Abstract
Objective
The present study planned to investigate the changes in the mRNA expression of inflammatory genes and their association with colorectal cancer (CRC). Our findings could be useful for noninvasive early screening of CRC patients.
Patients and methods
Venous blood of 20 CRC cases and 15 healthy controls was collected. The mRNA expression of COX-2, TNF-α, NF-κB and IL-6 genes was carried out by using real-time polymerase chain reaction. Relative quantification was done to find out the fold change of these genes.
Results
The mean age of cases and controls was 55 and 50 years, respectively. The ΔCt of COX-2, TNF-α, NF-κB and IL-6 genes was significantly (p < 0.05) lower in cases as compared to controls. Subsequently, the mRNA expression of these genes was, respectively, 3.56-, 3.4-, 1.71- and 3.86-fold higher in CRC cases as compared to controls. Positive correlation of ΔCt of COX-2 was found with ΔCt of TNF-α (r = 0.461, p = 0.041) and NF-κB (r = 0.536, p = 0.015) in CRC cases. The mRNA expression of COX-2 was significantly lower in T2 stage, while mRNA expression of NF-κB was significantly lower in both T2 and T3 stages of CRC as compared to T4 stage.
Conclusion
The increased mRNA expression of COX-2 along with the high mRNA expression of TNF-α, NF-κB and IL-6 genes may be associative risk factors for CRC. COX-2 and NF-κB genes were more expressed in advanced stages of CRC indicating their role in tumor progression. Our findings support the possible role of blood biomarker in the screening of CRC patients in the early stages.
Keywords: colorectal cancer, mRNA expression, COX-2, TNF-α, NF-κB, IL-6, blood biomarker
Introduction
Colorectal cancer (CRC) is the fourth leading cause of cancer-related mortality worldwide. It is the third most diagnosed cancer in males and second in females.1 The incidence of CRC is on the rise among the Saudi population, particularly in young females.2 The development of CRC is multifactorial which may include genetics, environmental factors, dietary habits, smoking, lifestyle and pathological conditions.3,4 In developed nations, early screening of CRC in asymptomatic population has led to a recent decline in its incidence. The relative survival rate of CRC patients was reported to be up to 5%, with the survival rate up to 65%, in the US population in the last 35 years.5
Presently, screening methods include fecal occult blood testing, flexible sigmoidoscopy, colonoscopy, barium enema, CT colonography and methylation changes in fecal DNA.6,7 However, these methods are not widely accepted for screening the general population, possibly because of patient discomfort and distaste for the procedures, high cost, low sensitivity/specificity, etc. Although the recent technological advancement in diagnosis clearly revealed the biology and genetics of CRC, its exact etiology is still under investigation. Noninvasive screening methods could be more comprehensive to start the early diagnosis, reducing disease incidence and mortality.
The blood-based assay is a noninvasive, easily accessible and early detection screening tool which could be used for detection of circulating tumor cells originated from the primary tumor site.8,9 Its accessibility is also because of higher compliance by the CRC patients, high feasibility, no requirement of colonoscopy, reduction of cost, etc.
Prostaglandins (PGs) play a predominant role in cancer development through promoting cell proliferation, invasion, angiogenesis and immunosuppression.10 The COX-2-regulated synthesis of PGs, particularly PGE-2, causes various types of cancers including CRC.11 COX-2 gene is highly expressed in 85% of adenocarcinoma cases with a poor survival rate.12
NF-κB, IL-6 and TNF-α have been shown to regulate COX-2 gene expression, and these genes were also reported to be more expressed in CRC cases.13,14 Thus, the regulated expression of COX-2 gene is important for minimizing the risk of CRC and normal physiology. Although many studies have been conducted for the identification of mRNA blood biomarker alternative to surgical and tissue biopsy,8,9,11,15,16 an established and conclusive blood marker does not exist. The mRNA expression in blood epithelial cells could be used as an alternative to surgical and tissue biopsy.
