Abstract
The essential bacteriophage T7-encoded single-stranded DNA binding protein is the nexus of T7 DNA metabolism. Multiple layers of macromolecular interactions mediate its function in replication, recombination, repair, and the maturation of viral genomes. In addition to binding ssDNA, the protein binds to DNA polymerase and DNA helicase, regulating their activities. The protein displays potent homologous DNA annealing activity, underscoring its role in recombination.
Keywords: Bacteriophage T7, Single-stranded DNA binding protein, DNA replication annealing, homologous, recombination
1. Introduction
The apparent simplicity of bacteriophages often belies the complexity of their molecular functions. Bacteriophage T7, a lytic phage of Escherichia coli, is a sterling example of this statement. Its ~40 kb linear, double-stranded DNA (dsDNA) encodes proteins necessary for transcription, replication, recombination, and virion packaging, while relying minimally on host functions, albeit with important exceptions (Figure 1) (1–3). The multifunctional single-stranded DNA (ssDNA) binding protein (gp2.5) of bacteriophage T7 is the hub linking all the major processes of T7 DNA metabolism (4–10).
Figure 1.
Genetic map of the bacteriophage T7 genome. Rectangles represent genes with the essential genes designated by numbers. The early Class I genes, including that for T7 RNA polymerase (gene 1), are transcribed by E. coli RNA polymerase. Afterwards, T7 RNA polymerase transcribes Class II gene, involved in DNA replication, and Class III genes, encoding phage structural proteins.
Two groups initially identified a ~ 25 kDa protein induced upon infection of E. coli with bacteriophage T7 that exhibited strong binding to ssDNA-affinity columns, and which enhanced the activity of T7 DNA polymerase (4–6). Pioneering work by Studier identified the majority of the genes essential for bacteriophage T7 growth by screening lethal amber mutants generated by chemical mutagenesis (7, 8). The gene encoding the T7 DNA-binding protein, however, was not among these essential genes since its nucleotide sequence does not encode suitable amber codons (9). The gene encoding the T7 DNA-binding protein was finally identified after an extensive screen of hundreds of strains isolated after heavy mutagenesis (9). Several strains were found that resulted in a T7 DNA-binding protein with altered electrophoretic mobility. The gene was mapped by genetic crosses coupled to electrophoretic analysis of protein patterns from the resulting recombinant strains. The results showed that the gene encoding the T7 DNA-binding protein was located between gene 2 and gene 3, hence the designation of gene 2.5. When the T7 genome was sequenced shortly after, the genomic location of gene 2.5 led to the identification of the coding sequence (1). Definitive proof of the essential nature of gene 2.5 was obtained form the inability of a recombinant T7 phage bearing a deletion of gene 2.5 to synthesize viral DNA after infecting E. coli (10). Rezende et al. identified residues essential for the function of gp2.5 by screening for lethal mutations in gene 2.5 using a genetic complementation assay with a library of randomly mutagenized plasmids encoding gp2.5 (11). Twenty distinct lethal mutations encoding single amino acid changes were identified in gp2.5 affecting protein dimerization, DNA binding, and homologous DNA annealing (11–13). These latter mutations are discussed in subsequent sections.
Expression of gene 2.5 is driven by T7 RNA polymerase-mediated transcription of class II genes that encode the T7 proteins of DNA replication. Among these gene products are a DNA polymerase, DNA helicase-primase, and both an exonuclease and an endonuclease. Gp2.5 is one of the most abundant T7 proteins and can be detected around 6 minutes after phage infection (1, 2, 9). Gp2.5 is a 25.7-kDa protein of 232 amino acid residues and an isoelectric point of 4. 7. A strong overall negative charge at physiological pH values is a feature shared by many of the T7-encoded proteins involved in nucleic acid metabolism. Homologs of gene 2.5 are present in numerous bacteriophage species and, interestingly, at least four families of nucleo-cytoplasmic large DNA viruses that infect eukaryotes (14, 15).
2. Structure of gp2.5, DNA Binding, and Homologous DNA Annealing
2. 1. Gp2.5 adopts an OB-fold structure
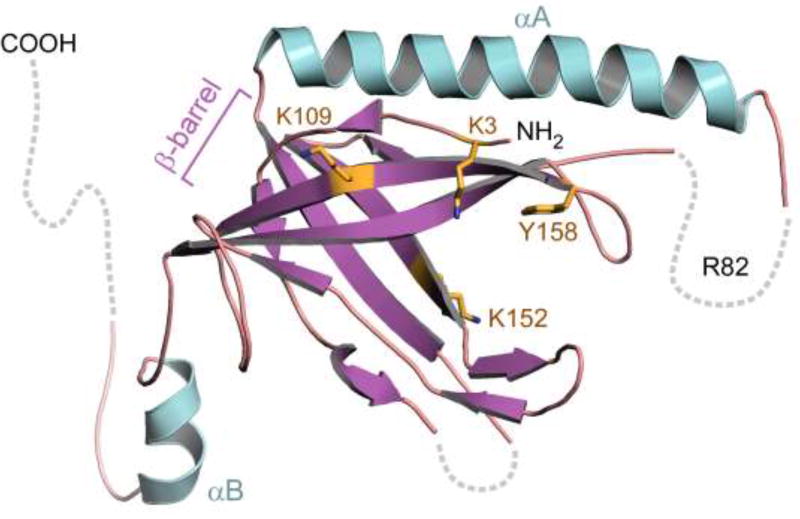
The crystal structure of gp2.5 shows a dimer in which each monomer adopts a conserved oligosaccharide-oligonucleotide binding fold (OB-fold) which consists of a five-stranded anti-parallel β barrel capped by an α-helix on one end (αA) (16) (Figure 2). The structural core of the β-barrel is hydrophobic and solvent-excluding, with loops extending from two β sheets forming a prominent groove on the surface of the fold, the major site of interaction with ssDNA (13). An alpha helix unique to gp2.5, αA, is located between strands β2 and β3 of the OB-fold. αA packs against strands β1 and β3, occluding the interior of the β-barrel. Other ssDNA binding proteins have similar OB-folds but they lack helix αA, instead having a short α-helix that interacts with β strands on the opposite face of the β-barrel (16–18).
Figure 2.
Crystal Structure of T7 gp2.5. Gp2.5 consists of a single OB-fold domain containing an anti-parallel five-stranded β-barrel (purple). Basic and aromatic acid residues implicated in DNA binding (K3, K109, K152, Y158) lie in the β-barrel and are indicated yellow. Alpha helix A (αA, turquoise) is located on the opposite face of the β-barrel relative to other prokaryotic ssDNA binding proteins. R84, a residue critical for the ability of gp2.5 to mediate homologous ssDNA annealing, is located in a disordered loop connecting αA to β3 (dashed line). The C-terminal tail, not present in the construct used for crystallization, is likely disordered and is shown protruding away from the β-barrel (dashed line).
A common strategy of binding by ssDNA binding proteins involves electrostatic and stacking interactions between ssDNA and the OB-fold, as observed in the co-crystal structures of other ssDNA binding proteins with ssDNA (17, 19–21). The structure of gp2.5 does not contain ssDNA and there is little amino acid sequence homology among ssDNA binding proteins members. However, superposition of the OB-folds of ssDNA binding proteins of various species with the OB-fold of gp2.5 reveals conserved aromatic residues that stack against the bases in ssDNA. This finding suggests that gp2.5 binds to ssDNA in a similar manner. Introduction of mutations into OB-fold residues results in decreased affinity for ssDNA and impaired annealing of homologous DNA (Figure 2) (11, 13, 16).
2. 2. An acidic C-terminal tail of gp2.5 and its terminal Phe residue are critical for its interaction with proteins
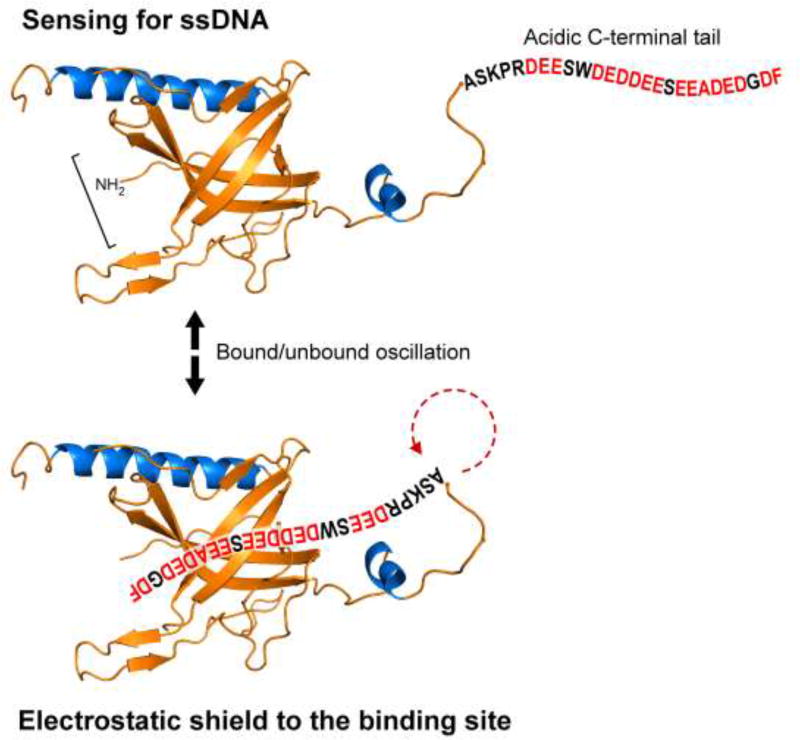
A key molecular feature of gp2.5 is an acidic C-terminal tail, 28 residues in length (Figure 3). While this tail was not present in the construct used for crystallization, there is an abundance of data underscoring its functional significance. The C-terminal tail is essential for phage growth; it modulates the oligomerization of gp2.5 and its interaction with ssDNA, T7 DNA polymerase, and helicase-primase (22–24). A variant of gp2.5 lacking the C-terminal tail (gp2.5Δ26) exhibits altered behavior in size-exclusion chromatography, and a loss of interaction with polymerase and helicase-primase (22). Similar results are obtained if the terminal phenylalanine residue is deleted or substituted with non-aromatic residues (23, 25). Substitution of the negatively charged residues with neutral residues results in alleles incapable of supporting T7 growth even when the terminal phenylalanine is present. Interestingly, the multifunctional gene 4 protein, the helicase-primase, also contains an acidic C-terminal tail with a terminal phenylalanine (26, 27). If either the C-terminal tail or the terminal phenylalanine are altered, the interaction of helicase-primase with DNA polymerase is eliminated, suggesting that gp2.5 and helicase-primase compete for the same binding site on T7 DNA polymerase (26). Chimeric proteins where the C-terminal tails of gp2.5 and the helicase-primase were interchanged support phage growth. Chimeric gp2.5 bearing the helicase-primase C-terminal tail supports wild-type levels of DNA synthesis in vivo but shows lower affinity for T7 DNA polymerase (23, 26). Interestingly, a chimeric gp2.5 containing the C terminus of E. coli SSB is also functional. It supports T7 growth, forms dimers, and interacts with T7 DNA polymerase, likely because E. coli SSB has a similar length, charge distribution, and a C-terminal phenylalanine (23, 28).
