Abstract
Developmental constraint and sexual conflict shape the evolution of heteromorphic sex chromosomes. These contrasting forces are perhaps strongest during spermatogenesis in species with XY males. In this review, we consider how the unique regulatory environment and selective pressures of spermatogenesis interact to impact sex chromosome evolution in mammals. We explore how each developmental phase of spermatogenesis influences sex chromosome gene content, structure, and rate of molecular evolution, and how these attributes may contribute to speciation. We argue that a developmental context is fundamental to understanding sex chromosome evolution and that an evolutionary perspective can shed new light on our understanding of sperm development.
Keywords: Genomic conflict, sex-biased expression, faster-X, recombination, hybrid male sterility, speciation
Constraint and conflict in sex chromosome evolution
The evolution of heteromorphic sex chromosomes (see Glossary) sets in motion unique evolutionary and developmental dynamics. As Ohno noted [1], ‘The so-called sex-linked genes are nothing more than the original Mendelian genes which were there when the X or the Z was an ordinary chromosome’. These ‘ordinary’ genes are under strong developmental constraint to maintain expression levels between males and females through dosage compensation [reviewed in 2], leading to the remarkable conservation of X chromosome gene content across placental mammals [3], known as Ohno’s law [see also 4]. But sex-linked genes are often more than just ordinary genes. For example, sex chromosomes tend to accumulate sexually antagonistic genes [5–7] and evolve functional specialization through the enrichment of sex-biased genes [8]. In this review, we argue that the dichotomy of developmental constraint and sexual conflict inherent to sex chromosomes has profound consequences for their evolution. We propose that to fully understand sex chromosome evolution these contrasting forces must be considered in a developmental context. In particular, we focus on how the unusual regulatory dynamics of heteromorphic sex chromosomes during spermatogenesis (Figure 1A, Key Figure) both constrains aspects of sex chromosome evolution and provides opportunities for rapid divergence.
Figure 1. Overview of mammalian spermatogenesis.
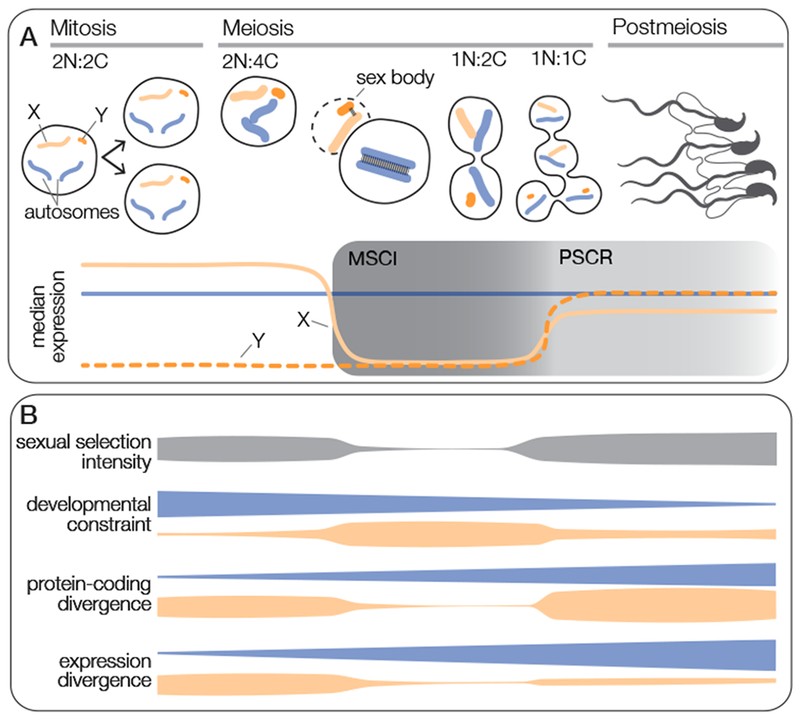
A. Spermatogenesis is a dynamic developmental process defined by reduction in ploidy and progressive specialization of germ cells. Spermatogenesis begins at the outside of the seminiferous tubule and proceeds inward towards the lumen. First, undifferentiated spermatogonia undergo multiple rounds of mitotic division. These cells then progress through meiosis, during which homologous chromosomes pair, crossing over takes place, and there are two subsequent rounds of cell division. The resulting haploid products undergo spermiogenesis to produce highly specialized spermatozoa. Spermatogenic genes on the autosomes (blue) are on average expressed throughout spermatogenesis, although there are groups of genes that are more highly expressed early (prior to MSCI) or late in spermatogenesis [31,48,71,102,103]. Genes on the X and Y chromosomes (orange) are silenced during meiosis (MSCI) and remain transcriptionally repressed in postmeiotic development (PSCR). Genes that escape PSCR tend to be highly expressed [3,73,74,76]. ‘N’ indicates autosomal ploidy, ‘C’ indicates autosomal copy number. B. Conceptual model for the molecular evolution of spermatogenic genes on and off the sex chromosomes. Patterns of protein-coding and gene expression divergence are influenced by variation in the intensity of sexual selection and the degree of developmental constraint across spermatogenesis. Protein-coding and expression divergence data summarized from [31].
Developmental constraint and sexual conflict are integral parts of spermatogenesis. The integrity of spermatogenesis is maintained through numerous developmental checkpoints [9,10], which may make aspects of spermatogenesis inherently sensitive to novel genetic variation introduced by mutation [11]. Over evolutionary timescales, selection against mutations that impact critical components of spermatogenesis slows the accumulation of genetic divergence between species. At the same time, other aspects of spermatogenesis may evolve rapidly in response to sexual selection or sexual conflict. For example, common forms of sexual selection, such as sperm competition [12] or cryptic female choice [13,14], can drive rapid divergence in sperm form, function, and rates of production [e.g. 15–19]. As a consequence, sperm are one of the most morphologically diverse cell types in animals [e.g. 20,21]. These themes are particularly germane to genes that reside on the sex chromosomes; X- and Y-linked genes experience additional developmental constraints during spermatogenesis (discussed below), while their sex-biased transmission creates opportunities for rapid evolution and sexual antagonism (Box 1).
