Abstract
Background
Knowledge of drug-sensitivity patterns of Mycobacterium tuberculosis complex (MTBC) strains isolated from patients is an important aspect of TB control strategy. This study was conducted to evaluate the drug sensitivity of MTBC isolates in South Omo, southern Ethiopia.
Materials and methods
A total of 161 MTBC isolates (153 from new cases and eight re-treatment TB cases) were isolated using Lowenstein Jensen medium of which 126 isolates were able to be tested for drug sensitivity by BACTEC™MGIT™ 960 system, while all the 161 isolates were tested by GenoType® MTBDRplus VER 2.0. Descriptive statistics and logistic regression were used to express and present results.
Results
On the basis of MGIT 960 system, the prevalence of mono-resistance was 9.2% (11/119) in the new cases, although neither poly-resistance nor multidrug resistance (MDR) was recorded in these cases. On the basis of GenoType MTBDRplus assay, two of the 153 isolates (1.3%) of the new cases were mono-resistant for rifampicin (RIF) and one of these isolates had known rpoB gene mutation (H526D). One of the eight (12.5%) isolates obtained from the re-treatment cases was MDR with rpoB gene mutation (D516V) and katG gene mutation (S315T2). Taking MGIT 960 system as a gold standard, the sensitivities of the MTBDRplus assay were 33.3%, 100% and 100% for detection of resistance to isoniazid, RIF and MDR, respectively. On the other hand, its specificities were 99.2%, 100% and 100% for detection of resistance to RIF, isoniazid and MDR, respectively.
Conclusion
The magnitude of drug resistance was relatively low in the new TB cases of South Omo as compared to the reports from the other regions of the country. This is encouraging and hence the TB Control Program in the Zone should strengthen its program so that the emergence of drug resistance is inhibited.
Keywords: drug resistance, MTBDRplus, MGIT 960
Introduction
Tuberculosis (TB) is a disease caused by Mycobacterium tuberculosis complex (MTBC). It is one of the leading causes of deaths from single infectious diseases and ninth from any cause of deaths worldwide.1 The therapeutic effectiveness of agents plays a very crucial role in the control of TB. The discovery of effective TB treatment, streptomycin (STR) in 1940s2,3 to rifampicin (RIF) in 1960s,4,5 made experts in the area to feel that it is time to end TB. But the emergence of resistant strains of M. tuberculosis has created a major threat to the control of TB.6 Multidrug resistant TB (MDR-TB) has reached to epidemic proportions in some countries.7
Globally, the proportions of MDR/RIF-resistant TB were estimated to be 4.1% in new TB cases and 19.0% in previously treated TB cases in 2016.1 About 6.2% MDR-TB cases were extensively drug resistant (XDR)-TB.1 Ethiopia stands 14th among the 30 high MDR-TB burden countries.1 Early detection of drug resistance is essential for appropriate treatment and prevention of further spread of resistant strains.8 However, drug sensitivity test (DST) is not accessible to all TB patients in Ethiopia due to inadequate laboratory capacity, inadequate transport network and resource limitation.9,10 Phenotypic DST is considered as the gold standard but it is done in limited laboratories in Ethiopia. Accordingly, 10.0% of pulmonary MDR-TB cases were detected and the patients received treatment.11 The remaining 90% were most likely undiagnosed and being a source of infection or started treatment based on patient history. Therefore, strengthening the DST in the country is important.
Liquid culture: BACTEC MGIT 960 system12 and molecular techniques including GenoTypeMTBDRplus13 and Gene Xpert MTB/RIF,14 can detect drug resistance easier and faster. The presence of a mutation in rpoB gene15 leads to resistance for RIF and mutations in inhA gene16 and/or in katG gene17 lead to resistance for isoniazid (INH). BACTEC MGIT System, GeneXpert MTB/RIF and GenoTypeMTB-DRplus assays are recently introduced in Ethiopia and are used for the detection of MDR-TB in selected health facilities and research centers. Although molecular assays can rapidly detect MTBC and mutation associated with drug resistance in a day(s), it should be complemented by phenotypic DST.18
Surveillance of drug-resistant TB in a population of specific localities such as South Omo Zone, which is a pastoral area and located at the periphery in the southern Ethiopia, has a crucial role to optimize drug treatment and to tackle the dissemination of resistant MTBC strains in the area. Therefore, this study was carried out with the objective of evaluating drug sensitivity of MTBC isolated from South Omo, southern Ethiopia using MGIT 960 system and GenoType MTBDRplus assay.
Materials and methods
Study setting
South Omo Zone is located in southern Ethiopia and borders Kenya and South Sudan19,20 and consists of eight districts and one city administration (Jinka). Zone is situated at 4.43°–6.46° latitude and 35.79°–36.06° longitude. According to the 2007 census, the total population of Zone was 577,673 (7.5% urban and 92.5% rural) of whom 50% were men.21 Jinka General Hospital (JGH) is the only Hospital in Zone and has been functional since April 2001.22 JGH provides health services for about 1,000,000 people from its catchment area in and outside the Zone. JGH was the main sample collec tion site. Human subject recruitment and sample collection were also conducted in 12 other health centers from different districts using community-based screening.
Ethical clearance
Ethical clearance was obtained from Aklilu Lemma Institute of Pathobiology, Addis Ababa University (ALIPB-AAU) Institutional Review Board (ref. no. ALIPB/IRB/22-B/2012/13) and the National Research Ethics Committee (ref no. 3.10/785/07). Permission to conduct the study was also obtained from South Omo Zone Health Department. Written informed consent was obtained from study participants.