Patients and methods
In the present study, a total of 20 CRC cases and 15 controls were recruited from the Department of Gastroenterology, Prince Sultan Military Medical City (PSMMC), Riyadh. The ethical clearance was obtained from the institutional ethical clearance committee, PSMMC, Riyadh. Written informed consent was taken from each of the participants in the study. Pathologically confirmed cases of adenocarcinoma were referred for surgery. The controls were age-matched and were free from any chronic, communicable or noncommunicable diseases.
RNA isolation and synthesis of cDNA
Venous blood (2 mL) was drawn in EDTA vials from each subject. The blood samples were fixed with TRI Reagent BD (Sigma-Aldrich, St. Louis, MO, USA), stored at −80°C and processed for total RNA isolation as per the manufacturer’s instructions (Sigma-Aldrich). The concentration and purity of total RNA were determined using NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). The quality and integrity of the RNA were determined by the ratio of absorbance at 260/280 nm, with values between 1.8 and 2.0 considered to indicate good quality. The quality of RNA was further checked by agarose gel electrophoresis by mixing with RNA gel loading dye (Thermo Fisher Scientific Inc.). The 28s rRNA band was found to be more intense than the 18s rRNA band, and there was no intense smearing down from the 28s and 18s bands. Any sample having degraded RNA was considered low quality and was excluded from the study. On the same day, total RNA (2 µg) was converted into single-strand cDNA using Maxima first-strand cDNA synthesis kit (Thermo Fisher Scientific Inc.) as per the manufacturer’s protocol. cDNA was further diluted four times in nuclease-free water (Thermo Fisher Scientific Inc.) before use in the real-time quantitative polymerase chain reaction (RT-qPCR). The volume of cDNA did not comprise >1/10 of the total PCR volume.
An RT-qPCR experiment was conducted to measure the mRNA expression of inflammatory genes. The reactions were performed on LightCycler 480 II, RT-qPCR system (Roche, Basel, Switzerland). Briefly, the PCR amplification master mix of 45 µL contained 6.75 µL of diluted cDNA, 10 µL of SYBR green master mix (Thermo Fisher Scientific Inc.), 10 pmol each of forward and reverse specific primer pairs and 4 µL of nuclease-free water. Out of 45 µL master mix, 20 µL was dispensed into two PCR tubes. Duplicate of each sample along with no-template control wells was kept for PCR amplification. The primer sequences and annealing temperatures used for COX-2, TNF-α, NF-κB and IL-6 genes are mentioned in Table 1. The thermal profile for COX-2 gene consisted of initial denaturation for 1 min at 95°C, 40 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 55°C and extension for 30 s at 72°C. In this analysis, gene expression normalization was done by using GAPDH, a constitutive gene, and its primer sequences are also shown in Table 1. The amplification program was similar for TNF-α, NF-κB, IL-6 and GAPDH genes except that annealing temperatures were kept at 50, 55, 50 and 55°C, respectively.
Table 1.
Primers sequence and annealing temperature used for RT-PCR amplification
| Genes | Primer sequence | Annealing temperature (°C) |
|---|---|---|
| COX-2 | Forward: 5′-CCGGGTACAATCGCACTTAT-3′ | 55 |
| Reverse: 5′-GGCGCTCAGCCATACAG-3′ | ||
| TNF-α | Forward: 5′-TCTGGGCAGGTCTACTTTGG-3′ | 50 |
| Reverse: 5′-TCTTCTCAAGTCCTGCAGCA-3′ | ||
| NF-κB | Forward: 5′-AAGACCCACCCCACCATCAA-3′ | 55 |
| Reverse: 5′-AAACTGTGGATGCAGCAGCGGTC-3′ | ||
| IL-6 | Forward: 5′-AAACAACCTGAACCTTCCAAAGA-3′ | 50 |
| Reverse: 5′-GCAAGTCTCCTCATTGAATCCA-3′ | ||
| GAPDH | Forward: 5′-CAGTGATGGCATGGACTGTG-3′ | 55 |
| Reverse: 5′-CACATCGCTGAGACACCA-3′ |
Abbreviation: RT-PCR, real-time polymerase chain reaction.
