used in this study
| Name | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| CD44 | 5′-GACACATAGCTCAATGCTTCAGC-3′ | 5′-GATGCCAAGATGATCAGCCATTCTGCAAT-3′ |
| β-Actin | 5′-CTCCATCCTGGCCTCGCTGT-3′ | 5′-GCTGTCACCTTCACCGTTCC -3′ |
Acknowledgments
This study was supported by funds from the National Natural Science Foundation of China (81702757, 81702346, 81600506, 81702927); the Medicine Science and Technology research project of Henan province (201602032, 201702001, 201702032); Youth Innovation Fund of the First Affiliated Hospital of Zhengzhou University (YNQN2017167, RRS, YNQN2017031, YNQN2017032); Foundation of Henan Educational Committee (18A320038); The Joint Research Fund of the First Affiliated Hospital of Zhengzhou University and Dalian Institute of Chemical Physics Chinese Academy of Sciences (RRS and ZGR). The funding body had no role in the design of the study, collection, analysis, and interpretation of the data, or manuscript writing.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Kanthan R, Senger JL, Ahmed S, Kanthan SC. Gallbladder Cancer in the 21st Century. J Oncol. 2015;2015:967472–26. doi: 10.1155/2015/967472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dwivedi AN, Jain S, Dixit R. Gall bladder carcinoma: Aggressive malignancy with protean loco-regional and distant spread. World J Clin Cases. 2015;3(3):231–244. doi: 10.12998/wjcc.v3.i3.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma A, Sharma KL, Gupta A, Yadav A, Kumar A. Gallbladder cancer epidemiology, pathogenesis and molecular genetics: Recent update. World J Gastroenterol. 2017;23(22):3978–3998. doi: 10.3748/wjg.v23.i22.3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hundal R, Shaffer EA. Gallbladder cancer: epidemiology and outcome. Clin Epidemiol. 2014;6:99. doi: 10.2147/CLEP.S37357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baig M, Guarino M, Petrelli N. Report on demographics of gall bladder cancer in Delaware and retrospective review of treatment strategies for gallbladder cancer in a large community cancer center. Surg Oncol. 2016;25(2):86–91. doi: 10.1016/j.suronc.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Hsu JM, Xia W, Hsu YH, et al. STT3-dependent PD-L1 accumulation on cancer stem cells promotes immune evasion. Nat Commun. 2018;9(1):9. doi: 10.1038/s41467-018-04313-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toh TB, Lim JJ, Chow EK. Epigenetics in cancer stem cells. Mol Cancer. 2017;16(1):29. doi: 10.1186/s12943-017-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mcgrail DJ, Lin CC, Dai H, et al. Defective Replication Stress Response Is Inherently Linked to the Cancer Stem Cell Phenotype. Cell Rep. 2018;23(7):2095–2106. doi: 10.1016/j.celrep.2018.04.068. [DOI] [PubMed] [Google Scholar]
- 9.Mayr C, Ocker M, Ritter M, Pichler M, Neureiter D, Kiesslich T. Biliary tract cancer stem cells - translational options and challenges. World J Gastroenterol. 2017;23(14):2470–2482. doi: 10.3748/wjg.v23.i14.2470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leon G, Macdonagh L, Finn SP, Cuffe S, Barr MP. Cancer stem cells in drug resistant lung cancer: Targeting cell surface markers and signaling pathways. Pharmacol Ther. 2016;158:71–90. doi: 10.1016/j.pharmthera.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 11.Li H, Jin Y, Hu Y, et al. The PLGF/c-MYC/miR-19a axis promotes metastasis and stemness in gallbladder cancer. Cancer Sci. 2018;109(5):1532–1544. doi: 10.1111/cas.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yadav A, Gupta A, Rastogi N, et al. Association of cancer stem cell markers genetic variants with gallbladder cancer susceptibility, prognosis, and survival. Tumour Biol. 2016;37(2):1835–1844. doi: 10.1007/s13277-015-3929-6. [DOI] [PubMed] [Google Scholar]
