Abstract
Interleukin (IL)-1 receptor-associated kinase (IRAK) 4 is a central enzyme of the Toll-like receptor (TLR) pathways. This study tested the hypothesis that IRAK4 kinase activity is prerequisite for regulating innate immunity during infections with intracellular bacteria. To this end, we analyzed responses of macrophages obtained from mice expressing wild-type IRAK4 or its kinase-inactive K213M mutant (IRAK4KI) upon infection with intracellular bacteria Listeria monocytogenes or Mycobacterium smegmatis. In contrast to robust induction of cytokines by macrophages expressing kinase-sufficient IRAK4, IRAK4KI macrophages expressed decreased tumor necrosis factor-α, IL-6, IL-1β, and C-C motif chemokine ligand 5 upon infection with L. monocytogenes or M. smegmatis. Bacterial infection of IRAK4KI macrophages led to attenuated activation of IRAK1, mitogen-activated protein kinases (MAPKs) and nuclear factor (NF)-κB, impaired induction of inducible nitric oxide (NO) synthase mRNA and secretion of NO, but resulted in elevated microbial burdens. Compared to wild-type animals, systemic infection of IRAK4KI mice with M. smegmatis or L. monocytogenes resulted in decreased levels of serum IL-6 and CXC motif ligand-1 but increased bacterial burdens in the spleen and liver. Thus, a loss of IRAK4 kinase activity underlies deficient cytokine and microbicidal responses during infection with intracellular bacteria L. monocytogenes or M. smegmatis via impaired activation of IRAK1, MAPKs, and NF-κB but increases bacterial burdens, correlating with decreased induction of NO.
Keywords: Inflammation, Innate Immunity, Infection, Toll-like Receptor, Macrophage
Introduction
Toll-like receptors (TLRs) sense microbes or host “alarmins” to activate innate immunity and orchestrate adaptive immune responses [1]. TLRs present on the plasma membrane (TLR1, TLR5, TLR6), endosomes (TLR3, TLR7, TLR9) or in both compartments (TLR2, TLR4) in myeloid cells express an ectodomain involved in ligand recognition, a transmembrane segment, and an intracellular Toll-IL-1R (TIR) signaling domain[1, 2]. Upon ligand recognition, the majority of TLRs recruit myeloid differentiation primary response protein (MyD) 88, followed by recruitment of interleukin (IL)-1R-associated kinase (IRAK) 4, IRAK1, IRAK2 and IRAK-M [2, 3]. Clustering of IRAK4 molecules induces IRAK4 kinase activity, leading to phosphorylation and activation of downstream IRAKs and engagement of tumor necrosis factor (TNF) receptor -associated factor (TRAF) 6 and transforming growth factor-β-activated kinase (TAK) 1 [3–5]. TAK1 phosphorylates and activates MAPKs and inhibitor of nuclear factor (NF)-κB kinase (IKK)-β to trigger activation and nuclear translocation of transcription factors, culminating in transcription of inflammatory cytokine genes [4]. TLR3 and TLR4 signal from endosomes via TIR domain-containing adapter inducing interferon (IFN)-β (TRIF) that recruits TRAF-associated NF-κB activator-binding kinase-1 and IKK-ε. These kinases activate IFN regulatory factor (IRF)-3, leading to its nuclear translocation and transcription of type I IFNs [2]. The TRIF pathway also mediates delayed activation of MAPKs and NF-κB via the receptor-interacting protein-1-TRAF6 signaling axis [2].
Studies with IL-1β and defined TLR agonists have revealed that activated IRAK4 signals via the TAK1-IKK-β axis to mediate classical activation of NF-κB, engages the MK2-dependent pathway to stabilize cytokine mRNAs [4, 6, 7], and elicits rapid activation of nucleotide-binding domain-like receptor containing pyrin domain 3 (NLRP3) inflammasome [8, 9]. Kinase-inactive (K213M) IRAK4 (IRAK4KI) fails to mediate these responses [10], but signals via MAPK-extracellular regulated kinase (ERK) kinase 3 (MEKK3)-IKK-γ to trigger non-classical activation of NF-κB. This pathway up-regulates cytokines (e.g. CC motif ligand (CCL) 2, CCL4, CCL7), signaling molecules (TLR2, IRAK2) and TLR inhibitors (A20, IRAK-M) whose expression is not regulated by mRNA stability [4, 6, 7, 10–13]. Macrophages and mice harboring IRAK4KI are deficient in mounting responses to and containing infection with Streptococcus pneumoniae [13], a highly invasive extracellular Gram positive bacterial pathogen. However, the role of IRAK4 kinase activity in regulating innate immune responses during infections with intracellular bacteria, e. g. Listeria monocytogenes or mycobacteria, is largely unknown. Furthermore, while studies in MyD88- and IRAK4-deficient mice showed their involvement in nitric oxide (NO) production [14, 15], the role of IRAK4 kinase activity in inducing macrophage microbicidal activity and NO secretion has not been elucidated.
In this paper, we demonstrate that infection of IRAK4KI macrophages with intracellular bacteria Listeria monocytogenes or Mycobacterium smegmatis led to deficient induction of TNF-α, IL-6, IL-1β, CCL5 and NO, inhibited activation of IRAK1, MAPKs and NF-κB, comparable phosphorylation of IRF3, but increased bacterial burdens. Furthermore, mice harboring kinase-dead IRAK4 exhibited decreased levels of serum IL-6 and CXCL1 but increased splenic and liver microbial burdens after systemic infection with M. smegmatis or L. monocytogenes. Thus, IRAK4 kinase activity is prerequisite for mounting innate immune responses and restraining intracellular bacterial survival during infection with L. monocytogenes or M. smegmatis.
Materials and Methods
Antibodies (Abs) and Reagents
The following Abs were used: anti-phospho (p)-IRAK1, anti-IRAK1, anti-p65 NF-κB (Santa Cruz Biotechnology, Santa Cruz, CA); anti-p-IRAK-4, anti-p-p38, anti-p-ERK1/2, anti-ERK1/2, anti-p38, anti-IκB-α, anti-p-p65, anti-p-IRF3, and anti-β-tubulin (Cell Signaling Technology, Inc., Danvers, MA). Pam3Cys, S-[2,3-bis(palmitoyloxy)-(2-RS)-propyl]-N-palmitoyl-(R)-Cys-Ser-Lys4-OH], was purchased from InvivoGen (San Diego, CA).
Mice, Infection and Macrophage Isolation
C57BL/6J mice were from the Jackson Laboratory (Bar Harbor, Maine, USA). IRAK4KI mice [7, 8, 10, 12, 13] were provided by Drs. Kirk Staschke (Eli Lilly, Indianapolis, IN) and Stefanie Vogel (University of Maryland School of Medicine, Baltimore, MD). Mice were housed in the University of Connecticut Health Center animal facility, and 6-week-old male and female mice were used. All animal procedures were approved by the Institutional Animal Care and Use Committee. For infections, wild-type (WT) or IRAK4KI mice were i.p. administered M. smegmatis (5 × 106 CFUs) or L. monocytogenes (2 × 104 CFU), similar to doses used in previous reports [16, 17]. Six hours post infection (hpi), blood samples were obtained by retroorbital punctures and serum was prepared to analyze cytokines. At 48 hpi, spleens and livers were homogenized in PBS containing protease and phosphatase inhibitors (ThermoFisher Scientific, Waltham, MA) and bacterial colony forming units (CFUs) were enumerated by plating organ homogenates on the respective agars [16, 18]. To generate bone marrow-derived macrophages (BMDMs), bone marrow cells from femurs and tibias were differentiated for 7 days in DMEM (Mediatech, Herndon, VA, USA), supplemented with 10% FBS (Atlanta Biologicals, Flowery Branch, GA), 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin (complete (c) RPMI) and 15% L929 cell-conditioned, macrophage-colony-stimulating factor-containing supernatant [6].
