Abstract
Genomic selection using high-density single-nucleotide polymorphism (SNP) markers is used in dairy and beef cattle breeds to accurately estimate genomic breeding values and accelerate genetic improvement by enabling selection of animals with high genetic merit. This genome-wide association study (GWAS) aimed to identify genetic variants associated with beef fatty-acid composition (FAC) traits and to evaluate the accuracy of genomic predictions (GPs) for those traits using genomic best linear unbiased prediction (GBLUP), pedigree BLUP (PBLUP), and BayesR models. Samples of the longissimus dorsi muscle of 965 thirty-month-old Hanwoo steers (progeny of 73 proven bulls) were used to investigate 14 FAC traits. Animals were genotyped or imputed using two bovine SNP platforms (50K and 777K), and after quality control, 38,715 (50K) and 633,448 (777K) SNPs were subjected to GWAS and GP study using a cross-validation scheme. SNP-based heritability estimates were moderate to high (0.25 to 0.47) for all studied traits, with some exceptions for polyunsaturated fatty acids. Association analysis revealed that 19 SNPs in BTA19 (98.7 kb) were significantly associated (P < 7.89 × 10−8) with C14:0 and C18:1n-9; these SNPs were in the fatty-acid synthase (FASN) and coiled-coil domain-containing 57 (CCDC57) genes. BayesR analysis revealed that 0.41 to 0.78% of the total SNPs (n = 2,571 to 4,904) explained almost all of the genetic variance; the majority of the SNPs (>99%) had negligible effects, suggesting that the FAC traits were polygenic. Genome partitioning analysis indicated mostly nonlinear and weak correlations between the variance explained by each chromosome and its length, which also reflected the considerable contributions of relatively few genes. The prediction accuracy of breeding values for FAC traits varied from low to high (0.25 to 0.57); the estimates using the GBLUP and BayesR methods were superior to those obtained by the PBLUP method. The BayesR method performed similarly to GBLUP for most of the studied traits but substantially better for those traits that were controlled by SNPs with large effects; this was supported by the GWAS results. In addition, the predictive abilities of the 50K and 777K SNP arrays were almost similar; thus, both are suitable for GP in Hanwoo cattle. In conclusion, this study provides important insight into the genetic architecture and predictive ability of FAC traits in Hanwoo cattle. Our findings could be used in selection and breeding programs to promote production of meat with enhanced nutritional value.
Keywords: fatty acid, genomic prediction, genome-wide association, Hanwoo cattle, high-density genotype
INTRODUCTION
The fatty-acid composition (FAC) of beef is important for its quality, palatability, and nutritional value. The amounts and types of fatty acids present in beef influence the meat quality, color, flavor, and fat firmness (Webb and O’Neill, 2008). Consumers are now concerned about the types and amounts of fat and fatty acids, particularly those of animal origin, that they consume. Hanwoo (Korean native cattle) beef is popular in Korea due to its marbling, juiciness, tenderness, and characteristic flavor (Hwang et al., 2010), but its cost is triple that of imported beef (KAFTC, 2015). Therefore, the Hanwoo beef fatty-acid profile needs to be improved to meet consumer demand for palatable, healthy, and high-quality meat. Genomic techniques facilitate determination of the underlying genetic architecture and causal variants of complex traits, such as FAC.
Genome-wide association studies (GWASs) using high-density single-nucleotide polymorphism (SNP) markers enable determination of the genetic basis of complex traits in dairy and beef cattle populations (Goddard et al., 2016). Genetic variants associated with FAC traits in various cattle breeds and their crosses have been reported (Kelly et al., 2014; Lemos et al., 2016). Genomic prediction (GP) is used to evaluate the genetic merit of animals based on genome-wide SNP information for selection of prospective candidates at a younger age. Generally, prediction equations are developed based on the genotype and phenotype information of a reference population, and are used to predict the breeding values of genotyped animals (validation population). Various analytical methods are used to estimate breeding values and prediction accuracies in diverse cattle breeds (Erbe et al., 2012; Mehrban et al., 2017). However, no GWAS or GP using high-density SNP data for FAC traits in Hanwoo cattle has been reported. Here, we performed GWAS using a high-density SNP array (777K) to detect genetic variants or candidate regions associated with FAC traits and to compare the GP performances of best linear unbiased prediction (BLUP), genomic BLUP (GBLUP), and BayesR methods for those traits in Korean Hanwoo cattle.
