Abstract
Multitrait meta-analyses are a strategy to produce more accurate genome-wide association studies, especially for complex phenotypes. We carried out a meta-analysis study for traits related to sexual precocity in tropical beef cattle (Nellore and Brahman) aiming to identify important genomic regions affecting these traits. The traits included in the analyses were age at first calving (AFC), early pregnancy (EP), age at first corpus luteum (AGECL), first postpartum anoestrus interval (PPAI), and scrotal circumference (SC). The traits AFC, EP, and SCN were measured in Nellore cattle, while AGECL, PPAI, and SCB were measured in Brahman cattle. Meta-analysis resulted in 108 significant single-nucleotide polymorphisms (SNPs), at an empirical threshold P-value of 1.39 × 10−5 (false discovery rate [FDR] < 0.05). Within 0.5 Mb of the significant SNP, candidate genes were annotated and analyzed for functional enrichment. Most of the closest genes to the SNP with higher significance in each chromosome have been associated with important roles in reproductive function. They are TSC22D2, KLF7, ARHGAP29, 7SK, MAP3K5, TLE3, WDR5, TAF3, TMEM68, PPP1R15B, NR2F2, GALR1, SUFU, and KCNU1. We did not observe any significant SNP in BTA5, BTA12, BTA17, BTA18, BTA19, BTA20, BTA22, BTA23, BTA25, and BTA28. Although the majority of significant SNPs are in BTA14, it was identified significant associations in multiple chromosomes (19 out of 29 autosomes), which is consistent with the postulation that reproductive traits are complex polygenic phenotypes. Five proposed association regions harbor the majority of the significant SNP (76%) and were distributed over four chromosomes (P < 1.39 × 10−5, FDR < 0.05): BTA2 (5.55%) from 95 to 96 Mb, BTA4 (5.55%) from 94.1 to 94.8 Mb, BTA14 (59.26%) from 24 to 25 Mb and 29 to 30 Mb, and BTA21 (5.55%) from 6.7 Mb to 11.4 Mb. These regions harbored key genes related to reproductive function. Moreover, these genes were enriched for functional groups associated with immune response, maternal–fetal tolerance, pregnancy maintenance, embryo development, fertility, and response to stress. Further studies including other breeds and precocity traits could confirm the importance of these regions and identify new candidate regions for sexual precocity in beef cattle.
Keywords: Bos indicus, Brahman, early puberty, GWAS, meta-analysis, Nellore
INTRODUCTION
Reproductive traits are important for economic success in beef cattle production (Formigoni et al., 2005; Brumatti et al., 2011). Despite its economic importance, genetic gains of reproductive traits are generally slow. These traits are considered polygenic and highly influenced by environmental factors (Cardoso et al., 2015). Genomic selection has allowed higher genetic gains for fertility traits in beef and dairy cattle (Zhang et al., 2014; Garcia-Ruiz et al., 2016). In parallel with genomic selection, genome-wide association studies (GWAS) have detected quantitative trait loci (QTL) affecting reproductive traits aiming to better understand their genetic mechanisms (Hawken et al., 2012; Irano et al., 2016; Melo et al., 2017). However, these traits are controlled by many QTL of small effect and GWAS can produce high false-positive rates. Validation for the identified QTL requires further investigation. To improve QTL detection, GWAS meta-analyses corroborate evidence from independent studies of related traits by identifying polymorphisms that produce variation in common among complex traits (Bolormaa et al., 2014; Ramayo-Caldas et al., 2016). The aim of this study was to carry out meta-analysis across two independent Bos indicus populations to improve QTL detection for reproductive traits. Specifically, we aimed to: 1) identify genomic regions associated with sexual precocity traits measured relatively early in a cow’s life and the related bull trait scrotal circumference (SC); 2) identify candidate genes in those genomic regions; and 3) investigate biological roles of the identified candidate genes. The traits measured in Nellore were age at first calving (AFC), early pregnancy (EP), and scrotal circumference (SCN) and in Brahman were age at the first corpus luteum (AGECL), first postpartum anoestrus interval (PPAI), and scrotal circumference (SCB).
MATERIALS AND METHODS
Nellore Phenotypes and Genotypes
Information of animals from Alliance Nellore dataset, born in eight different farms distributed in Northwest, Southwest, and Midwest of Brazil were used. Heifers were either artificially inseminated or naturally mated. Usually farmers apply two breeding seasons, in the early breeding season females are exposed to the first mating at around 16 mo of age and, those females that failed to conceive have a second chance in the later breeding season, at about 26 mo of age. Females that did not conceive in the first neither in the second chance were culled and were not considered in the analysis of AFC. More details about the dataset are described in Costa et al. (2015) and Irano et al. (2016).
Summary statistics of Nellore phenotypes—AFC, EP, and SCN—are presented in Table 1. The phenotype AFC was defined as the number of days from birth to first calving; EP was defined as success (1) for heifers that had the first calf with less than 31 mo of age, i.e., heifers that got pregnant in the first breeding season, or failure (0) for those heifers that had the first calf after 31 mo of age. In both populations, SC was measured at about 18 mo of age, at the widest point of the scrotum with a standard metal tape, in centimeter.
Table 1.
Summary statistics of the traits AFC, EP, and SCN in Nellore cattle and AGECL, PPAI, and SCB in Brahman cattle
| Trait | Number of observations1 | Mean ± SE | Minimum | Maximum |
|---|---|---|---|---|
| AFC, d | 1,796 | 1068.2 ± 118.5 | 778.9 | 1292.8 |
| EP, % | 1,849 | 28.3 | — | — |
| , cm | 4,248 | 26.57 ± 2.56 | 17.47 | 36.17 |
| AGECL, d | 1,007 | 750.6 ± 142.14 | 394 | 1211 |
| PPAI, d | 629 | 180.11 ± 108.71 | 17 | 484 |
| , cm | 1,203 | 26.51 ± 2.76 | 18.25 | 39.25 |
1Number of observations considered in analyses: animals with available genotypes and phenotypes.
2SCN measured at approximately 521 d of age.
3SCB measured at approximately 540 d of age.
The contemporary groups (CGs) for AFC, EP, and SCN were defined by concatenating the information of herd, year and season of birth, weaning, and yearling management groups. Groups with less than four animals for AFC and EP or less than three animals for SCN were excluded. For AFC and SCN, we considered only data in the interval of CG ± 3 SD from the mean of each group. For EP CG without variability, i.e., presenting only females with the same categorical response were excluded. For SC the linear effect of the age of the animal at the recording was included as a covariate in the model. It was considered in the analysis just animals presenting age at recording from 10 to 24 mo of age. It was excluded a total of 20 animals that presented ages out of this interval.
For Nellore traits, phenotypes were previously corrected for CG effects to avoid biased fixed effect estimates because the whole Nellore dataset contained both genotyped and non-genotyped animals. CG estimates used to pre-correct the phenotypes were obtained under a regular mixed animal model (Henderson, 1984). Then, these corrected phenotypic traits for genotyped animals were used in GWAS approach.
Animals were genotyped with high-density Illumina Bovine HD Assay (Illumina, San Diego, CA) and GeneSeek Genomic Profiler Indicus HD—GGP75Ki (Neogen Corporation, Lincoln, NE), which have 777,962 and 74,677 (73,941 in common with HD) SNP markers, respectively. Genotype imputation to Bovine HD was performed for animals genotyped with GGP75Ki using FImpute software (Sargolzaei et al., 2014), taking into account pedigree information. The imputation accuracy is expected to be higher than 0.98 (Carvalheiro et al., 2014). Quality control was performed, removing animals with call rate <0.90 and removing SNP with minor allele frequency (MAF) < 0.01, call rate < 0.95, GC score < 0.15, Hardy–Weinberg equilibrium test P-value < 10−5, SNP in non-autosomal regions, and unmapped SNP. After quality control 2,923 females with genotypes for 412,993 SNP and 5,078 males with genotypes for 477,317 SNP remained for the analyses. To construct the genomic relationship matrix (G), SNP with MAF >0.1 were considered.
Brahman Phenotypes and Genotypes
Data from the Cooperative Research Centre for Beef Genetic Technologies (Beef CRC) were used in this study. The Beef CRC data for male and female Brahman cattle were previously described (Johnston et al., 2009; Burns et al., 2013; Corbet et al., 2013). The number of observations and descriptive statistics for each trait are presented in Table 1. The traits evaluated were AGECL, PPAI, and SCB. When the heifers achieved the weight of 200 kg, an ovarian ultrasound was carried out at every 4 to 6 wk to detect the first corpus luteum (CL) and calculate AGECL. To get PPAI phenotypes, the number of days from calving to the first ovulation postpartum was observed. Ovarian ultrasound was used to observe CL presence after calving to indicate an ovulatory event.
For AGECL and PPAI, CGs were formed by concatenating information of cohort, year of birth, and management group. For SCB, CG included the effects of cohort and year of birth. Age of young bull at recording was used as a covariate for SCB.