Bacterial cultures and infection
Green fluorescent protein (GFP)-expressing M. smegmatis and L. monocytogenes were provided by Dr. Volker Briken (University of Maryland College Park, College Park, MD) and Dr. Kamal Khanna (New York University School of Medicine, New York, NY), respectively. M. smegmatis were cultured in 7H9 broth containing 0.5% glycerol, 0.05% Tween-80, 10% oleic albumin dextrose complex enrichment (OADC) and 40 μg/ml kanamycin. L. monocytogenes was cultured in BHI media containing 10 μg/ml chloramphenicol. Bacteria were grown to mid-log phase, washed in PBS, and added to cells cultured in antibiotic-free DMEM at the respective multiplicity of infections (MOI) for 2 h. Cells were washed in PBS and incubated in complete (c) DMEM supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin and 100 μg/ml gentamycin.
Assessment of bacterial CFUs
BMDMs were grown overnight in 24-well plates (106 cells/well), infected for 1 h with L. monocytogenes (MOI 1) or 2 h with M. smegmatis (MOI 10) in antibiotic-free DMEM, washed with PBS, incubated in cDMEM containing 100 μg/ml genatamycin for the indicated times, and lysed in PBS containing 0.05% Tween-80 [19]. Spleens and livers from M. smegmatis- or L. monocytogenes-infected mice were homogenized in PBS containing protease and phosphatase inhibitors (ThermoFisher Scientific).The number of CFUs per 106 cells or in organ homogenates was determined by plating 10-fold serial dilutions of lysates on Middlebrook 7H10 plates complemented with 0.5% glycerol, 10% OADC enrichment, 40 μg/ml kanamycin (M. smegmatis) or blood heart infusion agar plates containing 10 μg/ml chloramphenicol (L. monocytogenes) [19]. Plates were incubated at 37°C and CFUs were counted after 24–48 h.
Analyses of bacterial uptake by flow cytometry
BMDMs (106 cells) were treated for 1 h with GFP-expressing L. monocytogenes (multiplicity of infection (MOI) 1) or for 2 h with M. smegmatis (MOI 10) in antibiotic-free medium, washed, and incubated in cDMEM containing 100 μg/ml gentamycin on ice or at 37°C for the indicated times. Cells were detached, washed in ice-cold PBS containing 3% FBS, stained with Zombie® Aqua cell viability dye (Biolegend), fixed for 15 min in 3.7% formaldehyde, and analyzed by FACS on a LSRII Flow Cytometer (BD Biosciences) to measure GFP fluorescence. The data were analyzed using the FlowJo software (Tree Star). BMDMs were gated using forward vs side scatters, live cells were gated as Zombie® Aqua-negative events, and GFP mean fluorescence intensity (MFI) values were analyzed.
Isolation of RNA and Real-Time PCR
RNA was extracted with TRIzol (ThermoFisher Scientific, Waltham, MA), cDNA was prepared from 1 μg of total RNA using the reverse transcription system (Promega) and analyzed by real-time PCR on a MyIQ real-time PCR machine (Bio-Rad). The following primers were used - glyceraldehyde 3-phosphate dehydrogenase (Gapdh): 5′-GCTGACCTGCTGGATTACATT-3′ (forward), 5′-GTTGAGAGATCATCTCCACCA-3′ (reverse); Tnf-α: 5′-CCCAGGCAGTCAGATCATCTTC-3′ (forward), 5′-GCTTGAGGGTTTGCTACAACATG-3′ (reverse); pro-Il-1β: 5′-CTGGTGTGTGACGTTCCCAT-3′ (forward), 5′-GATTCTTTCCTTTGAGGCCCA-3′ (reverse); inducible NO synthase (iNos): 5′-CGCCTTCAACACCAAGGTTG-3′ (forward), 5′-TGGGGACAGTCTCCATTCCCA-3′; Il-6: 5′-TCAGGAAATTTGCCTATTGAAAATTT-3′ (forward), 5′-GCTTTGTCTTTCTTGTTATCTTTTAAGTTGT-3′ (reverse). The data were calculated as a ratio of expression of gene of interest normalized to that of Gapdh, normalized to medium treatments and expressed as folds induction [12].
Western blot analyses
Cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 5 mM NaF, 2 mM sodium orthovanadate, 1 mM PMSF, 1 × complete protease inhibitors), and resuspended in Laemmli buffer (50 mm Tris-Cl, pH 6.8, 10% glycerol, 2% SDS, 0.1% bromophenol blue, 5% 2-mercaptoethanol), boiled for 5 min, separated on 4–20% polyacrylamide gels (Fisher Scientific, Waltham, MA), transferred to polyvinylidene fluoride membranes (Bio-Rad), blocked, and probed with the respective Abs, as described [12].
Analyses of cytokine secretion and NO release
Organ homogenates and supernatants from cell stimulated with TLR agonists or infected with bacteria were cleared by centrifugation (400 g, 10 min), and cytokine levels were determined by ELISA using commercial kits (BioLegend, San Diego, CA) per manufacturer’s recommendations. The ELISA kits had the following limits of detection: 16 pg/ml (IL-1β), 4 pg/ml (TNF-α, IL-6 and CXCL1) and 31.20 pg/ml (CCL5). Nitrite (NO2−; product of NO breakdown) concentrations in the supernatants were examined by the Griess method [20]. NO2− was quantified using sodium nitrite (NaNO2) as a standard.
Statistical Analysis
Data were processed by the GraphPad Prism software package (GraphPad Software, San Diego, CA), using one-way ANOVA with the Tukey’s post-hoc test to compare multiple experimental groups, or using Student’s t-test to perform pair-way comparisons, and the data were expressed as mean ± standard deviation (SD).