Figure 3.
Model for C-terminal tail/DNA-binding switch of gp2.5. The C-terminal tail of gp2.5 binds to the positively-charged DNA-binding cleft, oscillating between bound and bound states. This oscillation allows gp2.5 to sense for the presence of ssDNA and prevents interaction with random negatively charged surfaces. For simplicity, we depict a gp2.5 monomer binding its C-terminal tail through its own DNA-binding surface.
2. 3. Binding to ssDNA
Purified gp2.5 has a lower affinity for ssDNA than other ssDNA binding proteins (19, 20, 29–31). Gp2.5 binds ssDNA with an equilibrium dissociation constant (Kd) of ~ 0. 8 µM as measured by fluorescent titration, a value at least an order of magnitude higher than that for E. coli SSB and T4 gp32. Gp2.5 binds ~7 nucleotides of ssDNA per monomer of protein as shown by (23, 29, 32). Binding does not show appreciable cooperative behavior, again in contrast to the binding behavior of other ssDNA binding proteins (23, 29, 33). The salt dependence of the dissociation constant suggests that gp2.5 utilizes cationic sites as well as aromatic residues for binding ssDNA. Gp2.5 binds dsDNA primarily through electrostatic interactions and diffuses one-dimensionally along dsDNA as shown by single-molecule analysis (34). Although this interaction with dsDNA is 10,000-fold weaker than the interaction with ssDNA, it may facilitate the search of gp2.5 for sites of ssDNA (32–34).
The dimer interface observed in the crystal structure is thought to be stabilized by the reciprocal binding of the C-terminal tail of a gp2.5 monomer to the OB-fold of another gp2.5 monomer, and vice versa (16). This model suggests that binding of the C-terminal tail constitutes a molecular switch between gp2.5 dimerization and ssDNA binding (33, 35, 36). In the absence of ssDNA, the C-terminal tail of one monomer would be bound by the DNA-binding cleft of the other monomer. ssDNA binding would lead to displacement of the C-terminal tail, freeing it for interaction with other replication proteins. Evidence suggests that the C-terminal tail and ssDNA compete for the same binding cleft in gp2.5 (35). A chemically synthesized C-terminal peptide tail can be cross-linked to gp2.5Δ26 (lacking the C-terminal tail). Cross-linking of gp2.5Δ26 to the C-terminal peptide prevents DNA binding, while DNA binding inhibits cross-linking.
Mapping of NMR chemical shifts upon titration of labeled gp2.5Δ26, lacking the C-terminal tail, with either ssDNA or the C-terminal tail peptide showed that both affect the same gp2.5 residues (35). These NMR studies support a model in which both ssDNA and the C-terminal tail bind to the same, or similar, surface of the protein. However, binding of ssDNA was more stable than was binding of the C-terminal peptide, evidenced by the different magnitude of the chemical shift changes. Analysis of the NMR data revealed a rapid dissociation of the C-terminal peptide from gp2.5Δ26 and a slower dissociation from ssDNA (35). Interestingly, addition of increasing amounts of ssDNA leads to a loss of visible spectra, perhaps due to the formation of higher-ordered gp2.5Δ26-ssDNA complexes.
Marintcheva et al. (35) proposed that the C-terminal tail of gp2.5 oscillates between being bound to the DNA-binding cleft and a free state (Figure 3). When gp2.5 binds ssDNA, additional binding energy derived from hydrophobic or stacking interactions between gp2.5 and unpaired bases in the ssDNA further stabilize the complex, leading to displacement of the C-terminal tail. The now free negatively charged C-terminal tail is free to interact with positively charged residues in the segment of T7 DNA polymerase that binds thioredoxin or with T7 helicase-primase (37–40). It is unclear if the active gp2.5 species is a monomer or a dimer, as either could be accommodated based on these observations. Other prokaryotic ssDNA binding proteins, such as E. coli SSB and T4 gp32 have acidic C-terminal tails that regulate their interactions with protein (23). However, the former is a tetramer and the latter is a monomer (19). Binding of gp2.5 induces striking structural rearrangements in ssDNA (12, 29, 41). The electron micrographs of nucleoprotein complexes of gp2.5 and M13 ssDNA in the presence of magnesium show a highly compact structure, in contrast to the open configuration observed with E. coli SSB. Electron micrographs of gp2.5-M13 complexes show indications of intrastrand interactions (12, 24, 29), suggesting that protein-protein interactions between ssDNA-bound gp2.5 could be responsible for placing homologous sequences in close proximity. Interestingly, single molecule force-extension experiments reveal a contour length of ssDNA saturated with gp2.5 that approaches the contour length of dsDNA (41). While these two observations appear contradictory, we envision that the extended structure adopted by ssDNA when bound by gp2.5 likely facilitates search for homologous sequences. Protein-protein interactions between ssDNA-bound gp2.5 observed by electron microscopy are likely transient, and could be potentially perturbed by the forces applied in single-molecule experiments.
2.4. Gp2.5 Facilitates Annealing of Homologous DNA Sequences
Gp2.5 facilitates the annealing of homologous ssDNA 3–4 orders of magnitude over the spontaneous rate and is more efficient than ssDNA binding proteins from other organisms by 1–3 orders of magnitude1 (42). While the molecular mechanism through which gp2.5 accelerates homologous base pairing of DNA is unclear, this process is not due to the elimination of ssDNA secondary structure. E. coli SSB and T4 gp32 bind more tightly to ssDNA and remove secondary structure more efficiently but do not promote annealing as proficiently as gp2.5 (19, 20, 31, 42–45). The annealing of complimentary strands by gp2.5 is specific for DNA; no annealing of complementary RNA is observed. While the annealing reaction is somewhat tolerant to internal mismatches, it is very permissive to internal, unpaired loops1. In contrast to E. coli recA, another protein that promotes ssDNA annealing, gp2.5 does not catalyze strand-invasion, and does not hydrolyze ATP (46).
A gp2.5 variant where arginine 82 is altered to a cysteine (gp2.5 R82C) shows no defect in gp2.5 function in vitro other than an inability to facilitate homologous ssDNA annealing (12). Yet, this altered protein cannot support T7 growth. This finding strongly suggests that the annealing activity of gp2.5 is essential for growth of bacteriophage T7. Arg-82 lies in a disordered loop connecting the characteristic α-helix present in gp2.5 and its homologs (αA) to the β-barrel portion of the structure (Figure 2) (12). It is tempting to speculate that this region is critical for the ability of gp2.5 to mediate homologous ssDNA annealing (14, 15).
The observation that homologous ssDNA annealing activity of gp2.5 is essential for T7 growth reflects the fact that this activity is required at multiple steps in the metabolism of T7 chromosomes (see below). Annealing by gp2.5 is required for the formation of concatemers through base-pairing of the terminal redundancy of two chromosome molecules. Homologous recombination and dsDNA break repair in T7 also are heavily dependent on annealing of homologous ssDNA, and surprisingly, packaging of unit-length T7 genomes into virions also requires the presence of gp2.5 (47–49).
We envision that gp2.5 facilitates annealing of homologous ssDNA through protein-protein interactions between ssDNA-bound gp2.5 molecules. Formation of such a complex would increase the local concentration of ssDNA and promote base-pair formation. The conservation of the αA helix and loop in gp2.5 homologs in phage and eukaryotic viruses suggests that this region is important, perhaps mediating homotypic gp2.5 protein-protein interactions underlying oligomerization-dependent annealing.
3. Role of gp2.5 in DNA replication
Bacteriophage T7 has evolved an efficient and economical mechanism for DNA replication. The relatively large size of the T7 chromosome enables it to encode all but one of its replication proteins (Figure 1) (3) and thus bypass the more complex host replication system upon which smaller phages, such as M13 and ϕX174, are dependent. However, essentially all the proteins and replication mechanisms uncovered in the T7 replication system are applicable to those of more complex organisms (36–38).