MATERIALS AND METHODS
Animals and Phenotypes
A total of 965 Hanwoo steers born between 2010 and 2012 were included in this study. This half-sib population comprises the progenies of 73 Korean proven bulls and unrelated dams of the Daegwallyeong Hanwoo Company, Pyeongchang, Gangwon Province, South Korea. The Animal Care and Use Committee of the National Institute of Animal Science (NIAS), Rural Development Administration (RDA), South Korea, approved the experimental procedures, and the appropriate animal health and welfare guidelines were followed. The feeding regimen was similar to feedlot conditions, and concentrate and rice straw–based rations were provided. The ratios of concentrate and roughage to total feed were approximately 2.5:1, 4.5:1, and 8:1 for early (12 to 15 mo), middle (16 to 21 mo), and final (22 to 29 mo) fattening periods, respectively. The concentrate mixtures used in the aforementioned three fattening ratios contained 71 and 14%, 72 and 13%, and 73 and 12% total digestible nutrients and crude protein (CP), respectively. All animals were slaughtered at around 30 mo of age. After storage for ~24 h at 4°C, samples of longissimus dorsi (LOD) muscle were collected from the junction between the 12th and 13th ribs and were stored at −20°C until further analysis. The intramuscular fatty-acid levels were determined according to Bhuiyan et al. (2017). Briefly, total lipids were extracted from 200 mg of intramuscular adipose tissue according to a modification of the method of Folch et al. (1957). Individual triglycerides and phospholipids classes were isolated from total lipids by thin-layer chromatography using Silica Gel H (Merck, Darmstadt, Germany) with a solvent of chloroform:methanol:water (45:35:10, v/v/v). The appropriate standards were visualized by exposure to iodine vapor. Fatty acids were analyzed by gas-liquid chromatography (model 437, Chrompack, NJ) using a Packard Chrompack stainless steel column (3 mm × 10 mL) containing Chromasorb WAW 80/100 (Supelco Inc., Bellefonte, PA). The flow rate of the carrier gas (nitrogen) was 22 mL/min. Lauric acid (C12:0) was added as an internal standard methyl ester. The quantities of the fatty acids were calculated from the respective fatty-acid peaks according to Slover and Lanza (1979). The following 10 individual fatty acids and 4 fatty-acid groups were analyzed: myristic (C14:0), palmitic (C16:0), palmitoleic (C16:1n-7), stearic (C18:0), oleic (C18:1n-9), linoleic (C18:2n-6), γ-linoleic (C18:3n-6), linolenic (C18:3n-3), eicosenoic (C20:1n-9), and arachidonic (C20:4n-6); and sum of saturated fatty acids (SFA) = C14:0 + C16:0 + C18:0; sum of monounsaturated fatty acids (MUFA) = C16:1n-7+ C18:1n-9 + C20:ln-9; sum of polyunsaturated fatty acids (PUFA) = C18:2n-6 + C18:3n-6 + C18:3n-3 + C20:4n-6; and sum of unsaturated fatty acids (UFA) = sum of MUFA + PUFA.
Genotyping and Quality Control
Genomic DNA was extracted from LOD muscle samples using a DNeasy Blood and Tissue Kit (Qiagen, CA). DNA concentration and purity were determined using a NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, DE). Animals were genotyped using the Illumina Bovine SNP50 BeadChip (50K) and Illumina Bovine HD BeadChip (777K) platforms. All 50K genotyped animals were imputed to a high-density level (671,902 SNPs) using 777K genotype data as a reference, with Beagle 3.2.2 (Browning and Browning, 2009). SNPs on the sex chromosomes were not included in the analysis. SNP filtering was performed using PLINK 1.9 software (Purcell et al., 2007) based on the following exclusion criteria: minor allele frequency (MAF) <0.01; call rate <0.10, and Hardy–Weinberg equilibrium <0.0001; this resulted in exclusion of 38,873 SNPs. After quality control, 633,029 SNPs were retained for further analyses.
Genomic Heritability Estimates
Variance components and genomic heritabilities were estimated using a univariate restricted maximum likelihood (REML) model implemented in ASReml 4.0 (Gilmour et al., 2015). For this, the genetic relationship matrix (GRM) for additive genetic effects was constructed between all pairs of individuals from genotyped autosomal SNPs using GCTA 1.26 (Yang et al., 2011). The animal model included fixed effects of growing sites (3 regions, 21 farms), birth year (3 levels), and season (10 levels). Slaughter age and beef marbling score (MS) were fitted as linear covariates in the following model:
where is the vector of phenotypes; and are the fixed effects of year, season, and animal growing sites; and are linear covariates for the slaughter age and MS of beef, respectively; is the random additive genetic effect distributed as N(0, A), where the additive GRM A is calculated based on either genotype information or pedigree and is the additive genetic variance; and is the random residual effects.
Genome-Wide Association, Linkage Disequilibrium Analysis, and Identification of Genes
The phenotypic data on fatty-acid traits were adjusted for fixed effects (growing sites, birth year and season) and covariates (slaughter age and beef MS) using a linear mixed model implemented in R software 3.3.1 (R Foundation for Statistical Computing, Vienna, Austria). The adjusted phenotypes and constructed GRM were subsequently used in a GWAS under a mixed linear model including all candidate SNPs in GCTA 1.26 using the following association analysis model:
where y is the phenotype; a is the mean; b is the additive effects (fixed effect) of the candidate SNP to be tested for association; x is the SNP genotype indicator variable coded as 0, 1, or 2; g is the accumulated effects of all considered SNPs; and e is random residual effects.
