Abstract
Breast cancer is one of the leading cancer type among women in worldwide. Many breast cancer patients die every year due to the late diagnosis and treatment. Thus, in recent years, early breast cancer detection systems based on patient’s imagery are in demand. Deep learning attracts many researchers recently and many computer vision applications have come out in various environments. Convolutional neural network (CNN) which is known as deep learning architecture, has achieved impressive results in many applications. CNNs generally suffer from tuning a huge number of parameters which bring a great amount of complexity to the system. In addition, the initialization of the weights of the CNN is another handicap that needs to be handle carefully. In this paper, transfer learning and deep feature extraction methods are used which adapt a pre-trained CNN model to the problem at hand. AlexNet and Vgg16 models are considered in the presented work for feature extraction and AlexNet is used for further fine-tuning. The obtained features are then classified by support vector machines (SVM). Extensive experiments on a publicly available histopathologic breast cancer dataset are carried out and the accuracy scores are calculated for performance evaluation. The evaluation results show that the transfer learning produced better result than deep feature extraction and SVM classification.
Keywords: Breast cancer detection, Histopathologic image, Convolutional neural networks, Transfer learning, Deep feature extraction
Introduction
The cancer is a disease in which cells multiply uncontrollably and these cell crowd out normal cells. Breast cancer (BC) is one of the most common types of cancer disease, which has a high death rate in women and is in second place after lung cancer. BC begins with uncontrolled proliferation of cells in the milk glands and ducts that carry milk to nipple [1]. According to the estimates of the American Cancer Society in 2018, there will be 268,670 (women 266,120) new breast cancer cases and 41,400 (40,920 women) will die from BC [2]. On the other hand, early detection and diagnosis of BC both greatly increases the chance of successful treatment of this disease and decreases mental and physical pains suffered by patients. These are many non-invasive biomedical imaging techniques for early detection of BC such as Contrast-Enhanced (CE) digital mammography, ultrasound and magnetic resonance imaging (MRI) computed tomography (CT), positron emission tomography (PET) [3, 4]. Imaging methods as CT, PET and MRI have serious disadvantages in terms of exposure to high radiation. Although the initial detection of BC can be performed using these techniques, they don’t guarantee that the detected abnormality in breast tissues is malignant. For this reason, the treatment of the disease does not begin until a biopsy is performed to confirm the malignancy.
A biopsy is an invasive diagnosis technique which uses the imaging tests to show that a breast change that may be cancer. In the surgical open biopsy method (SOB), which is one of the biopsy methods, it needs larger lesion parts to be removed. Breast biopsies provide histologically assessment of the microscopic structure of the tissue. In breast biopsies, pathologists provide histologically assessment of the microscopic structure of the tissue and they are make final BC diagnosis by applying visual inspection of histological samples under the microscope [5]. Histopathology aims to differentiate between normal tissue, benign and malignant cells. Histopathological analysis have some disadvantages such as the zooming, focusing and scrolling needed on the picture takes highly time-consuming, depend on experience, fatigue and attention of pathologists [6]. Therefore, computer-assisted diagnosis (CAD) systems are used to relieve the workload on pathologists by automatically classifying benign and malignant tissues and by filtering obviously benign areas in the field of BC histopathology image analysis [7], and provide a second opinion to doctors for higher accuracy.
The diagnosis of BC from histopathology images is excellent usage area for a CAD system based on the machine leaning. One of the most recent machine learning methods is Deep Learning. Deep Learning based systems outdid conventional machine learning methods in many image analysis. Deep learning aids in breast exams and help patients avoid unnecessary biopsies. Deep learning has emerged and developed because of the inadequate image recognition of machine learning algorithms. The most common of the deep learning algorithms used in the literature is the convolutional neural networks (CNN). One of the diagnosis studies of breast cancer performed using convulsive neural networks are that C. Pearce made. In the study conducted, tumor findings were classified by deep learning method. The FCN (Full Connected Network) structure was used a binary classifier to determine if the individual’s images are mitotic [8]. Selvathi et al. were used a deep learning technique with unsupervised using mammography. In the method used, dense mammography images was classified. Accuracy rate was calculated as 98.5% [9]. Geras et al. were used 886 thousand large-scale mammography-based images. These images were used to screening for breast cancer. In the study, multi-deep convolutional network structure was used. It has been observed that the performance increases as the training set increases. It has been observed that the best resolution values are the original resolution values [E]. Bayramoğlu et al. has proposed two different network structures for cancer diagnosis. They are intended to classify by using convolutional neural networks (CNN). The single task CNN was used to predict malignant tumors. The multi task CNN was used to simultaneously predict malignant tumor and image magnification [10].