Brahman animals were genotyped using the Illumina BovineSNP50 version 1 or 2, and posteriorly imputed for high-density panel using Beagle software (Browning and Browning, 2009). To allow imputation, representative animals of the Beef CRC population were genotyped using the HD chip from Illumina as described by Fortes et al. (2013). Quality control excluded animals with call rates < 98%, SNP with call rates < 85% and MAF < 0.02. After quality control remained 660,433 SNP for PPAI, 659,845 for AGECL and 465,644 for SCB.
Genome-Wide Association Methods and Meta-analyses
A single-trait single-marker model was used to perform GWAS for all traits using the genomic mixed model proposed by Kang et al. (2010), which accounts for population substructure, fitted as follows for Nellore (1) and Brahman (2):
| (1) |
| (2) |
where y is a vector containing the phenotypic information, corrected for the fixed effects in Nellore data (equation 1), is a vector of ones, is an overall mean, X is an incidence matrix relating fixed effects (CG) in β with the phenotypes in y, s is the vector containing the genotypes coded as 0, 1, or 2 according to the number of B allele copies, a is the vector with the SNP additive genetic substitution effects, Z is the incidence matrix of polygenic random effects of the animals in u and ε is the vector of residuals. u and ε followed normal distribution with u~N (0, G ) and ε~N (0, I ), respectively, where G is the genomic relationship matrix for all individuals and SNP (except the SNP considered in a), calculated as described in the first method proposed by VanRaden (2008), is the additive genetic variance, I is an identity matrix, and is the residual variance. The SNP effect estimates were computed using the SNP & Variation Suite v8.3.0 software (Golden Helix, Inc., Bozeman, MT) under the model previously described and the EMMAX method (Kang et al., 2010).
Multitrait meta-analyses were performed using the method described by Bolormaa et al. (2014). This method consists of a statistic test following a χ2 distribution with n degrees of freedom, where n is the number of traits included in meta-analysis, calculated as:
| (3) |
where ti is a vector 6 × 1 of the ith SNP effect estimates for the six traits divided by their respective standard errors, t′i is the transpose vector of ti, and V−1 is an inverse of the 6 × 6 correlation matrix of the correlations between the t-values of the six traits across the 387,971 SNP considered in the model. The t-values vector was computed as:
| (4) |
where SE(ai) is the SE of ai (SNP effect vector). Just common SNP in both Nellore and Brahman panels were considered in the analysis. False discovery rate (FDR) (f) was calculated as:
| (5) |
where m corresponds to the number of tests (markers) considered in the meta-analysis, α is the significant threshold, and s is the number of tests, where p < α, where p is the correspondent observed p-value for each marker. To select α values producing an FDR lower than 0.05, the p-value rank position procedure (Benjamini and Hochberg, 1995) was used. In summary, equation 5 was changed to obtain α =fs/m, and s was defined as the largest p-value rank position i that satisfies pi ≤ fi/m with f = 0.05 (Pereira et al., 2016).
Gene Annotation and Enrichment Analysis
The genes within 0.5 Mb of significant SNP (P < 1.39 × 10−5) were annotated as candidate genes. These candidate genes were used to perform gene ontology (GO) and enrichment analyses, using DAVID bioinformatics resources (Huang da et al., 2009).
RESULTS AND DISCUSSION
Significant SNP
Heritabilities were previously reported for the traits and populations studied here. They were 0.08 for AFC, 0.30 for EP, 0.41 for SCN, 0.57 for AGECL, 0.52 for PPAI, and 0.75 for SCB (Forni and Albuquerque, 2005; Johnston et al., 2009, 2010; Corbet et al., 2013; Irano et al., 2016).
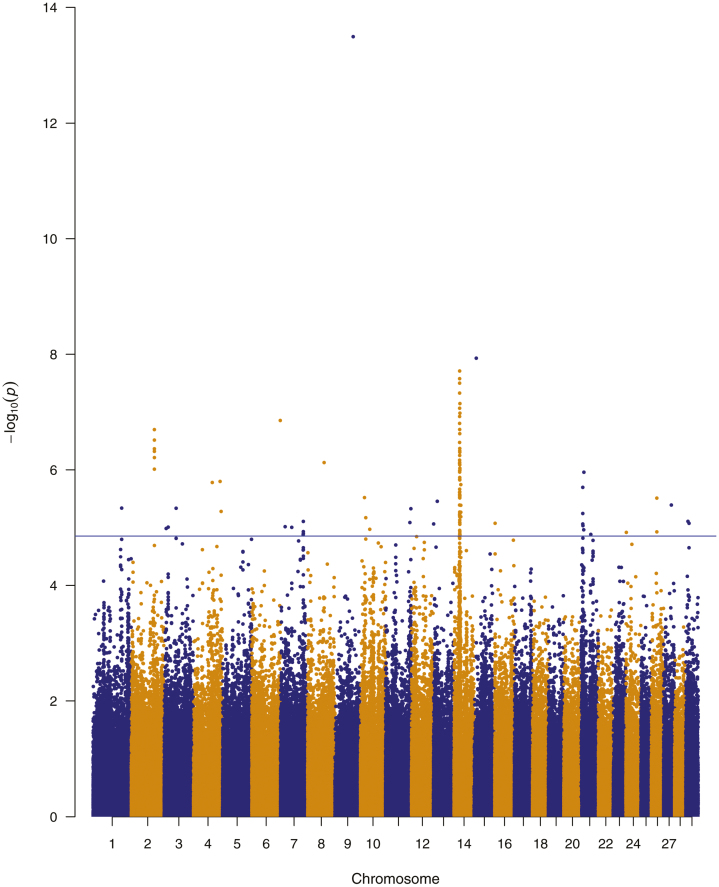
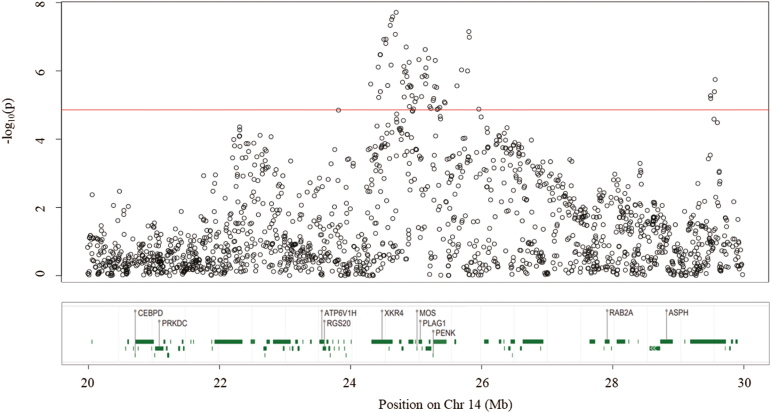
Meta-analysis resulted in 108 significant SNPs, at an empirical threshold P-value of 1.39 × 10−5 (Figure 1). This number of significant SNP was the maximum rank position, which P-value satisfied the condition P ≤ α with FDR = 0.05 (Supplementary Table S1). Most of the significant SNP (59.3%) were mapped to chromosome 14, producing a strong association peak between 24.3 and 26 Mb (Figure 2), in which 14 genes were harbored (XKR4, TRNAT-AUG, TMEM68, TGS1, LYN, RPS20, MOS, PLAG1, CHCHD7, SDR16C5, SDR16C6, PENK, LOC101907667, IMPAD1). Another region in BTA14, from 29.5 to 29.6 Mb presented a smaller peak, in which four SNPs were significant. All those SNPs (rs109465877, rs111023138, rs134875123, and rs42534153) were located in an intronic region of the gene NKAIN3, which was in a genomic window explaining 1% of the genetic variance of early pregnancy in Nellore heifers (Oliveira Junior et al., 2017).
Figure 1.
Manhattan plot of the meta-analysis for sexual precocity traits in Nellore and Brahman cattle. The y-axis represents the log inverse P-values for SNP associations and the x-axis represents the position in base pairs from chromosome 1 to 29. The blue line indicates genome-wide significance, a P-value cutoff of P < 1.39 × 10−5, which is equivalent of an FDR lower than 5%.
Figure 2.
Zoom in BTA14, region of the strongest peak. SNPs above the red line were significant (P < 1.39 × 10−5). In the bottom, genes mapped in this region (total = 87).
The peak observed in chromosome 14 is close to significant regions that were associated with Brahman and Nellore reproductive traits (Fortes et al., 2012; Fortes et al., 2013; Irano et al., 2016). The PLAG1 gene (mapped to 25,000,459 to 25,052,403 bp), a key gene associated with several traits of economic importance in different cattle breeds (Karim et al., 2011; Littlejohn et al., 2012; Nishimura et al., 2012; Fortes et al., 2013; Utsunomiya et al., 2013; Saatchi et al., 2014; Juma et al., 2016; Pereira et al., 2016) is located in this region. One of the most significant SNPs (P-value = 6.39 × 10−6), the rs109815800 was mapped in an intronic region of PLAG1.
Fortes et al. (2012) also found a region in chromosome 14 associated with AGECL and the age in which bull reach 26 cm of SC—phenotypes that represent the age at puberty for both females and males—for the same Brahman population used in this study. One of the most significant SNPs also was close to SDR16C6 and PLAG1 genes.