Cumulative fluorescence was measured at the end of the extension phase of each cycle. The amplified product was confirmed for specification by melt curve analysis using LC 480 II melt curve analysis software (Roche). The amplified product was also run on 2% agarose gel (Sigma-Aldrich) to check the specific amplification. Relative quantification was done to see the expression pattern of different genes. In RT-qPCR assay, a positive reaction is detected by the accumulation of fluorescent signals, and Ct (threshold cycle) is the number of cycles required for the fluorescent signal to cross the threshold (i.e. exceeds background level). Ct value is inversely proportional to the amount of target nucleic acid in the sample. Here, ΔCt = average Ct of target – average Ct of normalizer. The difference of mean Ct values between test and control was determined as ΔΔCt, where ΔΔCt = ΔCt control − ΔCt test. Further, true fold change (FC) was represented as follows: if FC > 1, true FC = FC and if FC < 1, true FC = −1/FC.17
Statistical analysis
Statistical software SSPS for Windows, version 16.0 (SPSS Inc., Chicago, IL, USA), was used for data presentation and statistical analysis. Unpaired Student’s t-test was applied to compare quantitative data. Spearman’s coefficient of correlation test was applied to find out correlation between ΔCt of COX-2 and ΔCt of TNF-α, NF-κB and IL-6 genes if any. p < 0.05 was considered as significant.
Results
The mean age of cases and controls was 55 and 50 years, respectively; 55% of the cases were <60 years old, and 45% were >60 years. The present study included equal number of males and females as controls. However, among CRC cases, the proportion of male patients was 35%, while that of female patients was 65%. In the present study, 70% of the tumors were localized in descending colon and 30% in ascending colon. The data stratification based on tumor stages revealed that 40% of the patients were in T3 stage of CRC followed by 30% each in T4 and T2 stages. When the patients were grouped based on differentiation grade into well, moderate and poor, 80% of the cases were found in moderate grade followed by well and poor grade of CRC. Lymph node metastasis in one to three regional lymph nodes (N1) was found in 35% of the cases, while metastasis in four or more lymph nodes (N2) was reported in 20% of the cases. Distant metastasis was reported in 35% of the CRC cases (Table 2).
Table 2.
clinical and pathological features of CRC cases
| Characteristics | Number of patients (%) |
|---|---|
| Gender | |
| Male | 7 (35) |
| Female | 13 (65) |
| Age | |
| <60 years | 11 (55) |
| >60 years | 9 (45) |
| Tumor localization | |
| Ascending colon | 6 (30) |
| Descending colon | 14 (70) |
| Tumor stage | |
| T2 | 6 (30) |
| T3 | 8 (40) |
| T4 | 6 (30) |
| Differentiation grade | |
| Well | 3 (15) |
| Moderate | 16 (80) |
| Poor | 1 (5) |
| Lymph node metastasis | 11 (55) |
| N1 | 7 (35) |
| N2 | 4 (20) |
| Distant metastasis (M1) | 7 (35) |
Note: T2: tumor growth in muscularis propria; T3: tumor growth in pericolorectal tissues; T4: tumor invaded to adjacent organ; N1: metastasis in one to three regional lymph nodes; N2: metastasis in four or more lymph nodes; M1: metastasis to distant site.
Abbreviation: CRC, colorectal cancer.