MTBC Isolates
The TB molecular epidemiological study was undertaken in South Omo Zone from September 2014 to May 2016. A total of 1200 sputum and fine needle aspirate samples were collected from TB cases and TB-suspected individuals in health centers through screening of the community. All 1200 samples were cultured on Lowenstein Jensen medium, yielding 201 acid fast bacilli (AFB) isolates of which 161 were part of the MTBC. Twenty-eight isolates were non-tuberculosis mycobacteria and 12 isolates were not members of the genus Mycobacterium. The AFB isolates were differentiated into three groups using region difference-based PCR9,23 and genus typing multiplex PCR.24 In addition, the species were identified based on the spoligotyping pattern according to Kamerbeek et al.25 MTBC isolates were stored in freezing medium and their DNA was extracted in dH2O. For DST analysis, the frozen suspension of the MTBC isolates were thawed and tested for DST using the BACTEC MGIT 960 system. Additionally, the frozen DNA of MTBC isolates were tested for DST using the GenoType MTBDRplus assay.
Phenotypic DST for first-line drugs using the BACTEC MGIT 960 system
Phenotypic DST of MTBC isolates to the first-line drugs was done using BD BACTEC™ MGIT™ 960 system in the Mycobacteriology Research Center at Jimma University (MRC-JU). The BACTEC MGIT™ (Becton Dickinson, Franklin Lakes, NJ, USA). Procedure was followed as described in its manual.26 The frozen MTBC isolates were thawed and then 100 µL of aliquots was added into 2 mL of phosphate-buffered saline and processed for decontamination using equal volume of 2.9% sodium citrate and 4% sodium hydroxide solution. Thereafter, the decontaminated suspension of isolates was cultured using the MGIT 960 system. Lyophilized preparations of the antibiotic drugs of MGIT 960 SIRE kit and pyrazinamide (PZA) kit were reconstituted in sterile dH2O. Final concentrations of STR 1.0 µg/mL, INH 0.1 µg/mL, RIF 1.0 µg/mL, ethambutol (EMB) 5.0 µg/mL and PZA 100.0 µg/mL were used. A growth control (GC) tube without drug was included for each isolate. The relative growth ratio between the drug-containing tube and GC tube was determined by the BD BACTEC™ MGIT™ 960 system’s software algorithm when 400 growth units for the GC tube were reached. DST results were reported qualitatively. Quality control was maintained by testing the batch of MGIT medium, SIRE Kit and PZA kit using the strain M. tuberculosis H37Rv.
Detection of RIF and INH resistance using GenoType® MTBDRplus assay
The identification of genotypic drug resistance to RIF (based on mutations the rpoB gene) and INH (based on mutations in katG and inhA genes) was carried out using the manufacturer’s instruction27 in the TB Laboratory at ALIPB-AAU. Accordingly, 50 µL of PCR mixture consisting of 10 µL AM-A GT MTBDRplus ver 2.0, 35 µL AM-B GT MTBDRplus ver 2.0 and 5µL DNA template from an isolate was used to perform this assay. M. tuberculosis H37Rv DNA template was used as a positive control. Water (Qiagen NV, Venlo, the Netherlands, product) served as a negative control. A thermal cycler (VWR, Leicestershire, UK) was programmed as follows: 15 min for enzyme activation at 95°C followed by 10 cycles of 30 sec denaturation at 95°C, 2 min annealing at 58°C, 20 cycles of 25 sec denaturation at 95°C, 40 sec annealing at 53°C, 40 sec elongation at 70°C, and finally elongation at 70°C for 8 min. Biotin-labeled amplicons were hybridized to DNA probes attached to a DNA strip®. Hybridization was done using the TwinCubator (Hain Lifescience, Nehren, Germany). Results were interpreted based on the presence and absence of wild-type (WT) and mutation (MUT) probes.
Data analysis
Data analysis was performed using IBM SPSS Statistics 20. Descriptive summaries are presented in Tables 1 and 2. Performance of the MTBDRplus assay in the diagnosis of RIF and INH resistance was evaluated taking results from the MGIT 960 test as gold standard. The association between drug resistance and other independent variables was evaluated using logistic regression models. P-values of less than 0.05 were considered as statistically significant.
Table 1.
Demographic and clinical characteristics of study participants
| Characteristics | Total N (%) | Number (%) of study participants
|
P-value | ||
|---|---|---|---|---|---|
| Non-pastoral | Pastoral | Unspecified | |||
| Sex | |||||
| Female | 64 (39.8) | 30 (42.9) | 26 (36.6) | 8 (40.0) | 0.751 |
| Male | 97 (60.2) | 40 (57.1) | 45 (63.4) | 12 (60.0) | |
| Age in years | |||||
| 15–29 | 72 (44.7) | 32 (45.7) | 34 (47.9) | 6 (30.0) | 0.017 |
| 30–44 | 60 (37.3) | 28 (40.0) | 19 (26.8) | 13 (65.0) | |
| 45+ | 29 (18.0) | 10 (14.3) | 18 (25.4) | 1 (5.0) | |
| Type of TB | |||||
| PTB+ | 110 (68.3) | 48 (68.6) | 49 (69.0) | 13 (65.0) | 0.005 |
| PTB− | 25 (15.5) | 5 (7.1) | 13 (18.3) | 7 (35.0) | |
| EPTB | 26 (16.1) | 17 (24.3) | 9 (12.7) | 0 (0.0) | |
| Treatment history | |||||
| New TB case | 153 (95.0) | 68 (97.1) | 65 (91.5) | 20 (100.0) | 0.171 |
| TB re-treatment case | 8 (5.0) | 2 (2.9) | 6 (8.5) | 0 (0.0) | |
| Total | 161 (100) | 70 (43.5) | 71 (44.1) | 20 (12.4) | |
Abbreviations: TB, tuberculosis; PTB+, smear-positive pulmonary TB; PTB–, smear-negative pulmonary; TB; EPTB, extra-pulmonary TB.