Manhattan plots of genome-wide P-values from the association analysis (−log10 transformed observed P-values) were produced using the “gap” package (Zhao, 2014) in R software 3.3.1. To determine the P-value threshold, Bonferroni adjustment was applied to correct for multiple hypotheses testing; the genome-wide suggestive and significant P-values were 1.58 × 10−6 (1.00/633,029) and 7.89 × 10−8 (0.05/633,029), respectively. Generally, GWAS does not identify causal variants of a trait from statistically significant SNPs within a strong linkage disequilibrium (LD) region. Therefore, LD analysis was performed to further locate the candidate regions or genes responsible for a trait. The LD coefficients (r2) were assessed with genome significant SNPs (P < 0.05) and the haplotype blocks were inferred using the so-called four-gamete rule (Wang et al., 2002). Analyses were carried out using Haploview 4.2 software (Barrett et al., 2005). We next functionally annotated the significant SNPs and searched for candidate genes in the blocks based on the Bos taurus (UMD 3.1) assembly, using the variant effect predictor (VEP) tools supported by Ensembl (McLaren et al., 2016).
Estimation of Variance Explained by SNPs and Chromosomes
To further decipher the genetic architecture of beef fatty-acid traits, we partitioned the cattle genome into 29 autosomes based on the SNPs on the respective chromosomes. The proportion of phenotypic variance explained by the autosomal SNPs on each chromosome was estimated by REML analysis in GCTA 1.26. A chromosome-wise GRM was constructed and the variance attributable to each chromosome was calculated by fitting the GRMs of all chromosomes (29) simultaneously in a joint analysis using the following model:
where y is the vector of phenotypes; β is the vector of fixed effects and covariates with its incidence matrix X; is the vector of genetic variance attributable to each chromosome; var() is from the joint analysis, where and are the SNP-derived GRM and additive genetic variance, respectively; and e is a random residual error. The proportion of variance captured by each chromosome was calculated as . The phenotypic variance explained by all autosomal SNPs is denoted by .
The Bayesian mixture model was used to estimate the genetic variance explained by individual SNPs that fits four posterior distributions simultaneously for each marker, and was implemented in BayesR (Erbe et al., 2012). This method assumes that the true SNP effects are derived from a combination of four distributions, with proportions of effect sizes of 0 to 1% (0.00, 0.0001, 0.001, and 0.01%) of the total genetic variance in each distribution using a single chain of 50,000 samples; the first 20,000 cycles were discarded as burn-in. The genetic contribution () of each SNP was calculated using the following formula:
where p and q are the allele frequencies for a given trait, is the additive effects of the SNPs, and is the additive genetic variance for a trait.
Genomic Prediction
The pedigree BLUP (PBLUP), GBLUP, and BayesR methods were used to evaluate the predictive ability of FAC traits. We employed the SNP genotype-based GBLUP and BayesR methods to calculate genomic estimated breeding values (GEBVs) and the pedigree-based BLUP method to estimate breeding values (estimated breeding value, EBV). A 10-fold cross-validation scheme was used to evaluate the accuracy of the GEBV and EBV estimates. The data set was divided into 10 equal-sized groups without overlapping of samples. In each cross-validation, nine groups were treated as the training population and the remaining group as the validation population. In the GBLUP method, a modified GRM was used to estimate GEBVs. Phenotypic values were deducted from the validation population and all individuals in the training and validation populations were combined to estimate the breeding values of the validation population using the following model:
where y is the vector of adjusted phenotypic values for fatty acids from the training population; X and Z are incidence matrices; b is the effects of fixed and covariate components; g is the vector of either GEBV or EBV distributed as g~(0, G), where is the additive genetic variance and G is the GRM (Yang et al., 2010) or numerator relationship matrix among animals (Henderson, 1975); and e is the vector of random residuals. All of these estimates were performed by ASReml v. 4.0. In addition, the BayesR method suggested by Erbe et al. (2012) was used to estimate the GEBVs of the validation animals. The statistical model similar to GBLUP was used to estimate the effects sizes of SNPs, and the resultant SNP effect sizes determined by the BayesR method were used to calculate the GEBVs for animals in the validation population. The accuracy of GP was calculated as the average of 10 cross-validation values by the following formula:
where r is the correlation between the estimated breeding values and the adjusted phenotypes of validation animals, and h2 is the heritability of the trait.
RESULTS
Phenotype and Heritability Estimates
Descriptive statistics and genomic heritability estimates () for 14 fatty-acid traits of LOD muscle are shown in Table 1. C18:1n-9 predominated (50.64%) among the intramuscular fatty acids analyzed, followed by palmitic and stearic acids (27.37 and 10.98%, respectively). The PUFA contents were low (0.04 to 0.51%), with the exception of the predominant PUFA, C18:2n-6 (1.81%). The estimates were high (>0.40) for C16:0 and C18:1n-9, as well as for SFA, UFA, and MUFA. The estimates for C14:0, C16:1n-7, C18:0, C18:2n-6, C20:1n-9, and PUFA varied from 0.25 ± 0.07 to 0.35 ± 0.08, compared with 0.04 ± 0.04 to 0.05 ± 0.04 for the fatty acids C18:3n-6, C18:3n-3, and C20:4n-6.
