Abstract
Mapping the paths that stem and progenitor cells take en route to differentiate and elucidating the underlying molecular controls are key goals in developmental and stem cell biology. However, with population level analyses it is difficult — if not impossible — to define the transition states and lineage trajectory branch points within complex developmental lineages. Single-cell RNA-sequencing analysis can discriminate heterogeneity in a population of cells and even identify rare or transient intermediates. In this review, we propose that using these data, one can infer the lineage trajectories of individual stem cells and identify putative branch points. Clonal lineage tracing of stem cells allows one to define the outcome of differentiation. Integrating these single cell-based approaches provides a robust strategy for establishing and testing models of how an individual stem cell changes through time to differentiate and self-renew.
Keywords: cell fate, lineage, scRNA-seq, single-cell RNA-sequencing, stem cells
1. Introduction
Many tissues continuously renew, incorporating new cells to maintain homeostasis, and some can also regenerate following injury. Both renewal and regenerative capacity rely upon the coordinated activity of stem cells to differentiate into a range of mature cell types and maintain a stem cell pool (Figure 1a). One of the fundamental challenges in developmental and stem cell biology is understanding how individual stem cells achieve this feat in vivo. To solve this problem, one must define how cells change through time and map the paths they take during differentiation. This knowledge is critical for unraveling the molecular mechanisms that control cell fate transitions and involves integrating a highly reductionist approach to discern the intrinsic molecular characteristics of each cell and a more systems level approach to characterize a cell’s progeny, derivatives, and environment. At a basic level, this is a fascinating problem at the core of how tissues and organisms develop and maintain homeostasis. From the translational perspective, detailed mapping of cell lineages lays the groundwork for reprogramming cells in vivo and designing strategies for cell replacement therapies.
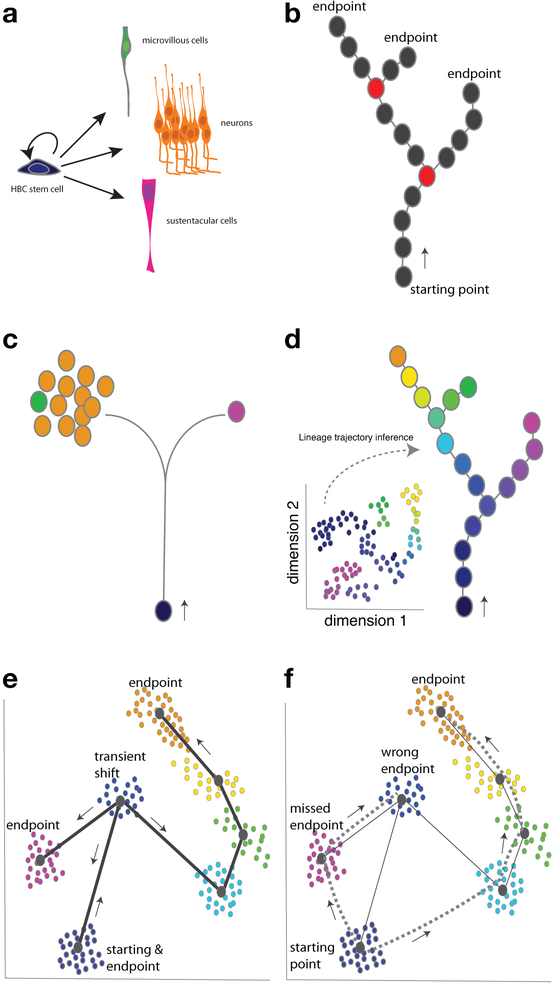
Figure 1.
Integrating single-cell RNA sequencing and lineage tracing can resolve complex cell populations and lineage trajectories. a) Multi potent stem cells can self-renew and give rise to a range of differentiated cell types. Shown are the horizontal basal cell (HBC) stem cells and differentiated cell types in the olfactory epithelium. In this review we focus on the two main lineages of the olfactory epithelium, sustentacular and neuronal, but microvillous cells are a third differentiated cell type. b–f) Circles represent individual cells or clusters of cells as indicated. b) Multipotent stem cells can give rise to multiple cell fate endpoints, and there are branch points along a lineage trajectory where one cell fate path is chosen. Circles colored red represent cell states that are branch points in this schematized lineage. c) Classic genetic lineage tracing, where a stem cell is labeled and its descendants are characterized, can provide insight into stem cell fate potential and the numbers of differentiated cells, different ones of which are indicated by the colored circles. d) scRNA-seq can discriminate the different cell types present in a heterogeneous population: here, we represent a hypothetical example of a reduced dimension plot of scRNA-seq data (inset), and this data can be used by lineage trajectory inference tools to predict the stem cell lineage trajectories, defining the cellular states and predicting lineage trajectory branch points. e) When the cell fate changes are associated with large shifts in gene expression, lineage trajectory inference can fail because the actual cell fate transition violates that assumption that cells that are more similar at the transcriptome level are closer together in the trajectory. Having time-stamping information can allow one to know that a transient state is early in the lineage and proceeds the appearance of later cell states. Incorporating this information into the model can lead to more accurate predictions. The predicted lineage is indicated by the solid lines and arrows. f) If a mature, differentiated cell type (an endpoint) is more similar in gene expression to the starting stem cell fate than an early, transient state, then lineage prediction inference will wrongly order the lineage trajectory progression, indicated here by the dashed lines.
A detailed understanding of how stem cells change through time has been elusive because one must define not only the endpoint but also all intermediate stages along the path of differentiation. Historically, fate maps have been produced by lineage tracing cells. While this has proven to be insightful, it usually provides data on the cell fate potential of labeled cells (i.e., what cell types the labeled cells can become) and not on the paths or molecular identity of cell states along the route. This is true whether the technique is applied at the population or clonal level. High-throughput sequencing approaches are excellent tools for defining the molecular status of cells, but population level analyses obscure the heterogeneity within tissues or cell lineages. However, single-cell RNA-sequencing (scRNA-seq) methods allow one to discriminate population level heterogeneity, overcoming the need for prior knowledge of the underlying cell types to fully interpret the data.