Table 2.
Phenotypic DST results for 126 MTBC isolates
| TB cases | No. of Isolates | First-line anti-TB drugs
|
PZA | DR type | % | |||
|---|---|---|---|---|---|---|---|---|
| STR | INH | RIF | EMB | |||||
| New (N=119) | 108 (90.8) | S | S | S | S | S | Pan-S | 90.8 |
| 3 (2.5) | R | S | S | S | S | Mono | 9.2 | |
| 1 (0.8) | S | S | S | R | S | |||
| 7 (5.9) | S | S | S | S | R | |||
|
| ||||||||
| Re-treatment (N=7) | 4 (57.1) | S | S | S | S | S | Pan-S | 57.1 |
| 2 (28.6) | S | R | S | S | R | Poly | 28.6 | |
| 1 (14.3) | R | R | R | R | R | MDR | 14.3 | |
|
| ||||||||
| Any resistance (N=126) | 14 (11.1) | 3.2 | 2.4 | 0.8 | 1.6 | 7.9 | MDR | 0.8 |
Abbreviations: DST, drug sensitivity test; MTBC, Mycobacterium tuberculosis complex; TB, tuberculosis; STR, streptomycin; INH, isoniazid; RIF, rifampicin; EMB, ethambutol; PZA, pyrazinamide; DR type, drug-resistance type; S, susceptible; Pan-S, pan-susceptible; R, resistant; MDR, multidrug resistance.
Results
Demographic and clinical characteristics of study participants
Demographic and clinical characteristics of the 161 study participants are summarized in Table 1. About 40% of the study participants were females. The age range of the study participants varied from 15 to 80 years with a mean and SD of 33.2 and 13.1 years, respectively. The majority (82%) of the participants were in the age range between 15 and 44 years. Age (P=0.017) and type of TB (P=0.005) were significantly associated with residential place.
Phenotypic drug sensitivity patterns as detected by MGIT 960 system
Phenotypic DST could be done on 78.3% (126/161) of the MTBC isolates (Figure 1) and the result is summarized in Table 2. About 91% of the isolates from the new TB case were pan-susceptible to anti-TB drugs, while four of the seven isolates from the re-treatment case isolates were pan-susceptible to anti-TB drugs. Mono-drug resistance was observed in 9.2% (11/119) of the isolates of the new TB cases. The highest incidence of mono-drug resistance was observed to PZA (5.9%) followed by STR (2.5%). Polydrug resistance and MDR were observed in two and one of the seven re-treatment cases, respectively. The results are summarized in Table 2.
Figure 1.
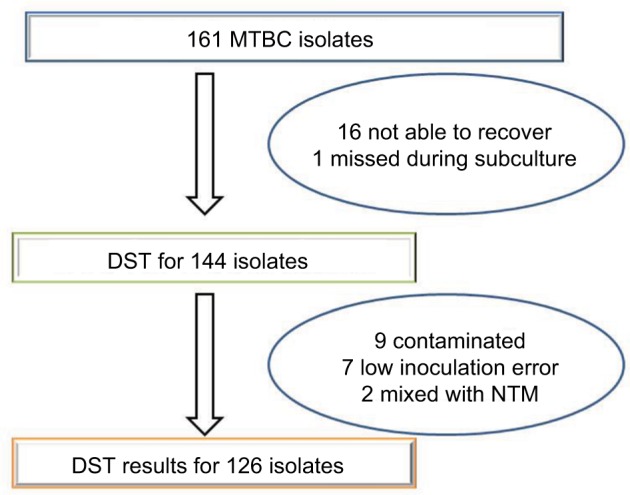
Total numbers of MTBC isolates and number of isolates with phenotypic DST result.
Abbreviations: MTBC, Mycobacterium tuberculosis complex; DST, drug sensitivity test; NTM, nontuberculous mycobacteria; NTM, nontuberculous mycobacteria.
Phenotypic drug resistance and its association with demographic characteristics
A summary result of the logistic regression indicating the association of phenotypic drug resistance and selected demographic characteristics is presented in Table 3. The re-treatment TB cases were significantly associated with increased odds of having drug-resistant isolates compared to the new TB cases (AOR=11.60, 95% CI: 1.70–78.92).
Table 3.