In this paper, a CAD system based on deep convolutional neural networks (CNN) is developed to help the pathologists classify the histopathological breast cancer images as benign and malignant. In this study, we are aimed to classify the histopathological breast cancer images taken from the BreaKHis database [11]. Different magnification rates of the images were taken in the classification made by using the CNN structure. At first, we used the BreakHis dataset for feature extraction. Then we used the data set for transfer learning. Feature extraction was done for classification using pre-trained neural network structure. The pre-trained network structure is AlexNet [12]. We have removed the last three layers of the network and added new layers to adopt the pre-trained network solve for our problem. In the last stage classification with support vector machine (SVM) has been completed.
The deep features that are considered in this work were extracted from the fully connected (fc) layers of the pre-trained AlexNet and VGG16 models. The fc6 and fc7 layers are considered in this work which produces 4096 dimensional feature vectors. These feature vectors either used by itself or used in a combination to augment the efficiency of the proposed work.
Related theories
In this section, we provide succinct descriptions of the related theories that are used in this work. Readers may refer to the related references for detailed information [13–15].
Convolutional neural networks
Convolutional neural networks (CNNs), which can be seen as one category of neural networks, are quite popular nowadays because of its high performance in a variety of image based object recognition applications [13, 14]. CNNs provide both feature extraction and classification with its end-to-end learning architecture. A general CNN architecture is composed of various layers such as convolution, pooling, normalization and fully connected layers. Convolution, pooling and normalization layers are embedded in a sequential way to construct the network. Performing convolution and pooling operations consecutively construct high level features on which classification is performed. The classification is performed in the fully connected layer of the CNN architecture. In CNN architecture, there are a huge number of parameters that need to be adjusted during CNN’s training. The training of CNN’s is generally carried out with the conventional back propagation algorithm.
Transfer learning
Transfer learning (TL) is defined as transferring knowledge, which was learned earlier in one domain, to another domain for classification and feature extraction purposes [15]. In the deep learning view, TL is carried out by using a deep CNN model which was trained earlier on a large dataset. The pre-trained CNN model is further trained (fine-tuning) on a new dataset with smaller numbers of training images comparable to the previously trained datasets. Recently, TL has been used in many deep learning applications because fine-tuning a pre-trained CNN model is usually much faster and easier than training a CNN model with randomly initialized weights from scratch. Generally, in the CNN models, edges, curves, corners and color blobs like features are learned in the initial layers, and the final layers of the CNN models represent abstract and specific features [13]. In practical applications, the last three layers of the pre-trained CNN model namely, fully connected layer, softmax layer and classification output layer, are discharged and the rest layers are transferred to the new classification task.
Deep feature extraction
Deep feature extraction can be seen as another type of transfer learning [15]. Instead of fine-tuning a pre-trained CNN model, the activation layers of the CNN model can be used to extract representative feature vectors. The activations of the earlier layers provide representations similar to low-level image features such as edges, while the deeper layers provide higher-level features salient for image classification. For example, in both ImageNets, the activations of the first and second fully connected layers (fc6 and fc7) are commonly used as a feature representation for image recognition tasks. The resulting feature vectors contain totally 4096 attributes.