We propose that by integrating clonal lineage tracing and single-cell RNA-sequencing analysis, one can build and test models of the cellular mechanisms that underlie stem cell development into tissues, organs, and organisms. Single-cell RNA-sequencing can molecularly define cell types — including intermediates within a developmental lineage — without a priori knowledge and be used to predict branching in lineage trajectories (Figure 1b), but it can only provide predictions that must be independently validated. Clonal lineage tracing data, which can identify descendants of labeled cells but by itself does not have the resolution to distinguish subtle differences in cells or identify branch points in lineage trajectories, can inform the interpretation of scRNA-seq data and be used to test predictions derived from scRNA-seq analysis. Ultimately, we view this integrative approach as a powerful method for identifying and validating intermediate cellular states that are often transient and for defining where lineages branch en route to forming diverse cell types. It can also help alleviate challenges to defining lineage trajectory maps caused by sudden, large changes in gene transcription or looping trajectories inherent in stem cell self-renewal. By applying this approach, we have been able to identify and validate transient stem cell states and demonstrate that they are critical windows of cell fate specification in vivo.
2. Lineage Tracing Defines Cell Fate Potential But Has Limitations
Lineage tracing, the technique of following a cell or group of cells and observing their descendants, is an important tool for defining the fate potential of cells and detecting the outcomes of differentiation.[1,2] The earliest lineage tracing was based on direct observation of cells as they divided and differentiated in transparent embryos.[3–5] In opaque embryos, early experimentalists labeled cells with vital dyes: to establish a fate map, the stained cells would be followed as they developed and the ultimate structures that they formed mapped back to the position in the embryo where the dye was applied.[6,7] A third approach was to distinguish the origin of cells based on unique pigmentation or cellular appearance. For example, the distinct pigmentation patterns of different newt species were utilized in organizer transplant experiments.[8]
These classical methods of lineage tracing served as a foundation for subsequent waves of technological innovations for labeling and tracing of individual cells in embryos and adult tissues in vivo. Microinjection of enzymes and fluorescent molecules,[9–13] the use of genetic modification to label cells, which was pioneered with viral transduction,[14] and co-opting bacterial and viral recombinases and inducible reporter proteins[15–17] were instrumental advancements. Subsequently, methods for sparsely labeling cells based on heritable reporters were developed for clonal lineage tracing by coupling cell division with interchromosomal recombination of reporter gene subunits[18] or by using a multi-fluorescent reporter transgenic strategy termed Brainbow,[19] which was then modified to create Confetti, a Cre recombinase-dependent, inducible reporter that facilitates clonal lineage tracing by exclusive expression of one of four possible fluorescent reporters.[20] Applying these tools, researchers have made great strides into the cell fate potential and specification of individual stem and progenitor cells in a range of tissues in developing and adult organisms.[18,20–23] More recently, methods using transposase or CRISPR-mediated modification of DNA coupled with high-throughput sequencing of the endpoint cells have allowed the creation of lineage hierarchies representing the relationships between the final differentiated cells.[24–27]
While powerful, current lineage tracing methods cannot identify intermediate stages along a lineage trajectory, and therefore they cannot pinpoint branch points in a trajectory. The main reason for these limitations is that most prospective in vivo lineage tracing involves labeling cells and then waiting for some duration before collecting, fixing, and observing the tissue to identify the endpoint (Figure 1c).[2] One then attempts to reconstruct the path that the cell took with only the endpoint data and without having observed the cell as it traversed the path. Therefore, while lineage tracing is a powerful tool for discerning fate potential of a labeled cell, the intermediate cellular states and branch points remain unresolved. Addressing this limitation are newly developed approaches where transposase or CRISPR/Cas9-mediated modification of DNA barcodes is coupled to scRNA-seq[28–31] that allows for the identification of lineage endpoints and the establishment of a lineage hierarchy or dendrogram. Modifications of this approach should provide a more facile way to combine the strengths of lineage tracing and scRNA-seq to define the cells that serve as lineage trajectory branch points, as we will discuss further below.
3. Single-Cell Transcriptomics Provides Exquisite Discriminatory Power
Tissues and organs are complex structures composed of multiple cell types, and even cells of the same type can display a range of phenotypic variation. Single-cell RNA-sequencing methods measure gene expression (i.e., the transcriptome) of individual cells. Thus, they allow one to distinguish cells from one another in a heterogeneous population based on their differences in gene expression. This powerful approach for detecting individual cellular variation within a complex population has been applied to identify a broad range of cell types and states from cells grown in vitro culture or isolated from tissues (Figure 1d).[32–50] Moreover, its discriminatory ability allows researchers to identify rare cell types in a larger population that would be obscured in bulk level analyses.[34,51–54] In addition to identifying rare cell types, collecting samples for scRNA-seq at a range of time-points during a complex biological process allows the identification of cell states that exist only transiently or during discrete time windows, cells that would most certainly be missed in bulk level analyses.[55–59] Furthermore, comparing scRNA-seq data between control or wild-type animals and genetic mutants can be a powerful way to understand the role of specific genes in developmental processes.[55,56,58]
In developing embryos and regenerative tissues, cells are usually asynchronous in their position along the axis of differentiation. After deconvolution of complex tissues or groups of cells into their constituent cell types based on scRNA-seq, the data can then be analyzed with lineage prediction tools to order cells along the axis of differentiation based on progressive changes in gene expression, and in some cases branch points in the lineage trajectories can be predicted as well[57,58,60–64] (Figure 1d). The first iterations of trajectory prediction algorithms were capable of ordering cells along a single trajectory but were largely unable to accommodate branching lineage trajectories — that is, cases where an immature progenitor cell gives rise to more than one lineage or cell type.[65–67] Subsequent methods attempted to predict where the branch points in trajectories occur.[55,56,59,61,68–72] These tools usually take as input reduced dimension gene expression data (e.g., after using principal component analysis) or nearest-neighbor graph representations and attempt to infer the branching lineage trajectory structure and order cells along the trajectories (Box 1, Figure 2). Regardless of the specific approach, these tools rely upon the assumption that cells that are more similar in gene expression are closer together on a lineage trajectory (Figure 1d). While this is a reasonable assumption, there are situations where cell fate transitions represent more saltatory changes in gene expression rather than subtle changes along a continuum (see below, Figure 1e,f). Furthermore, they also rely upon a second assumption that the paths are unidirectional, which presents difficulties for modeling stem cell self-renewal.
Box 1. scRNA-seq data analysis and inferring lineage trajectories.