Despite this evident peak in chromosome 14, the most significant SNP (P-value = 3.21 × 10−14) of the meta-analysis was located in chromosome 9 at 75.6 Mb. This SNP is in an intronic region of the MAP3K5 gene, which was identified as an estrogen receptor in human (Mirkin et al., 2005).
In Table 2 are presented the most significant SNP of each chromosome, its position and the closest gene. In BTA1, TSC22D2 emerged as a candidate gene in our meta-analysis study. This gene was high-expressed in human oocytes (Kakourou et al., 2013). In BTA2, KLF7 was the closest gene to the top SNP (rs134877457). It is associated with growth traits in Chinese cattle (Ma et al., 2011); presents an important role in mouse neurogenesis (Laub et al., 2001; Caiazzo et al., 2010) and olfactory neurogenesis (Kajimura et al., 2007); and plays a role in adipogenesis regulation in humans (Kawamura et al., 2006).
Table 2.
Number of significant SNP (P < 1.39 × 10−5) per Bos taurus autosome (BTA), the most significant SNP in each autosome (top SNP), its position, and its distance from the closest gene in base pairs
| BTA | Number of SNP | Top SNP | Position, bp | Gene Symbol | Distance, bp |
|---|---|---|---|---|---|
| 1 | 1 | rs110366479 | 118,606,312 | TSC22D2 | 5,852 |
| 2 | 6 | rs134877457 | 95,917,958 | KLF7 | 14,648 |
| 3 | 3 | rs109478958 | 49,436,731 | ARHGAP29 | 0 |
| 4 | 3 | rs136050748 | 110,440,534 |
Uncharacterized Protein
(ENSBTAG00000048097) |
530,702 |
| 6 | 1 | rs43492923 | 118,436,689 | 7SK – misc RNA 1 | 2,828 |
| 7 | 6 | rs135631400 | 94,710,749 | 7SK – misc RNA 1 | 510,435 |
| 8 | 1 | rs136590180 | 68,307,110 | Uncharacterized Protein (ENSBTAG00000012266) | 16,590 |
| 9 | 1 | rs110257163 | 75,613,158 | MAP3K5 | 0 |
| 10 | 3 | rs110458186 | 16,769,219 | TLE3 | 132,436 |
| 11 | 2 | rs108980439 | 104,936,387 | WDR5 | 0 |
| 13 | 2 | rs136854801 | 16,097,639 | TAF3 | 0 |
| 14 | 64 | rs109748092 | 24,710,609 | TMEM68 | 718 |
| 15 | 1 | rs110493922 | 9,064,376 | Uncharacterized Protein (ENSBTAG00000047425) | 141,535 |
| 16 | 1 | rs109871859 | 1,929,989 | PPP1R15B | 36,732 |
| 21 | 7 | rs110478544 | 11,435,365 | NR2F2 | 628,702 |
| 24 | 1 | rs133759831 | 2,276,768 | GALR1 | 129,893 |
| 26 | 2 | rs135708259 | 23,405,679 | SUFU | 47,301 |
| 27 | 1 | rs135961785 | 31,926,152 | KCNU1 | 0 |
| 29 | 2 | rs109184359 | 9,174,027 | Uncharacterized Protein (ENSBTAG00000046374) | 1,952 |
1miscRNA, miscellaneous RNA.
In BTA3 the top SNP (rs109478958) suggested ARHGAP29 as a candidate gene. Its protein was upregulated in bull semen with good quality, compared with bad-quality semen (Singh, et al., 2018). The top SNPs for chromosomes 6 (rs43492923) and 7 (rs135631400) were close to 7SK, a small nuclear ribonucleoprotein-coding gene (snRNA). This snRNA plays a role in DNA transcription and was found close to a significant SNP for bull fertility in chromosome 10 (Suchocki and Szyda, 2015); has a critical role in the control of primordial germ cell proliferation of mouse embryos (Okamura et al., 2012); and was detected in seminal plasma, ejaculated sperm, and epididymal sperm in pigs with a significant biological role in spermatogenesis (Chen et al., 2017).
In BTA9, the gene MAP3K5 was the closest to the top SNP (rs110257163) and was previously associated with regulation of apoptosis in porcine ovarian granulosa cells; the expression of MAP3K5 was increased when the levels of FSH hormone increased in those cells (Sirotkin et al., 2008). Also, this gene was a strong candidate associated with growth and reproduction traits in transcriptome analysis of bovine pituitary gland (Pareek et al., 2016), and it presented some interactions with estrogen receptors in rat primary cortical neurons (Singer et al., 1999).
In BTA10, TLE3 (closest to the top SNP rs110458186) was expressed in mouse mesoderm embryonic cells (Pfeffer et al., 2017). The expression of this gene was associated with FSH hormone increased levels in bovine granulosa cells (Nivet et al., 2018) and with estrogen receptors in human breast cancer cell line (Jangal et al., 2014). This gene was associated with mouse embryo survival. TLE3 mutants presented placenta defects (Gasperowicz et al., 2013) and this gene acted as a co-regulator of adipogenesis in mouse (Villanueva et al., 2011). This gene also affects the embryonic stem cell differentiation (Laing et al., 2015).
In BTA11, the candidate gene WDR5 presents an important role in porcine early embryo development (Ding et al., 2017) and was essential for vertebrate development (Wysocka et al., 2005; Gori et al., 2006). In BTA13, the most significant SNP (rs136854801) was located in an intronic region of the TAF3 gene. This gene harbors an SNP marker associated with the major histocompatibility complex (MHC) in an Italian human population (Pistis et al., 2013). The importance of the MHC for reproduction was previously discussed in Ziegler et al. (2010). Besides that, this gene was associated with endoderm differentiation in mouse embryonic stem cells (Liu et al., 2011), was significantly upregulated in human oocyte, and was enriched in the estrogen receptor signaling pathway (Kocabas et al., 2006).
In BTA14, TMEM68 was identified as a candidate gene. This gene was expressed in human epididymis (Belleannée et al., 2012), was upregulated in heifer blastocyst (Carter, et al., 2010) and was associated with feed intake and growth traits in cattle (Lindholm-Perry et al., 2012). In BTA16, PPP1R15B was the gene closest to the top SNP (rs109871859). It was expressed in bovine cumulus oocyte cells (Abd El Naby et al., 2013) and in bovine granulosa cells under FSH stimulation (Nivet et al., 2018). PPP1R15 gene knockout mouse embryos survived, but had several problems as grown retardation (Harding et al., 2009). In BTA21, the emergent candidate, NR2F2, was previously associated with Leydig cell steroidogenesis in mice testis cells (Mendoza-Villarroel et al., 2014). Bauersachs et al. (2006) concluded that this gene was associated with embryo implantation in cows. In BTA24 the gene closest to the top SNP (rs133759831) was GALR1. This gene was differentially expressed in ovine hypothalamus in pre-pubertal age compared with fetal and adult ages, suggesting a regulation role in the beginning of reproduction (Whitelaw et al., 2009). Also, it was located near a significant SNP associated with oligozoospermia and azoospermia in humans (Aston and Carrell, 2009), was associated with the regulation of ovarian steroids in rats (Mitchell et al., 2004), and interacted with testosterone in male rat brain (Bouret et al., 2000). In BTA26, SUFU was mapped close to the top SNP (rs135708259). This gene was a candidate gene associated with spermatogenesis in mouse, and it was found in spermatocytes (Szczepny et al., 2005). In BTA27, the KCNU1 was the closest gene to the top SNP (rs135961785). This gene presents an important role in semen quality (Santi et al., 2010), which could be observed in a knockout mouse experiment (SLO3, null mutation). Also Zeng et al. (2011) reported the importance of SLO3 for male fertility, by evaluating its role in the normal morphology and sperm motility in mouse. This gene was enriched in the pathway “Vascular smooth muscle contraction,” which was activated by progesterone treatment in neonatal mouse uterus, inhibiting its development (Filant et al., 2012).
On chromosomes 4, 8, 15, and 29, the top SNPs were close to uncharacterized proteins, and their identification is presented in Table 2. No significant SNPs were observed in BTA5, BTA12, BTA17, BTA18, BTA19, BTA20, BTA22, BTA23, BTA25, and BTA28. Evidence from the current meta-analysis seems to corroborate with previous knowledge of gene function for the discussed genes. Although the majority of the significant SNPs were located in chromosome 14, significant SNPs were also mapped in multiple chromosomes, which is consistent with the idea that reproductive traits are complex polygenic phenotypes.
Functional Annotation and Enrichment Analyses
A total of 389 genes were located within 0.5 Mb of the 108 significant SNP (P < 1.39 × 10−5). From these genes, functional annotation and enrichment analyses were performed and 333 genes were identified by DAVID software (Huang da et al., 2009), using bovine genome as a background gene list. Enrichment analyses identified 48 functional gene groups as significant in DAVID resources, using high astringency criterion. The top five functional gene groups, with higher enrichment score (ES), are presented in Table 3.
Table 3.