Proposed approach
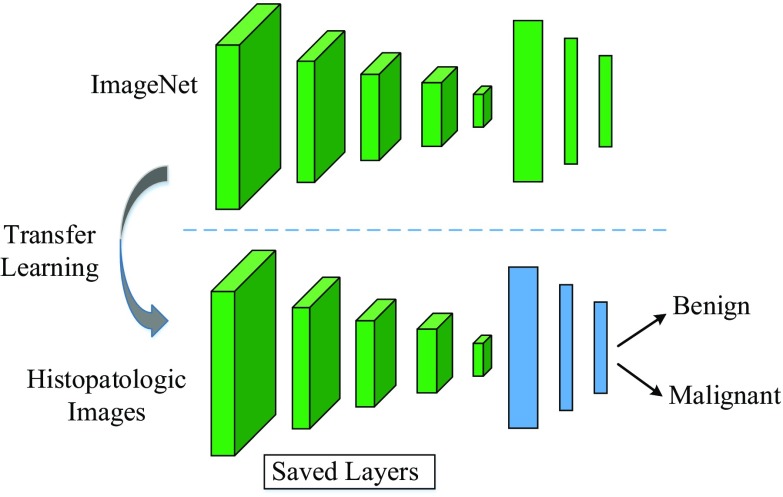
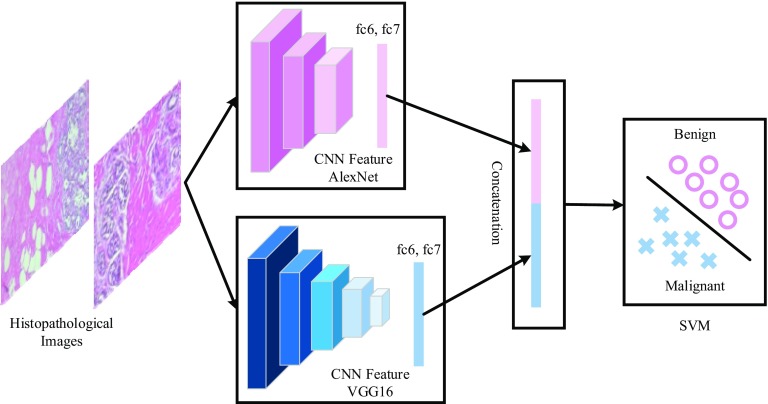
The proposed scheme and architecture of CNN are illustrated in Figs. 1 and 2, respectively. As seen in both figures, the input breast cancer histopathology images are fed into the pre-trained CNN models. For feature extraction, pre-trained AlexNet and VGG16 models are used [13, 14]. The obtained features vectors are then classified by SVM classifier to determine the class label of the input images. For fine-tuning only AlexNet model is considered.
Fig. 1.
An illustration for transfer learning. The last three layers of the pre-trained CNN model are discharged and the rest of layers are used for new classification task
Fig. 2.
An illustration for deep feature extraction and SVM classification. The fc6 and fc7 layers of the pre-trained CNN models (AlexNet, Vgg16) are used for deep feature extraction
Fine-tuning and feature extraction with pre-trained CNN models
For fine-tuning, the pre-trained AlexNet model is considered. AlexNet is known to be the first deep CNN model that was introduced by Krizhevsky et al. [13]. AlexNet model is totally comprised of 25 layers where 5 of them contain learnable weights and last three layers cover fully connected layers. In the AlexNet architecture, rectified linear units, normalization and max-pooling layers come after the convolutional layers and convolutional layers use varying kernel sizes. For TL adaptation, the last three layers of the AlexNet model are discharged because that layers were configured for 1000 classes ImageNet challenge [16]. These three layers are fine-tuned for the breast cancer detection problem where the number of classes is two (benign and malignant).
For deep feature extraction, AlexNet and VGG16 models are considered. VGG16 model is another deeper CNN model, which was proposed by Simonyan et al. [14]. VGG16 model contains totally 41 layers where 16 layers have learnable weights. 13 convolutional layers and 3 fully connected layers form the learnable layers. VGG16 model only uses small 3 × 3 kernels on all convolutional layers and similar to the AlexNet, max-pooling layers follow convolutional layers [13, 14]. The activations on the first and second fully connected layers (fc6, fc7) are used to extract the feature vectors. The resulting feature vectors from fc6 and fc7 contain totally 4096 attributes.