Demographic factors associated with phenotypic drug resistance of MTBC isolates
| Characteristics | Total N (%) | DR N (%) | Crude OR (95% CI) | Adjusted OR (95% CI) | P-value |
|---|---|---|---|---|---|
| Sex | |||||
| Female | 48 (38.1) | 7 (14.6) | 1.00 | 1.00 | |
| Male | 78 (61.9) | 7 (9.0) | 0.58 (0.19–1.76) | 0.55 (0.15–1.98) | 0.357 |
| Age in years | |||||
| 15–29 | 56 (44.4) | 6 (10.7) | 1.00 | 1.00 | |
| 30–44 | 48 (38.1) | 4 (8.3) | 0.76 (0.20–2.86) | 0.68 (0.16–2.86) | 0.601 |
| >=45 | 22 (17.5) | 4 (18.2) | 1.85 (0.47–7.33) | 0.11 (3.95) | 0.643 |
| TB type | |||||
| PTB+ | 84 (66.7) | 10 (11.9) | 1.00 | 1.00 | |
| PTB− | 19 (15.1) | 2 (10.5) | 0.87 (0.18–4.34) | 0.42 (0.04–4.77) | 0.483 |
| EPTB | 23 (18.3) | 2 (8.7) | 0.71 (0.14–3.47) | 0.50 (0.09–2.77) | 0.426 |
| Treatment history | |||||
| New TB cases | 119 (94.4) | 11 (9.2) | 1.00 | 1.00 | |
| Re-treatment TB cases | 7 (5.6) | 3 (42.9) | 7.36 (1.46–37.22) | 11.60 (1.70–78.92) | 0.012 |
| Residential areaa | |||||
| Non-pastoral | 54 (42.9) | 8 (14.8) | 1.00 | 1.00 | |
| Pastoral | 56 (44.4) | 5 (8.9) | 0.56 (0.17–1.85) | 0.46 (0.12–1.70) | 0.241 |
Note:
Participants with unknown residences were excluded from logistic regression analysis.
Abbreviations: MTBC, Mycobacterium tuberculosis complex; DR, drug resistance; TB, tuberculosis; PTB+, smear-positive pulmonary TB; PTB–, smear-negative pulmonary.
Genotypic drug resistance data
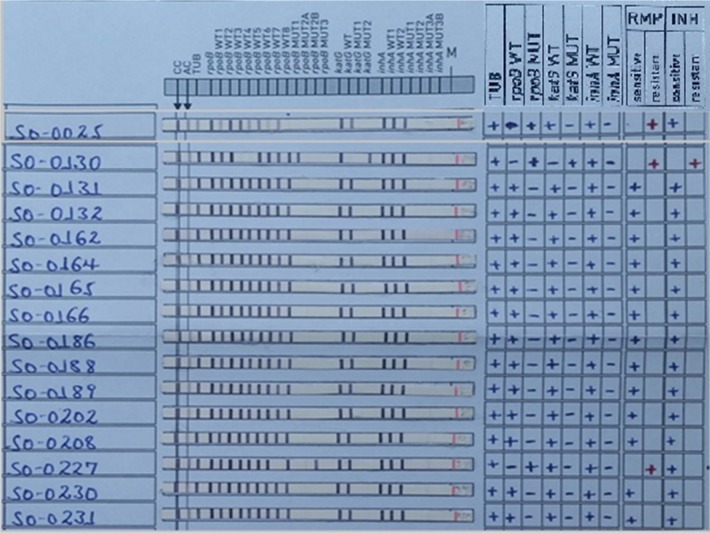
Genotypic DST results of selected 13 isolates out of 158 susceptible isolates and all the three isolates, which showed resistance, are depicted in Figure 2. As observed in Figure 2, RIF mono-drug resistance was 1.3% (2/153) in the new TB cases. One of these two isolates had known CAC→GAC mutation resulting in the amino acid substitution at H526D, while the other isolate had a missing rpoB-WT8 without a known mutation. One MDR-TB case was identified for eight TB re-treatment TB cases. The MDR isolate had a GAC→GTC mutation in rpoB (amino acid substitution D516V) and an AGC→ACA mutation in katG (amino acid substitution S315T2).
Figure 2.
Genotypic DST results of selected susceptible isolates and all resistant isolates.
Notes: Three isolates (SO-0025, SO-0130 and SO-0227) were resistant to RIF and/or INH and the rest 13 isolates were susceptible to both RIF and INH as representative for the 158 susceptible isolates to RIF and INH. In this illustration, isolate SO-0130 was from the re-treatment TB case and the remaining 15 isolates were from the new TB cases.
Abbreviations: DST, drug sensitivity test; RIF, rifampicin; INH, isoniazid; TB, tuberculosis; CC, conjugate control; AC, amplification control; WT, wild-type probe; MUT, mutation probe; TUB, M. tuberculosis complex.
Genotype MTBDRplus assay performance compared to MGIT 960 system
The diagnostic performance of the MTBDRplus assay was evaluated by the MGIT 960 system on 126 MTBC isolates. As summarized in Table 4, the sensitivities of the MTBDRplus assay for detection of resistance to INH, RIF and MDR were 33.3%, 100% and 100%, respectively. Its specificities for the detection of resistance to INH, RIF and MDR were 100%, 99.2% and 100%, respectively. One isolate showed RIF resistance to genotypic DST but not to phenotypic DST. On the contrary, two isolates showed INH resistance to phenotypic DST but not to genotypic DST.
Table 4.
GenoType MTBDRplus assay performance evaluation compared to MGIT 960 system
| MGIT 960 system
|
Total | Diagnostic performance of GenoType MTBDRplus assay
|
Level of agreement
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| R | S | Sensitivity | Specificity | PPV | NPV | k-value | ||||
| GenoType MTBDRplus assay | INH | R | 1 | 0 | 1 | 33.3 | 100 | 100 | 98.4 | 0.494 |
| S | 2 | 123 | 125 | |||||||
| RIF | R | 1 | 1 | 2 | 100 | 99.2 | 50 | 100 | 0.663 | |
| S | 0 | 124 | 124 | |||||||
| MDR | R | 1 | 0 | 1 | 100 | 100 | 100 | 100 | 1.00 | |
| S | 0 | 125 | 125 | |||||||
Abbreviations: R, resistant; S, susceptible; PPV, positive predictive value; NPV, negative predictive value; INH, isoniazid; RIF, rifampicin; MDR, multidrug resistance.