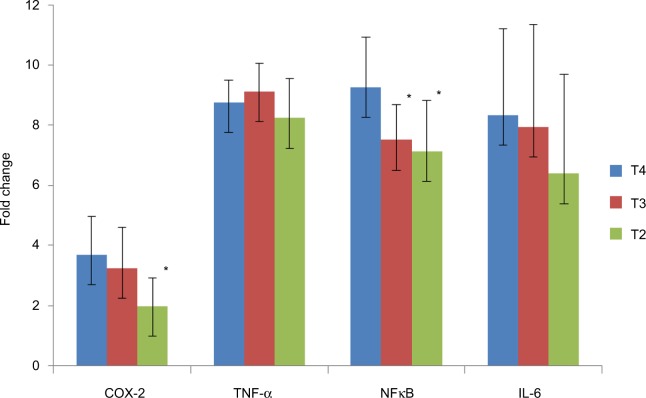
The ΔCt of COX-2, TNF-α, NF-κB and IL-6 genes was significantly (p < 0.05) lower in CRC cases as compared to controls. The mRNA expression of COX-2, TNF-α, NF-κB and IL-6 genes was, respectively, 3.56-, 3.40-, 1.71- and 3.86-fold higher in cases as compared to controls (Table 3). When the data were analyzed to find the correlation between ΔCt of COX-2 and ΔCt of TNF-α, NF-κB and IL-6 genes, we found significant positive correlation of ΔCt of COX-2 with ΔCt of TNF-α (r = 0.461, p = 0.041) and NF-κB (r = 0.536, p = 0.015) in CRC cases (Figures 1 and 2). We did not find any significant association of mRNA expression of COX-2, TNF-α, NF-κB and IL-6 genes with gender, age, tumor localization, lymph node metastasis and the presence of distance metastases. However, COX-2 mRNA expression was significantly (p < 0.05) lower in T2 stage of CRC, and NF-κB mRNA expression was significantly (p < 0.05) lower in both T2 and T3 stages of CRC as compared to T4 stage (Figure 3).
Table 3.
comparison of expression of COX-2, TNF-α, NF-κB and IL-6 genes in CRC cases and controls
| ΔCt of genes | Controls (mean ± SD) | Cases (mean ± SD) | p values | Fold change |
|---|---|---|---|---|
| COX-2 | 6.54 ± 0.487 | 2.98 ± 1.40 | 0.001* | ↑ 3.56 |
| TNF-α | 12.15 ± 0.936 | 8.75 ± 1.06 | 0.001* | ↑ 3.40 |
| NF-κB | 9.85 ± 1.23 | 7.92 ± 1.72 | 0.001* | ↑ 1.71 |
| IL-6 | 11.46 ± 1.47 | 7.60 ± 3.19 | 0.001* | ↑ 3.86 |
Note:
p < 0.05 was considered as significant.
Abbreviation: CRC, colorectal cancer.
Figure 1.
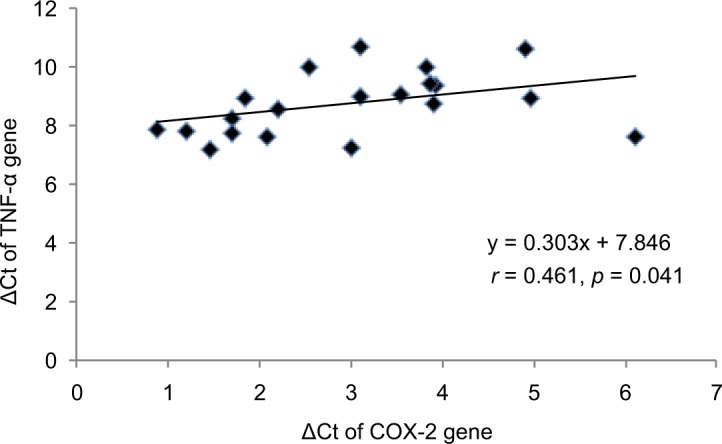
Correlation between ΔCt of COX-2 and TNF-α genes in CRC cases. spearman’s correlation (r) was applied to analyze the correlation data. p < 0.05 was considered as significant.
Abbreviation: CRC, colorectal cancer.
Figure 2.