Experimental results
As the proposed approach aims to detect breast cancer efficiently from histopathology images, BreaKHis dataset [6] is considered during experimental works. BreaKHis dataset covers totally 9109 microscopic images where 2480 of them are benign and the rest 5429 images are malignant samples. During the collection of the dataset, 82 patients’ breast tumor tissues were imagined by using different magnifying factors such as 40X, 100X, 200X, and 400X. All images are colored and of size 700 × 460 pixels. The dataset was arranged into five folds.
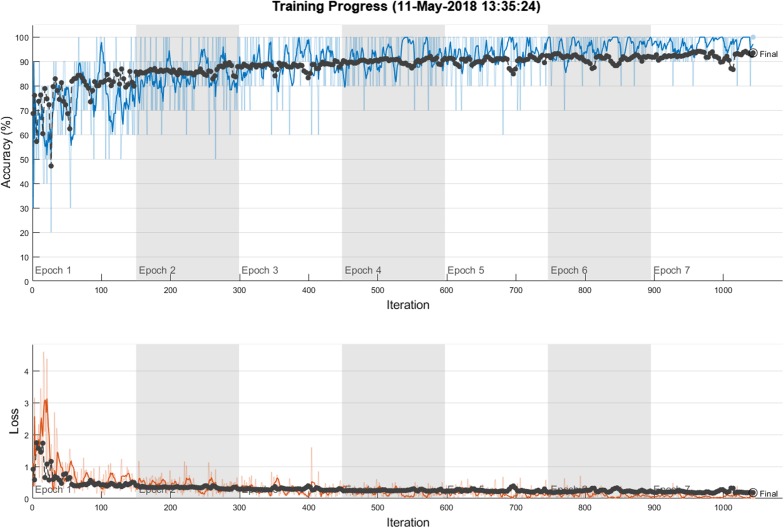
The experiment platform is configured with an Intel Core i7-4810 CPU and 32 GB memory. All input images were initially resized to sizes 227 × 227 and 224 × 224 for sake of convenience with AlexNet and VGG16 models, respectively. The detailed information about the network structures of AlexNet and VGG16 models can be viewed in [13, 14]. fc6 and fc7 activations were used for feature extraction. The extracted features were then concatenated accordingly. The SVM classifier with homogenous mapping and LIBLINEAR library with the L2-regularised L2-loss dual solver was considered because of its robustness to smaller amounts of training data [17, 18]. The SVM parameter C is searched in the range of [10−4–103]. The performance of the proposed method was scored using classification accuracy. For fine-tuning of the CNN model, the mini batch size was chosen as 10 and the initial learning rate was assigned as 0.0001. The initial learning rate was chosen small enough to slow down learning in the transferred layers. The maximum epoch number was set to 7 and the CNN model was trained by stochastic gradient descent with momentum. The training procedure is ended around 1100 iterations. Figure 3 shows the training progress of fine-tuned AlexNet model. While the first row shows the deviation of the accuracy against iterations, the second row shows the loss deviation against iterations.
Fig. 3.
The training progress of fine-tuned AlexNet model. The result is obtained for Fold 3, 40X magnification factor
The initial experiments were carried out with deep feature extraction and SVM classification. The obtained results were tabulated in Tables 1 and 2, respectively. While Table 1 shows the obtained results with the concatenated features from AlexNet-fc6 and Vgg16-fc6, Table 2 shows the produced accuracy scores with the concatenated deep features from AlexNet-fc7 and Vgg16-fc7. The rows of Tables 1 and 2 show the folds and the columns show the magnifying factors.
Table 1.