Box 1: The evolutionary dynamics of sex chromosomes.
Many of the unusual features of sex chromosomes are thought to reflect unique evolutionary dynamics related to their sex-biased transmission. The X and Y chromosomes are single copy, or hemizygous, in males and therefore sex-biased relative to the autosomes. Assuming a population with equal sex ratios, the X chromosome has 3/4 as many copies as a given autosome and spends 2/3 of its time in females while the Y chromosome is male-specific with 1/4 the autosomal population size. These deviations from autosomal conditions can impact two important aspects of sex chromosome evolution that are germane to spermatogenesis:
Genetic content
Selection in female and male environments can facilitate the accumulation of sexually antagonistic genes on the X chromosome that benefit one sex but are detrimental to the other [5,7]. The X chromosome spends most of its time in females, which can favor the fixation of X-linked mutations that have female-biased function [e.g. 30]. However, genes with at least partially recessive antagonistic effects can favor the evolution of male-biased functions on the X chromosome [e.g. 30,32]. The Y chromosome is only in males, thus mammalian Y-linked genes are largely testis-specific [34].
Rates of molecular evolution
Male hemizygosity results in immediate exposure of new recessive sex-linked alleles to selection. This can lead to more efficient purifying selection against deleterious mutations and the faster fixation of beneficial alleles relative to the autosomes, especially when considering genes that are primarily expressed in males [6,96]. Predictions on the relative efficacy of selection on and off the sex chromosomes depend on several factors, including sex ratios and variance in reproductive success, sex-specific mutation rates, and the average dominance and sex-specific fitness effects of mutations [36,96,97].
The amalgamation of rapidly evolving components of the sex chromosomes, embedded within a deeply conserved process, also appears to make spermatogenesis prone to disruption in hybrid progeny [22]. Reproductive isolation between species often arises as an indirect consequence of evolutionary divergence between populations. Given some evolutionary divergence, the combination of genomes from two different species can generate incompatible genetic interactions in hybrids [23]. Hybrid sterility (and inviability) usually first manifests in the heterogametic (XY or ZW) sex, a pattern known as Haldane’s rule [24, reviewed in 25]. In XY systems, hybrid male sterility tends to evolve very quickly and incompatibilities underlying hybrid male sterility are more common on the X chromosome [26], a phenomenon known as the Large X-effect [27, reviewed in 28].
Here, we will explore the evolution of sex chromosomes in the context of spermatogenesis. For simplicity, we focus on the mammalian X and Y chromosomes (female XX and male XY), where both the biology of spermatogenesis and patterns of sex chromosome evolution have been intensively studied. We examine how the three main phases of spermatogenesis — mitosis, meiosis, and the postmeiotic development of spermatozoa — impose specific developmental constraints and provide opportunities for evolutionary divergence. We further link evolutionary dynamics associated with each of these phases to the central role that the sex chromosomes play in the origin of species. Recent studies that have examined the evolutionary dynamics of sex chromosomes during spermatogenesis have shed new light on the mode and tempo of sex chromosome evolution, the regulatory underpinnings of X-linked hybrid male sterility, and the potential for antagonistic sex chromosome co-evolution. Synthesizing across these insights, we argue that the unique regulatory environments and selective pressures imposed by spermatogenesis are fundamental to understanding sex chromosome evolution and speciation. Likewise, a complete understanding of sperm development necessitates an evolutionary perspective.
Mitotic phase: Rapid evolution of the male-biased X chromosome
Testes are composed of interstitial cells and seminiferous tubules, which are the site of spermatogenesis. The first major phase of spermatogenesis is the proliferation of spermatogonia through mitotic division (Figure 1A). Undifferentiated spermatogonia are arrayed on the periphery of each tubule so that mitotic divisions proceed inwards (towards the tubular lumen) and in waves along each tubule. During this early proliferation phase, numerous mitotic cell divisions and the continuous production of sperm increase the likelihood of mutations, leading to a male-biased mutation rate in most mammals [reviewed in 29]. Mitotic germline mutations that are propagated in descendant cells create opportunities for heritable genetic novelty and adaptation. This also makes early spermatogenesis risky, and perhaps more prone to disruption by mutations than other phases of spermatogenesis [10].
The mammalian X chromosome is enriched relative to the autosomes for genes expressed in tissues with female-biased function(s), such as ovaries and placenta [30]. However, the X chromosome is also predicted to accumulate male-biased genes under some conditions (Box 1). Consistent with this prediction, the X chromosome is highly enriched for genes that are expressed early in spermatogenesis [30–32]. As discussed below, this prediction does not generally hold across spermatogenesis because other developmental constraints select against X-linkage during the later phases of spermatogenesis [30]. The lack of recombination between the X and Y chromosome fuels sex chromosome heteromorphy and is thought to have driven the loss of most genes from the non-recombinant portion of the Y [33]. Genes that are retained on the Y tend to be specific to the testis and are usually expressed in postmeiotic cells, where they likely have critical roles in sperm development [34, Figure 1A].
Genes expressed in spermatogonia on the X chromosome also tend to show faster protein sequence evolution relative to autosomal genes expressed during the same phase [31,35, Figure 1B]. Faster-X evolution is predicted to occur when beneficial mutations are on average recessive and exposed to selection on the X chromosome [6, reviewed in 36, Box 1]. Faster-X evolution is apparent across different functional classes of X-linked genes, but the effect appears to be the strongest for male-biased spermatogenic genes [e.g. 35]. In turn, faster-X evolution could drive the evolution of genetic incompatibilities that disrupt spermatogenesis in hybrid males [6]. Given the developmental sensitivity of the mitotic phase and the enrichment of rapidly evolving X-linked genes expressed in spermatogonia, we might further predict that the evolution of hybrid male sterility often involves disruption of early spermatogenesis. No such developmental bias has been established, although there is some evidence for disrupted gene regulation in hybrid males early in spermatogenesis [22,37]. Thus, the mitotic phase of spermatogenesis has had a big impact on the molecular evolution of the X chromosome, but it is unclear if these evolutionary dynamics also play an important role in speciation.
