Abstract
Lysophospholipids (LPLs) are metabolic intermediates in bacterial phospholipid turnover. Distinct from their diacyl counterparts, these inverted cone-shaped molecules share physical characteristics of detergents, enabling modification of local membrane properties such as curvature. The functions of LPLs as cellular growth factors or potent lipid mediators have been extensively demonstrated in eukaryotic cells but are still undefined in bacteria. In the envelope of Gram-negative bacteria, LPLs are derived from multiple endogenous and exogenous sources. Although several flippases that move non-glycerophospholipids across the bacterial inner membrane were characterized, lysophospholipid transporter LplT appears to be the first example of a bacterial protein capable of facilitating rapid retrograde translocation of lyso forms of glycerophospholipids across the cytoplasmic membrane in Gram-negative bacteria. LplT transports lyso forms of the three bacterial membrane phospholipids with comparable efficiency, but excludes other lysolipid species. Once a LPL is flipped by LplT to the cytoplasmic side of the inner membrane, its diacyl form is effectively regenerated by the action of a peripheral enzyme, acyl-ACP synthetase/LPL acyltransferase (Aas). LplT-Aas also mediates a novel cardiolipin remodeling by converting its two lyso derivatives, diacyl or deacylated cardiolipin, to a triacyl form. This coupled remodeling system provides a unique bacterial membrane phospholipid repair mechanism. Strict selectivity of LplT for lyso lipids allows this system to fulfill efficient lipid repair in an environment containing mostly diacyl phospholipids. A rocker-switch model engaged by a pair of symmetric ion-locks may facilitate alternating substrate access to drive LPL flipping into bacterial cells.
Keywords: lysophospholipid, LplT, membrane, flippase, bacterial lipid, remodeling
1. Introduction
The envelope of Gram-negative bacteria comprises two distinct membranes: an inner membrane (IM) and an outer membrane (OM) that are separated by a peptidoglycan-containing periplasmic space. These microorganisms maintain a relatively simple phospholipid composition. In Escherichia coli, the envelope mainly consists of ~70% phosphatidylethanolamine (PE), ~20% phosphatidylglycerol (PG) and ~5% cardiolipin (CL) with some minor phospholipid species: phosphatidylserine (PS), phosphatidic acid (PA), CDP-diacylglycerol (CDP-DAG) and lysophospholipids (LPLs) making up the rest ~5% (1). This phospholipid composition is established primarily by de novo synthesis on the IM via the intermediate PA which is then converted either to zwitterionic PE or anionic PG and CL (2).
LPLs are usually in a small fraction (≤ 1%) of the bacterial membrane, but may be accumulated in marine, pathogenic and stressed bacteria under special circumstances (3–5). LPLs are generated as metabolic intermediates in phospholipid synthesis or from membrane degradation. In bacteria, LPLs are mostly found in the form of lysophosphatidylethanolamine (LPE), which can be converted to dilysocardiolipin (diacyl-CL) and lysocardiohpin (triacyl-CL), at least in vitro (6). The role of LPLs remains poorly characterized in bacteria. But LPLs have been found to be essential in eukaryotic cells. Inflamed tissues secrete LPLs as potent chemotactic molecules (7,8) or stimulating factors (9), which are considered as potential biomarkers of certain diseases (10,11). Lysophosphatidic acid (LPA) and sphingosine-1-phosphate are potent signaling messengers and mitogens (12). Lysophosphatidylcholine (LPC) is a lipid precursor for biogenesis of docosahexaenoic acid, a primary structural component of the human brain, cerebral cortex and retina (13). The functional significance and mechanism of LPL turnover in the bacterial envelope remains obscure. Further investigation is needed (i) to correlate LPL turnover with bacterial growth, survival and pathogenic potential; and (ii) to understand the consequences of LPL turnover and its physiological significance during environmental adaptation in an infected host. Studying the metabolism of LPLs will help to address these fundamental questions in this underdeveloped research area.
LplT was originally identified as a LPE transporter in the IM of Gram-negative bacteria (14). A contradiction was whether LplT is able (or even necessary) to have a generalized LPL flippase function (15). Aas is a bifunctional enzyme catalyzing acyl transfer to LPL, generating diacyl form of the phospholipid. We recently reported that the LplT-Aas system facilitates remodeling of all three bacterial major phospholipids including PE, PG and CL with comparable translocation and remodeling efficiencies, and therefore is very important for maintaining stability and integrity of the membrane envelope (16). This review focuses on the biogenesis of LPLs from endogenous (intramembrane) and exogenous sources and provides a detailed characterization of the remodeling pathway controlled by the LplT-Aas system in Gram-negative bacteria. A new mechanistic model of LplT-mediated LPL transport is also proposed based on published biochemical data (14,16).
2. Origins of LPLs in the envelope of Gram-negative bacteria
2.1. LPLs generated by endogenous phospholipase A2
Many membrane-stressing events that compromise the integrity of the OM such as heat shock (17,18), T4 phage-induced lysis (19), colicin secretion (20), serum complement action (21), EDTA treatment (22), bactericidal-permeability increasing protein (BPI) action (23,24) and antimicrobial lipopeptide exposure (25) are accompanied by a drastic increase in the LPE level within the Gram-negative envelope.
The accumulation of LPE in the stressed envelope is mostly due to activation of PldA, a detergent-resistant phospholipase A on the OM (26) (Fig. 1-[1]). PldA remains dormant as an inactive monomer in normally growing cells (27). Although phospholipids are present in both the IM and the OM, their distribution on the OM is extremely asymmetric with lipopolysaccharides (LPS) residing in the outer leaflet and phospholipids (normally PE) in the inner leaflet. The phospholipid substrates lining the inner leaflet of the OM are normally inaccessible to the active site of PldA when the OM asymmetry is intact. However, under stressful conditions, disturbance of the OM integrity and asymmetry leads to activation of PldA via the Ca2+-dependent dimerization mechanism (28,29). The disturbed OM may further facilitate substrate accessibility to the PldA on the envelope surface (26,27,30–32).
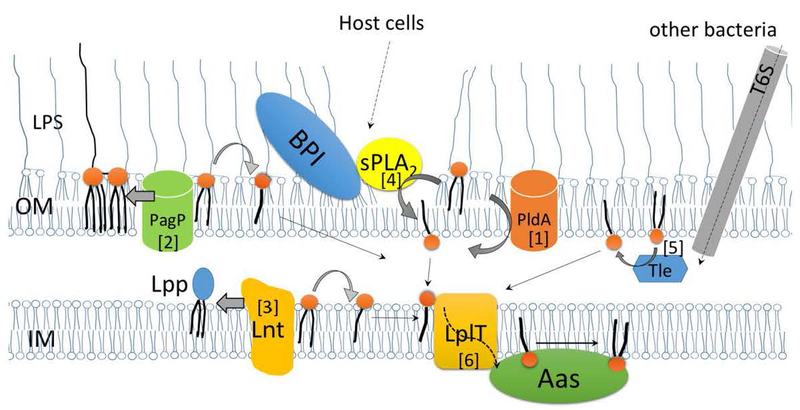
Fig. 1. Routes of lysophospholipid biogenesis and remodeling in the Gram-negative bacterial envelope.
Lysophospholipids are generated in the periplasm. In the OM, [1] activated PldA hydrolyzes a PE to generate LPE; [2] PagP catalyzes the transfer of a palmitoyl group from a PE to lipid A, generating hepta-acylated lipid A and LPE as by-product. In the IM, [3] Lnt transfers the fatty acid moiety from PE to the N terminus of a major outer membrane lipoprotein precursor (Lpp), generating a triacylated mature Lpp and releasing LPE as by-product. LPL can be generated by membrane degradation mediated by [4] exogenous sPLA2 and BPI from the host or by [5] the Tle protein delivered from invading bacteria via the type VI secretion system (T6S). The generated LPLs are translocated by LplT [6] from the outer leaflet to the inner leaflet of the IM where they can be reacylated by the coupled acyltransferase Aas.
In several pathogenic bacteria including Vibrio cholerae, Helicobacter pylori, Yersinia pseudotuberculosis, LPE is accumulated in accordance with their pathogenic or survival potential when confronting new hostile environmental conditions (3–5,33). V. cholerae remodels its lipid profile and accumulates LPE after exposure to bile salts (which may occur in the early stages of infection) or during growth in the presence of ocean sediment. Under both conditions, V. cholerae produces a large amount of LPE accounting for ~30% of the total lipid composition (34). Such abnormal LPE accumulation is unexpected given that no pldA homolog is encoded by this microorganism. It is worthwhile to note that this interesting phenomenon was confirmed not to be an artifact of lipid extraction but is a bona fide bacterial response to its natural environment (3). In addition, accumulation of LPE in the OM of H. pylori coincides with augmented invasive capacity accompanied by increased secretion of vacuolating cytotoxin A (a key toxin in this bacterium), urease release and better adherence to epithelial cells observed in vitro (4,35). Whether a high LPE/PE ratio stimulates bacterial virulence or is just a side effect of the overall bacterial metabolic response is still unknown. Accumulation of LPE has been implicated in stress resistance of H. pylori grown in acidified media (36).