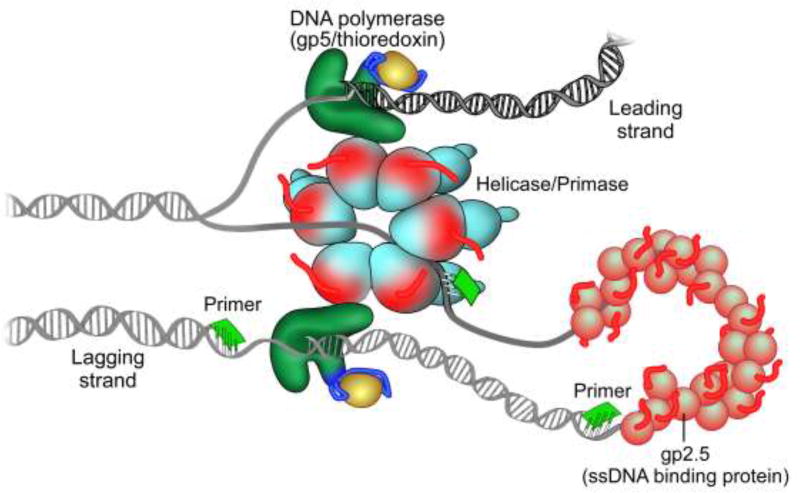
In vitro, only four proteins are required to reconstitute coordinated leading and lagging strand DNA synthesis (Figure 4) (36–38). The hexameric helicase-primase encoded by gene 4 unwinds duplex DNA to provide the ssDNA template for nucleotide polymerization by T7 DNA polymerase. The leading strand is copied continuously by T7 DNA polymerase, a 1:1 complex of gp5 and its processivity factor, E. coli thioredoxin. Thioredoxin binds to a unique segment, the thioredoxin binding domain (TBD), in the thumb domain of the polymerase and increases its affinity for the primer-template. The lagging strand is synthesized discontinuously in segments designated Okazaki fragments, which are later joined by DNA ligase. The primase domain of gp4 catalyzes the synthesis of tetraribonucleotides at specific DNA sequences that are transferred to DNA polymerase to initiate the synthesis of an Okazaki fragment. Finally, gp2.5 binds to any ssDNA at the replication fork and is essential for the coordination of events during synthesis of both strands (50–52). Most likely, its interaction with the polymerase-thioredoxin complex and the helicase-primase are important in establishing coordinated DNA synthesis.
Figure 4.
The T7 Replisome. The hexameric helicase/primase unwinds the DNA duplex to provide the template for DNA synthesis. The leading strand is continuously extended by T7 DNA polymerase in complex with its processivity factor, E. coli Trx, in association with the helicase. The primase synthesizes RNA primers on the lagging strand that are then extended by T7 DNA polymerase/thioredoxin leading to the formation of a replication loop containing a nascent Okazaki fragment. The T7 ssDNA-binding protein, gp2.5, stabilizes ssDNA replication intermediates, interacts with T7 DNA polymerase and helicase/primase, and is essential for the coordination of DNA synthesis on both strands.
3. 1. Gp2.5 interacts with T7 DNA polymerase
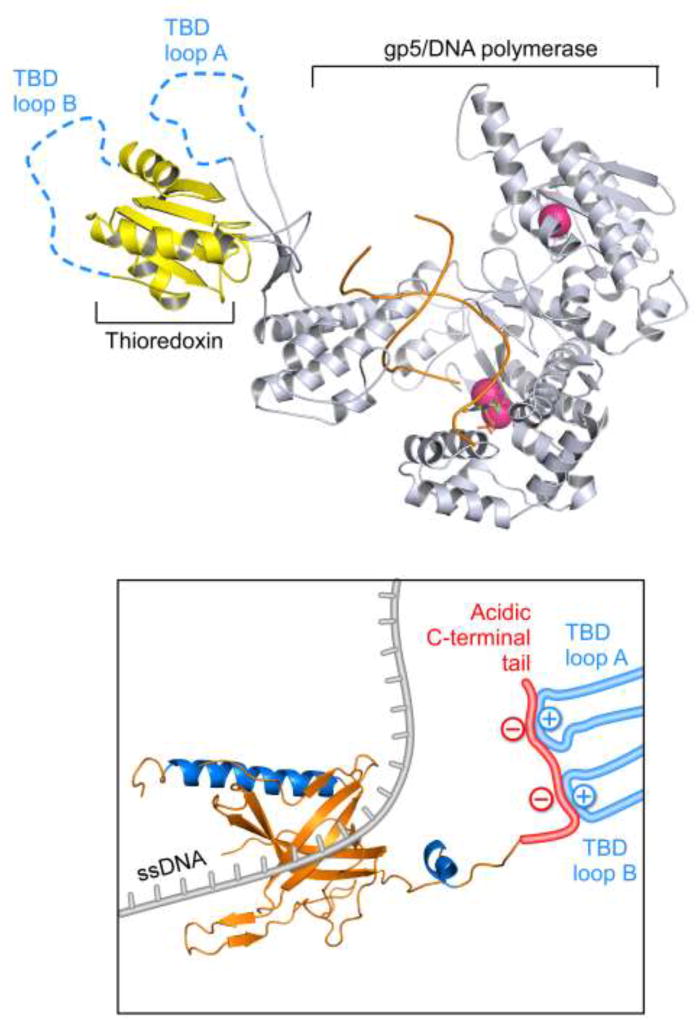
Gp2.5 binds directly to gp5, the polymerase subunit of T7 DNA polymerase to form a 1:1 complex with a Kd of 1 µM (53). Binding of gp2.5 to gp5 is mediated through the C-terminal tail of gp2.5 and the TBD of gp5 (22–24, 28, 39, 53). The interaction between gp2.5 and gp5 does not have an absolute requirement for thioredoxin but the presence of thioredoxin significantly enhances the affinity of gp5 for gp2.5 (39, 53). Thioredoxin interacts extremely weakly with gp2.5 (Kd of ~ 500 µM) suggesting that its effect on the affinity of gp5 for gp2.5 is not merely additive (39). Binding of thioredoxin to gp5 leads to the formation of two basically-charged loops, A and B, within the TBD (Figure 5) (54). The C-terminal tail of gp2.5 interacts with loops A and B electrostatically, as determined by surface plasmon resonance. Deletion of the C-terminal tail from gp2.5 eliminates its interaction with T7 DNA polymerase, as does the substitution of acidic residues with basic or neutral amino acids (22–24, 39, 40). The residues in gp5 that contact the C-terminal tail of gp2.5 also interact with the acidic C-terminal tail of the helicase-primase and are in close proximity to the gp5 DNA-binding groove (39, 55).
Figure 5.
Interaction between the TBD of gp5 and the C-terminal tail of gp2.5. Top panel: Binding of thioredoxin to gp5 leads to the formation of two positively-charged loops, TBD loops A and B (residues 275–285 and residues 299–314 of gp5, blue dashed lines). Bottom panel: When gp2.5 binds ssDNA (gray), its free acidic C-terminal tail (red) interacts electrostatically with TBD loops A and B (blue).
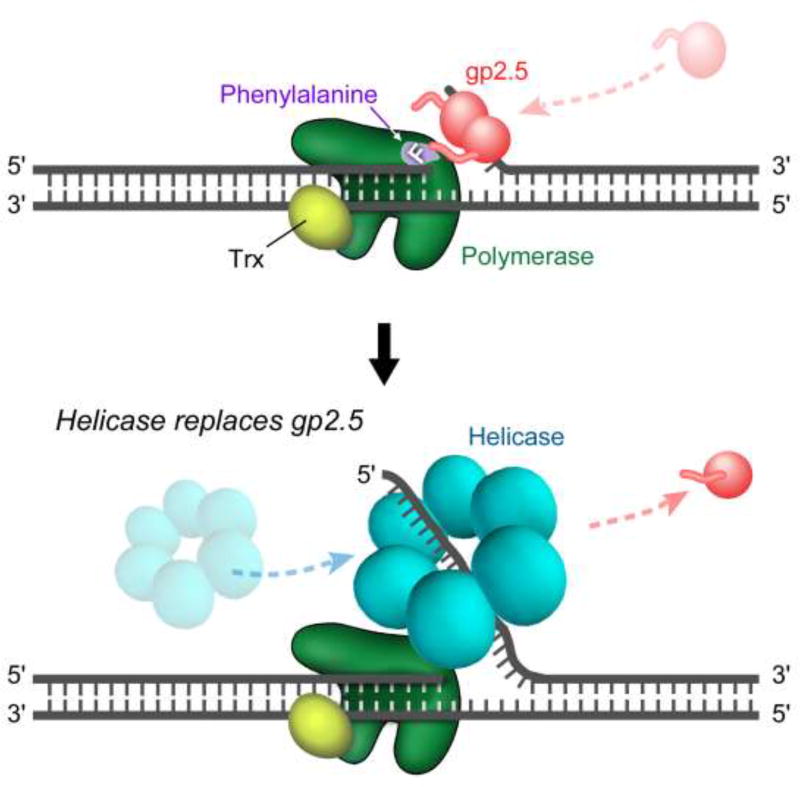
Interestingly, the C-terminal phenylalanine of gp2.5 is critical for interaction with T7 DNA polymerase. Removal of this phenylalanine or replacement with a non-aromatic residue is lethal in vivo and eliminates the interaction between gp2.5 and T7 DNA polymerase in vitro (23, 26). A genetic screen for second-site suppressors of a lethal variant of gp2.5, in which the C-terminal phenylalanine was placed in the penultimate position, identified two gp5 that suppress this lethal phenotype (56). The gp5 suppressor variants, bearing a G371K or an A411T/T258M alteration, are in close proximity to aromatic residues near the DNA binding site, suggesting these aromatic residues are potential sites of interaction between gp5 and the C-terminal phenylalanine of gp2.5 (57). The suppressor mutations identified likely introduce small changes in the hydrophobic binding pocket to accommodate binding of the artificially placed phenylalanine. The suppressor gp5 variants are comparable to wild-type gp5 in their ability to complement T7 phage lacking gene 5, and their polymerase activity. While gp2.5 lacking the C-terminal phenylalanine does not interact with wild-type T7 DNA polymerase, binding is restored by suppressor gp5 variants, indicating that the interaction between gp2.5 and T7 DNA polymerase is essential for phage growth (40, 56).