Meiotic phase: The constraint of silenced sex chromosomes
During meiosis, chromosomes condense, homologous chromosomes pair and recombine, and there are two rounds of cell division that produce haploid gametes (Figure 1A). Mammalian X and Y chromosomes typically only pair and recombine over a small region of homology called the pseudoautosomal region. Just prior to pairing and recombination, the X and Y chromosomes are transcriptionally silenced through meiotic sex chromosome inactivation (MSCI) and are sequestered in a cellular domain known as the sex body [38], where they remain inactivated throughout meiosis. MSCI originated soon after the origin of the mammalian sex chromosomes [39]. MSCI likely evolved from a more general mechanism in mammals, the meiotic silencing of unsynapsed chromatin (MSUC), which triggers the inactivation of autosomal regions that are not properly paired during meiosis [40]. This suggests that the silencing of the sex chromosomes is potentially a by-product of heteromorphy [11]. Alternatively, MSUC and MSCI may have evolved in response to genomic conflict as a way to prevent selfish genetic elements from being preferentially transmitted during meiosis [41,42]. Sex chromosomes may be silenced in response to genomic conflict because selfish genetic elements are predicted to arise more often and increase in frequency more easily on the sex chromosomes due to their sex-biased transmission [5,43,44, Box 1]. MSCI may also have evolved as a way of silencing X-linked female-biased genes that are toxic to spermatogenesis [11].
Regardless of the mechanistic underpinnings or evolutionary origins, the constraints imposed by MSCI have had profound impacts on the evolution of X chromosome gene content. For example, MSCI strongly selects against essential meiotic genes on the X chromosome. One apparent consequence of this is that many testis-biased genes have been copied from the sex chromosomes and inserted into the autosomes [45–47]. In at least one instance, meiotic expression of the autosomal copy appears to directly compensate for X-linked silencing [47]. Newly arisen male-biased genes are often X-linked, but over time the movement of these genes from the X to the autosomes has led to the de-enrichment of X-linked male meiotic genes [46]. Meiotic silencing may also select for X-linked genes that are critical in the later stages of meiosis to be highly expressed earlier in development (prior to meiosis or in early meiosis), so that transcripts persist through MSCI [48,49]. If true, then the constraints of MSCI may partially explain the enrichment for X-linked genes that are expressed earlier in spermatogenesis.
Sex chromosome evolution associated with the meiotic phase can contribute to speciation through at least two distinct mechanisms. First, recombination along the pseudoautosomal region can be essential for proper pairing and segregation of the sex chromosomes [50]. In mammals, this region of retained X-Y homology contains very few genes but their sequence and structure can evolve rapidly, likely as direct consequence of locally elevated recombination rates [e.g. 51,52]. Several studies have linked rapid pseudoautosomal divergence to the evolution of hybrid male sterility between closely related lineages of mice [53–55].
Second, regulation of MSCI may be prone to disruption in hybrids. When MSCI was first described by Lifschytz and Lindsley [56], they proposed that MSCI could be easily disrupted in hybrid genomes and that this could be a major developmental mechanism contributing to Haldane’s rule for hybrid male sterility. Nearly 40 years later, the first support for this hypothesis was demonstrated in mice [22,57–60] and likely in cats [61]. It remains to be seen if the disruption of MSCI or MSCI-like phenomena is a common mechanism underlying the rapid evolution of hybrid male sterility.
In mice, disruption of MSCI appears to be intimately tied to divergence of the autosomal encoded protein PRDM9 and its binding sites [59,62,63], which determine the location of double-strand breaks during recombination [64]. Divergence in PRDM9 binding sites can lead to the failure of PRDM9 to properly bind in hybrids [63,65], resulting in autosomal asynapsis [59,63,66], which in turn triggers MSUC and subsequent disruption of MSCI [22,59]. The mechanistic link between PRDM9 binding, autosomal asynapsis, and disrupted MSCI is not known. Unsynapsed autosomes are silenced using some of the same cellular components involved in MSCI. If this shared resource becomes depleted when MSUC is elevated [11], then the disruption of MSCI in hybrids may merely be a downstream consequence of widespread autosomal asynapsis and not associated with sex chromosome evolution per se [66]. However, Prdm9-associated hybrid sterility depends on an incompatible interaction with a specific region on the mouse X chromosome [60,67] that is also associated with X-linked control of global recombination rates [68,69]. These studies point towards a major role for X-linked control of meiotic recombination, MSCI, and the evolution of hybrid male sterility [22,69,70].
Postmeiotic phase: Conflict between the X and Y
The haploid cells produced during meiosis, spermatids, undergo dramatic differentiation during postmeiotic development. DNA becomes highly condensed and the morphological features of the mature spermatozoa take shape through the process of spermiogenesis (Figure 1A). Spermiogenesis is a highly specialized developmental process and many genes expressed during this phase are specific to postmeiotic cells [48,49,71,72]. Similar to MSCI, sex-linked genes are also partially silenced during postmeiotic development through a process known as postmeiotic sex chromosome repression (PSCR). It is unclear if PSCR is a continuation of MSCI or an independent repression of sex chromosome expression [73–75]. Unlike MSCI, many sex-linked genes escape PSCR and are highly expressed in round spermatids [31,34,73,74,76].
Genes expressed in postmeiotic cells tend to show rapid protein sequence evolution both on and off the sex chromosomes [31,35,74,77, Figure 1B]. This likely reflects relaxed evolutionary constraints typically associated with genes that are tissue or cell-specific [78], as well as positive selection on genes influencing sperm form and function across species [77]. The X-linked genes that escape PSCR tend to play critical roles in sperm development [79] but evolve even faster than postmeiotic genes on the autosomes [31]. Interestingly, a recent study found that postmeiotic gene expression divergence appears slower, not faster on the X-chromosome [31]. This result is based on a single pairwise comparison between closely related subspecies of mice, and other studies have produced conflicting results [e.g. 80].