While single-cell RNA-sequencing can be a powerful tool, there are some important challenges to consider in analyzing the resulting data. First, there is sparser coverage than in bulk level RNA-seq and hence an inflated number of drop-outs or “zeros” in the gene expression counts, more noise, and higher over dispersion. This means that it is imperative to use normalization strategies that minimize technical variation while maintaining biological variation.[78–80] It is also critical to collect biological replicates for specific experimental conditions to protect against batch effects. Because of the computational challenges of clustering cells in high-dimensional space, researchers often apply dimensionality reduction techniques (e.g., principal component analysis [PCA] and/or t-distributed stochastic neighbor embedding [t-SNE]). Then unsupervised clustering is applied to cluster cells by their gene expression. For a more detailed discussion and explanation of normalization strategies, dimensionality reduction techniques, and clustering, there are several excellent tools and reviews.[78–85]
We have used the lineage trajectory inference tool Slingshot to model branching lineage trajectories.[57,58] Slingshot first infers global lineage structure by using a cluster-based minimum spanning tree to define the number of lineages and where they branch, and then it infers the order or position of cells along each lineage, often referred to as pseudo time, by fitting smooth principal curves.[72] Slingshot was built to be flexible to the type of dimensionality reduction, normalization, and clustering procedures that have been applied to the data with the belief that there is no one-size-fits-all approach for analyzing single-cell sequencing data. Slingshot allows for semi-supervision of the model based on known biology: for example, one can specify known starting points and end-points, and this does not preclude the algorithm from inferring other end-points from the data. Differences and similarities among the different lineage trajectory prediction tools are detailed in a few studies.[72,78,86] Importantly, not all lineage trajectory prediction algorithms are equal, and in a side-by-side comparison of the same data sets across a range of parameters, Slingshot has been shown to outperform most.[87]
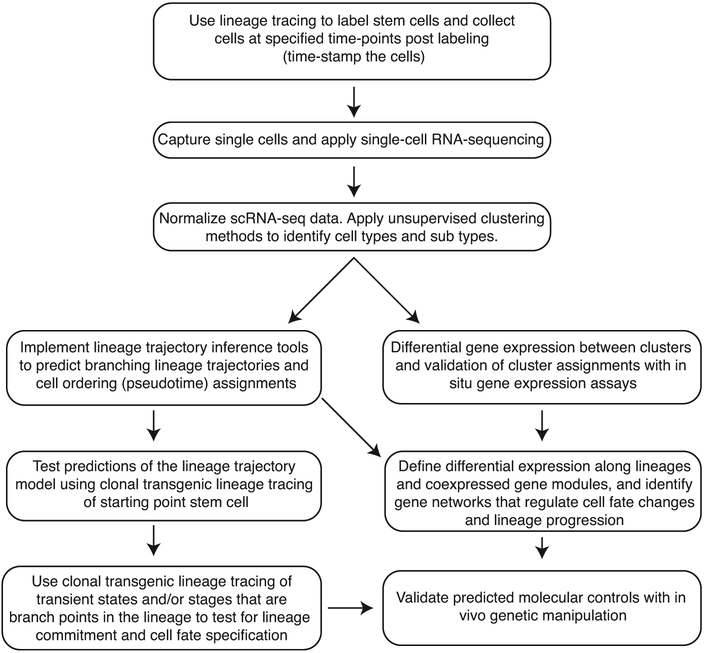
Figure 2.
Workflow for integrating single-cell RNA-sequencing and lineage tracing. We propose the indicated steps to integrate lineage tracing into the design and implementation of the scRNA-sequencing experiments. Clonal lineage tracing can be used to validate and test in silico predictions. By collecting cells at multiple time-points from lineage traced cells, transient states can be identified, and one knows that all cells are derived from the same cell type. scRNA-seq can be used to discriminate the different cell types present within a heterogeneous population. Following clustering of the data to identify cell fates/states, lineage trajectory inference tools can predict lineage trajectories including branch points. Gene expression differences and co-regulated gene expression along the lineage trajectories helps one identify the gene regulatory networks regulating cell fate changes. Predictions regarding lineage trajectory and genetic regulation can be tested with clonal lineage tracing and genetic manipulation.
Importantly, even the most sophisticated analysis of single-cell RNA-sequencing data and the ensuing models of lineage trajectories require experimental validation to demonstrate that cell types as defined transcriptomically do indeed exist in vivo and that the predicted trajectories represent reality. Validating cell types or transient states can be accomplished using RNA in situ hybridization to assess gene expression in the tissue based on genes or sets of genes measured as enriched in any given putative cellular type/state. Lineage trajectories must be validated by labeling individual progenitor cells and determining whether the output of differentiation is consistent with the inferred trajectory.
4. Integrating Lineage Tracing and Single-Cell RNA-Sequencing Helps Solve Key Challenges to Establishing Lineage Trajectories
We propose that integrating lineage tracing with scRNA-seq provides a robust framework for defining cell fate transitions, intermediate states, and stem cell branching lineage trajectories in vivo. Leveraging these two techniques together provides more resolution than either affords alone. In this framework, scRNA-seq of lineage traced cells provides the means to establish a model of stem cell lineage trajectories, whereas clonal lineage tracing provides a means to test and validate the model (Figure 2).