Wild-type T7 DNA polymerase is unable to initiate DNA synthesis at a nick in dsDNA. The presence of gp2.5, however, enables T7 DNA polymerase to catalyze limited strand-displacement synthesis at this site (Figure 5) (25, 58). Most likely gp2.5 binds to transiently ssDNA arising at the nick and stabilizes the ssDNA. This structure allows T7 DNA polymerase to catalyze strand-displacement synthesis to provide sufficient ssDNA onto which the helicase can load (58, 59). A lethal variant of gp2.5 where the conserved C-terminal phenylalanine is mutated to aspartate does not support initiation of strand displacement DNA synthesis (25). The inability of this variant to allow initiation of strand-displacement synthesis at a nick in DNA underscores the need for an interaction between gp2.5 and T7 DNA polymerase during this process and not merely a role of gp2.5 in stabilizing ssDNA.
The T7 helicase-primase also has an acidic C-terminal tail with a phenylalanine as the terminal residue (26, 27). Chimeric versions where C-terminal tails of gp2.5 and gp4 were swapped fully support T7 phage growth. The chimeric proteins bind to T7 DNA polymerase with similar affinities as the wild-type proteins, but the C-terminal tail of gp2.5 confers slightly increased affinity (23, 26). This observation suggest that the dynamic, mutually exclusive interactions of the helicase-primase and gp2.5 with T7 DNA polymerase, mediated through their C-terminal tails, are critical for coordination of their functions at the replication fork.
3. 2. Interaction of gp2.5 with T7 helicase-primase
The physical interaction between gp2.5 and T7 DNA polymerase is implied by experimental data, but the evidence for a direct interaction between the proteins is lacking. T7 helicase-primase binds to a gp2.5 affinity column under conditions of low ionic strength (53). The interaction between gp2.5 and the helicase-primase appears to require the acidic C-terminal tail of gp2.5. When gp2.5Δ26 is used as a ligand for affinity chromatography, the helicase-primase does not bind (22). Other physical methods to detect protein-protein interactions, such as surface plasmon resonance, have failed to measure an interaction between gp4 and gp2.5. Instead, the interaction between gp2.5 and the T7 helicase-primase has been inferred from the functional effects of gp2.5 on the enzymatic activities of the helicase-primase (24, 28, 51, 58, 60). While the underlying molecular mechanisms remain unknown, the interaction between gp2.5 and the T7 helicase-primase likely facilitates the association of the latter with ssDNA, an interaction that involves the C-terminal tail of gp2.5. For simplicity, in the following two subsections, we will refer to specific subdomains within the full-length T7 helicase-primase polypeptide, and their enzymatic activities, as “helicase” and “primase”.
3. 2. 1 Helicase
The T7 DNA helicase assembles as a hexamer onto linear or circular ssDNA in the presence of dTTP, without the need for loading factors (58, 61). In the absence of dTTP, a condition that does not permit binding of T7 helicase to ssDNA, gp2.5 can function as a helicase-loading factor. Under these conditions, T7 helicase associates with ssDNA coated with gp2.5, but not with ssDNA coated with E. coli SSB (58).
Wild-type gp2.5 modestly stimulates the dTTPase and helicase activities of T7 helicase. Elimination of the C-terminal tail as in gp2.5Δ26, abolishes both activities at concentrations where the ssDNA is saturated with gp2.5 (24).
3. 2. 2 Primase
Gp2.5 increases the initiation of RNA-primed DNA synthesis by T7 helicase-primase and T7 DNA polymerase using M13 ssDNA as a template (24, 58). Gp2.5 enhances the efficiency of oligoribonucleotide synthesis by T7 helicase-primase on ssDNA by reducing the dissociation of dimer intermediates from the helicase-primase, thereby increasing the proportion of full-length primers produced (52). Gp2.5 inhibits the proportion of primers extended approximately 2-fold. This inhibitory effect may be due to the sequestration of primase recognition sites by gp2.5 and/or the anchoring of the polymerase distal to the site of primer synthesis. However, fully-extended products accumulate with faster kinetics than in reactions lacking gp2.5, or those that contain either E. coli SSB or gp2.5Δ26 (52). A competition between the C-terminal tails of gp2.5 and the helicaseprimase for the TBD of the polymerase likely represents a molecular switch that enables the replisome to position the lagging strand polymerase in a configuration leading to formation of a priming complex.
3. 3. Gp2.5 coordinates DNA synthesis at the replication fork
Gp2.5 is indispensable for the coordination of leading- and lagging-strand DNA synthesis at the replication fork (50–52). In vitro analysis of the synthesis of leading-and lagging-strands occurring simultaneously can be accomplished using small, synthetic DNA minicircles of ~70 bp. The base sequence of DNA minicircles can be selectively designed to allow for the quantitative monitoring of DNA synthesis of each strand. For example by having guanosines only in one strand and cytosines in the other synthesis of leading and lagging strands can be monitored by the incorporation of radioactive dCMP or dGMP. In addition, the number and location of primase recognition sites can be specified.
Under conditions of coordinated DNA synthesis on the minicircle, the leading strand is extended continuously to sizes greater than 10 kb. The lagging strand is replicated as multiple Okazaki fragments with an average size of 1 kb (36–38). The consistency in Okazaki fragment size is remarkable because the replisome would encounter a primase recognition site on the minicircle on average, every 35 bp (50, 51). Okazaki fragments in the absence of gp2.5, or in the presence of gp2.5Δ26, show a distribution of sizes skewed towards species shorter than 0.5 kb (50, 51, 62). The periodicity in the extension pattern when reaction products are analyzed by denaturing PAGE strongly suggests that these products represent primer extension initiation/termination events, which are indicative of an over-utilization of primase recognition sites. Thus, the presence of gp2.5 tempers the levels of primer utilization, which leads to a bypass of priming events (52, 63). This bypass mechanism may allow leading- and lagging-strand DNA synthesis to proceed at comparable rates.
3. 4 Role of gp2.5 in initiation of DNA replication
The primary origin of initiation of DNA synthesis has been mapped to a 129 bp segment of DNA between positions 14.75 and 15.0, approximately 15% of the distance from the left end of the T7 DNA molecule (64). Sequence analysis (65) has shown that this region contains tandem promoters for T7 RNA polymerase that direct RNA synthesis rightward. No coding sequence is present between the promoters but an RNase III cleavage site is present in the RNA transcribed from this region. Immediately to the right of the promoters is a 61 bp AT-rich (79%) known to be essential for initiation at this origin (64).
Initiation of DNA replication can be reconstituted using T7 RNA polymerase, T7 DNA polymerase, T7 helicase-primase, and either T7 DNA or a T7 origin-containing plasmid DNAs (66, 67). On the origin-containing plasmid DNA synthesis is unidirectional proceeding in a rightward direction with respect to the T7 genetic map, the same direction as transcription by the T7 RNA polymerase (68). The product DNA has 10 to 60 ribonucleotides covalently linked to their 5’-termini. These RNA primers arise from each of the two promoters located in the origin region.
Initiation of DNA synthesis in the opposite direction requires the presence of a ssDNA binding protein (68). Both gp2.5 and E. coli SSB promote leftward synthesis but, interestingly, T4 gene 32 protein does not. Both T7 gp2.5 and E. coli SSB stimulate rightward synthesis. What is the role of ssDNA-binding protein in initiation of leftward synthesis? Transcription from the two promoters exposes sufficient ssDNA for the helicase to load and unwind DNA in the rightward direction. In order for helicase to load onto the opposite strand requires exposure of ssDNA to the left of the origin region. It has been proposed that ssDNA binding protein participates in this event.
3. 5. Gp2.5 is required for concatemer formation
Replication of T7 DNA proceeds via the formation of concatemers, head-to-tail arrangements of genome length DNA joined through a 160 bp region of complementarity called the terminal redundancy (3, 69). Since the T7 genome is linear, the first round of replication produces two DNA products containing 3’ ssDNA extensions. The 3’ ssDNA extensions of two molecules can anneal to form a linear concatemer (70, 71). The formation of concatemers requires gp2.5 and the 5’-3’ exonuclease (gp6) encoded by T7 (7). The direct role of gp2.5 in the annealing process is discussed below. Linear concatemers of increasing size can be detected early after infection, followed by the appearance of a nucleoprotein complex containing over 100 phage genome equivalents, with multiple regions of ssDNA which is thought to consist of actively replicating and recombining genomes (71–74). These recombination intermediates are likely converted into replication forks, and the complex DNA structure is eventually resolved by the T7 encoded endonuclease, gp3, to linear concatemers in preparation for packaging (75–77).