Results
Macrophages expressing IRAK4KI show reduced expression of cytokines upon infection with L. monocytogenes or M. smegmatis
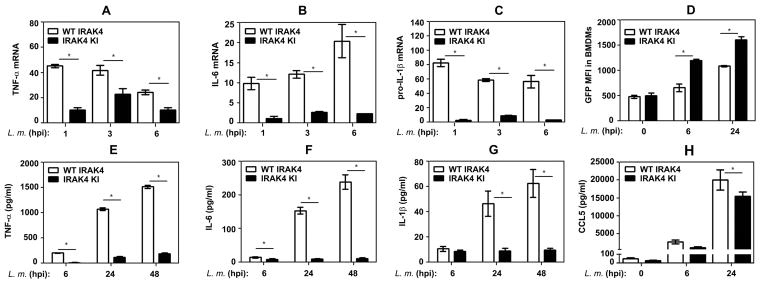
In response to TLR/IL-1R agonists, IRAK4 kinase activity is prerequisite for induction of inflammatory mediators whose expression is regulated by mRNA stability [4, 6, 7, 10–13] and for host resistance against S. pneumoniae [13], an extracellular bacterial pathogen. To explore the role of IRAK4 kinase activity in mounting macrophage cytokine responses during infection with intracellular bacteria, we used mice expressing WT or kinase-deficient (K213M) IRAK4. Infection of BMDMs from WT mice with L. monocytogenes up-regulated levels of TNF-α, IL-6 and pro-IL-1β mRNA by 25–43, 10–20 and 58–62 folds (Fig. 1A–C) and led to robust secretion of TNF-α, IL-6, IL-1β and CCL5 (Fig. 1E–H). In contrast, IRAK4KI BMDMs exhibited 62–91% inhibition of these responses (Fig. 1). Likewise, IRAK4KI BMDMs infected with M. smegmatis secreted markedly decreased (67–95% inhibition) levels of TNF-α, IL-1β, IL-6 and CCL5 compared to robust cytokine responses of macrophages expressing WT IRAK4 (Fig. 2). IRAK4KI BMDMs infected with GFP-expressing L. monocytogenes or M. smegmatis showed similar or higher levels of MFI for GFP compared to WT BMDMs (Fig. 1D, Fig. 2E), and exhibited similar bacterial CFUs immediately after infection (Fig. 5A, B). These data indicate that the differences in cytokine induction between WT and IRAK4KI BMDMs are not due to alterations in the bacterial uptake. Thus, a loss of IRAK4 kinase activity confers deficient expression of inflammatory cytokines by BMDMs infected with L. monocytogenes or M. smegmatis.
Figure 1. Macrophages expressing kinase-inactive IRAK4 exhibit deficient cytokine responses upon infection with L. monocytogenes.
BMDMs from mice expressing WT or IRAK4KI were infected with GFP-expressing L. monocytogenes (MOI 1), washed and incubated for the indicated times. A–C: RNA was isolated, reverse-transcribed and subjected to real-time qPCR with gene-specific primers. D: BMDMs were fixed and subjected to FACS analyses to determine MFI of GFP-positive cells. E–H: Cytokines levels in cell-free supernatants were examined by ELISA. The results (mean ± SD) of a representative experiment (n=3) are depicted. *p<0.05.
Figure 2. Infection with M. smegmatis results in attenuated cytokine responses in macrophages harboring kinase-inactive IRAK4.
After infection with GFP-expressing M. smegmatis (MOI 10), BMDMs were washed and incubated for the indicated times. A–D: Cytokines levels were analyzed in cell-free supernatants by ELISA. E: BMDMs were fixed and analyzed by FACS to determine MFI of GFP-positive cells. The summary data (mean ± SD) of 3 experiments (individual mice indicated as dots) are shown. *p<0.05.
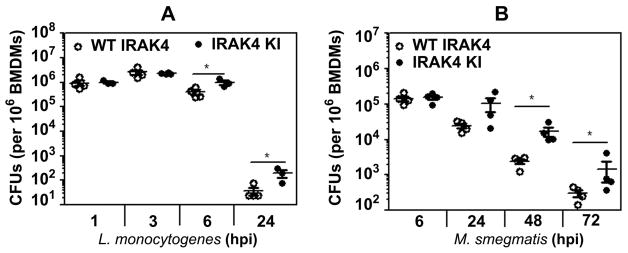
Figure 5. Macrophages harboring IRAK4KI exhibit increased microbial CFUs after infection with L. monocytogenes or M. smegmatis.
BMDMs expressing kinase-sufficient or kinase-deficient IRAK4 were infected with L. monocytogenes (MOI 1) or M. smegmatis (MOI 10). After washing, cells were incubated in gentamycin-containing medium for the indicated times, followed by lysing and plating cell lysates on agar plates to determine bacterial CFUs. The data (mean ± SD) represent a summary of three experiments, with individual mice shown in dots. *p<0.05.
A loss of IRAK4 kinase activity impairs activation of IRAK1, MAPK and NF-κB in macrophages infected with L. monocytogenes or M. smegmatis
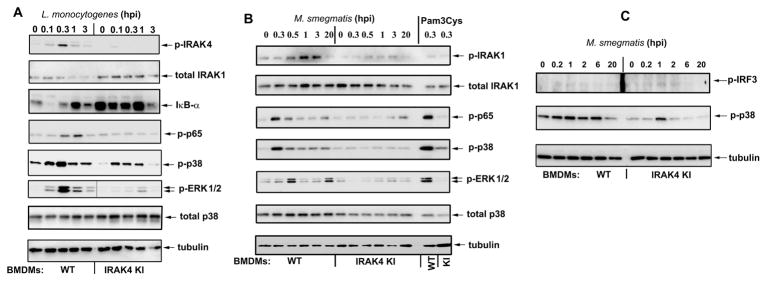
Next, we addressed the biochemical basis for attenuated responses of IRAK4KI macrophages during bacterial infection. WT BMDMs infected with L. monocytogenes showed robust phosphorylation of IRAK4, decreased levels of unmodified IRAK1, degradation of IκB-α, and phosphorylation of p38 MAPK and p65 NF-κB, whereas these responses were markedly attenuated in IRAK4KI BMDMs (Fig. 3A). During infection with M. smegmatis, WT, but not IRAK4KI, BMDMs exhibited efficient phosphorylation of IRAK1, ERK and p38 MAPKs and p65 NF-κB (Fig. 3B). Notably, IRAK4-independent, TRIF-driven [21] phosphorylation of IRF3 was induced comparably in WT and IRAK4KI BMDMs (Fig. 3C). These results indicate that deficiency in IRAK4 kinase activity underlies impaired phosphorylation and modifications of IRAK1, activation of ERK and p38 MAPK, degradation of IκB-α and phosphorylation of p65 NF-κB in BMDMs infected with L. monocytogenes or M. smegmatis but does not inhibit IRF3 activation.
Figure 3. IRAK-4 kinase activity is required for activation of IRAK1, MAPKs and NF-κB, but not IRF3, in BMDMs infected with L. monocytogenes or M. smegmatis.
BMDMs from WT or IRAK4KI mice were infected with L. monocytogenes (A) or M. smegmatis (B, C) (MOI 1 and 10, respectively). After washing, cells were incubated as shown, cell extracts were prepared and examined by Western blot analyses with the indicated Abs. The data of a representative (n=3) experiments are depicted.