- 13.Manhas J, Bhattacharya A, Agrawal SK, et al. Characterization of cancer stem cells from different grades of human colorectal cancer. Tumour Biol. 2016;37(10):14069–14081. doi: 10.1007/s13277-016-5232-6. [DOI] [PubMed] [Google Scholar]
- 14.Cho Y, Lee HW, Kang HG, Kim HY, Kim SJ, Chun KH. Cleaved CD44 intracellular domain supports activation of stemness factors and promotes tumorigenesis of breast cancer. Oncotarget. 2015;6(11):8709–8721. doi: 10.18632/oncotarget.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng Y, Wodzenski D, Gao D, et al. Stress-response protein RBM3 attenuates the stem-like properties of prostate cancer cells by interfering with CD44 variant splicing. Cancer Res. 2013;73(13):4123–4133. doi: 10.1158/0008-5472.CAN-12-1343. [DOI] [PubMed] [Google Scholar]
- 16.Xu H, Wu K, Tian Y, et al. CD44 correlates with clinicopathological characteristics and is upregulated by EGFR in breast cancer. Int J Oncol. 2016;49(4):1343–1350. doi: 10.3892/ijo.2016.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo JY, Hsu HS, Tyan SW, et al. Serglycin in tumor microenvironment promotes non-small cell lung cancer aggressiveness in a CD44-dependent manner. Oncogene. 2017;36(17):2457–2471. doi: 10.1038/onc.2016.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao S, Chen C, Chang K, et al. CD44 Expression Level and Isoform Contributes to Pancreatic Cancer Cell Plasticity, Invasiveness, and Response to Therapy. Clin Cancer Res. 2016;22(22):5592–5604. doi: 10.1158/1078-0432.CCR-15-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Y, Chen X, Yu Y, et al. LDHA is a direct target of miR-30d-5p and contributes to aggressive progression of gallbladder carcinoma. Mol Carcinog. 2018;57(6):772–783. doi: 10.1002/mc.22799. [DOI] [PubMed] [Google Scholar]
- 20.Randi G, Franceschi S, La Vecchia C. Gallbladder cancer worldwide: geographical distribution and risk factors. Int J Cancer. 2006;118(7):1591–1602. doi: 10.1002/ijc.21683. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S, Inigo JR, Kumar R, et al. Nimbolide reduces CD44 positive cell population and induces mitochondrial apoptosis in pancreatic cancer cells. Cancer Lett. 2018;413:82–93. doi: 10.1016/j.canlet.2017.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song JM, Molla K, Anandharaj A, et al. Triptolide suppresses the in vitro and in vivo growth of lung cancer cells by targeting hyaluronan-CD44/RHAMM signaling. Oncotarget. 2017;8(16):26927–26940. doi: 10.18632/oncotarget.15879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sawant S, Ahire C, Dongre H, et al. Prognostic significance of elevated serum CD44 levels in patients with oral squamous cell carcinoma. J Oral Pathol Med. 2018:665–673. doi: 10.1111/jop.12731. [DOI] [PubMed] [Google Scholar]
- 24.Puvanenthiran S, Essapen S, Haagsma B, et al. Co-expression and prognostic significance of the HER family members, EGFR-vIII, c-MET, CD44 in patients with ovarian cancer. Oncotarget. 2018;9(28):19662–19674. doi: 10.18632/oncotarget.24791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W, Jia H, Wang J, et al. A CD44-specific peptide, RP-1, exhibits capacities of assisting diagnosis and predicting prognosis of gastric cancer. Oncotarget. 2017;8(18):30063–30076. doi: 10.18632/oncotarget.16275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hu B, Ma Y, Yang Y, Zhang L, Han H, Chen J. CD44 promotes cell proliferation in non-small cell lung cancer. Oncol Lett. 2018;15(4):5627–5633. doi: 10.3892/ol.2018.8051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nam K, Oh S, Lee KM, Yoo SA, Shin I. CD44 regulates cell proliferation, migration, and invasion via modulation of c-Src transcription in human breast cancer cells. Cell Signal. 