Obtained classification accuracies when AlexNet-fc6 and Vgg16-fc6 deep features concatenation is used
| Accuracy (%) | ||||
|---|---|---|---|---|
| 40X | 100X | 200X | 400X | |
| Fold 1 | 86.31 | 89.34 | 88.58 | 88.40 |
| Fold 2 | 85.28 | 89.52 | 86.77 | 85.92 |
| Fold 3 | 83.71 | 90.73 | 93.02 | 90.71 |
| Fold 4 | 83.71 | 86.82 | 86.89 | 79.87 |
| Fold 5 | 85.42 | 89.65 | 88.31 | 88.84 |
The bold cases show the best accuracy values for each fold
Table 2.
Obtained classification accuracies when AlexNet-fc7 and Vgg16-fc7 deep features concatenation is used
| Accuracy (%) | ||||
|---|---|---|---|---|
| 40X | 100x | 200x | 400x | |
| Fold 1 | 85.10 | 89.34 | 87.77 | 87.94 |
| Fold 2 | 86.39 | 88.23 | 86.10 | 84.48 |
| Fold 3 | 83.41 | 91.18 | 93.78 | 90.71 |
| Fold 4 | 82.71 | 87.25 | 85.86 | 79.87 |
| Fold 5 | 85.29 | 89.16 | 88.05 | 87.03 |
The bold cases show the best accuracy values for each fold
As seen in Table 1, for Fold 1, the best classification accuracy 89.34% was produced for 100X magnification factor and the accuracy scores for 200X and 400X magnification factors were also 88.58% and 88.40%, respectively. The worse accuracy score for Fold 1 was obtained for 40X magnification factor. For Fold 2, the highest accuracy score 89.52% was also produced for 100X magnification factor. The other magnification factors also produced close accuracy scores to 100X magnification factor where the accuracy scores were 85.28, 86.77 and 85.92% respectively. Except 40X magnification factor, the obtained results for Fold 3 were better than the obtained results that were obtained for Folds 1, 2, 4 and 5. 93.02% accuracy score was recorded for 200X magnification factor. For Fold 4, 86.89% and 86.82% accuracy scores were obtained for 200X and 10X magnification factors. Finally, for Fold 5, 89.65% accuracy score was recorded for 100X magnification factor. An important observation from Table 1 is that the obtained accuracy scores for 100X magnification factor are better than the other magnification factors.
Table 2 represents the obtained results with concatenated AlexNet-fc7 and Vgg16-fc7 features. Before evaluating the obtained results fold by fold, it is worth to mentioning that 100X magnification factor outperformed by obtaining the best accuracy scores 89.34%, 88.23%, 87.25 and 89.16% for Folds 1, 2, 4 and 5, respectively. For Fold 3, the best accuracy 93.78% was produced for 200X magnification factor. In addition, any highest accuracy scores were not obtained for 40X and 400X magnification factors when all obtained results were observed as given in Tables 1 and 2.
Table 3 represents the obtained classification accuracy scores for fine-tuned AlexNet model. It is obvious that the fine-tuned AlexNet model achieved better results when Table 3 and other Tables 1 and 2 compared. Almost all obtained accuracy scores are above 90%. The highest accuracy score 93.57% was obtained on Fold 3 and 40X magnification factor. There is also another 93.24% accuracy score that was obtained in Fold 5 and 200X magnification factor. Folds 2 and 4 produced the highest accuracy scores 90.77% and 91.87% respectively for 400X magnification factor. Finally, Fold 1 produced 91.37% accuracy value for 40X magnification factor.
Table 3.
Obtained classification accuracies with fine-tuned AlexNet
| Accuracy (%) | ||||
|---|---|---|---|---|
| 40X | 100x | 200x | 400x | |
| Fold 1 | 91.37 | 91.35 | 88.87 | 90.77 |
| Fold 2 | 89.76 | 90.58 | 90.66 | 90.77 |
| Fold 3 | 93.57 | 92.50 | 92.64 | 90.77 |
| Fold 4 | 90.16 | 87.31 | 91.45 | 91.87 |
| Fold 5 | 89.96 | 91.15 | 93.24 | 92.31 |
The bold cases show the best accuracy values for each fold
A further comparison was carried out between the obtained results as given in Table 4. The comparisons were represented based on the calculated average accuracy score across the folds and theirs standard deviation. The columns of Table 4 show the magnification levels. As seen in Table 4, fine-tuned AlexNet model performed the highest average accuracy scores for all magnification levels and 90.96 ± 1.59% and the second best average accuracy score was produced by the feature set of AlexNet-fc6 + Vgg16-fc6. The worst average accuracy scores were obtained by the feature set of AlexNet-fc7 + Vgg16-fc7.