L. monocytogenes or M. smegmatis infection of macrophages harboring IRAK4KI leads to impaired NO production
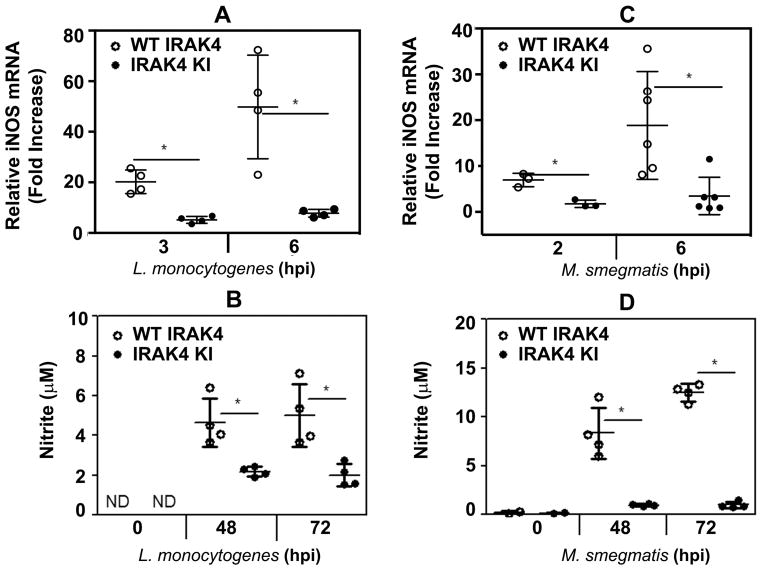
NO is critical for host defense against L. monocytogenes and mycobacteria [18, 20, 22–26], as evidenced by increased susceptibility to infections in mice subjected to pharmacological inhibition of inducible NO synthase or in carriers of polymorphisms rendering the enzyme dysfunctional [24, 27, 28]. While MyD88 is required for NO production [29], the involvement of IRAK4 in triggering NO secretion in response to bacterial infection is unknown. To delineate the role of IRAK4 kinase activity, we examined induction of iNOS mRNA and production of NO by macrophages derived from WT or IRAK4KI-expressing mice during infection with live intracellular bacteria. Figure 4 demonstrates that infection of WT BMDMs with L. monocytogenes (A, B) or M. smegmatis (C, D) markedly increased expression of iNOS mRNA (A, C) and induced NO-dependent nitrite production (B, D), while IRAK4KI macrophages had impaired iNOS mRNA induction and secreted significantly lower levels of NO. Thus, a loss of IRAK4 kinase activity impairs the capacity of macrophages to respond to infection with L. monocytogenes or M. smegmatis by production of NO, suggesting their deficient microbicidal functions.
Figure 4. Deficient induction of iNOS mRNA and secretion of NO in IRAK4KI-expressing macrophages infected with L. monocytogenes or M. smegmatis.
BMDMs obtained from WT or IRAK4KI mice were infected with L. monocytogenes (MOI 1, A, C) or M. smegmatis (MOI 10, B, D), washed and incubated for the indicated times. RNA samples were reverse-transcribed and subjected to real-time PCR analyses of iNOS mRNA expression (A, B). Cell-free supernatants were examined by the Griess assay to determine the levels of nitrite (B, D). The results (mean ± SD) or a representative experiment (n=3) are depicted. *p<0.05.
Increased microbial loads in IRAK4KI BMDMs infected with L. monocytogenes or M. smegmatis
In the next series of experiments, we examined whether deficient NO production by L. monocytogenes- or M. smegmatis-infected IRAK4KI macrophages affects intracellular bacterial replication. Infection of WT and IRAK4KI BMDMs with L. monocytogenes or M. smegmatis resulted in comparable bacterial CFUs immediately after infection and within the initial period of 3 hpi (L. monocytogenes) or 6 hpi (M. smegmatis) (Fig. 5A, B). However, compared to WT macrophages, significantly higher CFUs were recovered from IRAK4KI BMDMs at 6 and 24 hpi with L. monocytogenes (Fig. 5A) and at 24–72 hpi with M. smegmatis (Fig. 5B). These data indicate that deficiency of kinase activity of IRAK4 promotes bacterial replication and survival in macrophages.
Mice harboring IRAK4KI exhibit reduced serum IL-6 and CXCL1 but increased splenic and liver CFUs following systemic infection with M. smegmatis or L. monocytogenes
IRAK4 expression is prerequisite for immune defense against Gram-positive and some Gram-negative bacterial pathogens [15], and IRAK4 activity controls innate immune responses and susceptibility to infection with an extracellular bacterial pathogen S. pneumoniae in mice [13]. However, the role of IRAK4 kinase activity for host resistance against intracellular bacteria in vivo remains elusive. To address this question, we performed i.p. infection of WT and IRAK4KI mice with M. smegmatis or L. monocytogenes and determined levels of serum cytokines and microbial burdens in the spleen and liver. Figure 6 demonstrates that IRAK4KI mice had significantly lower expression of serum IL-6 and CXCL1 (66% and 77% reduction, respectively) compared to their levels observed in WT animals at 6 hpi with M. smegmatis (A). Notably, significantly higher CFUs were detected 48 hpi in the spleen and liver of M. smegmatis-infected IRAK4KI mice compared to WT mice (Fig. 6B). Compared to WT animals, i.p. infection of IRAK4KI mice with L. monocytogenes led to lower levels of CXCL1 (Fig. 6C) and IL6 (data not shown) in the serum, spleen and liver but yielded higher splenic and liver CFUs (Fig. 6D). Thus, during systemic infection with M. smegmatis or L. monocytogenes, a loss of IRAK4 kinase activity curtails induction of IL-6 and CXCL1 while increasing splenic and liver bacterial burdens, indicating a critical role for IRAK4 activity for host defense against intracellular bacteria.
Figure 6. Mice harboring kinase-dead IRAK4 exhibit deficient levels of IL-6 and CXCL1 but increased splenic and liver CFUs following systemic infection with M. smegmatis or L. monocytogenes.
WT or IRAK4KI mice were infected i.p. with M. smegmatis (5 × 106 CFUs, A and B) or L. monocytogenes (2 × 104 CFUs, C and D). A: Serum was obtained 6 hpi and analyzed by ELISA for IL-6 and CXCL1 levels. B and D: 48 hpi, spleens and livers were homogenized and serial dilutions of organ homogenates were plated on the respective agar plates to determine CFUs. C: Organ homogenates were analyzed by ELISA to determine levels of CXCL1. Shown are the results (mean ± SD) of 3 experiments with dots corresponding to individual mice. *p<0.05
Discussion
IRAK4 integrates signaling emanating from the majority of TLRs, except TLR3, to trigger induction of inflammatory cytokine genes, and mediates rapid, transcription-independent activation of NLRP 3 inflammasome [2, 4, 6–9]. Human IRAK4-deficient patients have recurrent pyogenic infection [30, 31], and IRAK4−/− mice succumb to infections with S. aureus or S. pneumoniae [13, 15, 32]. In response to defined IL-1R/TLR ligands, IRAK4 activity is required to induce cytokines whose expression is regulated by mRNA stability, and to optimally activate MAPKs and NF-κB via the TAK1-IKKβ pathway [4, 6, 7, 10–13]. Kinase-inactive IRAK4 adapter triggers MEKK3-dependent activation of IKK-α and IKK-γ, which initiates IκB-α phosphorylation without degradation and induce a subset of cytokines (CCL2, CCL4, CXCL10) and negative regulators (IRAK-M, A20) [4, 6, 7, 10–13]. While IRAK4 kinase deficiency renders mouse macrophages hyporesponsive to extracellular bacteria S. aureus or S. pneumoniae and conferred increased susceptibility to S. pneumoniae infection in mice [13, 15], the role of IRAK4 activity in host responses to intracellular bacteria L. monocytogenes and mycobacteria has been unknown.