4.1. Lineage Tracing Tests Predictions From Lineage Trajectory Inference Tools
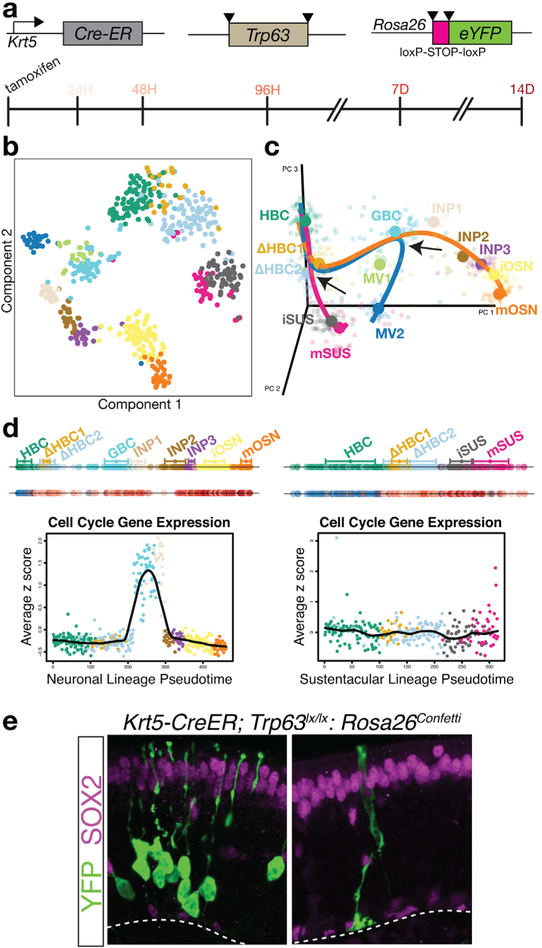
As an example of an integrated approach combining clonal lineage tracing in vivo with scRNA-seq, we identified and characterized the first transition states of the reserve stem cell of the olfactory epithelium — known as the horizontal basal cell (HBC) — upon its activation to differentiate and/or self-renew under various physiological conditions. In the absence of injury, HBC stem cells rarely differentiate; thus to increase the frequency of differentiation, we conditionally knocked out the transcription factor p63, which normally functions to repress their differentiation[57] (Figure 3a). This genetic manipulation causes the spontaneous differentiation of HBCs into multiple mature cell types of the olfactory epithelium. To identify the cell intermediates in the olfactory epithelial lineage, we performed scRNA-seq on fluorescence-activated cell sorting (FACS)-purified cells based either on expression of an olfactory progenitor-specific Sox2-eGFP tracer or YFP lineage-traced cells that were labeled using an HBC-specific Krt5-CreER driver and Rosa26-YFP reporter.[57] In addition to expected cell types, we identified new intermediate cell types (Figure 3b,c; ΔHBC1, ΔHBC2). Following normalization and clustering, we applied the lineage prediction algorithm Slingshot (Box 1). Slingshot predicted that the two main trajectories — the neuronal lineage trajectory and the sustentacular lineage trajectory — bifurcated early at a transitional intermediate (ΔHBC2) prior to the appearance of any proliferating cells (Figure 3c). Unlike the sustentacular lineage trajectory, cells of the neuronal lineage were predicted to then traverse two proliferative cell stages (Figure 3d). Therefore, one would predict that stem cell-derived neuronal clones would be multicellular while sustentacular clones would be small, perhaps even unicellular, clones and that clones should contain only cells of one cell type. To test this prediction, we scored differentiated cell clones derived from HBCs lineage traced in vivo using an HBC-specific inducible Cre recombinase driver coupled with the Cre-dependent Confetti reporter. The results from these in vivo clonal lineage tracing experiments confirmed the two lineage trajectories predicted by Slingshot from the single-cell RNA-sequencing data (Figure 3e). Importantly, clonal lineage tracing confirmed the prediction that sustentacular cells can form by direct cell fate conversion without cell division, demonstrating that cell fate changes from one cell type to another do not require cell division.[57]
Figure 3.
Lineage tracing validates lineage trajectory inference for the olfactory HBC stem cell during differentiation. a) To assess the behavior of olfactory HBC stem cells in uninjured tissue, we used an HBC stem cell specific Cre recombinase that coupled genetic ablation of Trp63 (p63), which induces more HBCs to differentiate, with transgenic lineage tracing, and collected cells in a time-course of differentiation. Triangles represent loxP sites that underlie the Cre recombinase-induced conditional knockout of p63 and conditional activation of the eYFP lineage reporter. b) Cells can be visualized in reduced dimension gene expression space. Here, we present a t-distributed stochastic neighbor embedding (t-SNE) plot, and cells are colored by cluster. c) After clustering the cells, we used Slingshot to infer the branching lineage trajectories. Slingshot predicted two bifurcations (arrows), an early bifurcation between the sustentacular and neuronal lineages followed by a second bifurcation of microvillous cells from the neuronal lineage. d) Cells can be ordered along their respective lineages. We present data for the neuronal (left) and sustentacular cell lineage (right). In the top line, cells are colored by their cluster assignment; in the bottom line, cells are colored by the time-point at which they were collected; blue cells are wild-type for p63 and remain in the resting state, and the shade of red represents the time-point (indicated in panel a) of collection after the cells are induced to differentiate. The plots represent the expression of a cell cycle gene set in the neuronal and sustentacular cell lineages. Two clusters in the neuronal lineage (globose basal cells, GBC; immediate neuronal precursors, INP1) show high expression of cell cycle genes, suggesting that the neuronal lineage involves transit through proliferative progenitor fates. e) Clonal lineage tracing of differentiating HBCs demonstrated that most clones were due to an early bifurcation, prior to cell division and included either neurons or sustentacular cells, and neuronal clones were multi-cellular and sustentacular cells could form without cell division. Neurons were distinguished from sustentacular cells by morphology and presence or absence of SOX2 protein expression by immunohistochemistry (magenta). These observations confirmed the main predictions from the branching lineage model derived from Slingshot. Panels a, b, c, and e were adapted with permission.[57] Copyright 2017, Elsevier.
4.2. Time-stamping Cells Helps Resolve Trajectories Confounded by Jumps in Gene Expression
In a second example of integrating clonal lineage tracing and scRNA-seq, we investigated the stem cell lineage of the olfactory epithelium during injury-induced regeneration. In this approach we labeled cells prior to inducing tissue regeneration and collected cells for scRNA-seq at defined time-points post injury,[58] in effect providing a time-stamp of the duration of regeneration in each cell. Time-stamping provides additional information with which to interpret the scRNA-seq data and further serves to constrain the lineage prediction analysis. After clustering and identifying the different cell types and applying lineage prediction algorithms, one can assess whether a given cell state exists in a brief time-window (i.e. is transient) or if it is composed of cells from a range of lineage traced time-points. Furthermore, time-stamping also allows one to identify the earliest stage in a lineage at which a given cell fate/state arises.