4. Role of gp2.5 in DNA Recombination, Repair, and Packaging
4. 1. Recombination
Homologous recombination in bacteria infected by bacteriophage T7 is extensive and extremely efficient (64–67). Although it is independent of host recombination pathways, the mechanism used in T7-infected cells is still unknown. Studies with extracts from T7 mutants show a requirement for T7 DNA polymerase, helicase-primase, gene 6 exonuclease, gene 3 endonuclease, and gp2.5 (78–80). Gp2.5 is likely critical for T7 recombination, as it efficiently mediates annealing of complementary ssDNA (see below) (26, 27. 42, 69). In conjunction with T7 helicase-primase the two proteins mediate DNA transfer between homologous strands (see below (70, 71).
4. 1. 2. Gp2.5 and T7 helicase-primase mediate homologous strand exchange
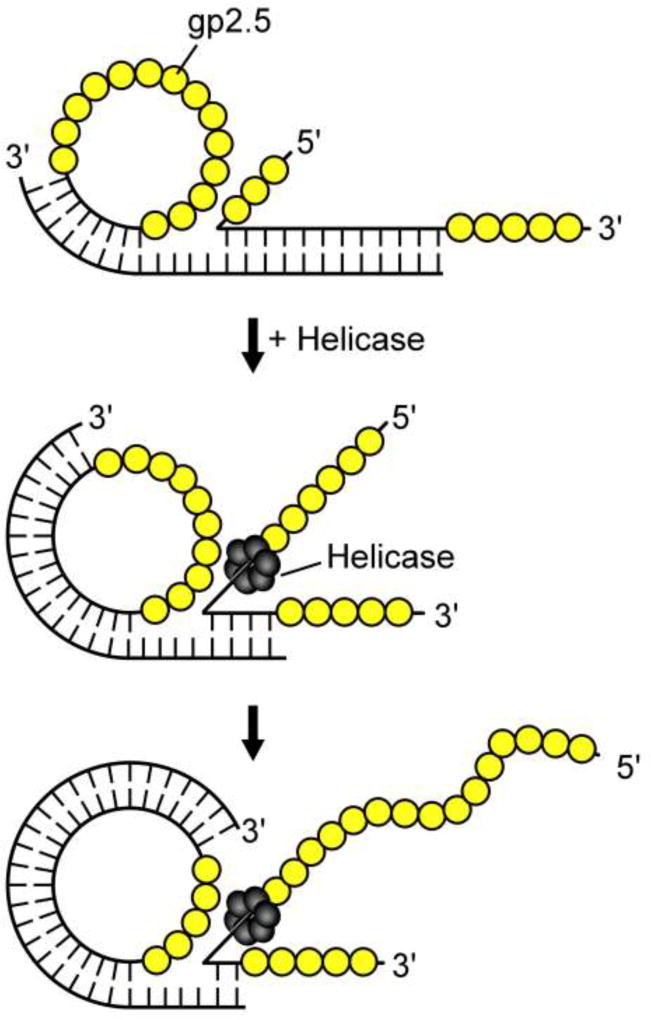
Gp2.5, in coordination with the T7 helicase-primase, mediates the transfer of strands between homologous DNA molecules (Figure 6) (81). In this process, gp2.5 stimulates the annealing of ssDNA between regions of homologous ssDNA, resulting in the formation of a hybrid molecule. T7 DNA helicase then unwinds the complementary strand to mediate strand transfer. This process requires an interaction between gp2.5 and DNA helicase, as gp2.5Δ26 cannot mediate strand transfer, even though it binds DNA and mediates annealing of homologous ssDNA. Strand transfer is tolerant to the presence of thymine dimers and short regions of non-homology (82, 83).
Figure 6.
Synthesis at a nick in dsDNA is facilitated by gp2.5. WT T7 DNA polymerase is unable to initiate DNA synthesis at a nick in dsDNA. Top panel: T7 DNA polymerase idles at the nick by alternating between 3’-5’ exonuclease and nucleotide polymerization activities. An interaction between gp2.5 and T7 DNA polymerase allows limited strand-displacement synthesis. This interaction is mediated, in part, by the C-terminal phenylalanine of gp2.5. In addition, gp2.5 likely stabilizes the ssDNA produced. Bottom panel: The helicase hexamer assembles on the displaced 5’-single-stranded tail when it is sufficiently long, and displaces gp2.5. Helicase and T7 DNA polymerase form a stable complex and continue DNA synthesis.
A possible mechanism for homologous recombination in T7-infected cells could involve a process similar to the recombination-dependent DNA replication occurring in phage T4 (3, 84–86). During this process, single-stranded 3’ ends generated by resection by T7 gene 6 5’-3’ exonuclease invade a double-stranded region and anneal to homologous strands, a process mediated by gp2.5 and T7 DNA helicase. After their replication, an enzyme such as T7 gp3 endonuclease resolves the complex recombination intermediates.
4. 2. Repair
4. 2. 1. dsDNA break repair
In prokaryotic systems, broken chromosome ends are predominantly repaired by homologous recombination (87). Repair of dsDNA breaks in T7-infected cells is quite efficient and proceeds through a recombination homologous-repair mechanism, which involves the incorporation of a donor DNA fragment into the damaged chromosome (88, 89). This process is independent of the E. coli RecA pathway and is carried out almost exclusively by T7 proteins, with gp2.5 being essential (47, 90, 91). A gene 2.5 deletion mutant blocks repair of dsDNA breaks, but inactivating mutations in T7 helicase-primase have no effect, suggesting that DNA strand transfer is not required (47).
Repair of a gap in dsDNA is accomplished by the formation of 3’ single-stranded tails through the 5’-3’ exonucleolytic digestion of double-stranded donor DNA and the broken acceptor DNAs by T7 exonuclease. Annealing of ssDNA segments bearing sequence homology by gp2.5 follows this event. T7 DNA polymerase (or E. coli DNA polymerase I) then fills unextended regions, followed by ligation, completing the repair (47, 88–92).
4. 2. 2. Nicks in dsDNA and lesions to DNA bases
T7 bacteriophage does not encode a specialized DNA base repair system and its growth is impaired in E. coli defective in DNA repair (93). However, mutagenized T7 genomes can produce successful infection when transfected into E. coli, indicating that it can utilize the host repair mechanisms (94–98). In addition, recent evidence indicates that some DNA lesions/adducts can be overcome by an intrinsic translesion synthesis ability of T7 DNA polymerase, when coupled to helicase, to replicate through sites of damage that would otherwise be inhibitory (99, 100). Helicase coupled-T7 DNA polymerase can bypass single-stranded nicks in dsDNA and the presence of gp2.5 enhances this activity (25, 100). Neither T7 DNA polymerase nor helicase can bypass a nick individually. Upon bypass, gp2.5 promotes initiation of strand-displacement DNA synthesis by binding to the displaced ssDNA generated by the helicase-polymerase complex (25, 100). Helicase coupled-T7 DNA polymerase can also tolerate and directly overcome leading-strand template lesions, which are thought to present the most potent barrier to replication fork progression, but any role gp2.5 may play in this process is yet to be examined. (99).
4. 3. Packaging
Phage capsid and maturation proteins functionally cooperate with DNA replication proteins to process concatemers and package genomes into virions (48, 101–105). Processing of concatemers involves the introduction of nicks at the terminal redundancy junction, strand displacement synthesis, and conversion of the single-stranded regions to dsDNA (3, 105). One can envision a role of gp2.5 in conjunction with T7 DNA polymerase and DNA helicase in this complex process. In support of this notion, in vitro packaging reactions that lack gp2.5 yield abnormally low titers of phage, underscoring its importance in this process (47, 49).
5. Homologs of gp2.5
Bioinformatic analyses indicate that a large number of bacteriophage genomes encode proteins with homology to gp2.5 (14, 15). These putative gp2.5 homologs share a similar gene location in the viral genome. Also, their sequence suggests the presence the distinctive αA between strands β2 and β3 of the OB-fold, as well as residues know to be functionally important for gp2.5 function, including highly acidic residues at the C-terminus of the polypeptide. Homologs of gp2.5 are not limited to phages related to T7, or even prokaryotic systems (14). Homologs of T7 gp2.5 are found in four families of nucleo-cytoplasmic large DNA viruses (NCLDVs): Phycodnaviridae, Mimiviridae, Iridoviridae and Marseillevirus. These viruses feature some of the largest viral genomes known and do not depend on host factors for DNA replication (106). This observation is perhaps not too surprising considering the proposal that eukaryotic dsDNA viruses are directly descended from bacteriophages (107, 108). In addition, ICP8 the major ssDNA binding protein from herpes simplex virus-1 is also distantly related to gp2.5, sharing a common ancestor (14). Like gp2.5, ICP8 can anneal ssDNA and it is essential for replication of leading- and lagging-strands using minicircle DNA in vitro (109). However, only the middle segment of ICP8 shares any significant homology to gp2.5 and the two differ in their oligomerization state.
6. Conclusion
Despite the progress made in elucidating the complex layers of interactions between gp2.5 and its macromolecular partners, much work remains in order to understand its various functions. How gp2.5 coordinates the synthesis of leading- and lagging-DNA strands at the replication fork is unknown. Given its central role in DNA synthesis, repair, recombination, and packaging, it is likely that gp2.5 regulates these processes as well, particularly in the higher-order nucleoprotein DNA factories observed in vivo. The prodigious acceleration of homologous DNA annealing by gp2.5 is essential to its functions, yet we still lack an adequate understanding of the molecular events occurring during this process. Likewise, the role of gp2.5 in the coordination of leading and lagging strand synthesis is critical but the molecular interactions involved are not known. For example, the role of the essential C-terminal phenylalanine of gp2.5 is yet to be elucidated. Finally, investigating the numerous gp2.5 homologs found in other organisms will shed light into the evolution and function of this ssDNA-binding protein family.