This paper shows that macrophages harboring kinase-inactive IRAK4 exhibited deficient activation of IRAK1, MAPKs and NF-κB, and reduced expression of TNF-α, IL-6, IL-1β and, to a lesser extent, CCL5 upon infection with L. monocytogenes or M. smegmatis. Compared to WT cells, IRAK4KI-expressing BMDMs showed equal infectivity at the start of infection and even higher bacterial uptake and survival at later times post infections, indicating that deficient cytokine responses cannot be explained by lower microbial burdens. L. monocytogenes and M. smegmatis express ligands activating TLR2 (mycobacterial lipoarabinomannan, listeria-derived lipoproteins), TLR7 (bacterial ssRNA) or TLR9 (bacterial DNA) [33–37]. These TLRs activate IRAK4 enzymatic activity to induce IRAK4-dependent phosphorylation and activation of IRAK1 [3–5]. In line with the requirement for IRAK4 to activate IRAK1 [3–5], we observed deficient IRAK4 and IRAK1 phosphorylation and reduced IRAK1 modifications (reflected in decreased levels of unmodified IRAK1) in IRAK4KI BMDMs infected with bacteria. Phosphorylated, activated IRAK1 engages TRAF6-TAK1 to interact and activate the IKK complex, mediating activation of MAPKs and NF-κB [4, 6, 7, 10–13]. We show that impaired auto-phosphorylation of IRAK4 and activation of IRAK1 in IRAK4KI BMDMs infected with L. monocytogenes or M. smegmatis was accompanied by deficient phosphorylation of ERK and p38 MAPK, p65 NF-κB and reduced IκB-α degradation.
Of note, WT and IRAK4KI-expressing BMDMs exhibited comparable phosphorylation of TRIF-dependent IRF3, indicating a tailored impact of IRAK4 kinase activity primarily on MyD88-dependent signaling cascades, in line with previous findings [12, 13]. We also demonstrate herein different dependence on IRAK4 kinase activity for the induction of various cytokines in infected macrophages, with strict dependence for TNF-α, IL-6, and IL-1β and lower dependency for CCL5. Sensing of mycobacterial species and L. monocytogenes involves several sensors, e.g. TLR2, TLR3, TLR4, TLR7 and TLR9 that engage MyD88-IRAK4-driven and the TRIF-dependent pathways [20, 22, 33, 37–39]. Since CCL5 can be induced via the TRIF pathway [21] that does not utilize IRAK4, while TNF-α, IL-6 and IL-1β rely primarily on the MyD88-IRAK4 module [40], this could explain the observed differences. Furthermore, IRAK4 kinase activity is required to stabilize mRNA for several cytokines, e.g. TNF-α, IL-6, IL-1β, while CCL5 induction could be less dependent on mRNA stabilization, analogous to other cytokines (CCL2, CCL4, CCL7) [6]. We also do not exclude that IRAK4 kinase activity could uniquely influence epigenetic regulation of various cytokine genes. Studies are ongoing to delineate the mechanisms responsible for differential dependence of various cytokines on IRAK4 kinase activity.
Our data obtained during in vitro infection of BMDMs with L. monocytogenes and M. smegmatis extend observations on the prerequisite role of IRAK4 activity for optimal activation of IRAK1, MAPKs and NF-κB in cells stimulated with TLR/IL-1R agonists [6, 7, 10–13]. However, they are in contrast to a report employing IRAK4 chemical inhibitor in human monocytes that showed a redundant role for IRAK4 kinase activity to mediate phosphorylation of IRAK1, ERK MAPK and p65 NF-κB [41]. It is plausible that differences in cell types (mouse vs human) and/or different inhibitory modes imparted by IRAK4 genetic mutations (K213M) vs. chemical inhibitors could be responsible for the observed differences. In addition, different engagement of TLRs and TLR cooperation with other receptors occurring during stimulation with defined ligands vs infection with bacteria could differentially affect signaling strengths. It is possible that strong TLR signaling could efficiently activate MEKK3 or engage additional pathways (e.g. phosphoinositide-3 kinase) by kinase-inactive IRAK4 adapter, accounting for MAPK and NF-κB activation. Under lower signaling strengths, MEKK3 engagement is impaired or does not occur and activation of the IRAK-1-TRAF6-TAK1 axis is required for optimal responses. Experiments are in progress to address this hypothesis.
Transcription of Tnf-α, Il-6 and Il-1β genes is governed by coordinated activation of MAPK-driven transcription factors (e.g. activator protein-1) and IKK-β-mediated activation of NF-κB and recruitment of transcription factors to the respective cytokine promoters [2]. In line with this notion, we demonstrate that a loss of IRAK4 kinase activity led to impaired activation of MAPK and NF-κB in L. monocytogenes- and M. smegmatis-infected BMDMs and significantly lowered expression of TNF-α, IL-6 and IL-1β. It is likely that during infection with L. monocytogenes or M. smegmatis, the presence of IRAK4KI in macrophages impairs both recruitment of transcription factors to promoters of the respective cytokine genes and activation of the MK2-dependent pathway of stabilization of cytokine mRNAs, as was shown for defined TLR agonists [4]. Reduced production of IL-1β by IRAK4KI-expressig macrophages upon infection with L. monocytogenes or M. smegmatis is likely to result from decreased transcription of pro-IL-1β mRNA. Notably, IRAK4 kinase activity is indispensable for rapid activation of NLRP3 inflammasome that produces active caspase-1 to generate mature IL-1β for secretion [8, 9]. Therefore, we cannot also exclude that impaired activation of NLRP3-driven caspase-1 in IRAK4KI macrophages contributes to reduced IL-1β secretion in response to bacterial infection. Experiments are ongoing to dissect the impact of deficiency in IRAK4 kinase activity on activation of NLRP3 and other inflammasomes during macrophage infection with L. monocytogenes or M. smegmatis.
NO is one of the major bactericidal mediators involved in host defense against L. monocytogenes and mycobacterial species [18, 20, 22-26], and genetic mutations in inducible NO synthase or its pharmacological inhibition have been linked to listeriosis and mycobacterial infections [24, 27, 28]. NO production requires MyD88 [29], but the role of IRAK4 kinase activity in inducing NO during bacterial infections has not been explored. We show significantly impaired capacities of macrophages harboring IRAK4KI to up-regulate iNOS mRNA and to induce NO upon infection with L. monocytogenes or M. smegmatis compared to robust responses exhibited by WT macrophages. To the best of our knowledge, our study is the first showing that IRAK4 kinase activity is prerequisite to NO production during macrophage sensing of bacterial pathogens. Deficient activation of NO production in IRAK4KI-expressing macrophages could be accounted by cell-intrinsic mechanisms. In this scenario, the alternative MEKK3-IKK-γ pathway induced by inactive IRAK4 adapter [4, 10, 11] cannot compensate for the loss of activity of the TAK1-IKK-β signaling axis that requires IRAK4 kinase activity [4, 6, 7, 10–13, 42]. Alternatively, mechanisms of indirect regulation could also play a role, as macrophages devoid of IRAK4 kinase activity show deficient induction of a variety of cytokines, including those implicated in up-regulation of NO production (IL-1β) via NF-κB- and STAT-1-dependent induction of iNOS transcription [43]. Additional studies are required to elucidate the exact molecular basis of impaired NO production in macrophages harboring kinase-inactive IRAK4.