Top five most significant functional gene groups enriched in DAVID v.6.7 bioinformatics resources (https://david.ncifcrf.gov/home.jsp)
| Functional groups | Top GO term, code ~ name | Ontology | ES | FDR | Genes |
|---|---|---|---|---|---|
| 1 | IPR001751~ S100/CaBP-9k-type, calcium binding |
7.06 | 5.2E-6 | S100A13, S100A1, S100A14, S100A16, S100A2, S100A3, S100A4, S100A7 | |
| 2 | UP_SEQ_FEATURE~ domain: EF-hand 1 |
2.79 | 1.2E-1 | S100A13, S100A1, S100A14, S100A16, S100A2, S100A4, CAPS, NCS1, S100A7 | |
| 3 | GO:0016477~ Cell migration |
Biological process | 2.59 | 2.2E0 |
FCER1G, cdk5, DBH, MP14,
NOS3, NR2F2, pex7, LOC515718 |
| 4 | GO:0019864~ IgG binding |
Molecular function | 1.99 | 1.7E0 | FCER1G, FCGR3, FCGR2B |
| 5 | GO:0051047~ Positive regulation of secretion |
Biological process | 1.65 | 1.6E1 | FCER1G, CHRNB2, cdk5, serp1 |
GO terms, ES, and FDR are reported for each functional group; genes in each group are also reported.
Groups 1 (S100/CaBP-9k-type, calcium binding) and 2 (domain: EF-hand 1) were related to calcium binding pathway. S100 proteins are a family protein expressed just in vertebrates. They participate in several biological events, as regulation of proliferation, differentiation, apoptosis, and inflammation (Donato et al., 2013). Calbindin-D9k (CaBP-9k-type) is a vitamin-D-dependent Ca2+ binding protein, member of the S100 family. This protein is mainly found in intestinal tissue, but was also found in other tissues as uterus and placenta (Mathieu et al., 1989; Krisinger et al., 1995; Emam et al., 2016). All the genes clustered in group 1 were members of S100A family. This gene family was expressed in ovary, prostate, and testis of human and rats (Wicki et al., 1996) and in thyroid gland, placenta, and prostate (Cannon et al., 2011). Their roles are generally associated with immunological function, promoting the sterility of reproductive tissues against pathogens (Germeyer et al., 2009; Teijeiro and Marini, 2015); however, they were also related to embryo implantation process (Gray et al., 2006; Smits et al., 2018), to response to temperature stress (Landriscina et al., 2001), cell proliferation, and apoptosis (Jin et al., 2011).
The majority of genes clustered in group 2 were also in group 1. This functional group, EF-hand 1, presents a group of proteins that share a similar structure, which is commonly found in calcium binding proteins. Besides of some genes in common with group 1, other two genes were enriched in this group, CAPS, NCS1. CAPS gene was expressed in human epididymis and endometrium during follicular phase (Thimon et al., 2007; Blockeel et al., 2011), while NCS1 was expressed in bovine blastocysts cultured in vitro under stimulation of thyroid hormones, presenting an important role in mammalian neuron-memory development (Ashkar et al., 2016).
Group 3 (Cell migration) is a fundamental process in the development and maintenance of multicellular organisms. Several reproductive events depend on this process, as embryo formation (Jovanović et al., 2010), immunological responses (Sánchez‐Madrid and del Pozo, 1999), spermatogenesis (Smith et al., 2012), and oogenesis (Rørth et al., 2000). Genes clustered in this group were associated with daughter pregnancy rate, cow conception rate (Ortega et al., 2016), development of nervous system (Kapur et al., 1991; Kanaani et al., 2005; Jessberger et al., 2009), embryo lethality (Nourizadeh-Lillabadi et al., 2010), postnatal mortality and growth retardation (Pallares and Gonzalez–Bulnes, 2010), immunity (Bogdan, 2015), embryo differentiation (Rosa and Brivanlou, 2011), and perinatal mortality (Brites et al., 2003).
The group 4 clustered genes associated with the immunoglobulin G (IgG). This is the most common isotype in human blood and extracellular fluid (Janeway et al., 2007), and the most abundant antibody class present in healthy individuals. This antibody was found in the mucus of woman reproductive tract in Fahrbach et al. (2013), which could suggest a role in the fortification of mucus barriers in the female reproductive tract. Besides that, mouse experiments were performed with a specific monoclonal IgM and demonstrated that females immunized with this antibody presented gestational failure (Sthoeger et al., 1993). A similar effect was observed in another mouse experiment, by Ornoy et al. (2003), where the administration of a purified IgG in pregnant mouse reduced the yolk sac and the embryo growth. This pathway enriched the genes FCER1G, FCGR3A, and FCGR2B, which were associated with daughter pregnancy rate, cow conception rate (Ortega et al., 2016), embryo–maternal recognition in cattle (Mamo et al., 2012), and infertility in female mouse (Wetendorf et al., 2017).
The fifth top group was “Positive regulation of secretion.” As group 3, this group is related to a large variety of biological mechanisms, mainly mediated by regulatory hormones. Genes clustered in this group were associated with positive regulation of immune response (Macen et al., 1993; Kiba, 2016), neurogenesis (Jessberger et al., 2009), and response to stress (Faria et al., 2012).
David results presented gene clusters that were mostly associated with immune events. Earlier studies with Bos indicus cattle have found genes playing roles in immune responses, adaptability, and reproduction (Bahbahani et al., 2018). Fayemi (2005) observed that the presence of antibodies (IgG) in bulls’ sperm was related with fertility. Our results are in agreement with previous studies with Bos indicus cattle, pointing the importance of the immune competence to guarantee reproductive success in Bos indicus cattle.
Besides most of significant SNPs were located in chromosome 14, our meta-analysis identified genomic regions harboring significant SNP associations in the majority of the autosome chromosomes (19 out of 29), as expected for polygenic traits. The genes mapped within these associated regions are plausible candidate genes because of their position and due to previous evidence suggesting their functional association with reproduction, as discussed above. The candidate genes identified could be grouped in functional categories that seem to affect immune system and consequently, mammalian reproduction.
In summary, five regions distributed over four chromosomes presented the majority of the most significant SNP (76%): BTA2 (5.55%) from 95 to 96 Mb, BTA4 (5.55%) from 94.1 to 94.8 Mb, BTA14 (59.26%) from 24 to 25 Mb and 29 to 30 Mb, and BTA21 (5.55%) from 6.7 Mb to 11.4 Mb. Two independent populations were studied here, animals from different breeds, born and bred in different geographic regions. The genes identified as candidates in the meta-analysis are probably affecting physiological mechanisms that control sexual precocity in both breeds, because we found key genes expressed in reproductive tissues and playing important roles in immune response (including in reproductive tract), maternal–fetal tolerance, pregnancy maintenance, embryo development, fertility, and response to stress, or were located in previously reported QTL regions for reproductive traits in bovine. Also, we found some candidate genes associated with important hormones for the regulation of puberty in mammalian, as the FSH, estrogens, and testosterone. The importance of these hormones for sexual precocity is expected to be conserved among mammalians.
In short, the genes presented here are both positional and functional candidates for sexual precocity in Bos indicus cattle. Future works could fine-map and validate the identified genomic regions and elucidate which genes are in fact harboring the mutations that could explain the proposed QTL.
SUPPLEMENTARY DATA
Supplementary data are available at Journal of Animal Science online.
Footnotes
The authors acknowledge the researcher mobility International Cooperation Program CAPES/COFECUB (88881.133149/2016-01) that facilitated the collaborations between UNESP and the University of Queensland. Financial support for this research was granted by Fundação de Amparo à Pesquisa (FAPESP—2009/16118-5), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq—559631/2009-0), and Coordenação de Aperfeiçoamento de pessoal de nível superior (CAPES). This work used the legacy dataset of the Cooperative Research Centre for Beef Genetic Technologies (www.beefcrc.com) and the Alliance Nellore dataset (www.gensys.com.br).