Discussion
In the present study, the DST was performed on 161 MTBC isolates, which were isolated from clinical cases in South Omo Zone, southern Ethiopia. The findings of this study are expected to be useful for the TB Control Program of Zone and would serve as baseline data for future studies.
Majority of the isolates were recovered from the economically productive age group, and this observation is similar with observation made by the previous studies.28–31 The reason could be due to the mobility of this age group for economy and social reasons. Similarly, relatively large number of isolates were recovered from males than females which is in agreement with the WHO report.1 This could be attributed to the difference between the two sex groups in biological, societal role and access to health facilities.32
In this study, 90.8% of the isolates of the new TB cases were phenotypically pan-susceptible and mono-drug resistance was observed relatively in lower percentage when compared with the percentage reported by the national survey.33 The highest mono-drug resistance was observed against PZA and similar observations were made in Quebec34 and People’s Republic of China.35 PZA is part of the first- and the second-line anti-TB drugs10 and helps to shorten the duration of TB treatment.36 PZA resistance is associated with less probable of treatment success.37 Hence, the existence of more PZA-resistant isolates in the study area could have an impact on TB Control Program of the area. Two of the PZA-resistant isolates in this study were M. bovis, which are naturally resistant to PZA.38
In this study, STR mono-drug-resistance in the new TB cases was the second higher proportion next to PZA. This finding is similar with those reported by the previous studies in Ethiopia29,33,39–41 and could be due to the widespread use of STR for treatment of the other bacterial infections.42,43 The least percentage to resistance was observed against EMB and this observation is in agreement with the results reported by earlier studies.29,33,39–41 No resistance was observed either against INH or against RIF in isolates of the new TB cases. But mono-resistance was observed against INH in the re-treatment cases and was associated with poor treatment outcomes.44 Hence, strengthening case management could help to avert the effect of INH resistance. The proportion of MDR-TB in the re-treatment TB cases was similar to that of one study reported earlier,41 while it was less than those reported by the other studies.29–31,40 However, it was difficult to make a reliable comparison because of the small sample size of the re-treatment cases in this study.
One of the isolates recovered from the re-treatment TB cases was resistant to both RIF and INH (MDR)and the majority (95%) of RIF resistance occurred within RIF resistance determining region with 81 bp length when there is a mutation in the rpoB gene.15 Mutations in the katG, inhA and ahpC genes lead to INH resistance, which does not occur in a specific region like that of RIF resistance.45 Mostly katG gene mutation is associated with high-level resistance.46 The MDR isolate missed rpoB WT3 and rpoB WT4 genes and had a mutation at rpoBMUT1 with an amino acid change of D516V (GAC→GTC),which is a synonym of D435V (GAC→GTC).47 This isolate, in addition, missed WT katG gene and developed katG MUT2 with an amino acid change of S315T2 (AGC→ACA). Similarly, such rpoB gene mutation types were observed at low frequency in studies conducted in Ethiopia48 and Pakistan,49 and at high frequency in a study conducted in East Hungary.50 The amino acid change of S315T2 (AGC→ACA) in this isolate was not previously reported from Ethiopia but the most common reported amino acid change in Ethiopia has been S315T1, which is in the range of 83%–100%.28,48,51–53
One of RIF mono-resistant isolates missed rpoB WT7 gene and developed mutation at rpoB MUT2B with an amino acid change of H526D, which is a synonym of H445D.47 Such type of mutation with an amino acid change of H526D was previously reported in Ethiopia in the proportion of 0%–22%.28,48,51,53 The other RIF mono-resistant isolate missed rpoB WT8 and had no known mutation, which may not indicate real resistance but rather it could be due to a silent mutation in rpoB gene.54 Similarly, such resistance was reported in previous studies of Ethiopia with various proportions including 11.8%53 and 44.4%.51 But most RIF-resistant isolates missing rpoB WT8 are accompanied by an S531L amino acid change.28,48,53,55 RIF mono-resistant isolates were identified from the new TB cases, which could be acquired from TB patients who had RIF-resistant TB or the patients could previously be treated with RIF for other bacterial infection.42,43 Molecular detection of RIF resistance is used as a surrogate marker for MDR-TB.11,13,28 Hence, the identification of RIF-resistant and MDR isolates in the present study indicated the existence of MDR-TB in the study area, which highlights the need for strengthening of the TB control program in the study area.
The sensitivity of GenoType MTBDRplus assay for detection of INH resistance in the present study was lower than those reported by previous studies in Ethiopia,48,51 Uganda55 and Pakistan.49 The MTBDRplus assay was not able to detect the INH resistance in the two isolates, which were detected by MGIT 960 system. This could be due to mutations in the inhA promoter and the ahpC-oxyR intergenic regions, which have not been included in the strips56 or it could also be due to unidentified mutation. The high specificity of the MTB-DRplus assay in detecting INH resistance, RIF resistance and MDR MTBC isolates agrees with the specificity of previous studies.48,51,55 RIF resistance in one isolate was detected by the MTBDRplus assay but not by MGIT 960 system. Being susceptible by phenotypic method but resistant by genotypic method may be associated with false RIF resistance due to a silent mutation which make the probe fails to hybridize on a strip and interpreted as RIF resistant.57 The three isolates that created discordance between the assays used need further analysis by DNA sequencing to fully understand reasons for the observed discordance.
Conclusion
The magnitude of drug resistance was relatively low in new TB cases of South Omo as compared to those reported from the other regions of the country. This is encouraging, and hence, the TB Control Program in Zone should strengthen its program and make continuous surveillance so that early detection enables the concerned bodies to take appropriate actions to inhibit the emergence of drug resistance in South Omo.