Many of the sex-linked genes that are expressed only in postmeiotic cells are also ampliconic genes [3,74,76,81]. The leading mechanistic hypothesis for this genomic pattern is that copy number expansions evolve essentially as dosage compensation responses that allow critical X-linked spermatogenic genes to overcome the repressive chromatin environment of PSCR [3,76,79]. This model posits a relationship between ampliconic expansions, overall expression levels, and sperm development. However, a recent study showed that deleting copies of ampliconic genes on the mouse X-chromosome did not reduce fertility, despite reducing gene expression levels by half [82]. Thus, the functional relationship between postmeiotic amplicon evolution and spermatogenesis is still unclear.
The evolution of ampliconic genes expressed during the postmeiotic phase of spermatogenesis appears to be a general feature of mammalian sex chromosomes. Sex-linked ampliconic gene families have been identified in several mammals, including mice [80], cattle [83], and humans [84]. Ampliconic gene regions vary dramatically in size and position, even among closely related mammals [3,4,81,85,86, Figure 2A]. In particular, ampliconic gene regions on the Y chromosome have been independently acquired and expanded multiple times in mammals (Table 1), with similar patterns of expression and high specificity to postmeiotic spermatogenesis [3,84]. Similarly, X chromosome gene content is largely conserved across mammals [1], but ampliconic genes are the striking exception [3, Figure 2B]. Sex-linked ampliconic gene evolution appears to be so rapid in mice that there can be extensive copy number variation within populations [80,87,88], and there are strong signatures of frequent positive selective sweeps near ampliconic genes in cats [89] and primates [90].
Figure 2. Ampliconic regions of the sex chromosomes evolve rapidly.

A. Cladogram and Y chromosome sketches showing that the structure and size of the Y chromosome is highly variable even between closely related species, largely because of variation in the ampliconic regions [4,81,85,86,89]. Chromosome sketches for species marked with (*) show only the male specific region of the Y chromosome. B. The number of ampliconic, multicopy, and single-copy X chromosome genes unique to human, unique to mouse, and shared between the two species [data from 3]. Gene content is highly conserved for single-copy and, to a lesser extent, multicopy genes. However, the ampliconic genes are highly divergent.
Table 1: Y Chromosome ampliconic gene families in mammals.
Ampliconic regions are very difficult to sequence and assemble accurately. Currently, the most accurate method is single-haplotype iterative mapping and sequencing (SHIMS), which uses bacterial artificial chromosome (BAC) cloning to amplify long reads and then identifies single-nucleotide differences among clones to identify different copies of a gene, find overlaps among copies, and properly order these copies [104]. BAC cloning combined with expression data can also be used to sequence ampliconic regions, although it is less accurate because it does not have a method for distinguishing among different copies of the same gene or ordering those copies [89]. Recently, a new method for ampliconic region sequencing was developed that uses flow sorting, droplet digital PCR, and bioinformatics to isolate a chromosome or region of interest for enrichment [105]. This method appears to be more accurate than BAC cloning and may provide an affordable alternative to SHIMS. Improved technology for sequencing highly repetitive regions will be important for understanding the acquisition and expansions of ampliconic gene families on the sex chromosomes.
| Organism | # | % of Chr Y | Co-amplified X/Y homologs | testis-biased expression | Sequencing method | References |
|---|---|---|---|---|---|---|
| Mouse (Mus musculus) | 5 | 98% | Slx/Slxl1, Sly; Sstx, Ssty1/Ssty2; Srsx, Srsy | Yes | SHIMS | [4,81] |
| Human (Homo sapiens) | 9 | 45% (of euchromatin) | VCX, VCY; HSFX1/HSFX2, HSFY | Yes | SHIMS | [4,85,106] |
| Chimpanzee (Pan troglodytes) | 6 | 57% (of euchromatin) | Unknown | Yes | SHIMS | [4,85] |
| Gorilla (Gorilla gorilla gorilla) | 8 | -- | Unknown | Unknown | Flow sorting, short and long read sequencing, gene expression, ddPCR | [105] |
| Rhesus (Macaca mulatta) | 5a | 4.5% (of euchromatin) | Unknown | Yes | SHIMS | [4,89,107] |
| Bull (Bos taurus) | ≥ 7 | -- | EGLX and EGLY | Yes | BAC cloning, gene expression | [4,83] |
| Dog (Canis lupus familiaris) | 2a | 9.7%b | Unknown | Yes | BAC cloning, cytogenic data, gene expression | [89] |
| Cat (Felis silvestris catus) | 9a | 80%b | Unknown | Yes | BAC cloning, cytogenic data, gene expression | [89] |
| Pig (Sus scrofa) | ~2–5 | -- | Unknown | Nod | BAC cloning, optical mapping | [86] |
| Opossum (Monodelphis domestica) | 4c | -- | Unknown | Unknown | SHIMS | [4] |
| Rat (Rattus norvegicus) | 8c | -- | Unknown | Yes | SHIMS | [4] |
| Marmoset (Callithrix jacchus) | 10c | -- | Unknown | Yes | SHIMS | [4] |
What drives the rapid and recurrent evolution of these sex-linked ampliconic gene families in mammals? Such dynamic evolutionary patterns would seem unlikely under a model of simple dosage compensation responses to PSCR. In mice, an ampliconic gene family is directly involved in the maintenance of PSCR [91] and this mechanistic connection may in turn drive an evolutionary arms-race in relative copy numbers of X- and Y-linked genes [92,93]. Expression of the ampliconic Y-linked Sly gene decreases postmeiotic sex chromosome expression while expression of the ampliconic X-linked Slx gene appears to increase sex-linked expression [91,94]. These antagonistic regulatory effects set the stage for intragenomic conflict driving co-evolved increases in Slx and Sly copy numbers and other co-regulated X, Y, and autosomal amplicons [80,92,93, Box 2]. These patterns demonstrate that intragenomic conflict can be a powerful force driving the rapid evolution and expansion ampliconic of sex-linked gene families. It remains to be seen if the antagonistic co-evolutionary dynamics of amplicons uncovered in mice are a general feature of heteromorphic sex chromosomes that are regulated by PSCR.
Box 2: Genomic conflict between mouse sex chromosomes.