Table 4.
Comparison of the obtained results for each magnification factor
| Method | Accuracy (%) | |||
|---|---|---|---|---|
| 40X | 100x | 200x | 400x | |
| AlexNet-fc6 + Vgg16-fc6 | 84.87 ± 1.14 | 89.21 ± 1.44 | 88.65 ± 2.41 | 86.75 ± 4.21 |
| AlexNet-fc7 + Vgg16-fc7 | 84.58 ± 1.49 | 89.03 ± 1.46 | 88.31 ± 3.20 | 86.00 ± 4.08 |
| Fine-tuned AlexNet | 90.96 ± 1.59 | 90.58 ± 1.96 | 91.37 ± 1.72 | 91.30 ± 0.74 |
The bold cases show the best accuracy values for each fold
We also compared the obtained results with the results that were published in [6, 19]. In [19], researchers applied end-to-end CNN approach and in [6], the authors used various classifiers such as neural networks (NN), quadratic discriminant analysis (QDA), SVM and random forest (RF) for breast cancer detection in breast histopathological images. Table 5 shows the related comparisons. As seen in Table 5, for each magnification factor, our proposed method achieved better accuracy scores than the compared scores. The CNN method achieved the second best accuracy scores for each magnification factor. Especially for 200X and 400X each magnification factors, the accuracy scores that were produced by our proposed method are significant.
Table 5.
Comparison of the proposed method with some of the published results
| Method | Accuracy (%) | |||
|---|---|---|---|---|
| 40X | 100x | 200x | 400x | |
| NN [6] | 80.9 ± 2.0 | 80.7 ± 2.4 | 81.5 ± 2.7 | 79.4 ± 3.9 |
| QDA [6] | 83.8 ± 4.1 | 82.1 ± 4.9 | 84.2 ± 4.1 | 82.0 ± 5.9 |
| RF [6] | 81.8 ± 2.0 | 81.3 ± 2.8 | 83.5 ± 2.3 | 81.0 ± 3.8 |
| SVM [6] | 81.6 ± 3.0 | 79.9 ± 5.4 | 85.1 ± 3.1 | 82.3 ± 3.8 |
| CNN [19] | 90.0 ± 6.7 | 88.4 ± 4.8 | 84.6 ± 4.2 | 86.1 ± 6.2 |
| Our work | 90.96 ± 1.59 | 90.58 ± 1.96 | 91.37 ± 1.72 | 91.30 ± 0.74 |
The bold cases show the best accuracy values for each fold
Conclusions
This paper compares the classification efforts of transfer learning and deep feature extraction on breast cancer detection based on the histopathological images. For deep feature extraction, two popular deep CNN architectures namely AlexNet and Vgg16 models are used and for transfer learning. The BreaKHis dataset is preferred in the experimental works due to huge number of sample images. Three different experimental works are considered. In the first one, the feature vectors from the fc6 layers of both AlexNet and Vgg16 models are extracted and then are concatenated. In the second one, the fc7 layers of AlexNet and Vgg16 models are used to feature extraction and obtained feature vectors are concatenated. An SVM classifier is used in the first and second experimental works for classification of the images into benign and malignant classes. In the third experiments, the pre-trained AlexNet model is further tuned on breast cancer images. Experimental results show that fine-tuned AlexNet outperformed and the first experiments results are better than the second one.
In the future works, we are planning to use other CNN models for improving the classification accuracy. In addition, the data augmentation will be considered in the future works for transfer learning.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Erkan Deniz, Email: edeniz@firat.edu.tr.