Acknowledgments
The research was funded by the National Institute of Health with grant number U01HG007472-01, the Addis Ababa University, the Arba Minch University and the Ethiopian Public Health Institute (EPHI). The authors would like to thank communities and health professionals in South Omo and staff members of ALIPB-AAU and MRC-JU for their unreserved support during the research work. The authors would like to thank Harar, Adama and Bahirdar Health Research and Regional Laboratories for the borrowing of MGIT DST set carriers. Finally, authors would like to thank Mr. Zelalem Yaregal and Mr. Bazezew Yenew (EPHI), Mr. Musse Brhane (Harar), Mr. Mohammed Suaudi (Adama) and Mr. Gashaw Yitayew (Bahirdar) for their help during phenotypic DST.
Footnotes
Author contributions
BW was principal investigator who conceived and designed the study and participated in sample collection, laboratory work, data analysis and interpretation and drafting of the manuscript. GM participated in the study design, data analysis and interpretation. G Abebe participated in phenotypic DST laboratory work. ST and TM participated in genotypic DST laboratory work. TT participated in sample collection. RP participated in the study conception and design. G Ameni participated in the study conception, design and supervision. GM, G Abebe, ST, TM, TT, RP and G Abebe also participated in critical review of the manuscript. All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work. All the authors read and approved the submitted manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.World Health Organization . Global Tuberculosis Report. Geneva: World Health Organization; 2017. [Accessed August 10, 2018]. Available from: http://www.who.int/tb/publications/global_report/en/ [Google Scholar]
- 2.Streptomycin treatment of pulmonary tuberculosis: a Medical Research Council investigation. BMJ. 1948;2:769–782. [PMC free article] [PubMed] [Google Scholar]
- 3.Schatz A, Bugle E, Waksman SA. Streptomycin, a substance exhibiting antibiotic activity against gram-positive and gram-negative bacteria. Proc Soc Exp Biol Med. 1944;55:66–69. doi: 10.1097/01.blo.0000175887.98112.fe. [DOI] [PubMed] [Google Scholar]
- 4.Calvori C, Frontali L, Leoni L, Tecce G. Effect of rifamycin on protein synthesis. Nature. 1965;207(995):417–418. doi: 10.1038/207417a0. [DOI] [PubMed] [Google Scholar]
- 5.Sensi P. History of the development of rifampin. Rev Infect Dis. 1983;5(Suppl 3):S402–S406. doi: 10.1093/clinids/5.supplement_3.s402. [DOI] [PubMed] [Google Scholar]
- 6.Cohn DL, Bustreo F, Raviglione MC. Drug-resistant tuberculosis: review of the worldwide situation and the WHO/IUATLD Global Surveillance Project. Clin Infect Dis. 1997;24(Suppl 1):S121–S130. doi: 10.1093/clinids/24.supplement_1.s121. [DOI] [PubMed] [Google Scholar]
- 7.World Health Organization . Drug-Resistant TB Surveillance and Response: Supplement Global Tuberculosis Report 2014. Geneva: World Health Organization; 2014. [Accessed August 10, 2018]. Available from: http://www.who.int/iris/handle/10665/137095. [Google Scholar]
- 8.World Health Organization . Guidelines for Surveillance of Drug Resistance in Tuberculosis. Geneva: World Health Organization; 2009. [Accessed August 10, 2018]. Available from: http://www.who.int/tb/publications/surveillance_guidelines/en/ [Google Scholar]
- 9.Biadglegne F, Sack U, Rodloff AC. Multidrug-resistant tuberculosis in Ethiopia: efforts to expand diagnostic services, treatment and care. Antimicrob Resist Infect Control. 2014;3(1):31. doi: 10.1186/2047-2994-3-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Federal Ministry of Health Ethiopia . Guideline for Program and Clinical Management of Drug Resistant Tuberculosis. Addis Ababa: FMOH; 2009. [Google Scholar]
- 11.Federal Ministry of Health Ethiopia and Ethiopian Public Health Institute . Implementation Guideline for GeneXpert MTB/RIF Assay in Ethiopia. Addis Ababa: Ethiopian Public Health Institute; 2014. [Accessed August 10, 2018]. Available from: https://www.medbox.org/et-guidelines-hiv-tb/implementation-guideline-for-gene-xpert-mtbrif-assay-in-ethiopia/preview? [Google Scholar]
- 12.Siddiqi S, Ahmed A, Asif S, et al. Direct drug susceptibility testing of Mycobacterium tuberculosis for rapid detection of multidrug resistance using the Bactec MGIT 960 system: a multicenter study. J Clin Micro-biol. 2012;50(2):435–440. doi: 10.1128/JCM.05188-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.World Health Organization . Molecular Line Probe Assay for Rapid Screening of Patients at Risk of Multidrug-resistant Tuberculosis (MDR-TB): Policy Statment. Geneva: World Health Organization; 2008. [Accessed August 10, 2018]. Available from: http://www.who.int/tb/laboratory/line_probe_assays/en/ [Google Scholar]
- 14.World Health Organization . Rapid implementation of the Xpert MTB/RIF diagnostic test. Geneva: World Health Organization; 2011. [Accessed August 10, 2018]. Available from: http://www.who.int/tb/publications/tb-amplificationtechnology-implementation/en/ [Google Scholar]