Table 1.
Summary statistics, additive genetic variance, and genomic heritability estimates of fatty acid composition traits in Korean Hanwoo cattle1
| Trait2 | Mean3 | SD | ± SE | |
|---|---|---|---|---|
| Myristic acid (C14:0) | 3.348 | 0.571 | 0.082 | 0.26 ± 0.08 |
| Palmitic acid (C16:0) | 27.37 | 1.911 | 1.617 | 0.44 ± 0.09 |
| Palmitoleic acid (C16:1n-7) | 4.754 | 0.866 | 0.212 | 0.34 ± 0.08 |
| Stearic acid (C18:0) | 10.98 | 1.360 | 0.420 | 0.25 ± 0.07 |
| Oleic acid (C18:1n-9) | 50.64 | 2.510 | 3.023 | 0.49 ± 0.09 |
| Linoleic acid (C18:2n-6) | 1.810 | 0.428 | 0.043 | 0.26 ± 0.08 |
| γ-Linoleic acid (C18:3n-6) | 0.039 | 0.013 | 6.9 × 10−6 | 0.04 ± 0.04 |
| Linolenic acid (C18:3n-3) | 0.084 | 0.042 | 5.6 × 10−5 | 0.04 ± 0.05 |
| Eicosenoic acid (C20:1n-9) | 0.509 | 0.122 | 0.005 | 0.35 ± 0.08 |
| Arachidonic acid (C20:4n-6) | 0.173 | 0.085 | 3.0 × 10−4 | 0.05 ± 0.04 |
| Saturated fatty acid (SFA) | 41.70 | 2.554 | 2.770 | 0.43 ± 0.09 |
| Unsaturated fatty acid (UFA) | 58.30 | 2.554 | 2.724 | 0.42 ± 0.09 |
| Monounsaturated fatty acid (MUFA) | 56.18 | 2.539 | 2.965 | 0.47 ± 0.09 |
| Polyunsaturated fatty acid (PUFA) | 2.106 | 0.482 | 0.052 | 0.25 ± 0.08 |
1Fatty acids concentrations were expressed as a percentage of the total fatty acid analyzed from longissimus dorsi muscle samples.
2SFA (sum of saturated fatty acid) = C14:0 + C16:0 + C18:0; MUFA (sum of monounsaturated fatty acid) = C16:1n-7+ C18:1n-9 + C20:ln-9; PUFA (sum of polyunsaturated fatty acid) = C18:2n-6 + C18:3n-6 + C18:3n-3 + C20:4n-6; UFA (sum of unsaturated fatty acid) = sum of MUFA + PUFA.
3A total of 965 samples investigated in this study.
Genome-Wide Association Study
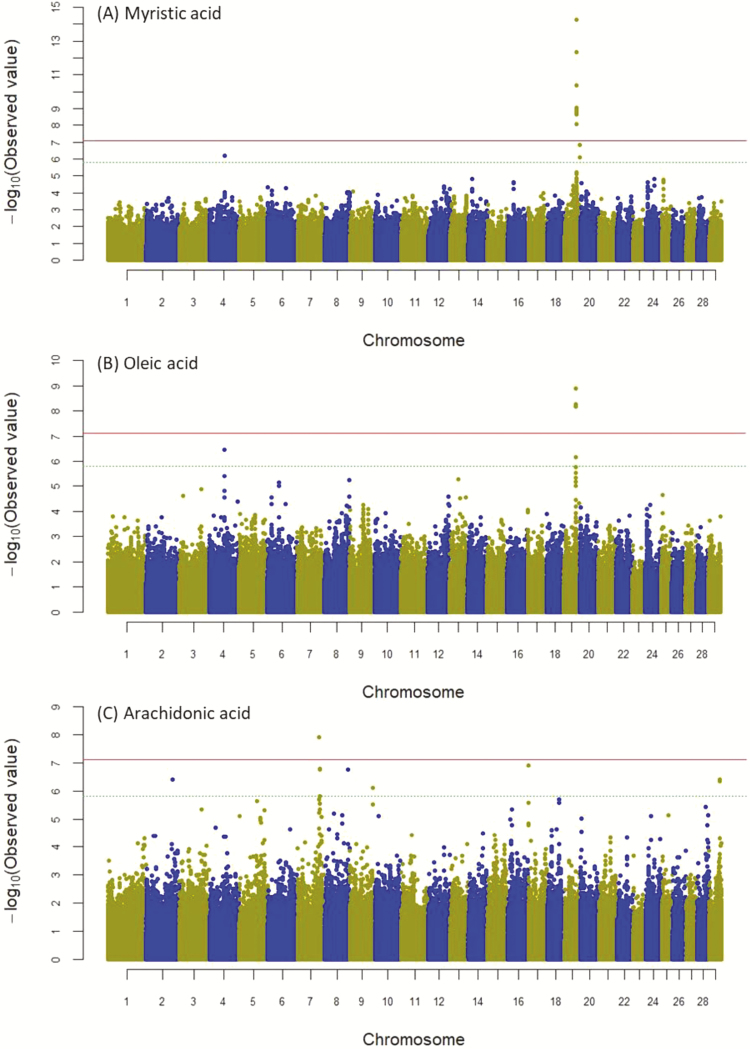
Manhattan plots of the GWAS results for significantly associated fatty-acid traits are presented in Figure 1, and the details of significant SNPs in Table 2. Twenty genome-wide significant SNPs located on chromosomes 7 and 19 were associated with the fatty acids C14:0, C18:1n-9, and C20:4n-6 (P < 7.89 × 10−8). Among them, 19 SNPs spanning a 51.3- to 51.4-Mb region in BTA19 had significant effects on C14:0. Moreover, five SNPs of the same chromosomal region of BTA19 were significantly associated with C18:1n-9 and C14:0. One intergenic SNP on BTA7 was significantly associated with C20:4n-6. Additionally, 13 SNPs located on BTA4, 7, 8, 9, 17, 19, and 29 had suggestive-level associations (P < 1.58 × 10−6) with the fatty acids C14:0, C18:1n-9, and C20:4n-6 (Figure 1). However, no associations were detected for the other seven individual fatty acids and four fatty-acid groups (Supplementary Figure S1).