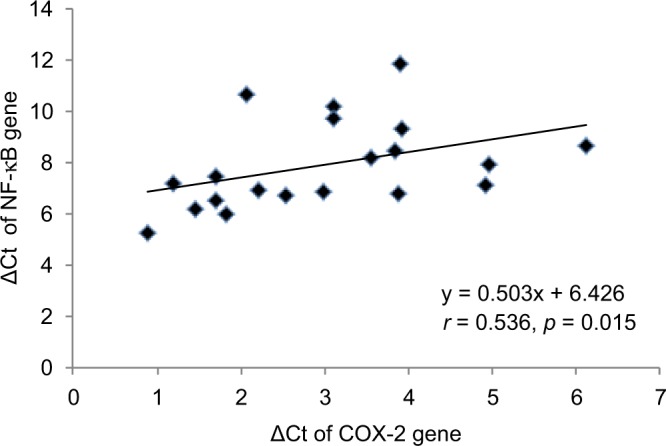
Correlation between ΔCt of COX-2 and NF-κB genes in CRC cases. Spearman’s correlation (r) was applied to analyze the correlation data. p < 0.05 was considered as significant.
Abbreviation: CRC, colorectal cancer.
Figure 3.
Comparison of mRNA expression of COX-2, TNF-α, NF-κB and IL-6 genes in T3 and T4 stages with mRNA expression of these genes in T2 stage of CRC. T2: tumor growth in muscularis propria; T3: tumor growth in pericolorectal tissues; T4: tumor invaded to adjacent organ. *p < 0.05 was considered as significant.
Abbreviation: CRC, colorectal cancer.
Discussion
Our study demonstrates the ΔCt of COX-2, TNF-α, NF-κB and IL-6 genes was significantly lower in peripheral blood of CRC cases as compared to healthy controls (Table 3). Accordingly, the FC of COX-2, TNF-α, NF-κB and IL-6 genes was higher in cases as compared to controls (Table 3). We also found significant positive correlation between ΔCt of COX-2 and ΔCt of TNF-α and NF-κB in CRC cases (Figures 1 and 2). There are numerous studies which revealed the role of COX-2 upregulation as a key setup in the colorectal tumorigenesis.15,18,19 Klampfer reported COX-2 gene expression was elevated in 50% of adenomas and in 85% of adenocarcinomas cases.20 The experimental studies also supported that overexpression of COX-2 causes tumorigenesis and its inhibition results in reduction in tumor incidence and progression.21 COX-2 enzyme acts as a link between inflammation and cancer through its involvement in the synthesis of prostaglandin, especially PGE2, which can promote cancer by stimulating angiogenesis, cell invasions, cell growth and cell survival.22 It may also change adhesion properties of extracellular matrix, and enhance apoptotic resistance and metastatic potential which could enhance cancer progression.23 The strong positive relation between COX-2 expression and production of PGE2 in tumor tissue has been reported earlier.24 Although its role in tumorigenesis is now well known, COX-2 could not be used as a gene therapy target and diagnostic marker because of conflicting reports, with some supporting its independent role in cancer while others did not.
The mechanism of cancer development is very complex which includes cell-to-cell interaction, cell signaling, environmental factors and gene–environment interaction. Therefore, we intended to evaluate the relationship between high COX-2 expression and other factors of known importance for tumor cell division, apoptosis and metastasis.