The Slx and Sly ampliconic gene families are found on the X and Y chromosomes, respectively, of house mice (Mus musculus) and have undergone rapid expansion, reaching sizes of over 100 gene copies [80,92]. Genomic conflict has been proposed to be the main evolutionary force driving this dramatic expansion [93]. Cocquet and colleagues [91,93,94] have performed experiments in male mice to knockdown the expression of Slx and Sly multicopy gene families (Figure I). When Slx transcription is knocked down, postmeiotic sex chromosome gene expression decreases slightly, sperm are mildly malformed, and males produce litters with male-biased sex ratios. When Sly transcription is knocked down, the sex chromosomes are overexpressed, sperm are severely abnormal, and males are mostly sterile. When Sly knockdowns do occasionally reproduce, they sire litters with sex ratios skewed towards females. Knocking down both Slx and Sly rescued male fertility and restored sex chromosome expression levels and offspring sex ratios. These experiments suggest that Slx and Sly are coevolving in an antagonistic arms-race to escape PSCR and bias their transmission to the next generation.
What is fascinating is that Slx and Sly have dramatically expanded their copy numbers since they arose in Mus and copy numbers vary among closely related subspecies [92] and within populations [80,87,88]. For example, M. m. musculus has ~100 copies of Slx and ~80 of Sly, while M. m. domesticus has ~50 copies of both Slx and Sly. These two subspecies form subfertile F1 hybrids in a natural hybrid zone in central Europe [98], leading to the intriguing possibility that imbalances in Slx and Sly copy number could contribute to hybrid sterility and speciation. Patterns of postmeiotic gene expression in reciprocal F1 hybrids are consistent with the Slx/Sly imbalance hypothesis: Slx excess hybrids are mostly sterile and overexpress the sex chromosomes, while Sly excess hybrids have only slight fertility problems and underexpress the sex chromosomes [22]. However, it is still unclear how much of the observed sterility in Slx excess hybrids is due to downstream effects of disrupted MSCI in the same cross (i.e. earlier disruption could have cascading effects on later cell stages), or other interactions between the X and Y chromosomes [99]. The phenotypic effects of Slx/Sly imbalance may also critically depend on interactions with other sex chromosome ampliconic regions and autosomal ampliconic gene regions that appear to be co-regulated with Sly [22,100,101].
The rapid evolution of postmeiotic genes could also lead to the evolution of hybrid male sterility. Similar to disruption of MSCI in hybrids, antagonistic arms races between X- and Y-linked genes provide an additional general regulatory mechanism through which reproductive isolation could rapidly evolve. Closely related species can have drastically different copy numbers of ampliconic genes [e.g. in mice 92]. Copy number imbalances in F1 hybrids could lead to one sex chromosome “winning” the arms-race and disrupting postmeiotic sex chromosome expression [93], resulting in hybrid male sterility (Box 2). Empirical support for this intriguing model remains tentative in mice [22]. However, ampliconic regions of the primate X chromosome with strong signatures of positive selection also show reduced gene flow between Neanderthals and anatomically modern humans, implying these regions could be barriers to gene flow between lineages [95].
Concluding Remarks
Sex chromosomes show extraordinary evolutionary dynamics shaped by their unique patterns of inheritance and by sexual selection. Sex chromosome evolution is also a product of strong developmental constraints imposed during male and female development. Here we focused on the interplay between sexual selection and constraints imposed during specific stages of spermatogenesis, arguing that this key developmental process plays a prominent role in shaping the evolution of mammalian sex chromosomes. This perspective affords additional insights into the evolution of sequences, gene contents, and structures of the sex chromosomes within and between mammalian species. One fundamental consequence of these processes is that sex chromosome evolution is intimately tied to the one of the most general patterns in the evolution of sexually-reproducing animals—the rapid evolution of hybrid male sterility. Linking sex chromosome evolution to the specific causes of male sterility is an ongoing challenge. However, it is becoming increasingly clear that the process of recombination and the epigenetic regulation of the sex chromosomes both seem especially prone to disruption in hybrids.
Spermatogenesis is a fundamental aspect of reproduction, but it is understood well in only a handful of model systems. Nonetheless, the hallmarks of sex chromosome evolution, such as sex-biased gene content, faster-X evolution, and the large role of the X chromosome in speciation is best understood in the context of a strong developmental framework. Future studies will benefit from incorporating and extending these perspectives across diverse mammalian systems (See Outstanding Questions).
Supplementary Material
Figure I.
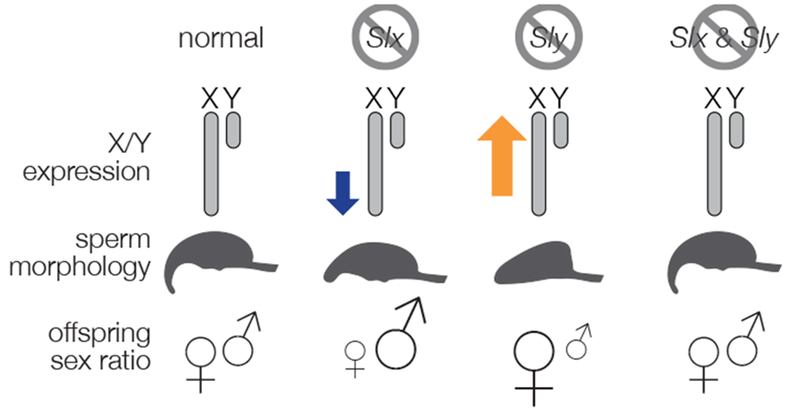
Consequence of Slx and Sly knockdown.
ACKNOWLEDGEMENTS
We would like to thank Julie Cocquet, Art Woods, Doug Emlen, Scott Taylor and three anonymous reviewers for their helpful feedback on this manuscript. EEKK was supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. (DGE-1313190). Research by JMG and ELL on sex chromosome evolution and reproduction was supported by grants from the Eunice Kennedy Shriver National Institute of Child Health and Human Development (R01-HD073439 and R01-HD094787) and the National Institute of General Medical Sciences (R01-GM098536). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation or the National Institutes of Health.