Figure 1.
Manhattan plot of GWAS for fatty acids myristic (A), oleic (B), and arachidonic (C) in Korean Hanwoo cattle. The y-axis represents −log10 (observed) P-values for genome-wide SNPs against their respective positions on each chromosome (x-axis). The horizontal solid and dot lines indicate the Bonferroni adjusted significant (7.89 × 10−8) and suggestive (1.58 × 10−6) threshold level, respectively.
Table 2.
Significant SNPs in genome-wide association study for myristic, oleic, and arachidonic acid in Korean Hanwoo cattle
| BTA | Position1 (bp) | Minor alleles | MAF2 | SNP effect | SNP location/ effect5 | Gene | |||
|---|---|---|---|---|---|---|---|---|---|
| SNP | β-value3 | SE | P-value4 | ||||||
| Myristic acid | |||||||||
| BovineHD1900014346 | 19 | 51303323 | T | 0.09 | 0.23 | 0.04 | 8.48E-09 | Intron | CCDC57 |
| BovineHD1900014348 | 19 | 51307828 | C | 0.11 | 0.24 | 0.04 | 2.04E-09 | Intron | CCDC57 |
| BovineHD1900014349 | 19 | 51312108 | A | 0.10 | 0.23 | 0.04 | 8.48E-09 | Intron | CCDC57 |
| BovineHD1900014350 | 19 | 51312886 | C | 0.11 | 0.24 | 0.04 | 2.04E-09 | Intron | CCDC57 |
| BovineHD1900014354 | 19 | 51319695 | T | 0.09 | 0.23 | 0.04 | 8.48E-09 | Intron | CCDC57 |
| BovineHD1900014355 | 19 | 51320976 | A | 0.10 | 0.23 | 0.04 | 8.48E-09 | Intron | CCDC57 |
| BovineHD1900014356 | 19 | 51322878 | T | 0.10 | 0.23 | 0.04 | 8.48E-09 | Intron | CCDC57 |
| BovineHD1900014357 | 19 | 51323848 | G | 0.11 | 0.24 | 0.04 | 1.23E-09 | Intron | CCDC57 |
| BovineHD1900014358 | 19 | 51325151 | A | 0.09 | 0.23 | 0.04 | 8.48E-09 | Intron | CCDC57 |
| ARS-BFGL-NGS-39328 | 19 | 51326750 | A | 0.09 | 0.23 | 0.04 | 8.48E-09 | Intron | CCDC57 |
| BovineHD1900014360 | 19 | 51333432 | T | 0.14 | 0.26 | 0.03 | 4.53E-13 | Intron | CCDC57 |
| BovineHD1900014361 | 19 | 51341014 | A | 0.13 | 0.26 | 0.03 | 4.67E-13 | Intron | CCDC57 |
| BovineHD1900014363 | 19 | 51343311 | A | 0.14 | 0.26 | 0.04 | 4.67E-13 | Intron | CCDC57 |
| BovineHD1900014364 | 19 | 51349695 | A | 0.14 | 0.26 | 0.03 | 4.67E-13 | Intron | CCDC57 |
| BovineHD1900014371 | 19 | 51380689 | T | 0.18 | 0.25 | 0.03 | 5.71E-15 | Upstream gene variant | FASN |
| ARS-BFGL-NGS-39983 | 19 | 51395684 | T | 0.15 | 0.24 | 0.04 | 4.20E-11 | Intron | FASN |
| BovineHD1900014376 | 19 | 51398083 | A | 0.15 | 0.22 | 0.03 | 1.40E-09 | Synonymous | FASN |
| BovineHD1900014377 | 19 | 51401022 | C | 0.16 | 0.22 | 0.04 | 8.74E-10 | Synonymous | FASN |
| BovineHD4100014275 | 19 | 51402032 | A | 0.15 | 0.21 | 0.04 | 2.29E-09 | Missense | FASN |
| Oleic acid | |||||||||
| BovineHD1900014360 | 19 | 51333432 | T | 0.14 | −0.94 | 0.16 | 6.80E-09 | Intron | CCDC57 |
| BovineHD1900014361 | 19 | 51341014 | A | 0.13 | −0.96 | 0.16 | 5.33E-09 | Intron | CCDC57 |
| BovineHD1900014363 | 19 | 51343311 | A | 0.14 | −0.96 | 0.16 | 5.33E-09 | Intron | CCDC57 |
| BovineHD1900014364 | 19 | 51349695 | A | 0.14 | −0.96 | 0.16 | 5.33E-09 | Intron | CCDC57 |
| BovineHD1900014371 | 19 | 51380689 | T | 0.18 | −0.90 | 0.15 | 1.31E-10 | Upstream gene variant | FASN |
| Arachidonic acid | |||||||||
| BovineHD0700026591 | 7 | 90802890 | T | 0.03 | 0.09 | 0.02 | 1.24E-08 | Intergenic | — |
1Based on reference NCBI Bos taurus sequence (genome assembly UMD 3.1).