LPE was also found to become dominant (12 %) in the OM of Y. pseudotuberculosis after shifting of the growth temperature from 8 °C to 37 °C or after exposure of the cells to heat shock (33). It is unclear why heat-stressed cells are able to retain LPE at such a high level without immediate degradation by PldB or remodeling by the LplT-Aas system in the IM despite the fact that homologs of aas and lplT genes are present in theYersinia genus.
2.2. LPLs as by-products of de novo biosynthetic pathways
The PhoPQ virulence two-component system is a major regulator of the OM lipid content and topography (externalization of CL) (37). This system modulates the glycerophospholipid (GPE) content within the bacterial envelope in response to phagosome acidification and the action of cationic antimicrobial peptides (CAMP). Several stress conditions (penetration attempts of CAMP, exposure to acidic pH, EDTA treatment, BPI binding, divalent cations limited growth conditions, the presence of bile acids or detergents etc.) displace LPS or calcium and magnesium ions from the outer leaflet and promote phospholipid externalization (31,38). In E. coli and Salmonella typhimurium, OM damage and spontaneous PhoPQ activation up-regulate the OM lipid A palmitoyltransfrase PagP (31,39). PagP transfers a palmitate chain from PE to hexaacylated lipid A or PG to form a hepta-acylated lipid A or triacylated PG, releasing LPE as byproduct (39,40) (Fig. 1-[2]). Biosynthesis of hepta-acylated LPS and triacylated PG fortifies the OM barrier to CAMP and promotes bacterial survival within acidified host phagosomes. In E.coli, triacylated PG is also synthesized by the addition of a fatty acid from LPE to PG by the lysophospholipase L2 PldB protein (41), which can therefore eliminate a “harmful” LPE from the IM. Triacylated PG synthesized by PagP and PldB can be distinguished because of the strict palmitate selectivity displayed by PagP in the OM compared to the more relaxed acyl chain specificity displayed by PldB in the IM.
Bacterial OM lipoprotein precursor (Lpp) undergoes a sequential maturation process mediated by three different membrane-bound enzymes on the outer leaflet of the IM (42,43). The final step is catalyzed by apolipoprotein N-acyltransferase (Lnt). Lnt transfers the sn-1 fatty acid moiety from PE to the N terminus of the Lpp precursor, generating mature triacylated lipoprotein and LPE as byproduct of the reaction (Fig. 1-[3]). LPL can also be generated during the synthesis of N-acylphosphatidylethanolamine (N-acyl-PE) (44). Although the catabolic pathway for N-acyl-PE remains uncharacterized in bacteria, it is synthesized in prokaryotes by transacylation of the sn-1 acyl chain of a phospholipid onto the amine of PE, with a LPL being simultaneously produced (45).
2.3. LPLs as result of exogenous phospholipase actions
The bacterial envelope establishes an essential compartmentalization of biochemical activities in cells. Its exterior accessibility also makes the envelope a common target of exogenous factors for antagonistic interactions between diverse organisms. Secretory phospholipases A2 (sPLA2)-mediated bacterial phospholipid degradation is a potent antibacterial mechanism in animal innate immune systems. Ten sPLA2 isoforms have been identified in human (46). They are expressed in different cell types including neutrophils and other phagocytic cells and are present in tears, plasma and many inflammatory fluids. Several isoforms exhibit their bactericidal activities towards both Gram-positive and Gram-negative bacteria. sPLA2 hydrolyzes membrane phospholipids of invading bacteria, producing the 1-acylated lyso form and free fatty acids that can cause bacterial membrane disorganization and eventual cell lysis (47).
Killing of Gram-positive bacteria by sPLA2 group IIA (sPLA2-GIIA) is very efficient. sPLA2-GIIA penetrates through the porous OM-lacking envelope to directly attack membrane phospholipids, with the preferred hydrolytic substrates PG and CL (47). The bactericidal activity of sPLA2-GIIA towards Gram-negative bacteria, such as E.coli and Pseudomonas aeruginosa, requires synergistic action of other innate immune antibacterial factors such as BPI or serum complementation membrane attack complex (MAC). These immune proteins disrupt the LPS-rich OM surface structure to assist sPLA2-GIIA access to PL components (48)(Fig. 1-[4]). The action of sPLA2-GIIA most likely takes place on the OM, at least at the initial stage. The damage may further then extend towards the IM, resulting in severe membrane degradation of >50% of the phospholipids (24). Disruption of E.coli spheroplasts by pancreatic sPLA2 produced multiple LPL species (16). Therefore, the bactericidal event is not limited to degradation of PE, but also affects PG and CL.
sPLA2-GIIA mediated phospholipid hydrolysis is substantially aggravated by activation of endogenous PldA in the OM. Incubation of rabbit polymorphonuclear leukocytes with an E.coli strain harboring a multicopy plasmid encoding pldA results in drastic degradation of up to 60% of bacterial PLs. The breakdown rate was reduced to 30% and 20% in the absence of the plasmid or in a ΔpldA strain, respectively (24). Activation of endogenous PldA may be triggered by the sPLA2 attack itself or by OM disruption caused by BPI or MAC either separately or synergistically. Why endogenous PldA assists the invading host sPLA2 in this antimicrobial event is still unknown and remains an interesting topic for study of bacterial host interaction in living organisms.
Gram-negative bacteria utilize their own phospholipases to achieve intra- and interspecies antagonistic interaction and competition. Activated bacteria deliver five divergent groups of lipase effectors (Tle1–5) to the recipient cells via the type VI secretion system (T6S) translocation apparatus (49). Tle1–5 possess phospholipase A1 or A2 activity and degrade PE in the periplasm of the recipient cells to generate LPE (Fig. 1-[5]). V. cholerae strains lacking tle2 exhibit impaired ability to kill E. coli. Remarkably, a type-specific immunity protein Tli is encoded adjacent to each tle gene. In the periplasm, Tli inactivates Tle via specific protein interaction, if both are from the same species, to ensure precise interspecies antagonistic action.
3. Function of LPLs in the bacterial envelope
The role of LPLs in bacteria remains poorly characterized. Although LPL-mediated signaling has not yet been discovered in bacteria, LPLs roles as potent messengers and mitogens have been demonstrated extensively in eukaryotes. Water-soluble LPA, once thought to be only a key intermediate of the intracellular de novo phospholipid biosynthesis pathway, has now been recognized as an important extracellular signaling molecule acting via a group of LPA specific G protein-coupled receptors (50,51). In bacteria, LPA’s only known role is as an intermediate in biosynthesis of PA, a key phospholipid precursor for membrane biogenesis (2).
LPLs may modulate mechanical properties of lipid bilayers in a specific manner. The adaptability and flexibility of the bacterial membrane structure necessitated by the environment is made possible only within a defined spectrum of bilayer and non-bilayer lipid mixtures (52). Harsh conditions are frequently encountered by bacteria surviving in changing environmental niches including an infected host. The structure and integrity of bacterial membranes must play a major role in the survival and propagation of bacteria in diverse environments. Various types of environmental stress, such as temperature, osmotic and pH stress, cause alterations in the physical properties of membrane lipids (53). When cone-shaped fully unsaturated PE is mixed with bilayer-prone lipids (PG), an elastic curvature stress is induced in the bilayer structure. Introduction of LPE reduces the stored curvature stress and therefore relaxes the “frustrated” bilayer due to its inverted cone-like geometrical shape (54). Although no gross defect in permeability of the bacterial cells upon increase of LPLs has been demonstrated, accumulation of LPLs may disrupt the membrane structure of mitochondria by increasing non-specific membrane permeability (55). LPL-induced changes in local membrane curvature can in turn modulate the function and conformation of embedded membrane proteins (56,57). However, it is not always clear how to translate these polymorphic phospholipid features into the biological functions of individual lipids or the collective physical properties of a lipid bilayer (58).