GLOSSARY
- Ampliconic gene
Highly similar multicopy gene families, usually in found in palindromic or tandem duplication arrays
- Autosomal asynapsis
The failure of autosomes to properly pair and recombine during meiosis. Unsynapsed autosomes trigger MSUC
- Cryptic female choice
A form of intersexual selection that arises when variation in the fertilization success among males is influenced by females
- Dosage compensation
Regulatory mechanisms that equalize the expression of sex-linked genes between males and females or relative to the autosomes. Dosage compensation can be chromosome-wide or gene-specific
- Faster-X evolution
The faster evolution of genes on the X chromosome relative to the autosomes. Originally predicted if beneficial mutations are on average recessive, allowing selection to act more efficiently on exposed X-linked mutations in hemizygous males
- Genomic conflict
When one part of the genome gains a reproductive advantage at the expense of another part[s] of the genome. Conflict can occur among genomes (e.g., maternal-offspring conflict during pregnancy) or within genomes (e.g., mitochondrial and nuclear genes, sexually antagonistic genes)
- Hemizygous
When only one copy of a chromosome is present in a diploid organism. The X and Y chromosomes are hemizygous in males (XY)
- Heterogametic
The sex that produces gametes with two different types of sex chromosomes (e.g., X and Y-bearing sperm). In mammals, males are the heterogametic sex
- Heteromorphic sex chromosomes
Sex chromosomes descended from homologous autosomes that differ in gene content, size, structure, and base composition
- Male-biased mutation
Higher mutation rates in the male germline caused by more cell divisions per generation in spermatogenesis relative to oogenesis
- Meiotic sex chromosome inactivation (MSCI)
Epigenetic silencing of sex chromosome gene expression during meiosis
- Meiotic silencing of unsynapsed chromatin (MSUC)
Silencing of gene expression in unsynapsed chromosomal regions during meiosis
- Postmeiotic sex chromosome repression (PSCR)
Transcriptional repression of the sex chromosomes during postmeiotic development
- Pseudoautosomal region
Region(s) of retained homology on heteromorphic sex chromosomes that pair and recombine in the heterogametic sex
- Sexual conflict
Differences in the evolutionary interests of males and females
- Spermatogenesis
The development of haploid gametes in the testes of males through mitosis, meiosis and postmeiotic development (i.e., spermiogenesis). See Figure 1A
- Spermatogonia
Undifferentiated diploid male germ cells. Spermatogonia undergo spermatogenesis to produce haploid spermatozoa
- Spermiogenesis
The final phase of spermatogenesis defined by the morphological development of round spermatids into spermatozoa
- Sperm competition
A form of intrasexual selection whereby the ejaculates of two or more males compete to fertilize a female’s eggs
- Sexually antagonistic gene
A gene showing sex-dependent fitness effects that are beneficial in one sex but harmful in the other
- Sex-biased gene
A gene that is expressed predominately or exclusively in one sex. Sex-biased genes are often expressed in sex-specific tissues (e.g., testes and accessory glands in males)
REFERENCES
- 1.Ohno S (1967) Sex chromosomes and sex-linked genes, Springer-Verlag. [Google Scholar]
- 2.Gu L and Walters JR (2017) Evolution of sex chromosome dosage compensation in animals: a beautiful theory, undermined by facts and bedeviled by details. Genome Biol Evol 9, 2461–2476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mueller JL et al. (2013) Independent specialization of the human and mouse X chromosomes for the male germ line. Nat Genet 45, 1083–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bellott DW et al. (2014) Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508, 494–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rice WR (1984) Sex chromosomes and the evolution of sexual dimorphism. Evolution 38, 735–742 [DOI] [PubMed] [Google Scholar]
- 6.Charlesworth B et al. (1987) The relative rates of evolution of sex chromosomes and autosomes. Am Nat 130, 113–146 [Google Scholar]
- 7.Connallon T and Clark AG (2010) Sex linkage, sex-specific selection, and the role of recombination in the evolution of sexually dimorphic gene expression. Evolution 64, 3417–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parsch J and Ellegren H (2013) The evolutionary causes and consequences of sex-biased gene expression. Nat Rev Genet 14, 83–87 [DOI] [PubMed] [Google Scholar]
- 9.Subramanian VV and Hochwagen A (2014) The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb Perspect Biol 6, a016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gunes S et al. (2015) Spermatogenesis, DNA damage and DNA repair mechanisms in male infertility. Reprod Biomed Online 31, 309–319 [DOI] [PubMed] [Google Scholar]
- 11.Turner JMA (2015) Meiotic silencing in mammals. Annu. Rev. Genet 49, 395–412 [DOI] [PubMed] [Google Scholar]
- 12.Parker GA (1970) Sperm competition and its evolutionary consequences in the insects. Biol Review 45, 525–567 [Google Scholar]
- 13.Eberhard W (1996) Female control: sexual selection by cryptic female choice, Princeton University Press. [Google Scholar]
- 14.Firman RC et al. (2017) Postmating female control: 20 years of cryptic female choice. Trends Ecol Evol 32, 368–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schärer L et al. (2011) Mating behavior and the evolution of sperm design. Proc Natl Acad Sci U S A 108, 1490–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higginson DM et al. (2012) Female reproductive tract form drives the evolution of complex sperm morphology. Proc Natl Acad Sci U S A 109, 4538–4543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rowe M et al. (2015) Postcopulatory sexual selection is associated with accelerated evolution of sperm morphology. Evolution 69, 1044–1052 [DOI] [PubMed] [Google Scholar]
- 18.Lüpold S et al. (2016) How sexual selection can drive the evolution of costly sperm ornamentation. Nature 533, 535–538 [DOI] [PubMed] [Google Scholar]
- 19.McLennan HJ et al. (2017) Greater sperm complexity in the Australasian old endemic rodents (Tribe: Hydromyini) is associated with increased levels of inter-male sperm competition. Reprod. Fertil. Dev 29, 921–930 [DOI] [PubMed] [Google Scholar]
- 20.Pitnick S et al. (2009) Sperm morphological diversity In Sperm biology an evolutionary perspective (Birkhead TR et al. , eds), pp. 69–149, Academic Press [Google Scholar]
- 21.Higginson DM et al. (2012) Convergence recurrence and diversification of complex sperm traits in diving beetles (Dytiscidae). Evolution 66, 1650–1661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larson EL et al. (2017) The composite regulatory basis of the large X-effect in mouse speciation. Mol Biol Evol 34, 282–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maheshwari S and Barbash DA (2011) The genetics of hybrid incompatibilities. Annu. Rev. Genet 45, 331–355 [DOI] [PubMed] [Google Scholar]
- 24.Haldane JBS (1922) Sex ratio and unisexual sterility in hybrid animals. J. Genet 12, 101–109 [Google Scholar]