LITERATURE CITED
- Abd El Naby W. A., Hagos T. H., Hossain M. M., Salilew-Wondim D., Gad A. Y., Rings F., Cinar M. U., Tholen E., Looft C., Schellander K.,. et al. 2013. Expression analysis of regulatory microRNAs in bovine cumulus oocyte complex and preimplantation embryos. Zygote 21:31–51. doi:10.1017/S0967199411000566 [DOI] [PubMed] [Google Scholar]
- Ashkar F. A., Revay T., Rho N., Madan P., Dufort I., Robert C., Favetta L. A., Schmidt C., and King W. A.. 2016. Thyroid hormones alter the transcriptome of in vitro-produced bovine blastocysts. Zygote 24:266–276. doi:10.1017/S0967199415000167 [DOI] [PubMed] [Google Scholar]
- Aston K. I., and Carrell D. T.. 2009. Genome-wide study of single-nucleotide polymorphisms associated with azoospermia and severe oligozoospermia. J. Androl. 30:711–725. doi:10.2164/jandrol.109.007971 [DOI] [PubMed] [Google Scholar]
- Bahbahani H., Salim B., Almathen F., Al Enezi F., Mwacharo J. M., and Hanotte O.. 2018. Signatures of positive selection in African Butana and Kenana Dairy Zebu cattle. Plos One 13:e0190446. doi:10.1371/journal.pone.0190446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauersachs S., Ulbrich S. E., Gross K., Schmidt S. E., Meyer H. H., Wenigerkind H., Vermehren M., Sinowatz F., Blum H., and Wolf E.. 2006. Embryo-induced transcriptome changes in bovine endometrium reveal species-specific and common molecular markers of uterine receptivity. Reproduction 132:319–331. doi:10.1530/rep.1.00996 [DOI] [PubMed] [Google Scholar]
- Belleannée C., Calvo E., Thimon V., Cyr D. G., Légaré C., Garneau L., and Sullivan R.. 2012. Role of microRNAs in controlling gene expression in different segments of the human epididymis. Plos One 7:e34996. doi:10.1371/journal.pone.0034996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., and Hochberg Y.. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B Stat. Methodol. 57:289–300. doi:10.2307/2346101 [Google Scholar]
- Blockeel C., Van Vaerenbergh I., Fatemi H. M., Van Lommel L., Devroey P., and Bourgain C.. 2011. Gene expression profile in the endometrium on the day of oocyte retrieval after ovarian stimulation with low-dose HCG in the follicular phase. Mol. Hum. Reprod. 17:33–41. doi:10.1093/molehr/gaq070 [DOI] [PubMed] [Google Scholar]
- Bogdan C. 2015. Nitric oxide synthase in innate and adaptive immunity: an update. Trends Immunol. 36:161–178. doi:10.1016/j.it.2015.01.003 [DOI] [PubMed] [Google Scholar]
- Bolormaa S., Pryce J. E., Reverter A., Zhang Y., Barendse W., Kemper K., Tier B., Savin K., Hayes B. J., and Goddard M. E.. 2014. A multi-trait, meta-analysis for detecting pleiotropic polymorphisms for stature, fatness and reproduction in beef cattle. Plos Genet. 10:e1004198. doi:10.1371/journal.pgen.1004198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouret S., Prevot V., Croix D., Howard A., Habert-Ortoli E., Jegou S., Vaudry H., Beauvillain J. C., and Mitchell V.. 2000. Expression of Galr1 and Galr2 galanin receptor messenger ribonucleic acid in proopiomelanocortin neurons of the rat arcuate nucleus: effect of testosterone. Endocrinology 141:1780–1794. doi:10.1210/endo.141.5.7469 [DOI] [PubMed] [Google Scholar]
- Brites P., Motley A. M., Gressens P., Mooyer P. A., Ploegaert I., Everts V., Evrard P., Carmeliet P., Dewerchin M., Schoonjans L.,. et al. 2003. Impaired neuronal migration and endochondral ossification in Pex7 knockout mice: a model for rhizomelic chondrodysplasia punctata. Hum. Mol. Genet. 12:2255–2267. doi:10.1093/hmg/ddg236 [DOI] [PubMed] [Google Scholar]
- Browning B. L., and Browning S. R.. 2009. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84:210–223. doi:10.1016/j.ajhg.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumatti R. C., Ferraz J. B. S., Eler J. P., and Formigonni I. B.. 2011. Development of selection index in beef cattle under focus of a bio-economic model. Arch. Zootec. 60:205–213. doi:10.4321/S0004-05922011000200005 [Google Scholar]
- Burns B. M., Corbet N. J., Corbet D. H., Crisp J. M., Venus B. K., Johnston D. J., Li Y., McGowan M. R., and Holroyd R. G.. 2013. Male traits and herd reproductive capability in tropical beef cattle. 1. Experimental design and animal measures. Anim. Prod. Sci. 53:87–100. doi:10.1071/AN12162 [Google Scholar]
- Caiazzo M., Colucci-D’Amato L., Esposito M. T., Parisi S., Stifani S., Ramirez F., and di Porzio U.. 2010. Transcription factor KLF7 regulates differentiation of neuroectodermal and mesodermal cell lineages. Exp. Cell Res. 316:2365–2376. doi:10.1016/j.yexcr.2010.05.021 [DOI] [PubMed] [Google Scholar]
- Cannon B. R., Zimmer D. B., and Weber D. J.. 2011. S100A1 (S100 calcium binding protein A1). Atlas Genet. Cytogenet. Oncol. Haematol. 15:873–876. doi:10.4267/2042/46035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso R. C., Alves B. R., Sharpton S. M., Williams G. L., and Amstalden M.. 2015. Nutritional programming of accelerated puberty in heifers: involvement of pro-opiomelanocortin neurones in the arcuate nucleus. J. Neuroendocrinol. 27:647–657. doi:10.1111/jne.12291 [DOI] [PubMed] [Google Scholar]
- Carter F., Rings F., Mamo S., Holker M., Kuzmany A., Besenfelder U., Havlicek V., Mehta J. P., Tesfaye D., Schellander K.,. et al. 2010. Effect of elevated circulating progesterone concentration on bovine blastocyst development and global transcriptome following endoscopic transfer of in vitro produced embryos to the bovine oviduct. Biol. Reprod. 83:707–719. doi:10.1095/biolreprod.109.082354 [DOI] [PubMed] [Google Scholar]
- Carvalheiro R., Boison S. A., Neves H. H., Sargolzaei M., Schenkel F. S., Utsunomiya Y. T., O’Brien A. M., Sölkner J., McEwan J. C., Van Tassell C. P.,. et al. 2014. Accuracy of genotype imputation in Nellore cattle. Genet. Sel. Evol. 46:69. doi:10.1186/s12711-014-0069-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Wu H., Shen D., Wang S., Zhang L., Wang X., Gao B., Wu T., Li B., Li K.,. et al. 2017. Comparative profiling of small RNAs of pig seminal plasma and ejaculated and epididymal sperm. Reproduction 153:785–796. doi:10.1530/REP-17-0014 [DOI] [PubMed] [Google Scholar]
- Corbet N. J., Burns B. M., Johnston D. J., Wolcott M. L., Corbet D. H., Venus B. K., Li Y., McGowan M. R., and Holroyd R. G.. 2013. Male traits and herd reproductive capability in tropical beef cattle. 2. Genetic parameters of bull traits. Anim. Prod. Sci. 53:101–113. doi:10.1071/AN12163 [Google Scholar]
- Costa R. B., Camargo G. M., Diaz I. D., Irano N., Dias M. M., Carvalheiro R., Boligon A. A., Baldi F., Oliveira H. N., Tonhati H.,. et al. 2015. Genome-wide association study of reproductive traits in Nellore heifers using Bayesian inference. Genet. Sel. Evol. 47:67. doi:10.1186/s12711-015-0146-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B., Cao Z., Hong R., Li H., Zuo X., Luo L., Li Y., Huang W., Li W., Zhang K.,. et al. 2017. WDR5 in porcine preimplantation embryos: expression, regulation of epigenetic modifications and requirement for early development. Biol. Reprod. 96:758–771. doi:10.1093/biolre/iox020 [DOI] [PubMed] [Google Scholar]
- Donato R., Cannon B. R., Sorci G., Riuzzi F., Hsu K., Weber D. J., and Geczy C. L.. 2013. Functions of S100 proteins. Curr. Mol. Med. 13:24–57. doi:10.2174/156652413804486214 [PMC free article] [PubMed] [Google Scholar]
- Emam M. A., Abouelroos M. E., and Gad F. A.. 2016. Expression of calbindin-D9K and vitamin D receptor in the uterus of Egyptian buffalo during follicular and luteal phases. Acta Histochem. 118:471–477. doi:10.1016/j.acthis.2016.04.009 [DOI] [PubMed] [Google Scholar]
- Fahrbach K. M., Malykhina O., Stieh D. J., and Hope T. J.. 2013. Differential binding of IgG and IgA to mucus of the female reproductive tract. Plos One 8:e76176. doi:10.1371/journal.pone.0076176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria D., Lentze N., Almaça J., Luz S., Alessio L., Tian Y., Martins J. P., Cruz P., Schreiber R., Rezwan M.,. et al. 2012. Regulation of ENaC biogenesis by the stress response protein SERP1. Pflugers Arch. 463:819–827. doi:10.1007/s00424-012-1091-1 [DOI] [PubMed] [Google Scholar]
- Fayemi O. 2005. Sperm antibodies and reproductive efficiency in the Zebu cattle in south-western Nigeria. Pakistan Vet. J. 25:111–114. [Google Scholar]