E. coli normally contains no major intracytoplasmic membrane structures or stores (lipid droplets) and all phospholipids are associated with the IM and the OM. It was suggested that the water solubility of a LPL is sufficient to permit its access not only to membranes but also to nonmembraneous cellular compartments such as the periplasm and the cytoplasm at micromolar concentrations (18). A mild heat shock from 30 to 42°C resulted in a 4-fold increase in LPE in less than 3 min, remaining constant for more than 1 hour in growing E.coli cells (17). LPE may exhibit chaperone-like properties in protein folding and protection against thermal denaturation. LPE, not PE or PC, promotes the functional folding of citrate synthase and glycosidase after urea denaturation and prevents the aggregation of citrate synthase at 42°C (18).
Accumulation of LPE is often accompanied by an increase in CL (21). The CL microdomain is localized in the septal region of dividing cells (59). Thus it is quite possible that LPE is present at the cell division site to relax bilayer stress induced by non-bilayer CL or to maintain an optimal membrane curvature. In accordance with this hypothesis, it has been suggested that PldA hydrolyzes phospholipids within the cell envelope during bacterial cell division (60). In support of this idea, an E.coli envC mutant defective in cell division exhibited a drastic increase in LPE level, whereas decreased cell length and separation of daughter cells were prompted by LPE in the septum division zone of the OM (21,61).
LPL may also regulate the function of membrane proteins associated with the bilayer. In the presence of LPC, the OmpF-like porin of Y. pseudotubeduclosis alters its ion channel activity and the large-conductance mechanosensitive channel MscL of E. coli is trapped in the fully open state (62,63). In both cases, the mechanisms are proposed to involve intrinsic membrane curvature stress and the resulting physical distortion of the lipid bilayer induced by incorporation of the LPL.
4. Retrograde LPL traffic routes within the envelope of Gram-negative bacteria
Obviously, LPLs as the products of phospholipid hydrolysis by endogenous PldA and exogenous PLA2 or as by-products of PagP and Lnt dependent enzymatic reactions should be cleared within the bacterial envelope under normal growth conditions in order to eliminate their potential membrane-destabilizing effect.
The asymmetric transmembrane arrangement in the OM is entropically disfavored and expected to be in a non-equilibrium thermodynamic state. Thus this asymmetry should be maintained in both normal and stressed cells (60). To preserve the OM integrity, LPE exhibits a retrograde movement from the OM to the IM, followed by a reacylation reaction to form diacyl PE (64–66). This remodeling occurs exclusively on the cytoplasmic side of the IM and is catalyzed by Aas (14) (Fig. 1[6]). The flipping of LPE from the periplasmic leaflet to the cytoplasmic surface of the IM is catalyzed by the generic flippase LplT, a dedicated retrograde transport system for LPL (14,16). Retrograde transport of diacyl phospholipids from the OM to the IM was shown to be mediated by the IM MlaFEDB complex via the periplasmic substrate binding protein MlaC (67). Utilization of this route has not yet been demonstrated for LPLs. Remarkably, functioning of the Mla system and PldA activity are interrelated via different mechanisms. When the Mla system is compromised, PldA degrades externalized diacyl phospholipids since the mutant cells fail to translocate the lipids back to the IM (60).
5. Scope for phospholipid flippase in Gram-negative bacteria
Transbilayer diffusion of amphipathic phospholipids in vitro is thermodynamically unfavorable and therefore is an extremely slow process. Thus phospholipid translocation in biological membranes is protein catalyzed (15). Lipid flippases mediate the net transfer of lipid molecules from one leaflet of a membrane to the other side often against a concentration gradient. These unique proteins therefore are able to either establish or annihilate/dissipate transmembrane lipid asymmetry. Several flippases that catalyze translocation of different classes of lipids have been identified in eukaryotes (13,68,69). MsbA is the only bacterial phospholipid flippase identified in E.coli by genetic approaches. The msbA conditional mutant accumulates phospholipids and LPS on the inner surface of the IM upon a shift to non-permissive conditions (70). Unfortunately, MsbA has never been demonstrated to promote translocation of phospholipids in vitro (71). Thus, it is unknown whether MsbA alone is necessary but not sufficient for flip-flop of phospholipids in vitro or whether any accessory proteins are required for MsbA to perform efficient phospholipid translocation (72). It is also possible that MsbA is only involved in lipid A transport but not required for phospholipid flip-flop across the IM (73,74). Other bacterial phospholipid flippases have been postulated based on partial purification and reconstituted activity of the detergent-extracted total membrane fraction (75). However it is difficult to extrapolate these in vitro results to the in vivo situation since the fluorescent phospholipid analogue substrates used are structurally different from their natural counterparts (15).
6. Identification of LplT in Gram-negative bacteria
LplT (TC subfamily 2.A.1.42) was first identified as a LPE transporter by the Rock and Saier groups (14). LplT consists of ten or twelve predicted transmembrane segments (Fig. 2b and 4a) and belongs to the Major Facilitator Superfamily (MFS). MFS members transport a great variety of small polar compounds including sugars, oligosaccharides, drugs, amino acids, nucleosides, organophosphate esters, Krebs cycle metabolites, and small inorganic ions (76). LplT is the only MFS member identified as a specific transporter of lipid thus far.
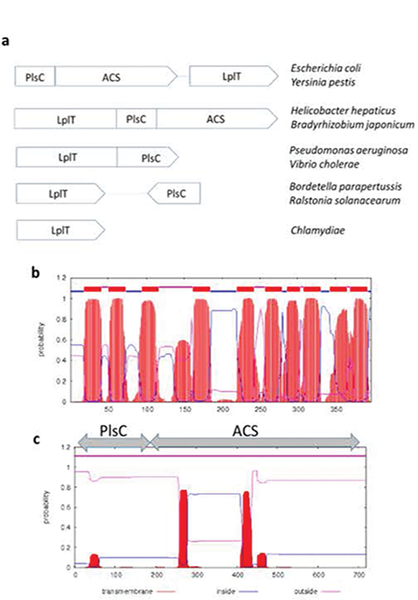
Fig. 2. Architectural scheme of the LplT-Aas system in Gram-negative bacteria.
Panel a, five classes of the LplT-Aas system in different Gram-negative bacterial genomes. Panel b & c, Membrane topology of LplT or Aas from Escherichia coli predicted by the TMHMM server (89). The LplT protein is predicted to have ten transmembrane helices. The Aas protein contains two tandem domains, the N-terminal acyltransferase (PlsC) domain and the C-terminal acyl-ACP synthetase (ACS) domain. Two hydrophobic segments were predicted in the ACS domain.
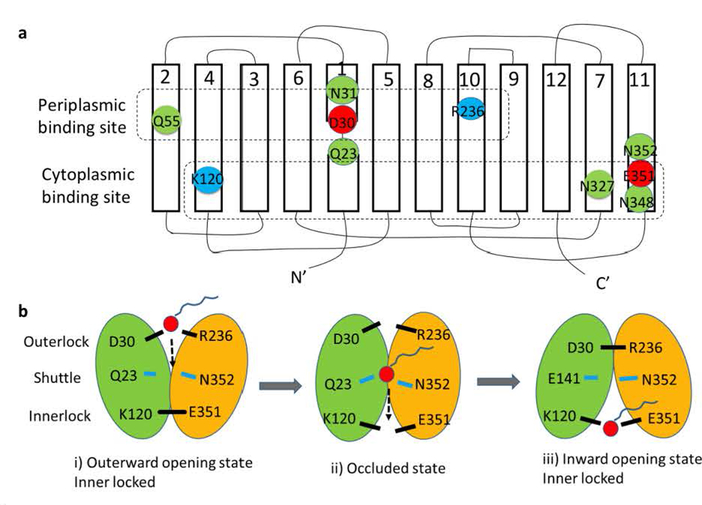
Fig. 4. Hypothetical model of the LplT-mediated lysophospholipid transport mechanism.
Panel a, Secondary structure of LplT showing the locations of the conserved residues in the putative substrate translocation pathway. The transmembrane topology diagram was rendered based on a structural homolog model of LplT from Klebsiella pneumonia generated previously (16), showing twelve transmembrane helices (1–12). Individual amino acids are represented by circles, with negatively charged residues in red, positively charged residues in blue, and other polar residues in green. The residues predicted to form the periplasmic binding site, the shuttle site and the cytoplasmic binding site are surrounded by dashed lines, respectively. Panel b, a hypothetical rocker-switch model for LplT-mediated LPL transport. The N-terminal domain formed by helices 1–6 and C-terminal domain formed by helices 7–12 of LplT are colored in green and yellow, respectively. The following three-step mechanism is proposed for LplT transport: i) an outward-facing conformation is stabilized by the outerlock formed by residues K120 and E351, which allows LPL access to the periplasmic binding site; ii) substrate binding induces a protein conformational change to open the innerlock to form an occluded state, allowing the substrate to pass via the shuttle site in the middle of the membrane towards the cytoplasmic binding site; iii) an inward-opening state is induced by outerlock formation of residues D30 and R236, which allows the substrate to be released from the cytoplasmic site to the inner leaflet of the membrane.