Abdulkadir Şengür, Email: ksengur@firat.edu.tr.
Zehra Kadiroğlu, Email: zehrakad@gmail.com.
Yanhui Guo, Email: yguo56@uis.edu.
Varun Bajaj, Email: varun@iiti.ac.in.
Ümit Budak, Email: ubudak@beu.edu.tr.
References
- 1.American Cancer Society. About breast cancer. https://www.cancer.org/content/dam/CRC/PDF/Public/8577.00.pdf.
- 2.American Cancer Society. Cancer facts & figures 2018. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2018/cancer-facts-and-figures-2018.pdf.
- 3.Wang L. Early diagnosis of breast cancer. Sensors. 2017;17–1572:1–20. doi: 10.3390/s17071572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dr. Filiz Yenicesu. Meme Kanserinde Görüntüleme Yöntemleri. https://www.duzen.com.tr/workshop/2011/Meme_Kanserinde_Goruntuleme_Y%C3%B6ntemleri_(Dr_Filiz_Yenicesu).pdf.
- 5.Joy JE, et al., editors. Saving women’s lives: strategies for improving breast cancer detection and diagnosis. Washington, DC: Natl Acad Press; 2005. [PubMed] [Google Scholar]
- 6.Spanhol FA, Oliveira LS, Petitjean C, Heutte L. A dataset for breast cancer histopathological image classification. IEEE Trans Biomed Eng. 2016;63(7):1455–1462. doi: 10.1109/TBME.2015.2496264. [DOI] [PubMed] [Google Scholar]
- 7.Gurcan MN, et al. Histopathological image analysis: a review. IEEE Rev Biomed Eng. 2009;2:147–171. doi: 10.1109/RBME.2009.2034865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pearce C. Convolutional neural networks and the analysis of cancer imagery. Stanford University; 2017.
- 9.Selvathi D, Poornila A. Aarthy, “Breast cancer detection ın mammogram ımages using deep learning technique” middle-east. J Sci Res. 2017;25(2):417–426. [Google Scholar]
- 10.Geras KJ, et al. High-resolution breast cancer screening with multi-view deep convolutional neural networks. 2017. arXiv preprint arXiv:1703.07047.
- 11.Bayramoglu N, Kannala J, Heikkilä J. Deep learning for magnification independent breast cancer histopathology image classification. In: 2016 23rd International conference on pattern recognition (ICPR), pp. 2440–2445. IEEE. 2016.
- 12.http://web.inf.ufpr.br/vri/breast-cancer-database.
- 13.Krizhevsky A, Sutskever I, Hinton GE. Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp. 1106–1114. 2012.
- 14.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. Computing research repository (CoRR), vol. abs/1409.1556. 2014.
- 15.Orenstein EC, Beijbom O. Transfer learning and deep feature extraction for planktonic image data sets. In: 2017 IEEE Winter conference on applications of computer vision (WACV). IEEE. 2017.
- 16.Russakovsky O, et al. Imagenet large scale visual recognition challenge. Int J Comput Vis. 2015;115(3):211–252. doi: 10.1007/s11263-015-0816-y. [DOI] [Google Scholar]
- 17.Hearst M.A., Dumais S.T., Osuna E., Platt J., Scholkopf B. Support vector machines. IEEE Intelligent Systems and their Applications. 1998;13(4):18–28. doi: 10.1109/5254.708428. [DOI] [Google Scholar]
- 18.Vedaldi Andrea, Zisserman Andrew. Efficient additive kernels via explicit feature maps. IEEE Trans Pattern Anal Mach İntell. 2012;34(3):480–492. doi: 10.1109/TPAMI.2011.153. [DOI] [PubMed] [Google Scholar]
- 19.Spanhol F, Oliveira LS, Petitjean C, Heutte L. Breast cancer histopathological image classification using convolutional neural network. In: International joint conference on neural networks (IJCNN 2016), Vancouver, Canada, 2016.