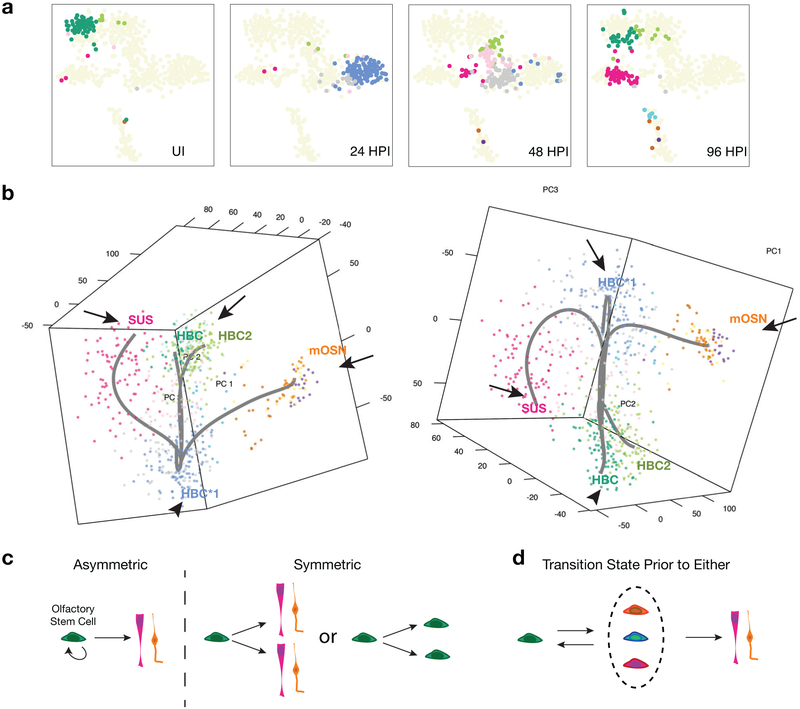
A key challenge that integrating clonal lineage tracing and time stamped scRNA-seq can address is the situation where the lineage trajectory is inconsistent with the expected assumption that cells that are closer together in a reduced dimension gene expression space are closer together in the developmental process (Figure 1e, f). For example, in our analysis of regeneration in the olfactory epithelium, all of the stem cells shift en masse to a proliferative stemcellstateat24 hr post in jury(24HPI)that is even further from the resting state in reduced dimension space (using PCA) than the sustentacular cells, which are a fully differentiated endpoint (Figure 4a,b). However, time-stamping reveals that this cluster contains only cells labeled at 24 hr, indicating that it represents a transient intermediate. This allowed us to constrain the parameters of the lineage trajectory prediction algorithm to be consistent with known biology by designating the activated state as the starting point to alleviate short circuiting the route (Figure 4b). If we had not done so, the transient, activated state intermediate would have been incorrectly predicted to be an endpoint. Importantly, clonal lineage tracing results were consistent with the prediction that all stem cells transit through this activated and proliferative state because all clones were multicellular, regardless of the cell type formed.[58]
Figure 4.
Activated state intermediates that are unique to tissue regeneration present challenges to lineage prediction. When gene expression shifts drastically between two cell fates, it contradicts an underlying assumption of all current lineage prediction algorithms that cell fate changes occur as gradual transitions along a continuum. Integrating a lineage tracing time-course into the scRNA-seq analysis can help overcome this obstacle. a) Olfactory HBC stem cell lineage cells. All cells shift away from the resting state (green) at 24-hr post injury (HPI), most to an activated state (blue and gray). The activated state is more distant from the resting state than the sustentacular support cells (magenta). This panel was adapted with permission.[58] Copyright 2017, Elsevier. b) Predicted branching lineage trajectory of the olfactory HBC stem cell during injury-induced regeneration, with the activated state as the starting point (left). If the activated state is not specified as the starting point in the lineage, then it will be incorrectly designated as an endpoint (right). The lineage tracing derived time-stamping allows us to choose the activated state as the starting point because all stem cells transit to this state upon injury. Clusters/cell types designated as the starting point for the lineage prediction tool, Slingshot, are indicated by an arrowhead; endpoints are indicated by the arrows. c) In traditional stem cell models, stem cells either asymmetrically divide at the individual level or adopt population asymmetry where individual cells either self-renew or differentiate. d) Based on the identification of an activated state that is unique to injury-induced regeneration and that occurs prior to olfactory stem cell self-renewal or differentiation, we propose a modified model of stem cell lineage determination during tissue regeneration.
4.3. Stem Cell Self-Renewal and Successive Rounds of Differentiation Pose Challenges to Lineage Trajectory Inference
Looping trajectories are lineage trajectories that are not unidirectional but instead also include a path back to an earlier cell state, posing a second related problem for lineage prediction algorithms. This is a central feature of stem cell self-renewal, which makes it difficult to find a unique solution for where cells reside in a lineage (Figures 1a, e, and f). However, by applying time-point specific lineage tracing, one can distinguish cells that cluster together as the ground or starting state based on their time of arrival and use this information to assist the placement of cells in the proper sequence of events. Cells in the lineage that correspond to the stem cell state at time zero (in our case, the uninjured [UI] stem cells, Figure 4a) are the earliest point in the lineage, and any cells that cluster in that group but are derived from lineage traced cells collected at later time-points must be the stem cell lineage that looped back to self-renew and reform the stem cell fate.
Another confounding problem for deconstructing lineage trajectories is that renewed stem cells can differentiate successive times.[58] If the route is the same upon each successive round, it should not alter lineage trajectory predictions. However, if the route is different, for example including different intermediates, then additional information — as for example afforded by time-stamping cells — would be required to assign cells to the appropriate position in their respective lineages. To confirm that the lineage trajectory prediction for the second phase of differentiation seeded from a renewed stem cell is correct, one could apply the same experimental approach and only label stem cells that had been renewed.
5. Activated, Heterogeneous States Are Developmental Windows for Cell Fate Determination
By integrating lineage tracing-based time-stamping in our scRNA-seq analysis of stem-cell mediated regeneration of the olfactory epithelium, we identified cell intermediates about which we had no prior knowledge. This approach also suggested that these early intermediate states are transient and heterogeneous and, therefore, likely to be the stage at which cell fates are specified (as predicted by the lineage prediction tool Slingshot). This prediction was validated using clonal lineage tracing, which revealed that a subset of cells were committed to differentiate and not self-renew, highlighting that this cellular state is a window along the axis of differentiation where cell fates become restricted.[58] Transcriptome heterogeneity has also been observed in other differentiating stem cells, especially at early stages of cell fate shifts, including early differentiating mouse embryonic stem cells, induced pluripotent stem cells, and early vertebrate development.[39,55,56,73,74]
These observations led us to propose a model for the way that stem cells respond to severe injury: following injury, stem cells shift to an activated state, where cell fate specification occurs, and then they either return to the stem cell fate (self-renewal) or go on to differentiate (Figure 4c,d). Thus in the olfactory epithelium, branching from this activated state can veer in at least three directions: backward to reform a stem cell or forward toward the two main differentiating lineages. The identification of these transient windows that form the branch points of lineage trajectories is consistent with other recent findings. During vertebrate embryonic development, there are cells at the predicted branch point for cell fate specification that express genes associated with more than one lineage.[55,56] The expression of genes for multiple lineages in a precursor that will ultimately commit to one path was reported years ago in the hematopoietic system.[75] More recently, in the hematopoietic dendritic lineage, Olsson et al.[52] observed a transient cell state (“mixed lineage intermediate”) that has features of both lineages, and they were able to trap cells in this intermediate and to push them toward one fate or the other. Consistent with this notion, others have argued that individual cell states along a lineage trajectory demonstrate an increased probability to respond to signals promoting a specific differentiation path.[76] In sum, we think that these activated, intermediate states represent windows along the developmental trajectory where gene regulatory networks are competing, and in the end, one transcriptional network will prevail and drive the cell toward the specified fate.