LplT is exclusively found in Gram-negative bacteria including Chlamydiae (Fig. 2a). In the genomes of most Gram-negative bacteria, the lplT gene is closely associated with aas, although their composition and relative orientation may be altered in different bacterial phyla. Aas is a bifunctional enzyme and consists of two tandem domains: the N-terminal acyltransferase (PlsC) domain and the C-terminal acyl-ACP synthetase (ACS) domain (Fig. 2c). In γ-proteobacteria E. coli and Yersinia pestis, the two genes are co-transcribed in one biscistronic operon. In the β-proteobacteria Bordetella and Ralstonia, the aas gene is short and only contains a PlsC domain. Any protein interaction between LplT and Aas on the membrane is still unknown, but remains as an attractive idea given their transcriptional synergy and functional coupling in the remodeling system (see below). Indeed, lplT and Aas exist as a fusion protein in several α-, and ε-proteobacteria including Bradyrhizobium japonicum and Helicobacter hepaticus (14). This integration may be efficient for proper lipid distribution in the bacterial envelope, as also seen in other lipid modification systems such as MprF in Gram-positive bacteria. MprF catalyzes synthesis of lysyl-PG on the cytoplasmic surface, which is subsequently flipped by its transporter domain to the outer leaflet of the membrane (77).
In E. coli, Aas performs LPL remodeling in two steps: 1) the ACS domain catalyzes ATP-dependent biosynthesis of acyl-ACP using acyl-CoA as acyl donor; and then 2) the acyl-ACP is utilized by the adjacent PlsC domain for acylation of LPL (Fig. 3)(78). Aas is considered a peripheral protein on the cytoplasmic membrane surface due to its two strong hydrophobic segments (Ile258-Ala277 and Phe409-Ala432) in the ACS domain (Fig. 2c). Its tight association with the membrane requires a high concentration of non-ionic detergent for extraction from the membrane fraction (data not shown). Interestingly, these two hydrophobic segments were predicted as transmembrane helices by other protein topology programs. LplT or Aas per se are functionally independent in E.coli cells since Aas activity is largely unaffected in the ΔlplT mutant (14). Although the amount of LPE increases by three fold in the E.coli ΔfadD, ΔlplT and Δaas triple mutant, it is still kept at a very low level (0.75%) under normal growth conditions (14). Physical proximity of the two proteins would greatly facilitate remodeling efficiency. The effectiveness of this coupling could become more significant when dealing with multiple exogenous PLA2 attacks that would generate a massive amount of LPLs (>20%) within the envelope (16).
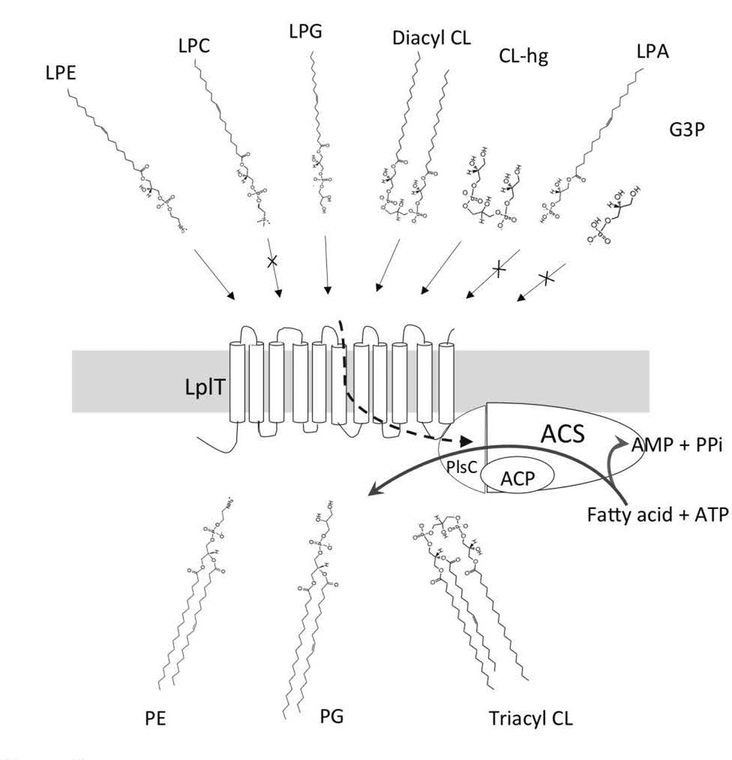
Fig. 3. Scheme of lysophospholipid remodeling mediated by the LplT-Aas system.
LplT substrates LPE, LPG, diacyl-CL and deacylated CL (CL-hg) are transported by LplT from the periplasm to the inner leaflet of the IM by an energy-independent mechanism. On the cytoplasmic surface, bifunctional Aas catalyzes acyl transfer to a flipped LPL using acyl-ACP as acyl donor, generating PE, PG or triacyl-CL respectively. Aas also catalyzes ATP-dependent synthesis of acyl-ACP using a fatty acid. LPA, LPC and glycerol-3-phosphate (G3P) are not LplT substrates.
It was previously suggested that LplT only transports LPE in E.coli (14). Recently, a more comprehensive study by our group of the LplT homolog from Klebsiella pneumonia reveals that LplT is a generalized bacterial LPL transporter with broad substrate specificity (16). Spontaneous flip/flopping of lipids, in particular polar LPLs, across the hydrophobic bilayer is usually a slow and energy unfavorable process. The membrane impermeability of LPL is apparently independent of the head group conformation (16). Lacking any lipid acylation activity in the periplasm establishes LplT as a critical mediator for the metabolism of LPLs. Several eukaryotic transporters utilize the energy of ATP hydrolysis or a counter electrochemical gradient to drive lipid translayer motion (13,69,79). Distinctly, LplT moves LPL using a passive diffusion mechanism which was not inhibited by arsenate blocking of ATP production or by carbonyl cyanide m-chlorophenylhydrazone, a protonophore that dissipates a proton gradient across the IM (14). MFS is also called the uniporter-symporter-antiporter family (76,80). A uniporter promotes equilibrium of substrate across the membrane simply by following a substrate concentration gradient. As discussed in Section 2, biogenesis of LPLs all takes place in the OM or the outer leaflet of the IM, whereas the remodeling of LPLs occurs on the cytoplasmic surface. The polarity of these metabolic activities on each side of the membrane consequently creates an inward substrate concentration gradient for LplT to move LPL into the cells.
7. Characterization of the LplT-Aas system
7.1. Transport of LPE by LplT
Intact bacterial cells are very inefficient in uptake of exogenous LPLs mainly due to the LPS barrier on the cell surface. The transport activity of LplT was measured using stabilized spheroplasts in which the OM had been removed (14,16). LPE is considered as a detergent-like lipid presumably capable of spontaneous partitioning into the lipid bilayer. However random insertion of a synthetic LPE fluorescent analog or 32P-labeled E.coli-derived LPE into spheroplasts was much less efficient in the absence of LplT. LPE uptake was significantly improved in the presence of LplT, yielding a μM substrate binding affinity. These studies conclude that lplT encodes a membrane transporter protein with a dedicated lipid flipping function.
7.2. Remodeling of LPE and LPG by the LplT-Aas system
Demonstration of direct LPE transport and LPE accumulation in a Δaas mutant raised several questions: 1) are LplT transport and acylation coupled or separable events? 2) to what extent does Aas-mediated acylation contribute to LPL remodeling? 3) is LplT transport facilitated by Aas-mediated continuous LPL reacylation potentially driving its downhill uptake? 4) is the LplT-Aas system specific for LPE or does it serve as a general machinery for LPL remodeling in bacterial cells? To answer these questions, LPL remodeling was directly monitored by thin-layer chromatography using various radioactive LPLs (16). 32P-labelled LPLs were generated by venom sPLA2-catalyzed lipolytic reactions of radiolabeled diacyl forms purified from a number of “lipid” mutant E.coli strains (81). These assays clearly show that both LPE and LPG are transported by LplT and then acylated by endogenous Aas (Fig. 3). No remodeling was detected in either the ΔlplT or Δaas mutant. E.coli cells apparently remodel LPG 3x faster than LPE. The higher remodeling rate may be attributed to the substrate selectivity of Aas towards LPG since LplT imports LPG and LPE with similar substrate binding affinities (~2 μM) and transport rates (16).