Figure 7.
Strand-exchange of homologous DNA facilitated by T7 helicase and gp2.5. Top panel: Gp2.5 (yellow) binds single-stranded regions of two partner DNA molecules and promotes their homologous base pairing to form a stable hybrid molecule. Middle panel: The helicase hexamer (dark gray) binds to the 5’-single-stranded tail, translocates in a 5’-3’ direction, and unwinds duplex DNA. Bottom panel: ssDNA formed behind the moving helicase anneals to the complementary strand on the circle as it is unwound.
Acknowledgments
We thank Steven Moskowitz (Advanced Medical Graphics) for illustrations. This work was supported by NIH F32GM101761 (A.J.H.), GM54397, and patent royalty (C.C.R.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Hernandez, A.J. and Richardson, C.C. Unpublished results.
Bibliography
- 1.Dunn JJ, Studier FW. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 2.Studier FW, Dunn JJ. Organization and expression of bacteriophage T7 DNA. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):999–1007. doi: 10.1101/sqb.1983.047.01.114. [DOI] [PubMed] [Google Scholar]
- 3.Calendar RL, Abedon ST. The Bacteriophages. Second. Oxford University Press; New York, New York: 2006. p. 768. [Google Scholar]
- 4.Reuben RC, Gefter ML. A DNA-binding protein induced by bacteriophage T7. Proc Natl Acad Sci U S A. 1973;70(6):1846–1850. doi: 10.1073/pnas.70.6.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scherzinger E, Litfin F, Jost E. Stimulation of T7 DNA polymerase by a new phage-coded protein. Mol Gen Genet. 1973;123(3):247–262. doi: 10.1007/BF00271243. [DOI] [PubMed] [Google Scholar]
- 6.Reuben RC, Gefter ML. A deoxyribonucleic acid-binding protein induced by bacteriophage T7. Purification and properties of the protein. J Biol Chem. 1974;249(12):3843–3850. [PubMed] [Google Scholar]
- 7.Studier FW. The genetics and physiology of bacteriophage T7. Virology. 1969;39(3):562–574. doi: 10.1016/0042-6822(69)90104-4. [DOI] [PubMed] [Google Scholar]
- 8.Studier FW. Bacteriophage T7. Science. 1972;176(4033):367–376. doi: 10.1126/science.176.4033.367. [DOI] [PubMed] [Google Scholar]
- 9.Studier FW. Identification and mapping of five new genes in bacteriophage T7. J Mol Biol. 1981;153(3):493–502. doi: 10.1016/0022-2836(81)90404-6. [DOI] [PubMed] [Google Scholar]
- 10.Kim YT, Richardson CC. Bacteriophage T7 gene 2.5 protein: an essential protein for DNA replication. Proc Natl Acad Sci U S A. 1993;90(21):10173–10177. doi: 10.1073/pnas.90.21.10173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rezende LF, Hollis T, Ellenberger T, Richardson CC. Essential amino acid residues in the single-stranded DNA-binding protein of bacteriophage T7. Identification of the dimer interface. J Biol Chem. 2002;277(52):50643–50653. doi: 10.1074/jbc.M207359200. [DOI] [PubMed] [Google Scholar]
- 12.Rezende LF, Willcox S, Griffith JD, Richardson CC. A single-stranded DNA-binding protein of bacteriophage T7 defective in DNA annealing. J Biol Chem. 2003;278(31):29098–29105. doi: 10.1074/jbc.M303374200. [DOI] [PubMed] [Google Scholar]
- 13.Hyland EM, Rezende LF, Richardson CC. The DNA binding domain of the gene 2.5 single-stranded DNA-binding protein of bacteriophage T7. J Biol Chem. 2003;278(9):7247–7256. doi: 10.1074/jbc.M210605200. [DOI] [PubMed] [Google Scholar]
- 14.Kazlauskas D, Venclovas C. Two distinct SSB protein families in nucleo-cytoplasmic large DNA viruses. Bioinformatics. 2012;28(24):3186–3190. doi: 10.1093/bioinformatics/bts626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopes A, Amarir-Bouhram J, Faure G, Petit MA, Guerois R. Detection of novel recombinases in bacteriophage genomes unveils Rad52, Rad51 and Gp2.5 remote homologs. Nucleic Acids Res. 2010;38(12):3952–3962. doi: 10.1093/nar/gkq096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hollis T, Stattel JM, Walther DS, Richardson CC, Ellenberger T. Structure of the gene 2.5 protein, a single-stranded DNA binding protein encoded by bacteriophage T7. Proc Natl Acad Sci U S A. 2001;98(17):9557–9562. doi: 10.1073/pnas.171317698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shamoo Y, Friedman AM, Parsons MR, Konigsberg WH, Steitz TA. Crystal structure of a replication fork single-stranded DNA binding protein (T4 gp32) complexed to DNA. Nature. 1995;376(6538):362–366. doi: 10.1038/376362a0. [DOI] [PubMed] [Google Scholar]
- 18.Raghunathan S, Ricard CS, Lohman TM, Waksman G. Crystal structure of the homo-tetrameric DNA binding domain of Escherichia coli single-stranded DNA-binding protein determined by multiwavelength x-ray diffraction on the selenomethionyl protein at 2.9-A resolution. Proc Natl Acad Sci U S A. 1997;94(13):6652–6657. doi: 10.1073/pnas.94.13.6652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chase JW, Williams KR. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- 20.Lohman TM, Ferrari ME. Escherichia coli single-stranded DNA-binding protein: multiple DNA-binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–570. doi: 10.1146/annurev.bi.63.070194.002523. [DOI] [PubMed] [Google Scholar]
- 21.Raghunathan S, Kozlov AG, Lohman TM, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7(8):648–652. doi: 10.1038/77943. [DOI] [PubMed] [Google Scholar]
- 22.Kim YT, Richardson CC. Acidic carboxyl-terminal domain of gene 2.5 protein of bacteriophage T7 is essential for protein-protein interactions. J Biol Chem. 1994;269(7):5270–5278. [PubMed] [Google Scholar]
- 23.Marintcheva B, Hamdan SM, Lee SJ, Richardson CC. Essential residues in the C terminus of the bacteriophage T7 gene 2.5 single-stranded DNA-binding protein. J Biol Chem. 2006;281(35):25831–25840. doi: 10.1074/jbc.M604601200. [DOI] [PubMed] [Google Scholar]
- 24.He ZG, Rezende LF, Willcox S, Griffith JD, Richardson CC. The carboxyl-terminal domain of bacteriophage T7 single-stranded DNA-binding protein modulates DNA binding and interaction with T7 DNA polymerase. J Biol Chem. 2003;278(32):29538–29545. doi: 10.1074/jbc.M304318200. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh S, Marintcheva B, Takahashi M, Richardson CC. C-terminal phenylalanine of bacteriophage T7 single-stranded DNA-binding protein is essential for strand displacement synthesis by T7 DNA polymerase at a nick in DNA. J Biol Chem. 2009;284(44):30339–30349. doi: 10.1074/jbc.M109.024059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee SJ, Marintcheva B, Hamdan SM, Richardson CC. The C-terminal residues of bacteriophage T7 gene 4 helicase-primase coordinate helicase and DNA polymerase activities. J Biol Chem. 2006;281(35):25841–25849. doi: 10.1074/jbc.M604602200. [DOI] [PubMed] [Google Scholar]
- 27.Notarnicola SM, Mulcahy HL, Lee J, Richardson CC. The acidic carboxyl terminus of the bacteriophage T7 gene 4 helicase/primase interacts with T7 DNA polymerase. J Biol Chem. 1997;272(29):18425–18433. doi: 10.1074/jbc.272.29.18425. [DOI] [PubMed] [Google Scholar]
- 28.Kong D, Richardson CC. Role of the acidic carboxyl-terminal domain of the single-stranded DNA-binding protein of bacteriophage T7 in specific protein-protein interactions. J Biol Chem. 1998;273(11):6556–6564. doi: 10.1074/jbc.273.11.6556. [DOI] [PubMed] [Google Scholar]
- 29.Kim YT, Tabor S, Bortner C, Griffith JD, Richardson CC. Purification and characterization of the bacteriophage T7 gene 2.5 protein. A single-stranded DNA-binding protein. J Biol Chem. 1992;267(21):15022–15031. [PubMed] [Google Scholar]
- 30.Ferrari ME, Bujalowski W, Lohman TM. Co-operative binding of Escherichia coli SSB tetramers to single-stranded DNA in the (SSB)35 binding mode. J Mol Biol. 1994;236(1):106–123. doi: 10.1006/jmbi.1994.1122. [DOI] [PubMed] [Google Scholar]
- 31.Kowalczykowski SC, Lonberg N, Newport JW, von Hippel PH. Interactions of bacteriophage T4-coded gene 32 protein with nucleic acids. I. Characterization of the binding interactions. J Mol Biol. 1981;145(1):75–104. doi: 10.1016/0022-2836(81)90335-1. [DOI] [PubMed] [Google Scholar]
- 32.Shokri L, Marintcheva B, Richardson CC, Rouzina I, Williams MC. Single molecule force spectroscopy of salt-dependent bacteriophage T7 gene 2.5 protein binding to single-stranded DNA. J Biol Chem. 2006;281(50):38689–38696. doi: 10.1074/jbc.M608460200. [DOI] [PubMed] [Google Scholar]