Last, it is important to note that after having applied clonal lineage tracing to test the predictions of branching lineage models, we found that in the early stages of the lineage, the inferred lineage trajectory should be thought of as the road most often taken, although there are alternate routes. For example, while stem cells usually commit to one lineage or the other, we found examples where they formed both lineages, suggesting there was either an asymmetric cell division and/or a stem cell that self-renewed then differentiated again into a different type of cell.[57,58]
6. Future Approaches and Considerations for Lineage Deconstruction
The strength of integrating lineage tracing with scRNA-seq at multiple time-points of a stem cell lineage is the power it gives one to constrain models of lineage trajectories and to deal with unexpected outcomes (Figure 1e,f). By sampling cells from multiple time-points along a lineage trajectory, transient intermediates and branch points can be identified. Combining scRNA-seq analysis with clonal lineage tracing allows one to test the predictions of the lineage models. The weaknesses are that it can be labor intensive to collect samples from transgenic animals for multiple time-points. Moreover, to validate the predictions of the model, one needs to identify genes that are specific to individual cellular states or time-windows along the developmental trajectories and design strategies to track the progeny of transient intermediates and cells that represent branch points in the trajectory. Despite these challenges, we deem the additional investment to be worthwhile because of the added resolution it provides for deconstructing lineage hierarchies. Moving forward, it would be powerful to apply this integrative approach to simultaneously label and collect multiple lineages or collect niche cells simultaneously with the lineage-traced cells, so that one could gain insight into how the niche and/or other lineages in the tissue behave in coordination with the stem cell lineage. This is especially relevant now that It is well-established that the niche plays a critical role in regulating the behavior of stem cells.[77]
New approaches promise to accelerate the ability to define multiple lineages simultaneously. For example, a CRISPR/Cas9-based method for modifying genomic DNA barcodes was used to establish lineage hierarchies in embryonic zebrafish.[25] This is similar to a strategy that uses transposon-based DNA modifications for lineage tracing to define the lineage relationships in the hematopoietic lineage.[24,26] More recent iterations now incorporate either CRISPR-mediated (scGESTALT, LINNAEUS, ScarTrace) or transposon-mediated (TracerSeq) modification of DNA barcodes coupled to scRNA-seq, allowing one to construct a lineage hierarchy or dendrogram and define the identities of the endpoint cells based on their transcriptomic profiles.[28–31]
In principle these methods could be applied at multiple time-points along a developmental process to capture intermediate cellular states, including lineage trajectory branch points, perhaps obviating the need for current lineage trajectory prediction algorithms. While such approaches have been applied mainly to early developmental processes, one group has developed an inducible system where barcode modification could be induced later in development,[28] an enhancement that provides a means to apply this strategy to juvenile and adult stem cell niches. Implementing these strategies in mice will be an important step toward developing an enhanced understanding of mammalian stem cell niches. One of the challenges going forward will be to develop methods for combining data from experiments performed on different animals, so that replicate samples with a range of different starting and ending points can be compared and aggregated.
7. Conclusions and Outlook
Understanding how stem cells give rise to a range of differentiated cell types and self-renew is a central problem in stem and developmental biology. We propose that by incorporating the classical approach of lineage tracing into scRNA-seq analysis at multiple time-points along development of the lineage allows one to identify transient intermediates and branch points in the lineage trajectory without prior knowledge. This type of insight is only possible by investigating the tissue at single cell resolution. The inferred lineage trajectories and branch points can be tested and validated by using clonal level lineage tracing. This integrative approach provides a robust framework for deconstructing how individual stem cells maintain tissues. Technological innovations like the application of DNA barcode modifications coupled with scRNA-seq that can be used to construct lineage hierarchies and define the transcriptional state of cells hold promise for a higher throughput approach for deconstructing lineage hierarchies. Major challenges and opportunities include the computational resources required to handle these ever larger data sets and validation of the predicted results. Ultimately, elucidation of these cellular mechanisms serves as the basis for understanding the molecular control of cell fate choice. The molecular identity data derived from scRNA-seq can be coupled to validated models of lineage trajectories to define the network of genes and signaling events that control how stem cells change through time.
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01DC007235, U01MH105979). D.D. is a fellow of the Berkeley Institute for Data Science, which is funded in part by the Gordon and Betty Moore Foundation (Grant GBMF3834) and the Alfred P. Sloan Foundation (Grant 2013–10–27).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
Contributor Information
Dr. Russell B. Fletcher, Department of Molecular and Cell Biology, University of California, 265 LSA, #3200, Berkeley, CA 94720,USA, rufletch@berkeley.edu
Dr. Diya Das, Department of Molecular and Cell Biology, University of California, 265 LSA, #3200, Berkeley, CA 94720,USA, rufletch@berkeley.edu Berkeley Institute for Data Science, University of California, Berkeley, CA 94720, USA.
Prof. John Ngai, Department of Molecular and Cell Biology, University of California, 265 LSA, #3200, Berkeley, CA 94720,USA, rufletch@berkeley.edu Helen Wills Neuroscience Institute, QB3 Functional Genomics Laboratory, University of California Berkeley, Berkeley, CA 94720, USA.