7.3. Remodeling of CL by the LplT-Aas system
Remodeling of CL has been previously demonstrated in eukaryotic cells to be mediated by a non-specific phospholipid transacylase, Tafazzin (82). Tafazzin catalyzes acyl transfer from CL to LPL in the mitochondrial membrane. Our recent study of LplT-Aas has revealed the first CL remodeling mechanism in bacteria (16).
Snake venom PLA2 degrades CL via a progressive deacylation reaction to produce diacyl-CL and subsequently monoacyl-CL, eventually generating a completely deacylated head group compound (16). To our knowledge, this compound has never been reported, thus was named CL-hg. LplT transports both diacyl-CL and CL-hg into spheroplasts with comparable kinetic parameters, despite their distinct chemical structures. Interestingly, both CL derivatives are remodeled by an Aas-dependent acylation reaction to generate tri-acyl CL (Fig. 3). No other CL intermediates were observed when CL-hg was used as substrate. Aas may implement unknown substrate binding and catalytic mechanisms to perform repetitive acyl transfers before releasing tri-acyl CL from the active site. Remodeling of monoacyl-CL has not yet been tested due to its poor production by venom sPLA2. It would not be surprising if the LplT-Aas system catalyzes this intermediate due to its analogous conformation with other two CL substrates. Remodeling to a triacyl form, not a tetra-acyl CL, may be a part of a membrane self-regulatory mechanism in bacteria. Phospholipid degradation is often compensated by the accumulation of CL and both events likely occur on the outer leaflet of the IM (21). Generation of a cylinder-like bilayer-prone triacyl-CL, instead of non-bilayer forming CL on the inner leaflet may help to reduce bilayer lateral tension to maintain maximal membrane stability under stressed conditions.
8. Substrate selectivity of LplT
8.1. Substrate head group
Several lipid permeases are known to transport multiple lipid types with a modest specificity (13,69,83). In contrast, LplT exhibits an unusual substrate selectivity (16). LplT is capable of transporting four substrates including LPE, LPG and two lyso forms of CL with nearly identical transport kinetics. But its substrate binding is strictly limited to LPLs derived from bacteria (Fig. 3). LPC is not a substrate of LplT despite the fact that it shares a similar zwitterionic head group structure with LPE. Therefore, LplT is expected to establish structural hindrance to prevent access of the bulky choline group to the binding site. Most bacteria including E.coli cannot synthesize PC but may live in a PC-rich environment such as the animal digestive tract. In the intestinal lumen, pancreatic PLA2 degrades food-derived PC to generate LPC (84). LPC is further catabolized to LPA and choline via autotaxin-dependent and autotaxin-independent reactions. LPA is also a phospholipid precursor in bacteria, but its metabolism occurs in the cytosol. Remarkably, LplT cannot transport LPA either (16). Exclusion of LPC and LPA prevents any incorporation of foreign lipids into the bacterial membrane, further supporting the notion that the function of LplT is membrane repair and possible involvement in modulation of bacterial membrane properties. The lack of binding of LPA also indicates that the head group moiety plays an imperative role in the substrate binding mechanism. Diacyl-CL may be viewed as LPG from each side of the molecule (Fig. 3). Additional study is needed to assess any role of the phosphate or glycerol backbone in substrate binding. Our current data support the conclusion that the ethanolamine or glycerol head group is the chemical determinant for substrate recognition (16).
8.2. Lyso vs diacyl phospholipids
Another unique feature of LplT is its high specificity for lyso lipids. LplT cannot be inhibited by PE, PG or CL (16). Occluding these membrane phospholipids enables LplT to perform properly in the dense diacyl lipid-packed bilayer. The effective transport of CL-hg minimizes any contribution of the acyl chain to substrate binding. Deletion of lplT also has no effect on fatty acid movement into the cells (14). These lines of evidence support a “credit card-swiping” model for LplT, which was previously proposed for lipid flippases (15). In this model, only the head group of the LPL slides along the polar translocation pathway while its hydrophobic fatty acid tail swings outside of the protein during translocation across the bilayer. To adapt this bipolar substrate orientation, LplT may have to establish a specific sliding tunnel on the protein-membrane interface, which perhaps only accommodates a slim LPL.
8.3. Acyl chain stereo-selectivity
Both 1-acyl and 2-acyl GPLs generated by venom PLA2 or R. arrhizus lipase have been used in LplT transport studies (14,16). These assays may be incapable of defining stereoselectivity of the acyl chain, since 2-acyl GPL is extremely unstable in biological solution and is quickly converted to the 2-acyl form by a spontaneous intra-molecular acyl migration, yielding a mixture mainly containing 1-acyl GPL (85,86). sPLA2s are a major exogenous threat for the bacterial membrane envelope. PLA1 from T. lanuginosus (Sigma) cannot degrade phospholipids in E.coli spheroplasts (data not shown). Probably, distinguishing between the two stereo-chemical configurations is not required for substrate recognition by LplT.
9. Putative mechanistic model for LplT transport
MFS members share a common substrate transport mechanism of domain rocker-switch and substrate alternating access (87). In the rocker-switch model, the two domains of a MFS protein undergo a rigid-body motion that facilitates the substrate-binding site being alternatively accessible from either side of the membrane. A structural model of LplT has been generated based on the crystal structure of the glycerol-3-phosphate transporter GlpT (16). The model shows an inward-facing conformation with a putative substrate translocation pathway opening towards the cytoplasmic surface. LplT catalyzes energy-independent LPL transport (14). The rocker switching may be triggered by substrate binding. Along the translocation pathway, several conserved charged or polar residues may assemble two symmetric substrate binding sites each localized on the periplasmic or cytoplasmic side of the membrane (Fig. 4a): a) the periplasmic binding site is formed by Asp30, Arg236, Asn31 and Gln55, and b) the cytoplasmic binding site is formed by Glu351, Lys120, Asn327 and Asn348. In the periplasmic site, Asp30 and Arg236 form an ionlock (the outerlock) via a salt bridge interaction, stabilizing the inward-facing conformation. In the cytoplasmic site, Glu351 and Lys120 separated apart in the current state may approach each other to form another ionlock (the innerlock) when the protein is switched to the outward-facing conformation. We also predict that Gln23 and Asn352 serve as a shuttle site in the middle of the pathway to mediate internal transfer of a substrate from the periplasmic site to the cytoplasmic site.
Based on this model, the transport mechanism of LplT is proposed to follow three steps (Fig. 4b): i) the outward-opening state is stabilized by the innerlock, allowing a LPL substrate access to the periplasmic binding site; ii) substrate binding induces conversion of an occluded state by opening the innerlock, which in turn helps the substrate move to the cytoplasmic binding site via the intermediate shuttle site; iii) the outerlock may be engaged to stabilize the protein in an inward-facing conformation which allows the substrate to be released from the cytoplasmic binding site to the inner leaflet of the membrane. A similar ionlock switching model has also been proposed for the LPC transporter protein MFSD2A (88), despite the lack of sequence homology between bacterial LplT and this mammalian transporter protein. These two proteins may utilize different mechanisms since 1) MFSD2A transports both LPC and LPE, while LplT excludes LPC; 2) LplT-mediated transport is energy independent, while MFSD2A utilizes a counter Na+ electrochemical gradient as energy driver; more importantly, 3) a fatty acyl chain is required for MFSD2A to mediate LPL transport while LplT recognizes the head group conformation exclusively. Further biochemical characterization will help to define the novel transport mechanism of LplT.
10. Physiological significance of the lplT-Aas system
Although multiple routes of LPL biogenesis have been reported, LplT-Aas is the only LPL remodeling system identified in Gram-negative bacteria thus far. The biological function of this novel system remains unidentified and our knowledge is limited to biochemical characterizations. Deletion of the lplT and/or aas gene shows no effect on bacterial growth and LPLs are still retained at a “safe” level (16). A combined study with endogenous LPL biogenesis pathways such as Lnt, PagP or PldA may help to delineate its housekeeping role in living bacteria.
The LplT-Aas system most likely contributes to the stability and functional integrity of the IM as a protective mechanism, in particular when cells confront an exogenous sPLA2 attack. Spheroplasts generated from ΔlplT or Δaas mutant cells exhibit much greater susceptibility to pancreatic sPLA2 coinciding with massive accumulation of multiple LPL species in the membrane in contrast to the high resistance of wild type (16). Although sPLA2-IIA kills both Gram-positive and Gram-negative bacteria in vitro, Gram-negative bacteria exhibit a greater resistance to the sPLA2. i.e. killing of Gram-negative bacteria, such as E.coli, requires a higher concentration of enzyme than present in the animal body even during inflammation (47). While the OM may act as a native barrier to the intruder, the LplT-Aas system that only exists in Gram-negative bacteria may serve as a bacterial defense system to counteract sPLA2-mediated attack by repairing the damaged membrane.