The COX-2 gene is located on chromosome 1 having NF-κB, IL-6 and TNF-α response elements on its promoter region. Therefore, it can be regulated by TNF-α, NF-κB and IL-6.25 Hence, targeting the expression of NF-κB, IL-6 and TNF-α is a matter of importance because it ultimately targets the expression of COX-2. Moreover, COX-2 inhibition may trigger apoptosis of tumor cells and/or sensitize them to cytotoxic treatments indicating that COX-2 could be used as a prognostic marker.26
NF-κB is an important pro-inflammatory transcription factor that regulates COX-2 gene expression.27,28 Yu et al29 reported the role of NF-κB in transition from colorectal adenoma with low-grade dysplasia to adenocarcinoma. The activated NF-κB translocates into the nucleus and may induce the expression of TNF-α and IL-6 and chemokines, all contributing to inflammation-related tissue damage. This could lead to aberrant upregulation of certain tumorigenic factors, adhesion proteins, chemokines and inhibitors of apoptosis that promote cell survival.29,30 The IL-6 is a multifunctional NF-κB-regulated cytokine that acts on epithelial and immune cells and regulates pre-neoplastic growth during CRC tumorigenesis.31 The effect of IL-6 in cancer is mediated by a transcription factor STAT3. NF-κB and STAT3 are constitutively activated in a variety of tumors, and their role in initiation and progression of the tumorigenic process has been revealed. Both can bind to the endogenous COX-2 promoter leading to the transcriptional regulation of the COX-2 gene.32 The increased gene expression of TNF-α may increase the production of reactive oxygen species leading to DNA damage and tumor initiation. The activated NF-κB may also induce TNF-α production leading to further increase of NF-κB activation after binding to TNF receptor.33
We also observed that the COX-2 mRNA expression was significantly (p < 0.05) lower in T2 stage, while NF-κB mRNA expression was significantly (p < 0.05) lower in both T2 and T3 stages of CRC as compared to T4 stage (Figure 3). Our findings of decreased expression of COX-2 and NF-κB genes in low-grade carcinomas suggested that the upregulation of these genes could occur at later stages of tumor progression from low- to high-grade tumors. Our findings are corroborated with previous studies showing high expression of COX-2 and NF-κB genes in higher stages of tumors.34
Conclusion
The present study demonstrated the upregulation of COX-2, TNF-α, NF-κB and IL-6 genes may be associated with the increased risk of CRC. Inhibition of COX-2 pathway could be a possible therapeutic target in the treatment of adenocarcinoma of the colon. The mRNA expression of these genes in blood epithelial cells could be used as a biomarker, an alternative to surgical and tissue biopsy. Further studies on the role of these genes in this pathway and other malignancies in the large blood samples are recommended to develop a blood-based test to screen asymptomatic CRC patients.
Acknowledgments
The authors are thankful to Mohammed Mahmiyaha Al-Asmari and S. Sadaf Rizvi for their help with laboratory work. No external grants or other support were received for conducting this study.
Footnotes
Author contributions
All authors contributed substantially to conception and design, acquisition of data, analysis and interpretation of data and drafting of the manuscript and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Alsanea N, Abduljabbar AS, Alhomoud S, Ashari LH, Hibbert D, Bazarbashi S. Colorectal cancer in Saudi Arabia: incidence, survival, demographics and implications for national policies. Ann Saudi Med. 2015;35(3):196–202. doi: 10.5144/0256-4947.2015.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arem H, Mayne ST, Sampson J, Risch H, Stolzenberg Solomon RZ. Dietary fat intake and risk of pancreatic cancer in the prostate, lung, colorectal and ovarian cancer screening trial. Ann Epidemiol. 2013;23(9):571–575. doi: 10.1016/j.annepidem.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Otani T, Iwasaki M, Yamamoto S, et al. Alcohol consumption, smoking, and subsequent risk of colorectal cancer in middle-aged and elderly Japanese men and women: Japan Public Health Center-based prospective study. Cancer Epidemiol Biomark Prev. 2003;12(12):1492–1500. [PubMed] [Google Scholar]