References
- [1].Kretzschmar K, Watt FM, Cell 2012, 148, 33. [DOI] [PubMed] [Google Scholar]
- [2].Woodworth MB, Girskis KM, Walsh CA, Nature Publishing Group 2017, 18, 230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Whitman CO, J. Morphol 1887, 1, 105. [Google Scholar]
- [4].Wilson EB, J. Morphol 1892, VI, 1. [Google Scholar]
- [5].Conklin EG, J. Acad. Nat. Sci. Phila 1905, 13, 1. [Google Scholar]
- [6].Vogt W, Roux Arch. Entw. Mech 1925, 106, 542. [DOI] [PubMed] [Google Scholar]
- [7].Vogt W, Roux Arch. Entw. Mech 1929, 120, 384. [DOI] [PubMed] [Google Scholar]
- [8].Spemann H, Mangold H, Roux Arch. Entw. Mech 1924, 100, 599. [Google Scholar]
- [9].Weisblat DA, Sawyer RT, Stent GS, Science 1978, 202, 1295. [DOI] [PubMed] [Google Scholar]
- [10].Weisblat DA, Zackson SL, Blair SS, Young JD, Science 1980, 209, 1538. [DOI] [PubMed] [Google Scholar]
- [11].Kimmel CB, Law RD, Dev. Biol 1985, 108, 94. [DOI] [PubMed] [Google Scholar]
- [12].Moody SA, Dev. Biol 1987, 119, 560. [DOI] [PubMed] [Google Scholar]
- [13].Bałakier H, Pedersen RA, Dev. Biol 1982, 90, 352. [DOI] [PubMed] [Google Scholar]
- [14].Price J, Turner D, Cepko C, Proc. Natl. Acad. Sci 1987, 84, 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Zinyk DL, Mercer EH, Harris E, Anderson DJ, Joyner AL, Curr. Biol 1998, 8, 665. [DOI] [PubMed] [Google Scholar]
- [16].Dymecki SM, Tomasiewicz H, Dev. Biol 1998, 201, 57. [DOI] [PubMed] [Google Scholar]
- [17].Soriano P, Nature Genet. 1999, 21, 70. [DOI] [PubMed] [Google Scholar]
- [18].Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L, Cell 2005, 121, 479. [DOI] [PubMed] [Google Scholar]
- [19].Livet J, Weissman TA, Kang H, Draft RW, Lu J, Bennis RA, Sanes JR, Lichtman JW, Nature 2007, 450, 56. [DOI] [PubMed] [Google Scholar]
- [20].Snippert HJ, van der Flier LG, Sato T, van Es JH, van den Born M, Kroon-Veenboer C, Barker N, Klein AM, van Rheenen J, Simons BD, Clevers H, Cell 2010, 143, 134. [DOI] [PubMed] [Google Scholar]
- [21].Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming G-L, Song H, Cell 2011, 145, 1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Clayton E, DoupE DP, Klein AM, Winton DJ, Simons BD, Jones PH, Nature 2007, 446, 185. [DOI] [PubMed] [Google Scholar]
- [23].Espinosa JS, Luo L, Neurosci J. 2008, 28, 2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Sun J, Ramos A, Chapman B, Johnnidis JB, Le L, Ho Y-J, Klein A, Hofmann O, Camargo FD, Nature 2014, 514, 322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, Shendure J, Science 2016, 353, aaf 7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Rodriguez-Fraticelli AE, Wolock SL, Weinreb CS, Panero R, Patel SH, Jankovic M, Sun J, Calogero RA, Klein AM, Camargo FD, Nature 2018, 553, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Siddiqi F, Chen F, Aron AW, Fiondella CG, Patel K, LoTurco JJ, Cereb Cortex 2014, 24, 508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Raj B, Wagner DE, McKenna A, Pandey S, Klein AM, Shendure J, Gagnon JA, Schier AF, Nature Biotechnol. 2018, 36, 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Spanjaard B, Hu B, Mitic N, Olivares-Chauvet P, Janjuha S, Ninov N, Junker JP, Nature Biotechnol. 2018, 36, 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, Klein AM, Science 2018, 36, eaar4362 http://science.sciencemag.org/content/early/2018/04/25/science.aar4362.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Alemany A, Florescu M, Baron CS, Peterson-Maduro J, van Oudenaarden A, Nature 2018, 556, 108. [DOI] [PubMed] [Google Scholar]
- [32].Deng Q, Ramskold D, Reinius B, Sandberg R, Science 2014, 343, 193. [DOI] [PubMed] [Google Scholar]
- [33].Islam S, Zeisel A, Joost S, La Manno G, Zajac P, Kasper M, Lönnerberg P, Linnarsson S, Nat. Meth 2013, 11, 163. [DOI] [PubMed] [Google Scholar]
- [34].Ramskold D, Luo S, Wang Y-C, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, Schroth GP, Sandberg R, Nature Biotechnol. 2012, 30, 777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA, Nat. Meth 2009, 6, 377. [DOI] [PubMed] [Google Scholar]
- [36].Furlan A, La Manno G, Lübke M, Häring M, Abdo H, Hochgerner H, Kupari J, Usoskin D, Airaksinen MS, Oliver G, Linnarsson S, Ernfors P, Nat. Neurosci 2016, 19, 1331. [DOI] [PubMed] [Google Scholar]
- [37].Gokce O, Stanley GM, Treutlein B, Neff NF, Camp JG, Malenka RC, Rothwell PE, Fuccillo MV, Südhof TC, Quake SR, Cell Rep. 2016, 16, 1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Jaitin DA, Kenigsberg E, Keren-Shaul H, Elefant N, Paul F, Zaretsky I, Mildner A, Cohen N, Jung S, Tanay A, Amit I, Science 2014, 343, 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kumar RM, Cahan P, Shalek AK, Satija R, Jay DaleyKeyser A, Li H, Zhang J, Pardee K, Gennert D, Trombetta JJ, Ferrante TC, Regev A, Daley GQ, Collins JJ, Nature 2014, 516, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, Lönnerberg P, Ryge J, Barker RA, Arenas E, Linnarsson S, Cell 2016, 167, 566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lake BB, Ai R, Kaeser GE, Salathia NS, Yung YC, Liu R, Wildberg A, Gao D, Fung H-L, Chen S, Vijayaraghavan R, Wong J, Chen A, Sheng X, Kaper F, Shen R, Ronaghi M, Fan J-B, Wang W, Chun J, Zhang K, Science 2016, 352, 1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA, Cell 2015, 161, 1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Patel AP, Tirosh I, Trombetta JJ, Shalek AK, Gillespie SM, Wakimoto H, Cahill DP, Nahed BV, Curry WT, Martuza RL, Louis DN, Rozenblatt-Rosen O, Suvà ML, Regev A, Bernstein BE, Science 2014, 344, 1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, Ramalingam N, Sun G, Thu M, Norris M, Lebofsky R, Toppani D, Kemp DW, Wong M, Clerkson B, Jones BN, Wu S, Knutsson L, Alvarado B, Wang J, Weaver LS, May AP, Jones RC, Unger MA, Kriegstein AR, West JAA, Nature Biotechnol. 2014, 32, 1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Poulin J-F, Zou J, Drouin-Ouellet J, Kim K-YA, Cicchetti F, Awatramani RB, Cell Rep. 2014, 9, 930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Usoskin D, Furlan A, Islam S, Abdo H, nnerberg PLO, Lou D, Hjerling-Leffler J, m JHO, Kharchenko O, Kharchenko PV, Linnarsson S, Ernfors P, Nat. Neurosci 2014, 18, 145. [DOI] [PubMed] [Google Scholar]