2Minor allele frequency.
3Regression coefficient.
4Significant threshold of genome-wide significance at 5% level for Bonferroni correction was P = 7.89 × 10−8.
5Location of SNP variants or genes was performed as per cattle genome reference sequence (UMD 3.1) using VEP tools (McLaren et al., 2016).
Allele substitution effects ranged between 0.24 and 0.29 (wt/wt%) in C14:0, whereas the effects were negative in C18:1n-9 and varied between −0.97 and −1.03. The Ensembl tools BioMart and VEP suggested that the significantly associated SNPs on BTA19 were harbored by the candidate genes fatty-acid synthase (FASN) and coiled-coil domain containing 57 (CCDC57). The most significant SNP for fatty acids C14:0 and C18:1n-9, rs41920007 (BTA19:51380689 bp), was located upstream of FASN (P < 1.18 × 10−15). In addition, two synonymous SNPs, rs41919992 and rs41919984 (BTA19:51398083 bp and BTA19:51401022 bp), and one missense SNP rs41919985 (BTA19:51402032 bp), which resulted in an amino acid change from threonine to alanine (T2263A), in FASN were significantly associated with C14:0 only (Table 2). All of the 15 intronic SNPs in CCDC57 were significantly associated with C14:0, and five were significantly associated with C18:1n-9. Haploview analysis of significant SNPs on BTA 19 revealed two LD blocks within which the SNP markers were tightly linked (Supplementary Figure S2). LD block 1 (77 kb) contained all 14 SNPs of CCDC57 and 1 SNP upstream of FASN, which exhibited the strongest association detected in this study. LD block 2 (6 kb) consisted of four significant SNPs in FASN.
Genome Partitioning of Genetic Variation
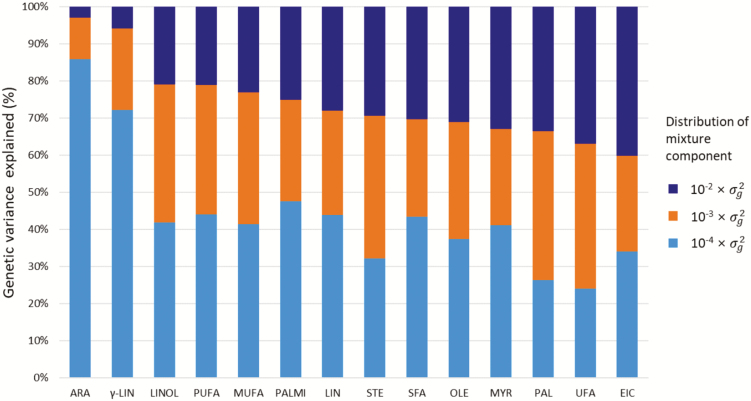
The total genetic variance was partitioned into individual chromosomes and SNPs to determine the genetic architecture underlying FAC. BayesR analysis revealed that individual SNPs generally made small contributions to the total genetic variance (Supplementary Figure S3). Specifically, 2,571 to 4,904 SNPs (0.41 to 0.78%) explained the vast majority of the genetic variance, whereas the remaining >99% of the SNPs had negligible effects (Supplementary Table S1), confirming the polygenic architecture of the study traits. Furthermore, the proportion of genetic variance explained by each mixture component differed markedly among the 14 fatty-acid traits (Figure 2). SNPs with larger effect sizes (10−2 × ) explained a substantial proportion of the total genetic variance of all traits except C18:3n-6 and C20:4n-6. Approximately 21 to 40% of the total genetic variance was explained by 35 to 61 SNPs, most of which were located on BTA3, 7, 8, 9, 13, 17, 19, and 23 (Figure 2; Supplementary Figure S3 and Table S1).
Figure 2.