Previous reports demonstrated the requirement of TLR2 and MyD88 for restricting intracellular replication of Coxiella burnetii [44, 45], and enhanced growth of Legionella pneumophilla in the lung of MyD88−/− mice [46]. Furthermore, downregulation of IRAK1 expression via micro-RNA 142-3p was shown to facilitate intracellular replication of M. bovis Bacillus Calmette–Guérin [47]. These findings highlight the importance of the MyD88-IRAK-1 axis in regulating survival of intracellular bacteria in the macrophage. However, the role of IRAK4 kinase activity in restraining intracellular bacteria is poorly understood. This study presents an important observation that deficiency in IRAK4 kinase activity leads to increased survival of intracellular L. monocytogenes and M. smegmatis in the macrophage. Our findings extend a previous publication that revealed higher lung microbial burdens in IRAK-4KI mice infected with extracellular bacterial pathogen S. aureus [13]. Collectively, they show that IRAK4 kinase activity is prerequisite to restrict survival of intracellular bacteria L. monocytogenes and M. smegmatis in the macrophage, possibly, due to its requirement for the induction of NO, one of the important microbicidal mediators. In addition, impaired production of IL-1β by macrophages harboring IRAK4KI could “report” impaired inflammasome activation, which could create an intracellular niche favoring bacterial replication. Finally, impaired MAPK and NF-κB activation by IRAK4KI macrophages could impair activation of autophagy, in turn leading to increased intracellular bacterial survival. Studies are in progress to address the role of these mechanisms in promoting intracellular growth of bacteria in IRAK4KI macrophages.
Pennini et al [13] revealed that IRAK4 kinase activity is prerequisite for innate immune defense against S. pneumoniae and clearance of infection. To uncover the role of IRAK4 kinase activity for infections with intracellular bacteria in vivo, we analyzed serum and organ cytokines and microbial burdens in WT and IRAK4KI mice after systemic infection with M. smegmatis or L. monocytogenes. This paper demonstrates for the first time that a loss of IRAK4 kinase activity underlies deficient induction of serum IL-6 and CXCL1 but increases bacterial CFUs in the spleen and liver following i.p. infection with M. smegmatis or L. monocytogenes.
Taken collectively, our paper shows that a loss of IRAK4 kinase activity leads to impaired production of TNF-α, IL-6, IL-1β, and CCL5 in macrophages infected with L. monocytogenes and M. smegmatis, under conditions of similar bacterial uptake immediately and at early times after infection. We demonstrate for the first time that in response to infection with L. monocytogenes or M. smegmatis, IRAK4KI macrophages exhibited low induction of NO, while showing increased intracellular bacterial survival. These results suggest that triggering of the non-classical pathway of NF-κB activation by kinase-inactive IRAK4 adapter in the macrophage is insufficient for mounting efficient cytokine responses and antibacterial resistance. Finally, we demonstrate deficient induction of serum cytokines and increased bacterial burdens in the spleen and liver in M. smegmatis- or L. monocytogenes-infected mice harboring kinase-dead IRAK4, indicating a critical role for IRAK4 activity for host defense against systemic infection with M. smegmatis or L. monocytogenes in vivo.
Acknowledgments
This study was supported by the National Institutes of Health grant R01 AI059524 (to AEM). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We are thankful to Drs. Kamal Khanna (New York University School of Medicine) and Volker Briken (University of Maryland College Park, College Park, MD) for kindly providing us with GFP-expressing L. monocytogenes and M. smegmatis, respectively. We are grateful to Drs. Kirk Staschke (Eli Lilly, Indianapolis, IN) and Stefanie Vogel (University of Maryland School of Medicine, Baltimore, MD) for providing us with IRAK4KI mice.
Abbreviations
- IL
interleukin
- TLR
Toll-like receptor
- IRAK
IL-1R-associated kinase
- MAPK
mitogen-activated protein kinase
- NF-κB
nuclear factor-κB
- NO
nitric oxide
- TIR
Toll-IL-1 receptor domain
- MyD88
myeloid differentiation primary response protein 88
- IRAK4KI
K213M kinase-inactive IRAK4
- TNF
tumor necrosis factor
- TRAF
TNFR-associated factor
- TAK
transforming growth factor-β-activated kinase
- IKK
inhibitor of NF-κB kinase
- TRIF
TIR domain-containing adapter inducing interferon-β
- IFN
interferon
- IRF
IFN regulatory factor
- ERK
extracellular regulated kinase
- MEKK
MAPK-ERK kinase
- CXCL
CXC motif ligand
- CCL
CC motif ligand
- NLRP
nucleotide-binding domain-like receptor containing pyrin domain
- Pam3Cys
S-[2,3-bis(palmitoyloxy)-(2-RS)-propyl]-N-palmitoyl-(R)-Cys-Ser-Lys4-OH
- GFP
green fluorescent protein
- BMDMs
bone marrow-derived macrophages
- c
complete
- MFI
mean fluorescent intensity
- MOI
multiplicity of infections
- hpi
hours post infection
- iNOS
inducible NO synthase
- CFUs
colony-forming units
- hpi
hours post infection
- WT
wild-type
- Abs
antibodies
- LPS
lipopolysaccharide
- MOI
multiplicity of infection
- MFI
mean fluorescence intensity
- Gapdh
glyceraldehyde 3-phosphate dehydrogenase
- SD
standard deviation
Footnotes
Authorship
GP conducted the experiments with M. smegmatis in vitro and in vivo, analyzed the results, and wrote the manuscript. MM performed macrophage in vitro experiments with L. monocytogenes and, in part, with M. smegmatis. AF and KK conducted experiments to analyze iNOS and cytokine mRNA expression in WT and IRAK4KI BMDMs infected with bacteria and performed infection of mice with L. monocytogenes. KK determined levels of serum and organ cytokines by ELISA. AEM conceived the idea for the project, conceptualized the research, analyzed the data, edited and finalized the manuscript.