- 33.Shokri L, Rouzina I, Williams MC. Interaction of bacteriophage T4 and T7 single-stranded DNA-binding proteins with DNA. Phys Biol. 2009;6(2):025002. doi: 10.1088/1478-3975/6/2/025002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shokri L, et al. Kinetics and thermodynamics of salt-dependent T7 gene 2.5 protein binding to single- and double-stranded DNA. Nucleic Acids Res. 2008;36(17):5668–5677. doi: 10.1093/nar/gkn551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marintcheva B, Marintchev A, Wagner G, Richardson CC. Acidic C-terminal tail of the ssDNA-binding protein of bacteriophage T7 and ssDNA compete for the same binding surface. Proc Natl Acad Sci U S A. 2008;105(6):1855–1860. doi: 10.1073/pnas.0711919105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamdan SM, Richardson CC. Motors, switches, and contacts in the replisome. Annu Rev Biochem. 2009;78:205–243. doi: 10.1146/annurev.biochem.78.072407.103248. [DOI] [PubMed] [Google Scholar]
- 37.Lee SJ, Richardson CC. Choreography of bacteriophage T7 DNA replication. Curr Opin Chem Biol. 2011;15(5):580–586. doi: 10.1016/j.cbpa.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kulczyk AW, Richardson CC. The Replication System of Bacteriophage T7. Enzymes. 2016;39:89–136. doi: 10.1016/bs.enz.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 39.Hamdan SM, et al. A unique loop in T7 DNA polymerase mediates the binding of helicase-primase, DNA binding protein, and processivity factor. Proc Natl Acad Sci U S A. 2005;102(14):5096–5101. doi: 10.1073/pnas.0501637102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosh S, Hamdan SM, Richardson CC. Two modes of interaction of the single-stranded DNA-binding protein of bacteriophage T7 with the DNA polymerase-thioredoxin complex. J Biol Chem. 2010;285(23):18103–18112. doi: 10.1074/jbc.M110.107656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hamdan SM, Loparo JJ, Takahashi M, Richardson CC, van Oijen AM. Dynamics of DNA replication loops reveal temporal control of lagging-strand synthesis. Nature. 2009;457(7227):336–339. doi: 10.1038/nature07512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabor S, Richardson CC. Method for nucleic acid hybridization using single-stranded DNA binding protein. 1996 (Google Patents) [Google Scholar]
- 43.Lu D, Myers AR, George NP, Keck JL. Mechanism of Exonuclease I stimulation by the single-stranded DNA-binding protein. Nucleic Acids Res. 2011;39(15):6536–6545. doi: 10.1093/nar/gkr315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bianco PR. The tale of SSB. Prog Biophys Mol Biol. 2017;127:111–118. doi: 10.1016/j.pbiomolbio.2016.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hernandez A, Jeong P, Lee SJ, Richardson CC. Annealing of homologous DNA by T7 SSB. (Unpublished.) [Google Scholar]
- 46.Bell JC, Kowalczykowski SC. RecA: Regulation and Mechanism of a Molecular Search Engine. Trends Biochem Sci. 2016;41(6):491–507. doi: 10.1016/j.tibs.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu M, Masker W. T7 single strand DNA binding protein but not T7 helicase is required for DNA double strand break repair. J Bacteriol. 2001;183(6):1862–1869. doi: 10.1128/JB.183.6.1862-1869.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Son M, Hayes SJ, Serwer P. Concatemerization and packaging of bacteriophage T7 DNA in vitro: determination of the concatemers' length and appearance kinetics by use of rotating gel electrophoresis. Virology. 1988;162(1):38–46. doi: 10.1016/0042-6822(88)90392-3. [DOI] [PubMed] [Google Scholar]
- 49.White JH. Ph.D. Dissertation. Harvard University; 1986. Formation and Processing of Concatemers of Bacteriophage T7, in Vitro. [Google Scholar]
- 50.Lee J, Chastain PD, 2nd, Kusakabe T, Griffith JD, Richardson CC. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol Cell. 1998;1(7):1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 51.Lee J, Chastain PD, 2nd, Griffith JD, Richardson CC. Lagging strand synthesis in coordinated DNA synthesis by bacteriophage t7 replication proteins. J Mol Biol. 2002;316(1):19–34. doi: 10.1006/jmbi.2001.5325. [DOI] [PubMed] [Google Scholar]
- 52.Hernandez AJ, Lee SJ, Richardson CC. Primer release is the rate-limiting event in lagging-strand synthesis mediated by the T7 replisome. Proc Natl Acad Sci U S A. 2016;113(21):5916–5921. doi: 10.1073/pnas.1604894113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim YT, Tabor S, Churchich JE, Richardson CC. Interactions of gene 2.5 protein and DNA polymerase of bacteriophage T7. J Biol Chem. 1992;267(21):15032–15040. [PubMed] [Google Scholar]
- 54.Ghosh S, Hamdan SM, Cook TE, Richardson CC. Interactions of Escherichia coli thioredoxin, the processivity factor, with bacteriophage T7 DNA polymerase and helicase. J Biol Chem. 2008;283(46):32077–32084. doi: 10.1074/jbc.M805062200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang H, et al. Helicase-DNA polymerase interaction is critical to initiate leading-strand DNA synthesis. Proc Natl Acad Sci U S A. 2011;108(23):9372–9377. doi: 10.1073/pnas.1106678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Marintcheva B, Qimron U, Yu Y, Tabor S, Richardson CC. Mutations in the gene 5 DNA polymerase of bacteriophage T7 suppress the dominant lethal phenotype of gene 2.5 ssDNA binding protein lacking the C-terminal phenylalanine. Mol Microbiol. 2009;72(4):869–880. doi: 10.1111/j.1365-2958.2009.06682.x. [DOI] [PubMed] [Google Scholar]
- 57.Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391(6664):251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- 58.Nakai H, Richardson CC. The effect of the T7 and Escherichia coli DNA-binding proteins at the replication fork of bacteriophage T7. J Biol Chem. 1988;263(20):9831–9839. [PubMed] [Google Scholar]
- 59.Lechner RL, Engler MJ, Richardson CC. Characterization of strand displacement synthesis catalyzed by bacteriophage T7 DNA polymerase. J Biol Chem. 1983;258(18):11174–11184. [PubMed] [Google Scholar]
- 60.He ZG, Richardson CC. Effect of single-stranded DNA-binding proteins on the helicase and primase activities of the bacteriophage T7 gene 4 protein. J Biol Chem. 2004;279(21):22190–22197. doi: 10.1074/jbc.M401100200. [DOI] [PubMed] [Google Scholar]
- 61.Ahnert P, Picha KM, Patel SS. A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. EMBO J. 2000;19(13):3418–3427. doi: 10.1093/emboj/19.13.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hernandez AJ, Richardson CC. Kinetics of Lagging-strand DNA Synthesis In Vitro by the Bacteriophage T7 Replication Proteins. J Vis Exp. 2017;120 doi: 10.3791/55312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Park K, Debyser Z, Tabor S, Richardson CC, Griffith JD. Formation of a DNA loop at the replication fork generated by bacteriophage T7 replication proteins. J Biol Chem. 1998;273(9):5260–5270. doi: 10.1074/jbc.273.9.5260. [DOI] [PubMed] [Google Scholar]
- 64.Tamanoi F, Saito H, Richardson CC. Physical mapping of primary and secondary origins of bacteriophage T7 DNA replication. Proc Natl Acad Sci U S A. 1980;77(5):2656–2660. doi: 10.1073/pnas.77.5.2656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saito H, Tabor S, Tamanoi F, Richardson CC. Nucleotide sequence of the primary origin of bacteriophage T7 DNA replication: relationship to adjacent genes and regulatory elements. Proc Natl Acad Sci U S A. 1980;77(7):3917–3921. doi: 10.1073/pnas.77.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fuller CW, et al. Mechanisms for the initiation of bacteriophage T7 DNA replication. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 2):669–679. doi: 10.1101/sqb.1983.047.01.078. [DOI] [PubMed] [Google Scholar]
- 67.Romano LJ, Tamanoi F, Richardson CC. Initiation of DNA replication at the primary origin of bacteriophage T7 by purified proteins: requirement for T7 RNA polymerase. Proc Natl Acad Sci U S A. 1981;78(7):4107–4111. doi: 10.1073/pnas.78.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fuller CW, Richardson CC. Initiation of DNA replication at the primary origin of bacteriophage T7 by purified proteins. Site and direction of initial DNA synthesis. J Biol Chem. 1985;260(5):3185–3196. [PubMed] [Google Scholar]
- 69.Dreiseikelmann B, Wackernagel W. The terminal redundant regions of bacteriophage T7 DNA: their necessity for phage production studied by the infectivity of T7 DNA after modification by various exonucleases. Mol Gen Genet. 1978;159(3):321–328. doi: 10.1007/BF00268269. [DOI] [PubMed] [Google Scholar]
- 70.Schlegel RA, Thomas CA., Jr Some special structural features of intracellular bacteriophage T7 concatemers. J Mol Biol. 1972;68(2):319–345. doi: 10.1016/0022-2836(72)90216-1. [DOI] [PubMed] [Google Scholar]