Genetic architecture of fatty-acid traits inferred by BayesR in Korean Hanwoo cattle. The colored bar partition indicates the proportion of genetic variation explained by the SNPs of respective mixture component. The proportion of variance was calculated as the sum of squares of SNP effects in each mixture component divided by the total genetic variance explained by SNPs. The fatty-acid ARA, arachidonic acid; γ-LIN, γ-linoleic acid; LINOL, linoleic acid; PUFA, polyunsaturated fatty acid; MUFA, monounsaturated fatty acid; PALMI, palmitoleic acid; LIN, linoleic acid; STE, stearic acid, SFA, saturated fatty acid; OLE, oleic acid; MYR, myristic acid; PAL, palmitic acid; UFA, unsaturated fatty acid and EIC, eicosenoic acid.
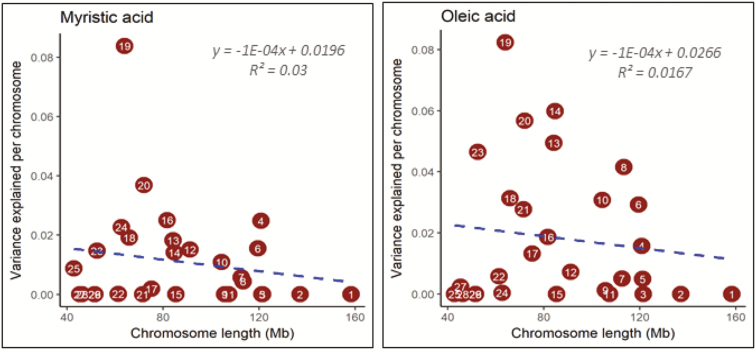
The proportion of variance explained by each chromosome is presented in Figure 3 and Supplementary Figure S4. The parameter estimates did not converge in the joint model for seven fatty-acid traits due to the relatively small data set and low heritability estimates for several traits. Among the converged traits, we found mostly nonlinear and weak relationships between the variance contributed by each chromosome and the chromosomal length. BTA19 alone explained 26.04 and 15.52% of the total phenotypic variance for fatty acids C14:0 and C18:1n-9, respectively, reflecting the considerable contribution of this chromosome segment (Figure 3). Remarkably, 19 significant SNPs accounted for 74.71% of the phenotypic variance in BTA19 for C14:0, whereas five significant SNPs in BTA19 contributed 32.83% of the phenotypic variance for C18:1n-9. BTA8, 13, 14, 20, and 23 contributed 7.83 to 11.29% of the total phenotypic variance for C18:1n-9.
Figure 3.
Genome partitioning for myristic and oleic acid in Korean Hanwoo cattle by joint analysis. The estimated proportion of variance explained by each chromosome against its length is presented in plots. The numbers in the circles represent chromosome number.
Genomic Prediction
The realized accuracies of GP (±SE) were estimated using the PBLUP, GBLUP, and BayesR methods (Table 3). The prediction accuracy varied from 0.25 to 0.57. Across all traits, the average prediction accuracies determined by PBLUP, GBLUP, and BayesR were 0.29, 0.38, and 0.41, respectively. The realized accuracies of the PBLUP method (0.25 to 0.34) were lower than those from the GBLUP and BayesR methods. The prediction accuracies of the GBLUP method using the 50K and 777K SNP data were moderate (>0.30) for all FAC traits, with the exception of C18:2n-6 and PUFA contents, the estimates of which were lower (0.26 to 0.28). The highest accuracy (0.57) was estimated for C14:0 using BayesR. The BayesR prediction accuracy for C14:0, C18:1n-9, C18:2n-6, and MUFA contents was 5 to 21% higher than that of the other two methods. However, both GBLUP and BayesR methods showed a similar prediction accuracy for C16:0, C18:0, SFA, UFA, and MUFA contents. In contrast, the prediction accuracy of GBLUP for C16:1n-7 and C20:1n-9 was superior to that of BayesR. There was no advantage to the 777K over the 50K SNP panel, as both showed similar prediction accuracies for most of the studied traits.
Table 3.
Realized accuracy (±SE) of predicted breeding value for fatty acid composition in Korean Hanwoo cattle
| GBLUP | BayesR | ||||
|---|---|---|---|---|---|
| Trait1 | PBLUP | 50K | 777K | 50K | 777K |
| C14:0 | 0.34 ± 0.07 | 0.34 ± 0.06 | 0.36 ± 0.06 | 0.57 ± 0.07 | 0.57 ± 0.07 |
| C16:0 | 0.32 ± 0.05 | 0.42 ± 0.05 | 0.42 ± 0.06 | 0.41 ± 0.06 | 0.41 ± 0.06 |
| C16:1n-7 | 0.32 ± 0.06 | 0.41 ± 0.04 | 0.43 ± 0.05 | 0.37 ± 0.07 | 0.35 ± 0.07 |
| C18:0 | 0.31 ± 0.08 | 0.41 ± 0.05 | 0.41 ± 0.05 | 0.45 ± 0.12 | 0.42 ± 0.11 |
| C18:1n-9 | 0.32 ± 0.06 | 0.40 ± 0.05 | 0.40 ± 0.05 | 0.45 ± 0.05 | 0.45 ± 0.06 |
| C18:2n-6 | 0.26 ± 0.07 | 0.28 ± 0.05 | 0.27 ± 0.05 | 0.36 ± 0.05 | 0.33 ± 0.07 |
| C20:1n-9 | 0.28 ± 0.04 | 0.41 ± 0.03 | 0.41 ± 0.03 | 0.36 ± 0.08 | 0.38 ± 0.08 |
| SFA | 0.30 ± 0.07 | 0.41 ± 0.05 | 0.41 ± 0.05 | 0.41 ± 0.07 | 0.42 ± 0.06 |
| UFA | 0.29 ± 0.07 | 0.41 ± 0.05 | 0.42 ± 0.05 | 0.42 ± 0.06 | 0.42 ± 0.07 |
| MUFA | 0.30 ± 0.07 | 0.42 ± 0.05 | 0.41 ± 0.05 | 0.41 ± 0.06 | 0.41 ± 0.06 |
| PUFA | 0.25 ± 0.05 | 0.28 ± 0.07 | 0.26 ± 0.06 | 0.35 ± 0.05 | 0.33 ± 0.07 |
1See Table 1 for trait abbreviations.