- Filant J., Zhou H., and Spencer T. E.. 2012. Progesterone inhibits uterine gland development in the neonatal mouse uterus. Biol. Reprod. 86:146, 1–146, 9. doi:10.1095/biolreprod.111.097089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formigoni I. B., Ferraz J. B. S., Silva J. A. I. I. V., Eler J. P., and Brumatti R. C.. 2005. Economic value for stayability and heifer pregnancy at 14 months in beef cattle herds. Arq. Bras. Med. Vet. Zootec. 57:220–226. doi:10.1590/S0102-09352005000800013 [Google Scholar]
- Forni S., and Albuquerque L. G.. 2005. Estimates of genetic correlations between days to calving and reproductive and weight traits in Nellore cattle. J. Anim. Sci. 83:1511–1515. doi:10.2527/2005.8371511x [DOI] [PubMed] [Google Scholar]
- Fortes M. R., Kemper K., Sasazaki S., Reverter A., Pryce J. E., Barendse W., Bunch R., McCulloch R., Harrison B., Bolormaa S.,. et al. 2013. Evidence for pleiotropism and recent selection in the plag1 region in Australian beef cattle. Anim. Genet. 44:636–647. doi:10.1111/age.12075 [DOI] [PubMed] [Google Scholar]
- Fortes M. R. S., Lehnert S. A., Bolormaa S., Reich C., Fordyce G., Corbet N. J., Whan V., Hawken R. J., and Reverter A.. 2012. Finding genes for economically important traits: Brahman cattle puberty. Anim. Prod. Sci. 52:143–150. doi:10.1071/AN11165 [Google Scholar]
- Garcia-Ruiz A., Cole J. B., VanRaden P. M., Wiggans G. R., Ruiz-López F. J., and Van Tassell C. P.. 2016. Changes in genetic selection differentials and generation intervals in US Holstein dairy cattle as a result of genomic selection. Proc. Natl. Acad. Sci. U. S. A. 113:E3995–E4004. doi:10.1073/pnas.1519061113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasperowicz M., Surmann-Schmitt C., Hamada Y., Otto F., and Cross J. C.. 2013. The transcriptional co-repressor TLE3 regulates development of trophoblast giant cells lining maternal blood spaces in the mouse placenta. Dev. Biol. 382:1–14. doi:10.1016/j.ydbio.2013.08.005 [DOI] [PubMed] [Google Scholar]
- Germeyer A., Sharkey A. M., Prasadajudio M., Sherwin R., Moffett A., Bieback K., Clausmeyer S., Masters L., Popovici R. M., Hess A. P.,. et al. 2009. Paracrine effects of uterine leucocytes on gene expression of human uterine stromal fibroblasts. Mol. Hum. Reprod. 15:39–48. doi:10.1093/molehr/gan075 [DOI] [PubMed] [Google Scholar]
- Gori F., Friedman L. G., and Demay M. B.. 2006. Wdr5, a WD-40 protein, regulates osteoblast differentiation during embryonic bone development. Dev. Biol. 295:498–506. doi:10.1016/j.ydbio.2006.02.031 [DOI] [PubMed] [Google Scholar]
- Gray C. A., Abbey C. A., Beremand P. D., Choi Y., Farmer J. L., Adelson D. L., Thomas T. L., Bazer F. W., and Spencer T. E.. 2006. Identification of endometrial genes regulated by early pregnancy, progesterone, and interferon tau in the ovine uterus. Biol. Reprod. 74:383–394. doi:10.1095/biolreprod.105.046656 [DOI] [PubMed] [Google Scholar]
- Harding H. P., Zhang Y., Scheuner D., Chen J. J., Kaufman R. J., and Ron D.. 2009. Ppp1r15 gene knockout reveals an essential role for translation initiation factor 2 alpha (eIF2α) dephosphorylation in mammalian development. Proc. Natl. Acad. Sci. USA. 106:1832–1837. doi:10.1073_pnas.0809632106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawken R. J., Zhang Y. D., Fortes M. R., Collis E., Barris W. C., Corbet N. J., Williams P. J., Fordyce G., Holroyd R. G., Walkley J. R.,. et al. 2012. Genome-wide association studies of female reproduction in tropically adapted beef cattle. J. Anim. Sci. 90:1398–1410. doi:10.2527/jas.2011-4410 [DOI] [PubMed] [Google Scholar]
- Henderson C. R. 1984. Applications of linear models in animal breeding. Guelph: University of Guelph. [Google Scholar]
- Huang da W., Sherman B. T., and Lempicki R. A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. doi:10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Irano N., de Camargo G. M., Costa R. B., Terakado A. P., Magalhães A. F., Silva R. M., Dias M. M., Bignardi A. B., Baldi F., Carvalheiro R.,. et al. 2016. Genome-wide association study for indicator traits of sexual precocity in Nellore cattle. Plos One 11:e0159502. doi:10.1371/journal.pone.0159502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway C. A., Travers P., Walport M., and Shlomchik M. J.. 2007. Imunobiologia: o sistema imune na saúde e na doença. 6th ed Sao Paulo: Artmed. [Google Scholar]
- Jangal M., Couture J. P., Bianco S., Magnani L., Mohammed H., and Gévry N.. 2014. The transcriptional co-repressor TLE3 suppresses basal signaling on a subset of estrogen receptor α target genes. Nucleic Acids Res. 42:11339–11348. doi:10.1093/nar/gku791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessberger S., Gage F. H., Eisch A. J., and Lagace D. C.. 2009. Making a neuron: Cdk5 in embryonic and adult neurogenesis. Trends Neurosci. 32:575–582. doi:10.1016/j.tins.2009.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Chen H., Luo A., Ding F., and Liu Z.. 2011. S100A14 stimulates cell proliferation and induces cell apoptosis at different concentrations via receptor for advanced glycation end products (RAGE). Plos One 6:e19375. doi:10.1371/journal.pone.0019375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston D. J., Barwick S. A., Corbet N. J., Fordyce G., Holroyd R. G., Williams P. J., and Burrow H. M.. 2009. Genetics of heifer puberty in two tropical beef genotypes in northern Australia and associations with heifer- and steer-production traits. Anim. Prod. Sci. 49:399–412. doi:10.1071/EA08276 [Google Scholar]
- Johnston D. J., Barwick S. A., Fordyce G., Holroyd R. G.. 2010. Understanding the genetics of lactation anoestrus in Brahman beef cattle to enhance genetic evaluation of female reproductive traits. Proceedings of the 9th World Congress on Genetics Applied to Livestock Production; 1-6 August, 2010; Leipzig, Germany: German Society for Animal Science. [Google Scholar]
- Jovanović M., Stefanoska I., Radojcić L., and Vićovac L.. 2010. Interleukin-8 (CXCL8) stimulates trophoblast cell migration and invasion by increasing levels of matrix metalloproteinase (MMP)2 and MMP9 and integrins alpha5 and beta1. Reproduction 139:789–798. doi:10.1530/REP-09-0341 [DOI] [PubMed] [Google Scholar]
- Juma A. R., Damdimopoulou P. E., Grommen S. V., Van de Ven W. J., and De Groef B.. 2016. Emerging role of PLAG1 as a regulator of growth and reproduction. J. Endocrinol. 228:R45–R56. doi:10.1530/JOE-15-0449 [DOI] [PubMed] [Google Scholar]
- Kajimura D., Dragomir C., Ramirez F., and Laub F.. 2007. Identification of genes regulated by transcription factor KLF7 in differentiating olfactory sensory neurons. Gene 388:34–42. doi:10.1016/j.gene.2006.09.027 [DOI] [PubMed] [Google Scholar]
- Kakourou G., Jaroudi S., Tulay P., Heath C., Serhal P., Harper J. C., and Sengupta S. B.. 2013. Investigation of gene expression profiles before and after embryonic genome activation and assessment of functional pathways at the human metaphase II oocyte and blastocyst stage. Fertil. Steril. 99:803–814.e23. doi:10.1016/j.fertnstert.2012.10.036 [DOI] [PubMed] [Google Scholar]
- Kanaani J., Prusiner S. B., Diacovo J., Baekkeskov S., and Legname G.. 2005. Recombinant prion protein induces rapid polarization and development of synapses in embryonic rat hippocampal neurons in vitro. J. Neurochem. 95:1373–1386. doi:10.1111/j.1471-4159.2005.03469.x [DOI] [PubMed] [Google Scholar]
- Kang H. M., Sul J. H., Service S. K., Zaitlen N. A., Kong S. Y., Freimer N. B., Sabatti C., and Eskin E.. 2010. Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42:348–354. doi:10.1038/ng.548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapur R. P., Hoyle G. W., Mercer E. H., Brinster R. L., and Palmiter R. D.. 1991. Some neuronal cell populations express human dopamine beta-hydroxylase-lacz transgenes transiently during embryonic development. Neuron 7:717–727. doi:10.1016/0896-6273(91)90275-5 [DOI] [PubMed] [Google Scholar]
- Karim L., Takeda H., Lin L., Druet T., Arias J. A., Baurain D., Cambisano N., Davis S. R., Farnir F., Grisart B.,. et al. 2011. Variants modulating the expression of a chromosome domain encompassing PLAG1 influence bovine stature. Nat. Genet. 43:405–413. doi:10.1038/ng.814 [DOI] [PubMed] [Google Scholar]