E.coli have additional mechanisms to facilitate LPL flipping in the IM. In ΔlplT mutants, the residual LPL transport activity (<20%) is not due to spontaneous lipid flipping (14,16). But current evidence supports LplT as the major pathway for LPL transport in E.coli. There are also multiple routes available for metabolism of LPLs (41). However, sensitive radioactive assay using TLC did not detect any products other than diacyl PE or PG or the triacyl form of CL or any LPL degradation in the LPL uptake and remodeling reaction (16). Therefore, Aas-catalyzed acylation is the major mechanism for processing LPLs that are imported by LplT. This evidence highlights the specificity of the LplT-Aas coupled LPL remodeling system.
11. Conclusions
Because newly generated or exogenous LPLs can potentially affect the integrity of the lipid bilayer, the destabilized membranes have to be repaired immediately in order to eliminate disruptive effects. The LplT/Aas translocation/acylation tandem is the universal and primary system for membrane phospholipid remodeling in Gram-negative bacteria. This coupled system facilitates uptake and specific rescuing of lyso forms of three Gram-negative bacterial major phospholipids with comparable kinetic and production yields, but excludes diacyl lipids access to the binding site and therefore represents a generalized membrane repairing mechanism. Bacteria have evolutionarily adapted a constant headgroup diacyl phospholipid composition on the membrane. Obviously, environmental and host effects induce perturbations of the bacterial phospholipid acylation-reacylation cycle, which would change the physical state of the OM and the whole envelope. The results presented in this review raise additional questions to be addressed in the future. 1) Is LplT-Aas mediated translocation and recycling limited only to rapid membrane repair or is it dedicated to fine tuning/determination of an optimal threshold level of LPE required for different physiological processes within envelope? 2) Does the LplT-Aas system function as a part of an acute membrane repair response in environmentally and host-stressed bacteria? 3) Does the LplT-Aas system act as a resistance mechanism that allows Gram-negative pathogens to tolerate the action of host sPLA2? Future structural and functional study may help to understand this novel transport mechanism and reveal the physiological role of this novel LPL remodeling system in Gram-negative bacteria.
Highlights.
Multiple routes of lysophospholipid biogenesis exist in Gram-negative bacteria.
LplT exhibits a broad substrate selectivity but only transports bacterial lipids.
LplT-Aas provides a unique bacterial membrane phospholipid repair mechanism.
A rocker-switch model for LplT-mediated lipid transport is proposed.
Acknowledgements:
we thank Drs. Shuilong Tong and Ziqiang Guan for contributions to the original work and Dr. Julia Lever for comments on manuscript.
Funding:
This work was supported by National Institute of Health grant R01GM098572 to LZ and H2020-MSCA-RISE EU to MB.
Abbreviations:
- PE:
phosphatidylethanolamine
- PG:
phosphatidylglycerol
- CL:
cardiolipin
- PS:
phosphatidylserine
- PA:
phosphatidic acid
- PC:
phosphatidylcholine
- CDP-DAG:
cytidine diphosphate diacylglycerol
- LPL:
lysophospholipid
- LPE:
lysophosphatidylethanolamine
- LPG:
lysophosphatidylglycerol
- LPC:
lysophosphatidylcholine
- LPA:
lysophosphatidic acid
- GPL:
glycerophospholipid
- G3P:
glycerol-3-phosphate
- IM:
inner membrane
- OM:
outer membrane
- BPI:
bactericidal-permeability-increasing protein
- PLA2:
phospholipase A2
- sPLA2-GIIA:
secretory phospholipase A2 group IIA
- MFS:
major facilitator superfamily
- LPS:
lipopolysaccharide
- Lpp:
lipoprotein
- T6S:
type VI secretion system
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- 1.Dowhan W (1997) Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem 66, 199–232 [DOI] [PubMed] [Google Scholar]
- 2.Zhang YM, and Rock CO (2008) Membrane lipid homeostasis in bacteria. Nature reviews. Microbiology 6, 222–233 [DOI] [PubMed] [Google Scholar]
- 3.Giles DK, Hankins JV, Guan Z, and Trent MS (2011) Remodelling of the Vibrio cholerae membrane by incorporation of exogenous fatty acids from host and aquatic environments. Mol Microbiol 79, 716–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tannaes T, Bukholm IK, and Bukholm G (2005) High relative content of lysophospholipids of Helicobacter pylori mediates increased risk for ulcer disease. FEMS Immunol Med Microbiol 44, 17–23 [DOI] [PubMed] [Google Scholar]
- 5.Taranto MP, Fernandez Murga ML, Lorca G, and de Valdez GF (2003) Bile salts and cholesterol induce changes in the lipid cell membrane of Lactobacillus reuteri. J Appl Microbiol 95, 86–91 [DOI] [PubMed] [Google Scholar]
- 6.Homma H, and Nojima S (1982) Synthesis of various phospholipids from 2-acyl lysophospholipids by Escherichia coli extract. Journal of biochemistry 91, 1103–1110 [DOI] [PubMed] [Google Scholar]
- 7.Whetton AD, Lu Y, Pierce A, Carney L, and Spooncer E (2003) Lysophospholipids synergistically promote primitive hematopoietic cell chemotaxis via a mechanism involving Vav 1. Blood 102, 2798–2802 [DOI] [PubMed] [Google Scholar]
- 8.Monet M, Gkika D, Lehen’kyi V, Pourtier A, Vanden Abeele F, Bidaux G, Juvin V, Rassendren F, Humez S, and Prevarsakaya N (2009) Lysophospholipids stimulate prostate cancer cell migration via TRPV2 channel activation. Biochimica et biophysica acta 1793, 528–539 [DOI] [PubMed] [Google Scholar]
- 9.Yamamoto N, Willett NP, and Lindsay DD (1994) Participation of serum proteins in the inflammation-primed activation of macrophages. Inflammation 18, 311–322 [DOI] [PubMed] [Google Scholar]
- 10.Murph M, Tanaka T, Pang J, Felix E, Liu S, Trost R, Godwin AK, Newman R, and Mills G (2007) Liquid chromatography mass spectrometry for quantifying plasma lysophospholipids: potential biomarkers for cancer diagnosis. Methods in enzymology 433, 1–25 [DOI] [PubMed] [Google Scholar]
- 11.Mills GB, Eder A, Fang X, Hasegawa Y, Mao M, Lu Y, Tanyi J, Tabassam FH, Wiener J, Lapushin R, Yu S, Parrott JA, Compton T, Tribley W, Fishman D, Stack MS, Gaudette D, Jaffe R, Furui T, Aoki J, and Erickson JR (2002) Critical role of lysophospholipids in the pathophysiology, diagnosis, and management of ovarian cancer. Cancer treatment and research 107, 259–283 [DOI] [PubMed] [Google Scholar]
- 12.Makide K, Uwamizu A, Shinjo Y, Ishiguro J, Okutani M, Inoue A, and Aoki J (2014) Novel lysophosphoplipid receptors: their structure and function. Journal of lipid research 55, 1986–1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen LN, Ma D, Shui G, Wong P, Cazenave-Gassiot A, Zhang X, Wenk MR, Goh ELK, and Silver DL (2014) Mfsd2a is a transporter for the essential omega-3 fatty acid docosahexaenoic acid. Nature 509, 503–506 [DOI] [PubMed] [Google Scholar]
- 14.Harvat EM, Zhang YM, Tran CV, Zhang Z, Frank MW, Rock CO, and Saier MH Jr. (2005) Lysophospholipid flipping across the Escherichia coli inner membrane catalyzed by a transporter (LplT) belonging to the major facilitator superfamily. The Journal of biological chemistry 280, 12028–12034 [DOI] [PubMed] [Google Scholar]
- 15.Sanyal S, and Menon AK (2009) Flipping lipids: why an’ what’s the reason for? ACS chemical biology 4, 895–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin Y, Bogdanov M, Tong S, Guan Z, and Zheng L (2016) Substrate Selectivity of Lysophospholipid Transporter LplT Involved in Membrane Phospholipid Remodeling in Escherichia coli. The Journal of biological chemistry 291, 2136–2149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Geus P, van Die I, Bergmans H, Tommassen J, and de Haas G (1983) Molecular cloning of pldA, the structural gene for outer membrane phospholipase of E. coli K12. Mol Gen Genet 190, 150–155 [DOI] [PubMed] [Google Scholar]