2015;27(9):1882–1894. doi: 10.1016/j.cellsig.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 28.Hao J, Madigan MC, Khatri A, et al. In vitro and in vivo prostate cancer metastasis and chemoresistance can be modulated by expression of either CD44 or CD147. PLoS One. 2012;7(8):e40716. doi: 10.1371/journal.pone.0040716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cortes-Dericks L, Schmid RA. CD44 and its ligand hyaluronan as potential biomarkers in malignant pleural mesothelioma: evidence and perspectives. Respir Res. 2017;18(1):58. doi: 10.1186/s12931-017-0546-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang SJ, Bourguignon LY. Role of hyaluronan-mediated CD44 signaling in head and neck squamous cell carcinoma progression and chemoresistance. Am J Pathol. 2011;178(3):956–963. doi: 10.1016/j.ajpath.2010.11.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaquero J, Lobe C, Tahraoui S, et al. The IGF2/IR/IGF1R Pathway in Tumor Cells and Myofibroblasts Mediates Resistance to EGFR Inhibition in Cholangiocarcinoma. Clin Cancer Res. 2018;24(17):4282–4296. doi: 10.1158/1078-0432.CCR-17-3725. [DOI] [PubMed] [Google Scholar]
- 32.Elaimy AL, Guru S, Chang C, et al. VEGF-neuropilin-2 signaling promotes stem-like traits in breast cancer cells by TAZ-mediated repression of the Rac GAP β2-chimaerin. Sci Signal. 2018;11(528):eaao6897. doi: 10.1126/scisignal.aao6897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin H, Wang B, Yu J, Wang J, Li Q, Cao B. Protein arginine methyltransferase 8 gene enhances the colon cancer stem cell (CSC) function by upregulating the pluripotency transcription factor. J Cancer. 2018;9(8):1394–1402. doi: 10.7150/jca.23835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang T, Xu J, Deng S, et al. Core signaling pathways in ovarian cancer stem cell revealed by integrative analysis of multi-marker genomics data. PLoS One. 2018;13(5):e0196351. doi: 10.1371/journal.pone.0196351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen J, Cao X, An Q, et al. Inhibition of cancer stem cell like cells by a synthetic retinoid. Nat Commun. 2018;9(1):1406. doi: 10.1038/s41467-018-03877-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi C, Tian R, Wang M, et al. CD44+ CD133+ population exhibits cancer stem cell-like characteristics in human gallbladder carcinoma. Cancer Biol Ther. 2010;10(11):1182–1190. doi: 10.4161/cbt.10.11.13664. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Cell lines used in this study
| Cell lines | Cell type | Source | Country |
|---|---|---|---|
| GBC-SD | Gallbladder carcinoma cell | Cell Bank of the Chinese Academy of Science | Shanghai, China |
| NOZ | Gallbladder carcinoma cell | Cell Bank of the Chinese Academy of Science | Shanghai, China |
Abbreviation: GBC, gallbladder cancer.
Table S2.
Information on antibodies used in this study
| Antibody | WB | IHC | Specificity | Company |
|---|---|---|---|---|
| β-Actin | 1:5,000 | / | Mouse monoclonal | Proteintech Group, Wuhan, China |
| CD44 | 1:2000 | 1:100 | Rabbit monoclonal | Proteintech Group, Wuhan, China |
Abbreviations: WB, western blotting; IHC, imunohistochemistry
Table S3.
siRNA sequence used in this study
| siRNA | Sense sequence | Anti-sense sequence |
|---|---|---|
| CD44 | 5′-UGCCUUUGAUGGACCAAUU-3′ | 5′-AUUGGUCCAUCAAAGGCAU-3′ |
| NC | 5′-UUCUCCGAACGUGUCACGU-3′ | 5′-CGUGACACGUUCGGAGAAU-3′ |
Abbreviation: NC, nonspecifc control siRNA.
Table S4.
qRT-PCR primer sequence used in this study
| Name | Forward primer sequence | Reverse primer sequence |
|---|---|---|
| CD44 | 5′-GACACATAGCTCAATGCTTCAGC-3′ | 5′-GATGCCAAGATGATCAGCCATTCTGCAAT-3′ |
| β-Actin | 5′-CTCCATCCTGGCCTCGCTGT-3′ | 5′-GCTGTCACCTTCACCGTTCC -3′ |