- 25.Delph LF and Demuth JP (2016) Haldane’s rule: genetic bases and their empirical support. J. Hered 107, 383–391 [DOI] [PubMed] [Google Scholar]
- 26.Masly JP and Presgraves DC (2007) High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS Biol 5, 1890–1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coyne JA and Orr HA (1989) Two rules of speciation In Speciation and its Consequences (Otte D and Endler JA, eds), pp. 180–207, Sinauer Associates [Google Scholar]
- 28.Presgraves DC (2008) Sex chromosomes and speciation in Drosophila. Trends Genet 24, 336–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson Sayres MA and Makova KD (2011) Genome analyses substantiate male mutation bias in many species. Bioessays 33, 938–945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khil PP et al. (2004) The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet 36, 642–646 [DOI] [PubMed] [Google Scholar]
- 31.Larson EL et al. (2016) Contrasting levels of molecular evolution on the mouse X chromosome. Genetics 203, 1841–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang PJ et al. (2001) An abundance of X-linked genes expressed in spermatogonia. Nat Genet 27, 422–426 [DOI] [PubMed] [Google Scholar]
- 33.Bachtrog D (2013) Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14, 113–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cortez D et al. (2015) Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488–493 [DOI] [PubMed] [Google Scholar]
- 35.Kousathanas A et al. (2014) Faster-X adaptive protein evolution in house mice. Genetics 196, 1131–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meisel RP and Connallon T (2013) The faster-X effect: integrating theory and data. Trends Genet 29, 537–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oka A et al. (2014) Evolutionarily diverged regulation of X-chromosomal genes as a primal event in mouse reproductive isolation. PLoS Genet 10, e1004301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Handel M (2004) The XY body: a specialized meiotic chromatin domain. Exp Cell Res 296, 57–63 [DOI] [PubMed] [Google Scholar]
- 39.Daish TJ et al. (2015) Lack of sex chromosome specific meiotic silencing in platypus reveals origin of MSCI in therian mammals. BMC Biology 13, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Turner JMA et al. (2005) Silencing of unsynapsed meiotic chromosomes in the mouse. Nat Genet 37, 41–47 [DOI] [PubMed] [Google Scholar]
- 41.Meiklejohn CD and Tao Y (2010) Genetic conflict and sex chromosome evolution. Trends Ecol Evol 25, 215–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rice WR (2013) Nothing in genetics makes sense except in light of genomic conflict. Annu. Rev. Ecol. Evol. Syst 44, 217–237 [Google Scholar]
- 43.Frank SA (1991) Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution 45, 262–267 [DOI] [PubMed] [Google Scholar]
- 44.Hurst LD and Pomiankowski A (1991) Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane’s rule and related phenomena. Genetics 128, 841–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Emerson JJ et al. (2004) Extensive gene traffic on the mammalian X chromosome. Science 303, 537–540 [DOI] [PubMed] [Google Scholar]
- 46.Zhang YE et al. (2010) Chromosomal redistribution of male-biased genes in mammalian evolution with two bursts of gene gain on the X chromosome. PLoS Biol 8, e1000494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang L et al. (2017) RPL10L is required for male meiotic division by compensating for RPL10 during meiotic sex chromosome inactivation in mice. Curr Biol 27, 1498–1505 [DOI] [PubMed] [Google Scholar]
- 48.da Cruz I et al. (2016) Transcriptome analysis of highly purified mouse spermatogenic cell populations: gene expression signatures switch from meiotic-to postmeiotic-related processes at pachytene stage. BMC Genomics 17, 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jan SZ et al. (2017) Unraveling transcriptome dynamics in human spermatogenesis. Development 144, 3659–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otto SP et al. (2011) About PAR: The distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet 27, 358–367 [DOI] [PubMed] [Google Scholar]
- 51.White MA et al. (2012) A pronounced evolutionary shift of the pseudoautosomal region boundary in house mice. Mamm Genome 23, 454–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hinch AG et al. (2014) Recombination in the human pseudoautosomal region PAR1. PLoS Genet 10, e1004503–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White MA et al. (2012) Genetics and evolution of hybrid male sterility in house mice. Genetics 191, 917–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dumont BL (2017) Meiotic consequences of genetic divergence across the murine pseudoautosomal region. Genetics 205, 1089–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hale DW et al. (1993) Meiotic abnormalities in hybrid mice of the C57BL/6J× Mus spretus cross suggest a cytogenetic basis for Haldane’s rule of hybrid sterility. Cytogenet Genome Res 63, 221–234 [DOI] [PubMed] [Google Scholar]
- 56.Lifschytz E and Lindsley DL (1972) The role of X-chromosome inactivation during spermatogenesis. Proc Natl Acad Sci U S A 69, 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Good JM et al. (2010) Widespread over-expression of the X chromosome in sterile F1 hybrid mice. PLoS Genet 6, e1001148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campbell P et al. (2013) Meiotic sex chromosome inactivation is disrupted in sterile hybrid male house mice. Genetics 193, 819–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhattacharyya T et al. (2013) Mechanistic basis of infertility of mouse intersubspecific hybrids. Proc Natl Acad Sci U S A 110, E468–E477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turner LM et al. (2014) Genomic networks of hybrid sterility. PLoS Genet 10, e1004162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davis BW et al. (2015) Mechanisms underlying mammalian hybrid sterility in two feline interspecies models. Mol Biol Evol 32, 2534–2546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mihola O et al. (2009) A mouse speciation gene encodes a meiotic histone H3 methyltransferase. Science 323, 373–375 [DOI] [PubMed] [Google Scholar]
- 63.Davies B et al. (2016) Re-engineering the zinc fingers of PRDM9 reverses hybrid sterility in mice. Nature 530, 171–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baudat F et al. (2010) PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327, 836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smagulova F et al. (2016) The evolutionary turnover of recombination hot spots contributes to speciation in mice. Genes Dev 30, 266–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gregorova S et al. (2018) Modulation of Prdm9-controlled meiotic chromosome asynapsis overrides hybrid sterility in mice. eLife 7, e34282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhattacharyya T et al. (2014) X chromosome control of meiotic chromosome synapsis in mouse inter-subspecific hybrids. PLoS Genet 10, e1004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dumont BL and Payseur BA (2011) Genetic analysis of genome-scale recombination rate evolution in house mice. PLoS Genet 7, e1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Balcova M et al. (2016) Hybrid sterility locus on chromosome X controls meiotic recombination rate in mouse. PLoS Genet 12, e1005906–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Payseur BA (2016) Genetic links between recombination and speciation. PLoS Genet 12, e1006066–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Margolin G et al. (2014) Integrated transcriptome analysis of mouse spermatogenesis. BMC Genomics 15, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Soumillon M et al. (2013) Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. CellReports 3, 2179–2190 [DOI] [PubMed] [Google Scholar]