- 18.Kern R, Joseleau-Petit D, Chattopadhyay MK, and Richarme G (2001) Chaperone-like properties of lysophospholipids. Biochem Biophys Res Commun 289, 1268–1274 [DOI] [PubMed] [Google Scholar]
- 19.Cronan JE Jr., and Wulff DL (1969) A role for phospholipid hydrolysis in the lysis of Escherichia coli infected with bacteriophage T4. Virology 38, 241–246 [DOI] [PubMed] [Google Scholar]
- 20.Pugsley AP, and Schwartz M (1984) Colicin E2 release: lysis, leakage or secretion? Possible role of a phospholipase. EMBO J 3, 2393–2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michel G (1979) The changes in cell size and phospholipid composition during growth of a chain-forming envC mutant of Escherichia coli. FEBS Lett 100, 258–260 [DOI] [PubMed] [Google Scholar]
- 22.Hardaway KL, and Buller CS (1979) Effect of ethylenediaminetetraacetate on phospholipids and outer membrane function in Escherichia coli. J Bacteriol 137, 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elsbach P, Weiss J, Franson RC, Beckerdite-Quagliata S, Schneider A, and Harris L (1979) Separation and purification of a potent bactericidal/permeability-increasing protein and a closely associated phospholipase A2 from rabbit polymorphonuclear leukocytes. Observations on their relationship. J Biol Chem 254, 11000–11009 [PubMed] [Google Scholar]
- 24.Wright GC, Weiss J, Kim KS, Verheij H, and Elsbach P (1990) Bacterial phospholipid hydrolysis enhances the destruction of Escherichia coli ingested by rabbit neutrophils. Role of cellular and extracellular phospholipases. The Journal of clinical investigation 85, 1925–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss J, Beckerdite-Quagliata S, and Elsbach P (1979) Determinants of the action of phospholipases A on the envelope phospholipids of Escherichia coli. J Biol Chem 254, 11010–11014 [PubMed] [Google Scholar]
- 26.Dekker N (2000) Outer-membrane phospholipase A: known structure, unknown biological function. Mol Microbiol 35, 711–717 [DOI] [PubMed] [Google Scholar]
- 27.Snijder HJ, and Dijkstra BW (2000) Bacterial phospholipase A: structure and function of an integral membrane phospholipase. Biochimica et biophysica acta 1488, 91–101 [DOI] [PubMed] [Google Scholar]
- 28.Dekker N, Tommassen J, Lustig A, Rosenbusch JP, and Verheij HM (1997) Dimerization regulates the enzymatic activity of Escherichia coli outer membrane phospholipase A. J Biol Chem 272, 3179–3184 [DOI] [PubMed] [Google Scholar]
- 29.Snijder HJ, Ubarretxena-Belandia I, Blaauw M, Kalk KH, Verheij HM, Egmond MR, Dekker N, and Dijkstra BW (1999) Structural evidence for dimerization-regulated activation of an integral membrane phospholipase. Nature 401, 717–721 [DOI] [PubMed] [Google Scholar]
- 30.Audet A, Nantel G, and Proulx P (1974) Phospholipase A activity in growing Escherichia coli cells. Biochimica et biophysica acta 348, 334–343 [DOI] [PubMed] [Google Scholar]
- 31.Jia W, El Zoeiby A, Petruzziello TN, Jayabalasingham B, Seyedirashti S, and Bishop RE (2004) Lipid trafficking controls endotoxin acylation in outer membranes of Escherichia coli. J Biol Chem 279, 44966–44975 [DOI] [PubMed] [Google Scholar]
- 32.Istivan TS, and Coloe PJ (2006) Phospholipase A in Gram-negative bacteria and its role in pathogenesis. Microbiology 152, 1263–1274 [DOI] [PubMed] [Google Scholar]
- 33.Davydova L, Bakholdina S, Barkina M, Velansky P, Bogdanov M, and Sanina N (2016) Effects of elevated growth temperature and heat shock on the lipid composition of the inner and outer membranes of Yersinia pseudotuberculosis. Biochimie 123, 103–109 [DOI] [PubMed] [Google Scholar]
- 34.Oliver JD, and Colwell RR (1973) Extractable lipids of gram-negative marine bacteria: phospholipid composition. J Bacteriol 114, 897–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bukholm G, Tannaes T, Nedenskov P, Esbensen Y, Grav HJ, Hovig T, Ariansen S, and Guldvog I (1997) Colony variation of Helicobacter pylori: pathogenic potential is correlated to cell wall lipid composition. Scand J Gastroenterol 32, 445–454 [DOI] [PubMed] [Google Scholar]
- 36.Tannaes T, Dekker N, Bukholm G, Bijlsma JJ, and Appelmelk BJ (2001) Phase variation in the Helicobacter pylori phospholipase A gene and its role in acid adaptation. Infect Immun 69, 7334–7340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dalebroux ZD, Edrozo MB, Pfuetzner RA, Ressl S, Kulasekara BR, Blanc MP, and Miller SI (2015) Delivery of cardiolipins to the Salmonella outer membrane is necessary for survival within host tissues and virulence. Cell Host Microbe 17, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leive L (1965) Release of lipopolysaccharide by EDTA treatment of E. coli. Biochem Biophys Res Commun 21, 290–296 [DOI] [PubMed] [Google Scholar]
- 39.Bishop RE, Gibbons HS, Guina T, Trent MS, Miller SI, and Raetz CR (2000) Transfer of palmitate from phospholipids to lipid A in outer membranes of gram-negative bacteria. EMBO J 19, 5071–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dalebroux ZD, Matamouros S, Whittington D, Bishop RE, and Miller SI (2014) PhoPQ regulates acidic glycerophospholipid content of the Salmonella Typhimurium outer membrane. Proc Natl Acad Sci U S A 111, 1963–1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hsu L, Jackowski S, and Rock CO (1991) Isolation and characterization of Escherichia coli K-12 mutants lacking both 2-acyl-glycerophosphoethanolamine acyltransferase and acyl-acyl carrier protein synthetase activity. The Journal of biological chemistry 266, 13783–13788 [PubMed] [Google Scholar]
- 42.Jackowski S, and Rock CO (1986) Transfer of fatty acids from the 1-position of phosphatidylethanolamine to the major outer membrane lipoprotein of Escherichia coli. J Biol Chem 261, 11328–11333 [PubMed] [Google Scholar]
- 43.Hillmann F, Argentini M, and Buddelmeijer N (2011) Kinetics and phospholipid specificity of apolipoprotein N-acyltransferase. J Biol Chem 286, 27936–27946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mileykovskaya E, Ryan AC, Mo X, Lin C-C, Khalaf KI, Dowhan W, and Garrett TA (2009) Phosphatidic Acid and N-Acylphosphatidylethanolamine Form Membrane Domains in Escherichia coli Mutant Lacking Cardiolipin and Phosphatidylglycerol. Journal of Biological Chemistry 284, 2990–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Merkel O, Schmid PC, Paltauf F, and Schmid HH (2005) Presence and potential signaling function of N-acylethanolamines and their phospholipid precursors in the yeast Saccharomyces cerevisiae. Biochimica et biophysica acta 1734, 215–219 [DOI] [PubMed] [Google Scholar]
- 46.Nevalainen TJ, Graham GG, and Scott KF (2008) Antibacterial actions of secreted phospholipases A2. Review. Biochimica et biophysica acta 1781, 1–9 [DOI] [PubMed] [Google Scholar]
- 47.Weiss JP (2015) Molecular determinants of bacterial sensitivity and resistance to mammalian Group IIA phospholipase A2. Biochimica et biophysica acta 1848, 3072–3077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Elsbach P, and Weiss J (1998) Role of the bactericidal/permeability-increasing protein in host defence. Current Opinion in Immunology 10, 45–49 [DOI] [PubMed] [Google Scholar]
- 49.Russell AB, LeRoux M, Hathazi K, Agnello DM, Ishikawa T, Wiggins PA, Wai SN, and Mougous JD (2013) Diverse type VI secretion phospholipases are functionally plastic antibacterial effectors. Nature 496, 508–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ishii I, Fukushima N, Ye X, and Chun J (2004) Lysophospholipid receptors: signaling and biology. Annu Rev Biochem 73, 321–354 [DOI] [PubMed] [Google Scholar]
- 51.Fukushima N, Ishii S, Tsujiuchi T, Kagawa N, and Katoh K (2015) Comparative analyses of lysophosphatidic acid receptor-mediated signaling. Cell Mol Life Sci 72, 2377–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morein S, Andersson A, Rilfors L, and Lindblom G (1996) Wild-type Escherichia coli cells regulate the membrane lipid composition in a “window” between gel and nonlamellar structures. J Biol Chem 271, 6801–6809 [DOI] [PubMed] [Google Scholar]