Conflict of interest. The authors declare that they have no conflicts of interest with the contents of this article
References
- 1.Medzhitov R, Janeway CA., Jr Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997;91:295–8. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 2.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annual review of immunology. 2015;33:257–90. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ferrao R, Zhou H, Shan Y, Liu Q, Li Q, Shaw DE, Li X, Wu H. IRAK4 dimerization and trans-autophosphorylation are induced by Myddosome assembly. Molecular cell. 2014;55:891–903. doi: 10.1016/j.molcel.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li X. IRAK4 in TLR/IL-1R signaling: possible clinical applications. European journal of immunology. 2008;38:614–8. doi: 10.1002/eji.200838161. [DOI] [PubMed] [Google Scholar]
- 5.Lin SC, Lo YC, Wu H. Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signalling. Nature. 2010;465:885–90. doi: 10.1038/nature09121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraczek J, Kim TW, Xiao H, Yao J, Wen Q, Li Y, Casanova JL, Pryjma J, Li X. The kinase activity of IL-1 receptor-associated kinase 4 is required for interleukin-1 receptor/toll-like receptor-induced TAK1-dependent NFkappaB activation. J Biol Chem. 2008;283:31697–31705. doi: 10.1074/jbc.M804779200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim TW, Staschke K, Bulek K, Yao J, Peters K, Oh KH, Vandenburg Y, Xiao H, Qian W, Hamilton T, Min B, Sen G, Gilmour R, Li X. A critical role for IRAK4 kinase activity in Toll-like receptor-mediated innate immunity. The Journal of experimental medicine. 2007;204:1025–36. doi: 10.1084/jem.20061825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin KM, Hu W, Troutman TD, Jennings M, Brewer T, Li X, Nanda S, Cohen P, Thomas JA, Pasare C. IRAK-1 bypasses priming and directly links TLRs to rapid NLRP3 inflammasome activation. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:775–80. doi: 10.1073/pnas.1320294111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fernandes-Alnemri T, Kang S, Anderson C, Sagara J, Fitzgerald KA, Alnemri ES. Cutting edge: TLR signaling licenses IRAK1 for rapid activation of the NLRP3 inflammasome. Journal of immunology. 2013;191:3995–9. doi: 10.4049/jimmunol.1301681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qin J, Yao J, Cui G, Xiao H, Kim TW, Fraczek J, Wightman P, Sato S, Akira S, Puel A, Casanova JL, Su B, Li X. TLR8-mediated NF-kappaB and JNK activation are TAK1-independent and MEKK3-dependent. J Biol Chem. 2006;281:21013–21. doi: 10.1074/jbc.M512908200. [DOI] [PubMed] [Google Scholar]
- 11.Yao J, Kim TW, Qin J, Jiang Z, Qian Y, Xiao H, Lu Y, Qian W, Gulen MF, Sizemore N, DiDonato J, Sato S, Akira S, Su B, Li X. Interleukin-1 (IL-1)-induced TAK1-dependent Versus MEKK3-dependent NFkappaB activation pathways bifurcate at IL-1 receptor-associated kinase modification. J Biol Chem. 2007;282:6075–6089. doi: 10.1074/jbc.M609039200. [DOI] [PubMed] [Google Scholar]
- 12.Xiong Y, Pennini M, Vogel SN, Medvedev AE. IRAK4 kinase activity is not required for induction of endotoxin tolerance but contributes to TLR2-mediated tolerance. J Leukoc Biol. 2013;94:291–300. doi: 10.1189/jlb.0812401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pennini ME, Perkins DJ, Salazar AM, Lipsky M, Vogel SN. Complete dependence on IRAK4 kinase activity in TLR2, but not TLR4, signaling pathways underlies decreased cytokine production and increased susceptibility to Streptococcus pneumoniae infection in IRAK4 kinase-inactive mice. J Immunol. 2013;190:307–316. doi: 10.4049/jimmunol.1201644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson DL, Ma C, Bergstrom KS, Huang JT, Man C, Vallance BA. MyD88 signalling plays a critical role in host defence by controlling pathogen burden and promoting epithelial cell homeostasis during Citrobacter rodentium-induced colitis. Cellular microbiology. 2008;10:618–31. doi: 10.1111/j.1462-5822.2007.01071.x. [DOI] [PubMed] [Google Scholar]
- 15.Suzuki N, Suzuki S, Duncan GS, Millar DG, Wada T, Mirtsos C, Takada H, Wakeham A, Itie A, Li S, Penninger JM, Wesche H, Ohashi PS, Mak TW, Yeh WC. Severe impairment of interleukin-1 and Toll-like receptor signalling in mice lacking IRAK-4. Nature. 2002;416:750–6. doi: 10.1038/nature736. [DOI] [PubMed] [Google Scholar]
- 16.Pattabiraman G, Panchal R, Medvedev AE. The R753Q polymorphism in Toll-like receptor 2 (TLR2) attenuates innate immune responses to mycobacteria and impairs MyD88 adapter recruitment to TLR2. J Biol Chem. 2017;292:10685–10695. doi: 10.1074/jbc.M117.784470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim YG, Park JH, Shaw MH, Franchi L, Inohara N, Nunez G. The cytosolic sensors Nod1 and Nod2 are critical for bacterial recognition and host defense after exposure to Toll-like receptor ligands. Immunity. 2008;28:246–57. doi: 10.1016/j.immuni.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 18.Beckerman KP, Rogers HW, Corbett JA, Schreiber RD, McDaniel ML, Unanue ER. Release of nitric oxide during the T cell-independent pathway of macrophage activation. Its role in resistance to Listeria monocytogenes. Journal of immunology. 1993;150:888–95. [PubMed] [Google Scholar]
- 19.Anes E, Peyron P, Staali L, Jordao L, Gutierrez MG, Kress H, Hagedorn M, Maridonneau-Parini I, Skinner MA, Wildeman AG, Kalamidas SA, Kuehnel M, Griffiths G. Dynamic life and death interactions between Mycobacterium smegmatis and J774 macrophages. Cellular microbiology. 2006;8:939–60. doi: 10.1111/j.1462-5822.2005.00675.x. [DOI] [PubMed] [Google Scholar]
- 20.Torres D, Barrier M, Bihl F, Quesniaux VJ, Maillet I, Akira S, Ryffel B, Erard F. Toll-like receptor 2 is required for optimal control of Listeria monocytogenes infection. Infect Immun. 2004;72:2131–2139. doi: 10.1128/IAI.72.4.2131-2139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamamoto M, Sato S, Hemmi H, Hoshino K, Kaisho T, Sanjo H, Takeuchi O, Sugiyama M, Okabe M, Takeda K, Akira S. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–3. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 22.Janot L, Secher T, Torres D, Maillet I, Pfeilschifter J, Quesniaux VF, Landmann R, Ryffel B, Erard F. CD14 works with toll-like receptor 2 to contribute to recognition and control of Listeria monocytogenes infection. The Journal of infectious diseases. 2008;198:115–24. doi: 10.1086/588815. [DOI] [PubMed] [Google Scholar]
- 23.Serbina NV, Salazar-Mather TP, Biron CA, Kuziel WA, Pamer EG. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 2003;19:59–70. doi: 10.1016/s1074-7613(03)00171-7. [DOI] [PubMed] [Google Scholar]