- Kawamura Y., Tanaka Y., Kawamori R., and Maeda S.. 2006. Overexpression of kruppel-like factor 7 regulates adipocytokine gene expressions in human adipocytes and inhibits glucose-induced insulin secretion in pancreatic beta-cell line. Mol. Endocrinol. 20:844–856. doi:10.1210/me.2005-0138 [DOI] [PubMed] [Google Scholar]
- Kiba T. 2016. Ventromedial hypothalamic lesions downregulate multiple immune signaling pathways in rat pancreatic islets. Neurosci. Lett. 610:177-181. doi:10.1016/j.neulet.2015.11.007 [DOI] [PubMed] [Google Scholar]
- Kocabas A. M., Crosby J., Ross P. J., Otu H. H., Beyhan Z., Can H., Tam W. L., Rosa G. J., Halgren R. G., Lim B.,. et al. 2006. The transcriptome of human oocytes. Proc. Natl. Acad. Sci. USA. 103:14027–14032. doi:10.1073/pnas.0603227103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisinger J., Jeung E. B., Simmen R. C., and Leung P. C.. 1995. Porcine calbindin-D9K gene: expression in endometrium, myometrium, and placenta in the absence of a functional estrogen response element in intron A. Biol. Reprod. 52:115–123. doi:10.1095/biolreprod52.1.115 [DOI] [PubMed] [Google Scholar]
- Laing A. F., Lowell S., and Brickman J. M.. 2015. Gro/TLE enables embryonic stem cell differentiation by repressing pluripotent gene expression. Dev. Biol. 397:56–66. doi:10.1016/j.ydbio.2014.10.007 [DOI] [PubMed] [Google Scholar]
- Landriscina M., Soldi R., Bagalá C., Micucci I., Bellum S., Tarantini F., Prudovsky I., and Maciag T.. 2001. S100A13 participates in the release of fibroblast growth factor 1 in response to heat shock in vitro. J. Biol. Chem. 276:22544–22552. doi:10.1074/jbc.M100546200 [DOI] [PubMed] [Google Scholar]
- Laub F., Aldabe R., Friedrich V. Jr, Ohnishi S., Yoshida T., and Ramirez F.. 2001. Developmental expression of mouse krüppel-like transcription factor KLF7 suggests a potential role in neurogenesis. Dev. Biol. 233:305–318. doi:10.1006/dbio.2001.0243 [DOI] [PubMed] [Google Scholar]
- Lindholm‐Perry A. K., Kuehn L. A., Smith T. P. L., Ferrell C. L., Jenkins T. G., Freetly H. C., and Snelling W. M.. 2012. A region on BTA14 that includes the positional candidate genes LYPLA1, XKR4 and TMEM68 is associated with feed intake and growth phenotypes in cattle 1. Anim. Genet. 43:216–219. doi:10.1111/j.1365-2052.2011.02232.x [DOI] [PubMed] [Google Scholar]
- Littlejohn M., Grala T., Sanders K., Walker C., Waghorn G., Macdonald K., Coppieters W., Georges M., Spelman R., Hillerton E.,. et al. 2012. Genetic variation in PLAG1 associates with early life body weight and peripubertal weight and growth in Bos taurus. Anim. Genet. 43:591–594. doi:10.1111/j.1365-2052.2011.02293.x [DOI] [PubMed] [Google Scholar]
- Liu Z., Scannell D. R., Eisen M. B., and Tjian R.. 2011. Control of embryonic stem cell lineage commitment by core promoter factor, TAF3. Cell 146:720–731. doi:10.1016/j.cell.2011.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Qu Y. J., Huai Y. T., Li Z. J., Wang J., Lan X. Y., Zhang C. L., Wang J. Q., and Chen H.. 2011. Polymorphisms identification and associations of KLF7 gene with cattle growth traits. Livest. Sci. 135:1–7. doi:10.1016/j.livsci.2010.04.014 [Google Scholar]
- Macen J. L., Upton C., Nation N., and McFadden G.. 1993. SERP1, a serine proteinase inhibitor encoded by myxoma virus, is a secreted glycoprotein that interferes with inflammation. Virology 195:348–363. doi:10.1006/viro.1993.1385 [DOI] [PubMed] [Google Scholar]
- Mamo S., Mehta J. P., Forde N., McGettigan P., and Lonergan P.. 2012. Conceptus-endometrium crosstalk during maternal recognition of pregnancy in cattle. Biol. Reprod. 87:6, 1–6, 9. doi:10.1095/biolreprod.112.099945 [DOI] [PubMed] [Google Scholar]
- Mathieu C. L., Burnett S. H., Mills S. E., Overpeck J. G., Bruns D. E., and Bruns M. E.. 1989. Gestational changes in calbindin-D9K in rat uterus, yolk sac, and placenta: implications for maternal-fetal calcium transport and uterine muscle function. Proc. Natl. Acad. Sci. USA. 86:3433–3437. doi:10.1073/pnas.86.9.3433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo T. P., de Camargo G. M. F., de Albuquerque L. G., and Carvalheiro R.. 2017. Genome-wide association study provides strong evidence of genes affecting the reproductive performance of Nellore beef cows. Plos One 12:e0178551. doi:10.1371/journal.pone.0178551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Villarroel R. E., Robert N. M., Martin L. J., Brousseau C., and Tremblay J. J.. 2014. The nuclear receptor NR2F2 activates star expression and steroidogenesis in mouse MA-10 and MLTC-1 leydig cells. Biol. Reprod. 91:26. doi:10.1095/biolreprod.113.115790 [DOI] [PubMed] [Google Scholar]
- Mirkin S., Arslan M., Churikov D., Corica A., Diaz J. I., Williams S., Bocca S., and Oehninger S.. 2005. In search of candidate genes critically expressed in the human endometrium during the window of implantation. Hum. Reprod. 20:2104–2117. doi:10.1093/humrep/dei051 [DOI] [PubMed] [Google Scholar]
- Mitchell V., Lecompte F., and Beauvillain J. C.. 2004. Regulation of galanin receptor GalR1 mRNA expression by ovarian steroids in oestrogen receptor alpha-immunoreactive neurones: identification of distinct populations of neurones in the preoptic area. J. Neuroendocrinol. 16:138–145. doi:10.1111/j.0953-8194.2004.01147.x [DOI] [PubMed] [Google Scholar]
- Nishimura S., Watanabe T., Mizoshita K., Tatsuda K., Fujita T., Watanabe N., Sugimoto Y., and Takasuga A.. 2012. Genome-wide association study identified three major QTL for carcass weight including the PLAG1-CHCHD7 QTN for stature in Japanese black cattle. BMC Genet. 13:40. doi:10.1186/1471-2156-13-40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivet A. L., Dufort I., Gilbert I., and Sirard M. A.. 2018. Short-term effect of FSH on gene expression in bovine granulosa cells in vitro. Reprod. Fertil. Dev. 30:1154–1160. doi:10.1071/RD17469 [DOI] [PubMed] [Google Scholar]
- Nourizadeh-Lillabadi R., Seilø Torgersen J., Vestrheim O., König M., Aleström P., and Syed M.. 2010. Early embryonic gene expression profiling of zebrafish prion protein (Prp2) morphants. Plos One 5:e13573. doi:10.1371/journal.pone.0013573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura D., Maeda I., Taniguchi H., Tokitake Y., Ikeda M., Ozato K., Mise N., Abe K., Noce T., Izpisua Belmonte J. C.,. et al. 2012. Cell cycle gene-specific control of transcription has a critical role in proliferation of primordial germ cells. Genes Dev. 26:2477–2482. doi:10.1101/gad.202242.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira Junior G. A., Perez B. C., Cole J. B., Santana M. H. A., Silveira J., Mazzoni G., Ventura R. V., Júnior M. L. S., Kadarmideen H. N., Garrick D. J.,. et al. 2017. Genomic study and medical subject headings enrichment analysis of early pregnancy rate and antral follicle numbers in Nelore heifers. J. Anim. Sci. 95:4796–4812. doi:10.2527/jas2017.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornoy A., Yacobi S., Matalon S. T., Blank M., Blumenfeld Z., Miller R. K., and Shoenfeld Y.. 2003. The effects of antiphospholipid antibodies obtained from women with SLE/APS and associated pregnancy loss on rat embryos and placental explants in culture. Lupus 12:573–578. doi:10.1191/0961203303lu405oa [DOI] [PubMed] [Google Scholar]
- Ortega M. S., Denicol A. C., Cole J. B., Null D. J., and Hansen P. J.. 2016. Use of single nucleotide polymorphisms in candidate genes associated with daughter pregnancy rate for prediction of genetic merit for reproduction in Holstein cows. Anim. Genet. 47:288–297. doi:10.1111/age.12420 [DOI] [PubMed] [Google Scholar]
- Pallares P., and Gonzalez-Bulnes A.. 2010. The effect of embryo and maternal genotypes on prolificacy, intrauterine growth retardation and postnatal development of nos3-knockout mice. Reprod. Biol. 10:241–248. doi:10.1016/S1642-431X(12)60044-8 [PubMed] [Google Scholar]
- Pareek C. S., Smoczyński R., Kadarmideen H. N., Dziuba P., Błaszczyk P., Sikora M., Walendzik P., Grzybowski T., Pierzchała M., Horbańczuk J.,. et al. 2016. Single nucleotide polymorphism discovery in bovine pituitary gland using RNA-seq technology. Plos One 11:e0161370. doi:10.1371/journal.pone.0161370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira A. G. T., Utsunomiya Y. T., Milanesi M., Torrecilha R. B., Carmo A. S., Neves H. H., Carvalheiro R., Ajmone-Marsan P., Sonstegard T. S., Sölkner J.,. et al. 2016. Pleiotropic genes affecting carcass traits in Bos indicus (Nellore) cattle are modulators of growth. Plos One 11:e0158165. doi:10.1371/journal.pone.0158165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer P. L., Smith C. S., Maclean P., and Berg D. K.. 2017. Gene expression analysis of bovine embryonic disc, trophoblast and parietal hypoblast at the start of gastrulation. Zygote 25:265–278. doi:10.1017/S0967199417000090 [DOI] [PubMed] [Google Scholar]