- 53.Los DA, and Murata N (2004) Membrane fluidity and its roles in the perception of environmental signals. Biochimica et biophysica acta 1666, 142–157 [DOI] [PubMed] [Google Scholar]
- 54.Ben-Zeev G, Telias M, and Nussinovitch I (2010) Lysophospholipids modulate voltage-gated calcium channel currents in pituitary cells; effects of lipid stress. Cell Calcium 47, 514–524 [DOI] [PubMed] [Google Scholar]
- 55.Hollie NI, Cash JG, Matlib MA, Wortman M, Basford JE, Abplanalp W, and Hui DY (2014) Micromolar changes in lysophosphatidylcholine concentration cause minor effects on mitochondrial permeability but major alterations in function. Biochimica et biophysica acta 1841, 888–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vasquez V, Sotomayor M, Cordero-Morales J, Schulten K, and Perozo E (2008) A structural mechanism for MscS gating in lipid bilayers. Science 321, 1210–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bavi O, Vossoughi M, Naghdabadi R, and Jamali Y (2014) The effect of local bending on gating of MscL using a representative volume element and finite element simulation. Channels (Austin) 8, 344–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dowhan W (2009) Molecular genetic approaches to defining lipid function. Journal of lipid research 50 Suppl, S305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mileykovskaya E, and Dowhan W (2000) Visualization of phospholipid domains in Escherichia coli by using the cardiolipin-specific fluorescent dye 10-N-nonyl acridine orange. J Bacteriol 182, 1172–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sutterlin HA, Shi H, May KL, Miguel A, Khare S, Huang KC, and Silhavy TJ (2016) Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc Natl Acad Sci U S A 113, E1565–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Michel GP, and Starka J (1984) Origin and fate of the lysophosphatidylethanolamine in a chain-forming mutant (envC) of Escherichia coli. Journal of general microbiology 130, 1391–1398 [DOI] [PubMed] [Google Scholar]
- 62.Rokitskaya TI, Kotova EA, Naberezhnykh GA, Khomenko VA, Gorbach VI, Firsov AM, Zelepuga EA, Antonenko YN, and Novikova OD (2016) Single channel activity of OmpF-like porin from Yersinia pseudotuberculosis. Biochimica et biophysica acta 1858, 883–891 [DOI] [PubMed] [Google Scholar]
- 63.Perozo E, Cortes DM, Sompornpisut P, Kloda A, and Martinac B (2002) Open channel structure of MscL and the gating mechanism of mechanosensitive channels. Nature 418, 942–948 [DOI] [PubMed] [Google Scholar]
- 64.McIntyre TM, and Bell RM (1978) Escherichia coli mutants defective in membrane phospholipid synthesis: binding and metabolism of 1-oleoylglycerol 3-phosphate by a plsB deep rough mutant. J Bacteriol 135, 215–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Homma H, Nishijima M, Kobayashi T, Okuyama H, and Nojima S (1981) Incorporation and metabolism of 2-acyl lysophospholipids by Escherichia coli. Biochimica et biophysica acta 663, 1–13 [DOI] [PubMed] [Google Scholar]
- 66.Hsu L, Jackowski S, and Rock CO (1989) Uptake and acylation of 2-acyllysophospholipids by Escherichia coli. J Bacteriol 171, 1203–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Malinverni JC, and Silhavy TJ (2009) An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci U S A 106, 8009–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharom FJ (2011) Flipping and flopping--lipids on the move. IUBMB Life 63, 736–746 [DOI] [PubMed] [Google Scholar]
- 69.Lopez-Marques RL, Theorin L, Palmgren MG, and Pomorski TG (2014) P4-ATPases: lipid flippases in cell membranes. Pflugers Arch 466, 1227–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doerrler WT, Gibbons HS, and Raetz CR (2004) MsbA-dependent translocation of lipids across the inner membrane of Escherichia coli. J Biol Chem 279, 45102–45109 [DOI] [PubMed] [Google Scholar]
- 71.Kol MA, van Dalen A, de Kroon AI, and de Kruijff B (2003) Translocation of phospholipids is facilitated by a subset of membrane-spanning proteins of the bacterial cytoplasmic membrane. J Biol Chem 278, 24586–24593 [DOI] [PubMed] [Google Scholar]
- 72.Doerrler WT (2006) Lipid trafficking to the outer membrane of Gram-negative bacteria. Mol Microbiol 60, 542–552 [DOI] [PubMed] [Google Scholar]
- 73.Tefsen B, Bos MP, Beckers F, Tommassen J, and de Cock H (2005) MsbA is not required for phospholipid transport in Neisseria meningitidis. J Biol Chem 280, 35961–35966 [DOI] [PubMed] [Google Scholar]
- 74.Tefsen B, Geurtsen J, Beckers F, Tommassen J, and de Cock H (2005) Lipopolysaccharide transport to the bacterial outer membrane in spheroplasts. J Biol Chem 280, 4504–4509 [DOI] [PubMed] [Google Scholar]
- 75.Kubelt J, Menon AK, Muller P, and Herrmann A (2002) Transbilayer movement of fluorescent phospholipid analogues in the cytoplasmic membrane of Escherichia coli. Biochemistry 41, 5605–5612 [DOI] [PubMed] [Google Scholar]
- 76.Pao SS, Paulsen IT, and Saier MH (1998) Major Facilitator Superfamily. Microbiology and Molecular Biology Reviews 62, 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, and Peschel A (2009) The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS pathogens 5, e1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jackowski S, Jackson PD, and Rock CO (1994) Sequence and function of the aas gene in Escherichia coli. The Journal of biological chemistry 269, 2921–2928 [PubMed] [Google Scholar]
- 79.Lopez-Marques RL, Poulsen LR, Bailly A, Geisler M, Pomorski TG, and Palmgren MG (2015) Structure and mechanism of ATP-dependent phospholipid transporters. Biochimica et biophysica acta 1850, 461–475 [DOI] [PubMed] [Google Scholar]
- 80.Goswitz VC, and Brooker RJ (1995) Structural features of the uniporter/symporter/antiporter superfamily. Protein science: a publication of the Protein Society 4, 534–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bogdanov M, Mileykovskaya E, and Dowhan W (2008) Lipids in the assembly of membrane proteins and organization of protein supercomplexes: implications for lipid-linked disorders. Sub-cellular biochemistry 49, 197–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schlame M (2013) Cardiolipin remodeling and the function of tafazzin. Biochimica et biophysica acta 1831, 582–588 [DOI] [PubMed] [Google Scholar]
- 83.Brunner JD, Lim NK, Schenck S, Duerst A, and Dutzler R (2014) X-ray structure of a calcium-activated TMEM16 lipid scramblase. Nature 516, 207–212 [DOI] [PubMed] [Google Scholar]
- 84.Hui DY (2016) Intestinal phospholipid and lysophospholipid metabolism in cardiometabolic disease. Current opinion in lipidology 27, 507–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dennis EA, Darke PL, Deems RA, Kensil CR, and Pluckthun A (1981) Cobra venom phospholipase A2: a review of its action toward lipid/water interfaces. Molecular and cellular biochemistry 36, 37–45 [DOI] [PubMed] [Google Scholar]
- 86.Okudaira M, Inoue A, Shuto A, Nakanaga K, Kano K, Makide K, Saigusa D, Tomioka Y, and Aoki J (2014) Separation and quantification of 2-acyl-1-lysophospholipids and 1-acyl-2-lysophospholipids in biological samples by LC-MS/MS. J Lipid Res 55, 2178–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Quistgaard EM, Low C, Guettou F, and Nordlund P (2016) Understanding transport by the major facilitator superfamily (MFS): structures pave the way. Nat Rev Mol Cell Biol 17, 123–132 [DOI] [PubMed] [Google Scholar]
- 88.Quek DQY, Nguyen LN, Fan H, and Silver DL (2016) Structural Insights into the Transport Mechanism of the Human Sodium-dependent Lysophosphatidylcholine Transporter MFSD2A. Journal of Biological Chemistry 291, 9383–9394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Krogh A, Larsson B, von Heijne G, and Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. Journal of molecular biology 305, 567–580 [DOI] [PubMed] [Google Scholar]