- [47].Villani A-C, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, Griesbeck M, Butler A, Zheng S, Lazo S, Jardine L, Dixon D, Stephenson E, Nilsson E, Grundberg I, McDonald D, Filby A, Li W, De Jager PL, Rozenblatt-Rosen O, Lane AA, Haniffa M, Regev A, Hacohen N, Science 2017, 356, eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, David E, Baruch K, Lara-Astaiso D, Toth B, Itzkovitz S, Colonna M, Schwartz M, Amit I, Cell 2017, 169, 1276. [DOI] [PubMed] [Google Scholar]
- [49].Vanlandewijck M, He L, Mäe MA, Andrae J, Ando K, Del Gaudio F, Nahar K, Lebouvier T, Laviña B, Gouveia L, Sun Y, Raschperger E, Räsänen M, Zarb Y, Mochizuki N, Keller A, Lendahl U, Betsholtz C, Nature 2018, 554, 475. [DOI] [PubMed] [Google Scholar]
- [50].Hashimshony T, Wagner F, Sher N, Yanai I, Cell Rep. 2012, 2, 666. [DOI] [PubMed] [Google Scholar]
- [51].Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N, Clevers H, van Oudenaarden A, Nature 2015, 525, 251. [DOI] [PubMed] [Google Scholar]
- [52].Olsson A, Venkatasubramanian M, Chaudhri VK, Aronow BJ, Salomonis N, Singh H, Grimes HL, Nature 2016, 537, 698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, Schwartz S, Fowler B, Weaver S, Wang J, Wang X, Ding R, Raychowdhury R, Friedman N, Hacohen N, Park H, May AP, Regev A, Nature 2014, 510, 363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Cao J, Packer JS, Ramani V, Cusanovich DA, Huynh C, Daza R, Qiu X, Lee C, Furlan SN, Steemers FJ, Adey A, Waterston RH, Trapnell C, Shendure J, Science 2017, 357, 661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF, Science 2018, 360, eaar3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW, Klein AM, Science 2018, 360, eaar5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Fletcher RB, Das D, Gadye L, Street KN, Baudhuin A, Wagner A, Cole MB, Flores Q, Choi YG, Yosef N, Purdom E, Dudoit S, Risso D, Ngai J, Cell Stem Cell 2017, 20, 817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Gadye L, Das D, Sanchez MA, Street K, Baudhuin A, Wagner A, Cole MB, Choi YG, Yosef N, Purdom E, Dudoit S, Risso D, Ngai J, Fletcher RB, Cell Stem Cell 2017, 21, 775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Plass M, Solana J, Wolf FA, Ayoub S, Misios A, Glažar P, Obermayer B, Theis FJ, Kocks C, Rajewsky N, Science 2018, 360, eaaq1723. [DOI] [PubMed] [Google Scholar]
- [60].Nowakowski TJ, Bhaduri A, Pollen AA, Alvarado B, Mostajo-Radji MA, Di Lullo E, Haeussler M, Sandoval-Espinosa C, Liu SJ, Velmeshev D, Ounadjela JR, Shuga J, Wang X, Lim DA, West JA, Leyrat AA, Kent WJ, Kriegstein AR, Science 2017, 358, 1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming G-L, Song H, Cell Stem Cell 2015, 17, 360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Treutlein B, Lee QY, Camp JG, Mall M, Koh W, Shariati SAM, Sim S, Neff NF, Skotheim JM, Wernig M, Quake SR, Nature 2016, 534, 391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Petropoulos S, Edsgärd D, Reinius B, Deng Q, Panula SP, Codeluppi S, Plaza Reyes A, Linnarsson S, Sandberg R, Lanner F, Cell 2016, 165, 1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].St evant I, Neirijnck Y, Borel C, Escoffier J, Smith LB, Antonarakis SE, Dermitzakis ET, Nef S, Cell Rep. 2018, 22, 1589. [DOI] [PubMed] [Google Scholar]
- [65].Bendall SC, Davis KL, Amir E-AD, Tadmor MD, Simonds EF, Chen TJ, Shenfeld DK, Nolan GP, Pe’er D, Cell 2014, 157, 714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL, Nature Biotechnol. 2014, 32, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Campbell K, Ponting CP, Webber C, BioRxiv 2015, 1 http://biorxiv.org/content/early/2015/09/18/027219.abstract [Google Scholar]
- [68].Setty M, Tadmor MD, Reich-Zeliger S, Angel O, Salame TM, Kathail P, Choi K, Bendall S, Friedman N, Pe’er D, Nature Biotechnol. 2016, 34, 637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Haghverdi L, Buettner F, Theis FJ, Bioinformatics 2015, 31, 2989. [DOI] [PubMed] [Google Scholar]
- [70].Ji Z, Ji H, Nucleic Acids Res. 2016, 44, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C, Nat. Meth 2017, 14, 979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S, BioRxiv 2017, 1. doi: 10.1101/128843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Klein AM, Mazutis L, Akartuna I, Tallapragada N, Veres A, Li V, Peshkin L, Weitz DA, Kirschner MW, Cell 2015, 161, 1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Thomson M, Liu SJ, Zou L-N, Smith Z, Meissner A, Ramanathan S, Cell 2011, 145, 875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T, Genes Dev. 1997, 11, 774. [DOI] [PubMed] [Google Scholar]
- [76].Jang S, Choubey S, Furchtgott L, Zou L-N, Doyle A, Menon V, Loew EB, Krostag A-R, Martinez RA, Madisen L, Levi BP, Ramanathan S, eLife 2017, 6, 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Gonzales KAU, Fuchs E, Dev. Cell 2017, 43, 387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bacher R, Kendziorski C, Genome Biol. 2016, 17, 405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Wagner A, Regev A, Yosef N, Nature Biotechnol. 2016, 34, 1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kolodziejczyk AA, Kim JK, Svensson V, Marioni JC, Teichmann SA, Mol. Cell 2015, 58, 610. [DOI] [PubMed] [Google Scholar]
- [81].Perraudeau F, Risso D, Street K, Purdom E, Dudoit S, F1000Res 2017, 6, 1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Risso D, Purvis L, Fletcher R, Das D, Ngai J, Dudoit S, Purdom E, BioRxiv 2018, 1. doi: 10.1101/280545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Cole MB, Risso D, Wagner A, DeTomaso D, Ngai J, Purdom E, Dudoit S, Yosef N, BioRxiv 2018, 1. doi: 10.1101/235382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].DeTomaso D, Yosef N, BMC Bioinformatics 2016, 17, 315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Risso D, Perraudeau F, Gribkova S, Dudoit S, Vert J-P, Nat. Commun 2018, 9, 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kester L, van Oudenaarden A, Cell Stem Cell 2018, 1 10.1016/j.stem.2018.04.014 [DOI] [PubMed] [Google Scholar]
- [87].Saelens W, Cannoodt R, Todorov H, Saeys Y, 2018, 1. doi: 10.1101/276907 [DOI] [Google Scholar]