- 24.Boockvar KS, Granger DL, Poston RM, Maybodi M, Washington MK, Hibbs JB, Kurlander RL. Nitric-Oxide Produced during Murine Listeriosis Is Protective. Infect Immun. 1994;62:1089–1100. doi: 10.1128/iai.62.3.1089-1100.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weiss G, Schaible UE. Macrophage defense mechanisms against intracellular bacteria. Immunological reviews. 2015;264:182–203. doi: 10.1111/imr.12266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mishra BB, Rathinam VA, Martens GW, Martinot AJ, Kornfeld H, Fitzgerald KA, Sassetti CM. Nitric oxide controls the immunopathology of tuberculosis by inhibiting NLRP3 inflammasome-dependent processing of IL-1beta. Nature immunology. 2013;14:52–60. doi: 10.1038/ni.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gómez LM, Anaya JM, Vilchez JR, Cadena J, Hinojosa R, Vélez L, Lopez-Nevot MA, Martín J. A polymorphism in the inducible nitric oxide synthase gene is associated with tuberculosis. Tuberculosis (Edinb) 2007;87:288–294. doi: 10.1016/j.tube.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 28.Scanga CA, Mohan VP, Tanaka K, Alland D, Flynn JL, Chan J. The inducible nitric oxide synthase locus confers protection against aerogenic challenge of both clinical and laboratory strains of Mycobacterium tuberculosis in mice. Infect Immun. 2001;69:7711–7. doi: 10.1128/IAI.69.12.7711-7717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scanga CA, Bafica A, Feng CG, Cheever AW, Hieny S, Sher A. MyD88-deficient mice display a profound loss in resistance to Mycobacterium tuberculosis associated with partially impaired Th1 cytokine and nitric oxide synthase 2 expression. Infect Immun. 2004;72:2400–4. doi: 10.1128/IAI.72.4.2400-2404.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Picard C, Puel A, Bonnet M, Ku CL, Bustamante J, Yang K, Soudais C, Dupuis S, Feinberg J, Fieschi C, Elbim C, Hitchcock R, Lammas D, Davies G, Al-Ghonaium A, Al-Rayes H, Al-Jumaah S, Al-Hajjar S, Al-Mohsen IZ, Frayha HH, Rucker R, Hawn TR, Aderem A, Tufenkeji H, Haraguchi S, Day NK, Good RA, Gougerot-Pocidalo MA, Ozinsky A, Casanova JL. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299:2076–9. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 31.Medvedev AE, Lentschat A, Kuhns DB, Blanco JC, Salkowski C, Zhang S, Arditi M, Gallin JI, Vogel SN. Distinct mutations in IRAK-4 confer hyporesponsiveness to lipopolysaccharide and interleukin-1 in a patient with recurrent bacterial infections. The Journal of experimental medicine. 2003;198:521–31. doi: 10.1084/jem.20030701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.von Bernuth H, Picard C, Puel A, Casanova JL. Experimental and natural infections in MyD88- and IRAK-4-deficient mice and humans. European journal of immunology. 2012;42:3126–35. doi: 10.1002/eji.201242683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Machata S, Tchatalbachev S, Mohamed W, Jansch L, Hain T, Chakraborty T. Lipoproteins of Listeria monocytogenes are critical for virulence and TLR2-mediated immune activation. Journal of immunology. 2008;181:2028–35. doi: 10.4049/jimmunol.181.3.2028. [DOI] [PubMed] [Google Scholar]
- 34.Stockinger S, Kastner R, Kernbauer E, Pilz A, Westermayer S, Reutterer B, Soulat D, Stengl G, Vogl C, Frenz T, Waibler Z, Taniguchi T, Rulicke T, Kalinke U, Muller M, Decker T. Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes. PLoS pathogens. 2009;5:e1000355. doi: 10.1371/journal.ppat.1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujiwara N, Naka T, Ogawa M, Yamamoto R, Ogura H, Taniguchi H. Characteristics of Mycobacterium smegmatis J15cs strain lipids. Tuberculosis. 2012;92:187–92. doi: 10.1016/j.tube.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Wieland CW, Knapp S, Florquin S, de Vos AF, Takeda K, Akira S, Golenbock DT, Verbon A, van der Poll T. Non-mannose-capped lipoarabinomannan induces lung inflammation via toll-like receptor 2. American journal of respiratory and critical care medicine. 2004;170:1367–74. doi: 10.1164/rccm.200404-525OC. [DOI] [PubMed] [Google Scholar]
- 37.Stamm CE, Collins AC, Shiloh MU. Sensing of Mycobacterium tuberculosis and consequences to both host and bacillus. Immunological reviews. 2015;264:204–19. doi: 10.1111/imr.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bai W, Liu H, Ji Q, Zhou Y, Liang L, Zheng R, Chen J, Liu Z, Yang H, Zhang P, Kaufmann SH, Ge B. TLR3 regulates mycobacterial RNA-induced IL-10 production through the PI3K/AKT signaling pathway. Cell Signal. 2014;26:942–950. doi: 10.1016/j.cellsig.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 39.Aubry C, Corr SC, Wienerroither S, Goulard C, Jones R, Jamieson AM, Decker T, O’Neill LA, Dussurget O, Cossart P. Both TLR2 and TRIF contribute to interferon-β production during Listeria infection. PLoS One. 2012;7:e33299. doi: 10.1371/journal.pone.0033299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kawai T, Adachi O, Ogawa T, Takeda K, Akira S. Unresponsiveness of MyD88-deficient mice to endotoxin. Immunity. 1999;11:115–22. doi: 10.1016/s1074-7613(00)80086-2. [DOI] [PubMed] [Google Scholar]
- 41.Cushing L, Stochaj W, Siegel M, Czerwinski R, Dower K, Wright Q, Hirschfield M, Casanova JL, Picard C, Puel A, Lin LL, Rao VR. Interleukin 1/Toll-like receptor-induced autophosphorylation activates interleukin 1 receptor-associated kinase 4 and controls cytokine induction in a cell type-specific manner. J Biol Chem. 2014;289:10865–75. doi: 10.1074/jbc.M113.544809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conze DB, Wu CJ, Thomas JA, Landstrom A, Ashwell JD. Lys63-linked polyubiquitination of IRAK-1 is required for interleukin-1 receptor- and toll-like receptor-mediated NF-kappaB activation. Mol Cell Biol. 2008;28:3538–3547. doi: 10.1128/MCB.02098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bogdan C. Nitric oxide synthase in innate and adaptive immunity: an update. Trends in immunology. 2015;36:161–78. doi: 10.1016/j.it.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 44.Bradley WP, Boyer MA, Nguyen HT, Birdwell LD, Yu J, Ribeiro JM, Weiss SR, Zamboni DS, Roy CR, Shin S. Primary Role for Toll-Like Receptor-Driven Tumor Necrosis Factor Rather than Cytosolic Immune Detection in Restricting Coxiella burnetii Phase II Replication within Mouse Macrophages. Infect Immun. 2016;84:998–1015. doi: 10.1128/IAI.01536-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zamboni DS, Campos MA, Torrecilhas AC, Kiss K, Samuel JE, Golenbock DT, Lauw FN, Roy CR, Almeida IC, Gazzinelli RT. Stimulation of toll-like receptor 2 by Coxiella burnetii is required for macrophage production of pro-inflammatory cytokines and resistance to infection. J Biol Chem. 2004;279:54405–15. doi: 10.1074/jbc.M410340200. [DOI] [PubMed] [Google Scholar]
- 46.Archer KA, Roy CR. MyD88-dependent responses involving toll-like receptor 2 are important for protection and clearance of Legionella pneumophila in a mouse model of Legionnaires’ disease. Infect Immun. 2006;74:3325–33. doi: 10.1128/IAI.02049-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu G, Zhang Z, Wei J, Zhang Y, Zhang Y, Guo L, Liu X. microR-142-3p down-regulates IRAK-1 in response to Mycobacterium bovis BCG infection in macrophages. Tuberculosis. 2013;93:606–11. doi: 10.1016/j.tube.2013.08.006. [DOI] [PubMed] [Google Scholar]