- 15.Telenti A, Imboden P, Marchesi F, et al. Detection of rifampicin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341(8846):647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 16.Banerjee A, Dubnau E, Quemard A, et al. inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science. 1994;263(5144):227–230. doi: 10.1126/science.8284673. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y, Heym B, Allen B, Young D, Cole S. The catalase-peroxidase gene and isoniazid resistance of Mycobacterium tuberculosis. Nature. 1992;358(6387):591–593. doi: 10.1038/358591a0. [DOI] [PubMed] [Google Scholar]
- 18.CLSI . Susceptibility Testing of Mycobacteria, Nocardia, and Other Aerobic Actinomycetes; Approved Standard. 2nd ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2011. pp. 1–64. [PubMed] [Google Scholar]
- 19.Southern Nations, Nationalities and Peoples Region Culture and Tourism Bureau . South Omo Zone Administrative Zone, Administrative Map. Hawassa: SNNPR; 2016. [Google Scholar]
- 20.United Nations Development Programme . Regional Map of Ethiopia. Addis Ababa: UNDP; 1996. [Google Scholar]
- 21.Central Statistical Agency . Population Size by Age and Sex: Federal Democratic Republic of Ethiopia, Population Census Commission, with support from UNFPA. Addis Ababa: Federal Democratic Republic of Ethiopia PCC; 2008. Summary and Statistical Report of the 2007 Population and Housing Census. [Google Scholar]
- 22.Wondale B, Medihn G, Teklu T, Mersha W, Tamirat M, Ameni G. A retrospective study on tuberculosis treatment outcomes at Jinka General Hospital, southern Ethiopia. BMC Res Notes. 2017;10(1):680. doi: 10.1186/s13104-017-3020-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brosch R, Gordon SV, Marmiesse M, et al. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci USA. 2002;99(6):3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wilton S, Cousins D. Detection and identification of multiple mycobacterial pathogens by DNA amplification in a single tube. PCR Methods Appl. 1992;1(4):269–273. doi: 10.1101/gr.1.4.269. [DOI] [PubMed] [Google Scholar]
- 25.Kamerbeek J, Schouls L, Kolk A, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35(4):907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siddiqi S, Rusch-Gerdes S. MGIT™ Procedure Manual For BACTEC™ MGIT 960™ TB System: Mycobacteria Growth Indicator Tube (MGIT) Culture and Drug Susceptibility Demonstration Projects. Geneva: FiND; 2006. pp. 1–52. [Google Scholar]
- 27.Hain Lifescience GenoType MTBDRplus VER 20: Molecular Genetic Assay for Identification of the M tuberculosis Complex and its Resistance to Rifampicin and Isoniazid from Clinical Specimens and Cultivated Samples. Germany: Hain Lifescience GmbH; 2012. [Accessed August 10, 2018]. pp. 1–13. Available from: https://www.ghdonline.org/uploads/MTBDRplusV2_0212_304A-02-02.pdf. [Google Scholar]
- 28.Brhane M, Kebede A, Petros Y. Molecular detection of multidrug-resistant tuberculosis among smear-positive pulmonary tuberculosis patients in Jigjiga town, Ethiopia. Infect Drug Resist. 2017;10:75–83. doi: 10.2147/IDR.S127903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agonafir M, Lemma E, Wolde-Meskel D, et al. Phenotypic and genotypic analysis of multidrug-resistant tuberculosis in Ethiopia. Int J Tuberc Lung Dis. 2010;14(10):1259–1265. [PubMed] [Google Scholar]
- 30.Abdella K, Abdissa K, Kebede W, Abebe G. Drug resistance patterns of Mycobacterium tuberculosis complex and associated factors among retreatment cases around Jimma, Southwest Ethiopia. BMC Public Health. 2015;15:599. doi: 10.1186/s12889-015-1955-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abate D, Taye B, Abseno M, Biadgilign S. Epidemiology of anti-tuberculosis drug resistance patterns and trends in tuberculosis referral hospital in Addis Ababa, Ethiopia. BMC Res Notes. 2012;5:462. doi: 10.1186/1756-0500-5-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borgdorff MW, Nagelkerke NJ, Dye C, Nunn P. Gender and tuberculosis: a comparison of prevalence surveys with notification data to explore sex differences in case detection. Int J Tuberc Lung Dis. 2000;4(2):123–132. [PubMed] [Google Scholar]
- 33.Getahun M, Ameni G, Kebede A, et al. Molecular typing and drug sensitivity testing of Mycobacterium tuberculosis isolated by a community-based survey in Ethiopia. BMC Public Health. 2015;15:751. doi: 10.1186/s12889-015-2105-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nguyen D, Brassard P, Westley J, et al. Widespread pyrazinamide-resistant Mycobacterium tuberculosis family in a low-incidence setting. J Clin Microbiol. 2003;41:2878–2883. doi: 10.1128/JCM.41.7.2878-2883.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D, Hu Y, Werngren J, et al. Multicenter study of the emergence and genetic characteristics of pyrazinamide-resistant tuberculosis in China. Antimicrob Agents Chemother. 2016;60(9):5159–5166. doi: 10.1128/AAC.02687-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Steele MA, Des Prez RM. The role of pyrazinamide in tuberculosis chemotherapy. Chest. 1988;94(4):845–850. doi: 10.1378/chest.94.4.845. [DOI] [PubMed] [Google Scholar]
- 37.Yee DP, Menzies D, Brassard P. Clinical outcomes of pyrazinamide-monoresistant Mycobacterium tuberculosis in Quebec. Int J Tuberc Lung Dis. 2012;16(5):604–609. doi: 10.5588/ijtld.11.0376. [DOI] [PubMed] [Google Scholar]