- 71.Frohlich B, Powling A, Knippers R. Formation of concatemeric DNA in bacteriophage T7-infected bacteria. Virology. 1975;65(2):455–468. doi: 10.1016/0042-6822(75)90051-3. [DOI] [PubMed] [Google Scholar]
- 72.Paetkau V, Langman L, Bradley R, Scraba D, Miller RC., Jr Folded, concatenated genomes as replication intermediates of bacteriophage T7 DNA. J Virol. 1977;22(1):130–141. doi: 10.1128/jvi.22.1.130-141.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kelly TJ, Jr, Thomas CA., Jr An intermediate in the replication of bacteriophage T7 DNA molecules. J Mol Biol. 1969;44(3):459–475. doi: 10.1016/0022-2836(69)90373-8. [DOI] [PubMed] [Google Scholar]
- 74.Langman L, Paetkau V. Purification and structures of recombining and replicating bacteriophage T7 DNA. J Virol. 1978;25(2):562–569. doi: 10.1128/jvi.25.2.562-569.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Stratling W, Ferdinand FJ, Krause E, Knippers R. Bacteriophage T7-DNA replication in vitro: an experimental system. Eur J Biochem. 1973;38(1):160–169. doi: 10.1111/j.1432-1033.1973.tb03046.x. [DOI] [PubMed] [Google Scholar]
- 76.Center MS, Richardson CC. An endonuclease induced after infection of Escherichia coli with bacteriophage T7. I. Purification and properties of the enzyme. J Biol Chem. 1970;245(23):6285–6291. [PubMed] [Google Scholar]
- 77.Center MS, Studier FW, Richardson CC. The structural gene for a T7 endonuclease essential for phage DNA synthesis. Proc Natl Acad Sci U S A. 1970;65(1):242–248. doi: 10.1073/pnas.65.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Powling A, Knippers R. Recombination of bacteriophage T7 in vivo. Mol Gen Genet. 1976;149(1):63–71. doi: 10.1007/BF00275961. [DOI] [PubMed] [Google Scholar]
- 79.Kerr C, Sadowski PD. The involvement of genes 3,4,5 and 6 in genetic recombination in bacteriophage T7. Virology. 1975;65(1):281–285. doi: 10.1016/0042-6822(75)90031-8. [DOI] [PubMed] [Google Scholar]
- 80.Araki H, Ogawa H. The participation of T7 DNA-binding protein in T7 genetic recombination. Virology. 1981;111(2):509–515. doi: 10.1016/0042-6822(81)90353-6. [DOI] [PubMed] [Google Scholar]
- 81.Kong D, Richardson CC. Single-stranded DNA binding protein and DNA helicase of bacteriophage T7 mediate homologous DNA strand exchange. EMBO J. 1996;15(8):2010–2019. [PMC free article] [PubMed] [Google Scholar]
- 82.Kong D, Griffith JD, Richardson CC. Gene 4 helicase of bacteriophage T7 mediates strand transfer through pyrimidine dimers, mismatches, and nonhomologous regions. Proc Natl Acad Sci U S A. 1997;94(7):2987–2992. doi: 10.1073/pnas.94.7.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kong D, Nossal NG, Richardson CC. Role of the bacteriophage T7 and T4 single-stranded DNA-binding proteins in the formation of joint molecules and DNA helicase-catalyzed polar branch migration. J Biol Chem. 1997;272(13):8380–8387. doi: 10.1074/jbc.272.13.8380. [DOI] [PubMed] [Google Scholar]
- 84.Weigel C, Seitz H. Bacteriophage replication modules. FEMS Microbiol Rev. 2006;30(3):321–381. doi: 10.1111/j.1574-6976.2006.00015.x. [DOI] [PubMed] [Google Scholar]
- 85.Mueser TC, Hinerman JM, Devos JM, Boyer RA, Williams KJ. Structural analysis of bacteriophage T4 DNA replication: a review in the Virology Journal series on bacteriophage T4 and its relatives. Virol J. 2010;7:359. doi: 10.1186/1743-422X-7-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem Sci. 2000;25(4):156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 87.Hiom K. DNA repair: common approaches to fixing double-strand breaks. Curr Biol. 2009;19(13):R523–525. doi: 10.1016/j.cub.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 88.Lai YT, Masker W. Repair of double-strand breaks by incorporation of a molecule of homologous DNA. Mol Microbiol. 2000;36(2):437–446. doi: 10.1046/j.1365-2958.2000.01861.x. [DOI] [PubMed] [Google Scholar]
- 89.Masker W. In vitro repair of double-strand breaks accompanied by recombination in bacteriophage T7 DNA. J Bacteriol. 1992;174(1):155–160. doi: 10.1128/jb.174.1.155-160.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Scearce LM, Masker W. Deletion between direct repeats in bacteriophage T7 gene 1.2. Mutat Res. 1993;288(2):301–310. doi: 10.1016/0027-5107(93)90098-z. [DOI] [PubMed] [Google Scholar]
- 91.Lai YT, Masker W. In vitro repair of gaps in bacteriophage T7 DNA. J Bacteriol. 1998;180(23):6193–6202. doi: 10.1128/jb.180.23.6193-6202.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lai YT, Masker W. Visualization of repair of double-strand breaks in the bacteriophage T7 genome without normal DNA replication. J Bacteriol. 2000;182(2):327–336. doi: 10.1128/jb.182.2.327-336.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sanchez G, Mamet-Bratley MD. Development of T7 phage and T7 phage containing apurinic sites in an exonuclease III, endonuclease IV double mutant of Escherichia coli. Biochem Cell Biol. 1992;70(7):605–608. doi: 10.1139/o92-092. [DOI] [PubMed] [Google Scholar]
- 94.Meyer M, Witte W. Mutagenesis in bacteriophage T7. II. UV induced mutagenesis. Z Allg Mikrobiol. 1976;16(4):283–287. doi: 10.1002/jobm.3630160408. [DOI] [PubMed] [Google Scholar]
- 95.Meyer M. Mutagenesis in bacteriophage T7. I. Chemically induced mutagenesis. Z Allg Mikrobiol. 1976;16(4):279–282. doi: 10.1002/jobm.3630160407. [DOI] [PubMed] [Google Scholar]
- 96.Lane D, Mamet-Bratley MD, Karska-Wysocki B. Host-cell reactivation of alkylated T7 bacteriophage. Biochim Biophys Acta. 1979;564(3):495–506. doi: 10.1016/0005-2787(79)90039-x. [DOI] [PubMed] [Google Scholar]
- 97.Bauer J, Krammer G, Knippers R. Asymmetric repair of bacteriophage T7 heteroduplex DNA. Mol Gen Genet. 1981;181(4):541–547. doi: 10.1007/BF00428750. [DOI] [PubMed] [Google Scholar]
- 98.Masker W. In vitro packaging of heteroduplex bacteriophage T7 DNA: evidence for repair of mismatched bases. J Bacteriol. 1986;168(2):762–768. doi: 10.1128/jb.168.2.762-768.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sun B, et al. T7 replisome directly overcomes DNA damage. Nat Commun. 2015;6:10260. doi: 10.1038/ncomms10260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhu B, Lee SJ, Richardson CC. Bypass of a nick by the replisome of bacteriophage t7. The Journal of biological chemistry. 2011;286(32):28488–28497. doi: 10.1074/jbc.M111.252023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.White JH, Richardson CC. Processing of concatemers of bacteriophage T7 DNA in vitro. J Biol Chem. 1987;262(18):8851–8860. [PubMed] [Google Scholar]
- 102.Sun M, Louie D, Serwer P. Single-event analysis of the packaging of bacteriophage T7 DNA concatemers in vitro. Biophys J. 1999;77(3):1627–1637. doi: 10.1016/S0006-3495(99)77011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chung YB, Hinkle DC. Bacteriophage T7 DNA packaging. I. Plasmids containing a T7 replication origin and the T7 concatemer junction are packaged into transducing particles during phage infection. J Mol Biol. 1990;216(4):911–926. doi: 10.1016/S0022-2836(99)80010-2. [DOI] [PubMed] [Google Scholar]
- 104.Cerritelli ME, Conway JF, Cheng N, Trus BL, Steven AC. Molecular mechanisms in bacteriophage T7 procapsid assembly, maturation, and DNA containment. Adv Protein Chem. 2003;64:301–323. doi: 10.1016/s0065-3233(03)01008-8. [DOI] [PubMed] [Google Scholar]
- 105.Fujisawa H, Morita M. Phage DNA packaging. Genes Cells. 1997;2(9):537–545. doi: 10.1046/j.1365-2443.1997.1450343.x. [DOI] [PubMed] [Google Scholar]
- 106.Colson P, et al. "Megavirales", a proposed new order for eukaryotic nucleocytoplasmic large DNA viruses. Arch Virol. 2013;158(12):2517–2521. doi: 10.1007/s00705-013-1768-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yutin N, Wolf YI, Koonin EV. Origin of giant viruses from smaller DNA viruses not from a fourth domain of cellular life. Virology. 2014;466–467:38–52. doi: 10.1016/j.virol.2014.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Forterre P, Gaia M. Giant viruses and the origin of modern eukaryotes. Curr Opin Microbiol. 2016;31:44–49. doi: 10.1016/j.mib.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 109.Bermek O, Willcox S, Griffith JD. DNA replication catalyzed by herpes simplex virus type 1 proteins reveals trombone loops at the fork. J Biol Chem. 2015;290(5):2539–2545. doi: 10.1074/jbc.M114.623009. [DOI] [PMC free article] [PubMed] [Google Scholar]