- 73.Namekawa SH et al. (2006) Postmeiotic sex chromatin in the male germline of mice. Curr Biol 16, 660–667 [DOI] [PubMed] [Google Scholar]
- 74.Sin HS et al. (2012) Human postmeiotic sex chromatin and its impact on sex chromosome evolution. Genome Res 22, 827–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Federici F et al. (2015) Incomplete meiotic sex chromosome inactivation in the domestic dog. BMC Genomics 16, 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mueller JL et al. (2008) The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet 40, 794–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Good JM and Nachman MW (2005) Rates of protein evolution are positively correlated with developmental timing of expression during mouse spermatogenesis. Mol Biol Evol 22, 1044–1052 [DOI] [PubMed] [Google Scholar]
- 78.Meisel RP (2011) Towards a more nuanced understanding of the relationship between sex-biased gene expression and rates of protein-coding sequence evolution. Mol Biol Evol 28, 1893–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sin H-S and Namekawa SH (2013) The great escape. Epigenetics 8, 887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Morgan AP and Pardo-Manuel de Villena F (2017) Sequence and structural diversity of mouse Y chromosomes. Mol Biol Evol 34, 3186–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soh YQS et al. (2014) Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159, 800–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kruger AN et al. (2018) Male mice with large inversions or deletions of X-chromosome palindrome arms are fertile and express their associated genes during post-meiosis. Sci Rep 8, 8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chang T-C et al. (2013) Male-specific region of the bovine Y chromosome is gene rich with a high transcriptomic activity in testis development. Proc Natl Acad Sci U S A 110, 12373–12378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hughes JF and Page DC (2015) The biology and evolution of mammalian Y chromosomes. Annu. Rev. Genet 49, 507–527 [DOI] [PubMed] [Google Scholar]
- 85.Hughes JF et al. (2010) Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 463, 536–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Skinner BM et al. (2016) The pig X and Y Chromosomes: structure, sequence, and evolution. Genome Res 26, 130–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Case LK et al. (2015) Copy number variation in Y chromosome multicopy genes is linked to a paternal parent-of-origin effect on CNS autoimmune disease in female offspring. Genome Biol 16, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pezer Ž et al. (2015) Divergence patterns of genic copy number variation in natural populations of the house mouse (Mus musculus domesticus) reveal three conserved genes with major population-specific expansions. Genome Res 25, 1114–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li G et al. (2013) Comparative analysis of mammalian Y chromosomes illuminates ancestral structure and lineage-specific evolution. Genome Res 23, 1486–1495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nam K et al. (2015) Extreme selective sweeps independently targeted the X chromosomes of the great apes. Proc Natl Acad Sci U S A 112, 6413–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cocquet J et al. (2009) The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol 7, e1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ellis PJI et al. (2011) Association of Sly with sex-linked gene amplification during mouse evolution: a side effect of genomic conflict in spermatids? Hum Mol Genet 20, 3010–3021 [DOI] [PubMed] [Google Scholar]
- 93.Cocquet J et al. (2012) A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet 8, e1002900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cocquet J et al. (2010) Deficiency in the multicopy Sycp3-like X-linked genes Slx and Slxl1 causes major defects in spermatid differentiation. Mol. Biol. Cell 21, 3497–3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dutheil JY et al. (2015) Strong selective sweeps on the X chromosome in the human-chimpanzee ancestor explain its low divergence. PLoS Genet 11, e1005451–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vicoso B and Charlesworth B (2009) Effective population size and the faster-X effect: an extended model. Evolution 63, 2413–2426 [DOI] [PubMed] [Google Scholar]
- 97.Vicoso B and Charlesworth B (2006) Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet 7, 645–653 [DOI] [PubMed] [Google Scholar]
- 98.Turner LM et al. (2012) Reduced male fertility is common but highly variable in form and severity in a natural house mouse hybrid zone. Evolution 66, 443–458 [DOI] [PubMed] [Google Scholar]
- 99.Campbell P and Nachman MW (2014) X-Y interactions underlie sperm head abnormality in hybrid male house mice. Genetics 196, 1231–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Moretti C et al. (2016) Expression and epigenomic landscape of the sex chromosomes in mouse post-meiotic male germ cells. Epigenetics & Chromatin 9, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Moretti C et al. (2017) SLY regulates genes involved in chromatin remodeling and interacts with TBL1XR1 during sperm differentiation. Cell Death Differ 24, 1029–1044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Shima JE et al. (2004) The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 71, 319–330 [DOI] [PubMed] [Google Scholar]
- 103.Chalmel F et al. (2007) The conserved transcriptome in human and rodent male gametogenesis. Proc Natl Acad Sci U S A 104, 8346–8351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bellott DW et al. (2017) Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nat Genet 49, 387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tomaszkiewicz M et al. (2016) A time- and cost-effective strategy to sequence mammalian Y Chromosomes: an application to the de novo assembly of gorilla Y. Genome Res 26, 530–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Skaletsky H et al. (2003) The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825–837 [DOI] [PubMed] [Google Scholar]
- 107.Hughes JF et al. (2012) Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 482, 82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