- 38.de Jong BC, Onipede A, Pym AS, et al. Does resistance to pyrazinamide accurately indicate the presence of Mycobacterium bovis? J Clin Microbiol. 2005;43(7):3530–3532. doi: 10.1128/JCM.43.7.3530-3532.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Biadglegne F, Tessema B, Sack U, Rodloff AC. Drug resistance of Mycobacterium tuberculosis isolates from tuberculosis lymphadenitis patients in Ethiopia. Indian J Med Res. 2014;140(1):116–122. [PMC free article] [PubMed] [Google Scholar]
- 40.Esmael A, Ali I, Agonafir M, et al. Drug resistance pattern of Mycobacterium tuberculosis in Eastern Amhara regional state Ethiopia. J Microb Biochem Technol. 2014;6:75–79. [Google Scholar]
- 41.Hamusse SD, Teshome D, Hussen MS, Demissie M, Lindtjørn B. Primary and secondary anti-tuberculosis drug resistance in Hitossa District of Arsi Zone, Oromia Regional State, Central Ethiopia. BMC Public Health. 2016;16:593. doi: 10.1186/s12889-016-3210-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Drug Administration and Control Authority of Ethiopia . Standard Treatment Guideline for Primary Hospitals. Addis Ababa: DACA; 2010. [Google Scholar]
- 43.Grouzard V, Rigal J, Sutton M. Clinical Guidelines Diagnosis and Treatment Manual. 2016 ed. Geneva: Medecins Sans Frontieres; 2016. [Google Scholar]
- 44.Gegia M, Cohen T, Kalandadze I, Vashakidze L, Furin J. Outcomes among tuberculosis patients with isoniazid resistance in Georgia, 2007–2009. Int J Tuberc Lung Dis. 2012;16(6):812–816. doi: 10.5588/ijtld.11.0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hillemann D, Rüsch-Gerdes S, Richter E. Evaluation of the GenoType MTBDRplus assay for rifampin and isoniazid susceptibility testing of Mycobacterium tuberculosis strains and clinical specimens. J Clin Microbiol. 2007;45(8):2635–2640. doi: 10.1128/JCM.00521-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gillespie SH. Evolution of drug resistance in Mycobacterium tuberculosis: clinical and molecular perspective. Antimicrob Agents Chemother. 2002;46(2):267–274. doi: 10.1128/AAC.46.2.267-274.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andre E, Goeminne L, Cabibbe A, et al. Consensus numbering system for the rifampicin resistance-associated rpoB gene mutations in pathogenic mycobacteria. Clin Microbiol Infect. 2017;23(3):167–172. doi: 10.1016/j.cmi.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Kebede A, Demisse D, Assefa M, et al. Performance of MTBDRplus assay in detecting multidrug resistant tuberculosis at hospital level. BMC Res Notes. 2017;10(1):661. doi: 10.1186/s13104-017-2989-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Javaid M, Ahmed A, Asif S, Raza A. Diagnostic plausibility of MTB-DRplus and MTBDRsl line probe assays for rapid drug susceptibility testing of drug resistant Mycobacterium tuberculosis strains in Pakistan. Int J Infect. 2016;3:e34903. [Google Scholar]
- 50.Bártfai Z, Somoskövi A, Ködmön C, et al. Molecular characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Hungary by DNA sequencing and the line probe assay. J Clin Microbiol. 2001;39(10):3736–3739. doi: 10.1128/JCM.39.10.3736-3739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bedewi Omer Z, Mekonnen Y, Worku A, et al. Evaluation of the Geno-Type MTBDRplus assay for detection of rifampicin- and isoniazid-resistant Mycobacterium tuberculosis isolates in central Ethiopia. Int J Mycobacteriol. 2016;5(4):475–481. doi: 10.1016/j.ijmyco.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 52.Biadglegne F, Tessema B, Rodloff AC, Sack U. Magnitude of gene mutations conferring drug resistance in Mycobacterium tuberculosis isolates from lymph node aspirates in ethiopia. Int J Med Sci. 2013;10(11):1589–1594. doi: 10.7150/ijms.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tadesse M, Aragaw D, Dimah B, et al. Drug resistance-conferring mutations in Mycobacterium tuberculosis from pulmonary tuberculosis patients in Southwest Ethiopia. Int J Mycobacteriol. 2016;5(2):185–191. doi: 10.1016/j.ijmyco.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 54.Alonso M, Palacios JJ, Herranz M, et al. Isolation of Mycobacterium tuberculosis strains with a silent mutation in rpoB leading to potential misassignment of resistance category. J Clin Microbiol. 2011;49(7):2688–2690. doi: 10.1128/JCM.00659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Albert H, Bwanga F, Mukkada S, et al. Rapid screening of MDR-TB using molecular Line Probe Assay is feasible in Uganda. BMC Infect Dis. 2010;10:41. doi: 10.1186/1471-2334-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seifert M, Catanzaro D, Catanzaro A, Rodwell TC. Genetic mutations associated with isoniazid resistance in Mycobacterium tuberculosis: a systematic review. PLoS One. 2015;10(3):e0119628. doi: 10.1371/journal.pone.0119628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mathys V, van de Vyvere M, de Droogh E, Soetaert K, Groenen G. False-positive rifampicin resistance on Xpert® MTB/RIF caused by a silent mutation in the rpoB gene. Int J Tuberc Lung Dis. 2014;18(10):1255–1257. doi: 10.5588/ijtld.14.0297. [DOI] [PubMed] [Google Scholar]