- Pistis G., Okonkwo S. U., Traglia M., Sala C., Shin S. Y., Masciullo C., Buetti I., Massacane R., Mangino M., Thein S. L.,. et al. ; CHARGE Consortium Hematology Working. 2013. Genome wide association analysis of a founder population identified TAF3 as a gene for MCHC in humans. Plos One 8:e69206. doi:10.1371/journal.pone.0069206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramayo-Caldas Y., Renand G., Ballester M., Saintilan R., and Rocha D.. 2016. Multi-breed and multi-trait co-association analysis of meat tenderness and other meat quality traits in three French beef cattle breeds. Genet. Sel. Evol. 48:37. doi:10.1186/s12711-016-0216-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rørth P., Szabo K., and Texido G.. 2000. The level of C/EBP protein is critical for cell migration during Drosophila oogenesis and is tightly controlled by regulated degradation. Mol. Cell 6:23–30. doi:10.1016/S1097-2765(05)00008-0 [DOI] [PubMed] [Google Scholar]
- Rosa A., and Brivanlou A. H.. 2011. A regulatory circuitry comprised of miR-302 and the transcription factors OCT4 and NR2F2 regulates human embryonic stem cell differentiation. Embo J. 30:237–248. doi:10.1038/emboj.2010.319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatchi M., Schnabel R. D., Taylor J. F., and Garrick D. J.. 2014. Large-effect pleiotropic or closely linked QTL segregate within and across ten us cattle breeds. BMC Genomics 15:16. doi:10.1186/1471-2164-15-442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez‐Madrid F., and del Pozo M. A.. 1999. Leukocyte polarization in cell migration and immune interactions. EMBO J. 18:501–511. doi:10.1093/emboj/18.3.501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi C. M., Martínez-López P., de la Vega-Beltrán J. L., Butler A., Alisio A., Darszon A., and Salkoff L.. 2010. The SLO3 sperm-specific potassium channel plays a vital role in male fertility. FEBS Lett. 584:1041–1046. doi:10.1016/j.febslet.2010.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargolzaei M., Chesnais J. P., and Schenkel F. S.. 2014. A new approach for efficient genotype imputation using information from relatives. BMC Genomics 15:478. doi:10.1186/1471-2164-15-478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer C. A., Figueroa-Masot X. A., Batchelor R. H., and Dorsa D. M.. 1999. The mitogen-activated protein kinase pathway mediates estrogen neuroprotection after glutamate toxicity in primary cortical neurons. J. Neurosci. 19:2455–2463. doi:10.1523/JNEUROSCI.19-07-02455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Sengar G. S., Singh U., Deb R., Junghare V., Hazra S., Kumar S., Tyagi S., Das A. K., Raja T. V.,. et al. 2018. Functional proteomic analysis of crossbred (holstein Friesian × sahiwal) bull spermatozoa. Reprod. Domest. Anim. 53:588–608. doi:10.1111/rda.13146 [DOI] [PubMed] [Google Scholar]
- Sirotkin A. V., Benco A., Tandlmajerova A., Vasícek D., Kotwica J., Darlak K., and Valenzuela F.. 2008. Transcription factor p53 can regulate proliferation, apoptosis and secretory activity of luteinizing porcine ovarian granulosa cell cultured with and without ghrelin and FSH. Reproduction 136:611–618. doi:10.1530/REP-08-0229 [DOI] [PubMed] [Google Scholar]
- Smith B. E., and Braun R. E.. 2012. Germ cell migration across sertoli cell tight junctions. Science 338:798–802. doi:10.1126/science.1219969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits K., Willems S., Van Steendam K., Van De Velde M., De Lange V., Ververs C., Roels K., Govaere J., Van Nieuwerburgh F., Peelman L.,. et al. 2018. Proteins involved in embryo-maternal interaction around the signalling of maternal recognition of pregnancy in the horse. Sci. Rep. 8:5249. doi:10.1038/s41598-018-23537-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SNP & Variation Suite ™ (Version 8.3.0) [Software]. Bozeman, MT: Golden Helix, Inc.-[accessed 13 August 2018]. http://www.goldenhelix.com. [Google Scholar]
- Sthoeger Z. M., Mozes E., and Tartakovsky B.. 1993. Anti-cardiolipin antibodies induce pregnancy failure by impairing embryonic implantation. Proc. Natl. Acad. Sci. USA. 90:6464–6467. doi:10.1073/pnas.90.14.6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchocki T., and Szyda J.. 2015. Genome-wide association study for semen production traits in Holstein–Friesian bulls. J. Dairy Sci. 98:5774–5780. doi:10.3168/jds.2014-8951 [DOI] [PubMed] [Google Scholar]
- Szczepny A., Jans D. A., Hime G., and Loveland K. L.. 2005. 247. Expression of components of the hedgehog signalling pathway during murine spermatogenesis. Reprod. Fertil. Dev. 17:98. doi:10.1071/SRB05Abs247 [Google Scholar]
- Teijeiro J. M., and Marini P. E.. 2015. S100A7 in the fallopian tube: a comparative study. Zygote 23:229–236. doi:10.1017/S0967199413000464 [DOI] [PubMed] [Google Scholar]
- Thimon V., Koukoui O., Calvo E., and Sullivan R.. 2007. Region-specific gene expression profiling along the human epididymis. Mol. Hum. Reprod. 13:691–704. doi:10.1093/molehr/gam051 [DOI] [PubMed] [Google Scholar]
- Utsunomiya Y. T., do Carmo A. S., Carvalheiro R., Neves H. H., Matos M. C., Zavarez L. B., Pérez O’Brien A. M., Sölkner J., McEwan J. C., Cole J. B.,. et al. 2013. Genome-wide association study for birth weight in Nellore cattle points to previously described orthologous genes affecting human and bovine height. BMC Genet. 14:52. doi:10.1186/1471-2156-14-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaden P. M. 2008. Efficient methods to compute genomic predictions. J. Dairy Sci. 91:4414–4423. doi:10.3168/jds.2007-0980 [DOI] [PubMed] [Google Scholar]
- Villanueva C. J., Waki H., Godio C., Nielsen R., Chou W. L., Vargas L., Wroblewski K., Schmedt C., Chao L. C., Boyadjian R.,. et al. 2011. TLE3 is a dual-function transcriptional coregulator of adipogenesis. Cell Metab. 13:413–427. doi:10.1016/j.cmet.2011.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetendorf M., Wu S. P., Wang X., Creighton C. J., Wang T., Lanz R. B., Blok L., Tsai S. Y., Tsai M. J., Lydon J. P.,. et al. 2017. Decreased epithelial progesterone receptor A at the window of receptivity is required for preparation of the endometrium for embryo attachment. Biol. Reprod. 96:313–326. doi:10.1095/biolreprod.116.144410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelaw C. M., Robinson J. E., Chambers G. B., Hastie P., Padmanabhan V., Thompson R. C., and Evans N. P.. 2009. Expression of mRNA for galanin, galanin-like peptide and galanin receptors 1-3 in the ovine hypothalamus and pituitary gland: effects of age and gender. Reproduction 137:141–150. doi:10.1530/REP-08-0266 [DOI] [PubMed] [Google Scholar]
- Wicki R., Schäfer B. W., Erne P., and Heizmann C. W.. 1996. Characterization of the human and mouse cDNAs coding for S100A13, a new member of the S100 protein family. Biochem. Biophys. Res. Commun. 227:594–599. doi:10.1006/bbrc.1996.1551 [DOI] [PubMed] [Google Scholar]
- Wysocka J., Swigut T., Milne T. A., Dou Y., Zhang X., Burlingame A. L., Roeder R. G., Brivanlou A. H., and Allis C. D.. 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 H4 methylation and vertebrate development. Cell 121:859–872. doi:10.1016/j.cell.2005.03.036 [DOI] [PubMed] [Google Scholar]
- Zeng X. H., Yang C., Kim S. T., Lingle C. J., and Xia X. M.. 2011. Deletion of the Slo3 gene abolishes alkalization-activated K+ current in mouse spermatozoa. Proc. Natl. Acad. Sci. USA. 108:5879–5884. doi:10.1073/pnas.1100240108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. D., Johnston D. J., Bolormaa S., Hawken R. J., and Tier B.. 2014. Genomic selection for female reproduction in Australian tropically adapted beef cattle. Anim. Prod. Sci. 54:16–24. doi:10.1071/AN13016 [Google Scholar]
- Ziegler A., Santos P. S., Kellermann T., and Uchanska-Ziegler B.. 2010. Self/nonself perception, reproduction and the extended MHC. Self. Nonself. 1:176–191. doi:10.4161/self.1.3.12736 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.