- 5.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 6.Nicholson FB, Barro JL, Atkin W, et al. Population screening for colorec-tal cancer. Aliment Pharmacol Ther. 2005;22(11–12):1069–1077. doi: 10.1111/j.1365-2036.2005.02695.x. [DOI] [PubMed] [Google Scholar]
- 7.Muller HM, Oberwalder M, Fiegl H, et al. Methylation changes in faecal DNA: a marker for colorectal cancer screening? Lancet. 2004;363(9417):1283–1285. doi: 10.1016/S0140-6736(04)16002-9. [DOI] [PubMed] [Google Scholar]
- 8.Solmi R, Ugolini G, Rosati G, et al. Microarray-based identification and RT-PCR test screening for epithelial-specific mRNAs in peripheral blood of patients with colon cancer. BMC Cancer. 2006;6:250. doi: 10.1186/1471-2407-6-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lauriola M, Ugolini G, Rosati G, et al. Identification by a Digital Gene Expression Displayer (DGED) and test by RT-PCR analysis of new mRNA candidate markers for colorectal cancer in peripheral blood. Int J Oncol. 2010;37(2):519–625. doi: 10.3892/ijo_00000701. [DOI] [PubMed] [Google Scholar]
- 10.Elzagheid A, Emaetig F, Elsaghayer W, et al. Neurofibromin expression is associated with aggressive disease and poor outcome in colorectal carcinoma. Anticancer Res. 2016;36(10):5301–5306. doi: 10.21873/anticanres.11102. [DOI] [PubMed] [Google Scholar]
- 11.Martin KJ, Graner E, Li Y, et al. High-sensitivity array analysis of gene expression for the early detection of disseminated breast tumor cells in peripheral blood. Proc Natl Acad Sci. 2001;98:2646–2651. doi: 10.1073/pnas.041622398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta RA, Dubois RN. Colorectal cancer prevention and treatment by inhibition of cyclooxygenase-2. Nat Rev Cancer. 2001;1(1):11–21. doi: 10.1038/35094017. [DOI] [PubMed] [Google Scholar]
- 13.Ke J, Long X, Liu Y. Role of NF-kappaB in TNF-alpha-induced COX-2 expression in synovial fibroblasts from human TMJ. J Dent Res. 2007;86(4):363–367. doi: 10.1177/154405910708600412. [DOI] [PubMed] [Google Scholar]
- 14.Asting AG, Carén H, Andersson M, Lönnroth C, Lagerstedt K, Lund-holm K. COX-2 gene expression in colon cancer tissue related to regulating factors and promoter methylation status. BMC Cancer. 2011;11:238. doi: 10.1186/1471-2407-11-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elzagheid D, Fatma Emaetig F, Alkikhiahigh L, et al. Cyclooxygenase-2 expression is associated with advanced stages in colorectal cancer. Anticancer Res. 2013;33(8):3137–3144. [PubMed] [Google Scholar]
- 16.Hatakeyama K, Yamakawa Y, Fukuda Y, et al. A novel splice variant of XIAP-associated factor 1 (XAF1) is expressed in peripheral blood containing gastric cancer-derived circulating tumor cells. Gastric Cancer. 2015;18(4):751–761. doi: 10.1007/s10120-014-0426-3. [DOI] [PubMed] [Google Scholar]
- 17.Enquobahrie DA, Williams MA, Qiu C, et al. Early pregnancy peripheral blood gene expression and risk of preterm delivery: a nested case control study. BMC Pregnancy Childbirth. 2009;9:56. doi: 10.1186/1471-2393-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdullah M, Rani AA, Sudoyo AW, Makmun D, Handjari DR, Hernowo BS. Expression of NF-kB and COX2 in colorectal cancer among native Indonesians: the role of inflammation in colorectal carcinogenesis. Acta Med Indones. 2013;45(3):187–192. [PubMed] [Google Scholar]
- 19.Rothwell PM, Fowkes FG, Belch JF, Ogawa H, Warlow CP, Meade TW. Effect of daily aspirin on long-term risk of death due to cancer: analysis of individual patient data from randomised trials. Lancet. 2011;377(9759):31–41. doi: 10.1016/S0140-6736(10)62110-1. [DOI] [PubMed] [Google Scholar]
- 20.Klampfer L. Cytokines, inflammation and colon cancer. Curr Cancer Drug Targets. 2011;11(4):451–464. doi: 10.2174/156800911795538066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Y, Prescott SM. Many actions of cyclooxygenase-2 in cellular dynamics and in cancer. J Cell Physiol. 2002;190:279–286. doi: 10.1002/jcp.10068. [DOI] [PubMed] [Google Scholar]
- 22.Wang D, Wang H, Brown J, et al. CXCL1 induced by prostaglandin E2 promotes angiogenesis in colorectal cancer. J Exp Med. 2006;203:941–951. doi: 10.1084/jem.20052124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cahlin C, Lonnroth C, Arvidsson A, Nordgren S, Lundholm K. Growth associated proteins in tumor cells and stroma related to disease progression of colon cancer accounting for tumor tissue PGE2 content. Int J Oncol. 2008;32(4):909–918. [PubMed] [Google Scholar]
- 24.Soumaoro LT, Uetake H, Higuchi T, Takagi Y, Enomoto M, Sugihara K. Cyclooxygenase-2 expression: a significant prognostic indicator for patients with colorectal cancer. Clin Cancer Res. 2004;10(24):8465–8471. doi: 10.1158/1078-0432.CCR-04-0653. [DOI] [PubMed] [Google Scholar]
- 25.Ramsay RG, Ciznadija D, Vanevski M, Mantamadiotis T. Transcriptional regulation of cyclooxygenase expression: three pillars of control. Int J Immunopathol Pharmacol. 2003;16(2):59–67. [PubMed] [Google Scholar]
- 26.Peng L, Zhou Y, Wang Y, Mou H, Zhao Q. Prognostic significance of COX-2 immunohistochemical expression in colorectal cancer: a meta-analysis of the literature. PLoS One. 2013;8(3):e58891. doi: 10.1371/journal.pone.0058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakamoto K, Maeda S, Hikiba Y, et al. Constitutive NF-kappaB activation in colorectal carcinoma plays a key role in angiogenesis, promoting tumor growth. Clin Cancer Res. 2009;15(7):2248–2258. doi: 10.1158/1078-0432.CCR-08-1383. [DOI] [PubMed] [Google Scholar]
- 28.Wang S, Liu Z, Wang L, Zhang X. NF-kappaB signaling pathway, inflammation and colorectal cancer. Cell Mol Immunol. 2009;6(5):327–334. doi: 10.1038/cmi.2009.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu HG, Yu LL, Yang Y, et al. Increased expression of RelA/nuclear factor-kappa B protein correlates with colorectal tumorigenesis. Oncology. 2003;65(1):37–45. doi: 10.1159/000071203. [DOI] [PubMed] [Google Scholar]
- 30.Karin M, Cao Y, Greten FR, Li ZW. NF-κB in cancer: from innocent bystander to major culprit. Nat Rev Cancer. 2002;2(4):301–310. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 31.Greten FR, Eckmann L, Greten TF, et al. IKK®. links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell. 2004;118(3):285–296. doi: 10.1016/j.cell.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 32.Gong J, Xie J, Bedolla R, Rivas P, Chakravarthy D, Freeman JW. Combined targeting of STAT3/NF-κB/COX-2/EP4 for effective management of pancreatic cancer. Clin Cancer Res. 2014;20(5):1259–1273. doi: 10.1158/1078-0432.CCR-13-1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O’Connor PM, Lapointe TK, Beck PL, Buret AG. Mechanisms by which inflammation may increase intestinal cancer risk in inflammatory bowel disease. Inflamm Bowel Dis. 2010;16(8):1411–1420. doi: 10.1002/ibd.21217. [DOI] [PubMed] [Google Scholar]
- 34.Kömhoff M, Guan Y, Shappell HW, Davis L, Jack G, Shyr Y, et al. Enhanced expression of cyclooxygenase-2 in high grade human transitional cell bladder carcinomas. Am J Pathol. 2000;157(1):29–35. doi: 10.1016/S0002-9440(10)64513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]