DISCUSSION
In previous candidate gene analysis and GWASs have suggested potential genetic variants or chromosomal regions associated with beef FAC traits in various cattle breeds, including Hanwoo. Here, we conducted a GWAS and GP of breeding values for 14 FAC traits using high-density SNP markers to identify associated loci and assess the genetic architecture of the FAC traits.
Fatty-Acid Measurements and Heritability Estimates
In general, our fatty-acid results are in line with the findings of Jung et al. (2013) and Choi et al. (2016) in Hanwoo cattle, Chen et al. (2015) in Angus cattle, and Sasago et al. (2017) in Japanese Black cattle. Notably, the intramuscular FAC differs according to the cattle breed, sex, fatty-acid assessment method, sample type, feedlot condition, and uptake of exogenous fatty acids (Smet et al., 2004). The genomic heritability estimates of this study are in accordance with those reported by Chen et al. (2015) in Angus and admixed genetic background Angus cattle. However, Saatchi et al. (2013) reported higher estimates in Angus cattle (0.49 to 0.57 for C14:0, C14:1, C16:0, C16:ln-7, C18:0, C18:1n-9, SFA, and MUFA; and 0.06 to 0.22 for 18:2n-6, 18:3n-3, 18:3n-6, 20:3n-3, 20:3n-6, 20:4n-6, and PUFA). Compared with our results, Lemos et al. (2016) obtained similar estimates for individual saturated and monounsaturated fatty acids (0.21 to 0.48) in indicine Nellore cattle. Conversely, they reported estimates of 0.31 to 0.38 for PUFAs (C18:2n-6, C18:3n-3, C20:3n-6, and C20:4n-6), suggesting use of different feedlot conditions and feeding regimes, as beef PUFA contents are dependent on the type of forage. Moreover, Onogi et al. (2015) reported high estimates for seven fatty-acid traits (0.55 to 0.98) in Japanese Black cattle. These discrepancies among studies may be associated with the cattle breed, type of meat sample, population size, and statistical methods used.
The pedigree-based heritability estimates in this study (data not shown) were higher than those of the genomic estimates (0.32 ± 0.11 to 0.56 ± 0.14 for C14:0, C16:0, C16:1n-7, C18:0, C18:1n-9, SFA, UFA, and MUFA; and 0.03 ± 0.05 to 0.25 ± 0.09 for PUFAs). Earlier studies reported 8 to 17% higher pedigree derived h2 than genomic estimates for reproductive and milk yield traits in Holstein–Friesian cattle populations (Veerkamp et al., 2011) and is consistent with our findings. The lower estimates may be due to the lack of complete LD between SNPs and quantitative trait loci (QTLs) associated with the trait (Goddard et al., 2011). SNP markers may not capture all of the genetic variance for most of the investigated traits in dairy cattle, resulting in lower estimates (Christensen and Lund, 2010; Liu et al., 2011), which also supports our results. Overall, the moderate-to-high genomic heritability estimates for the majority of fatty-acid traits suggest host genetic effects, and so genomic selection could be applied to improve those traits. In contrast, the low estimates of PUFAs indicate poor marker prediction and suggest that non-genetic factors make considerable contributions to these traits. The estimates for fatty-acid traits were subsequently used to estimate the realized accuracy of GP.
Genome-Wide Association Study
Similar to our findings, Bouwman et al. (2014) identified 10 SNPs in FASN and CCDC57 significantly associated with C14:0 in bovine milk; of them, rs41920007 had the most significant association. Sasago et al. (2017) and Chen et al. (2015) reported that the rs41921177 SNP (BTA19:51326750 bp) exhibited the most significant association with several FAC traits, including C14:0 and C18:1n-9, in Japanese Black and Canadian Angus cattle, respectively. This intronic SNP marker in CCDC57 was also significantly associated with C14:0 in our study. Ishii et al. (2013) and Kelly et al. (2014) identified a number of SNPs in a 7.0-Mb region (49 to 55 Mb) of BTA19 centered around FASN that were significantly associated with C14:0, C14:1, C16:0, C16:1, and C18:1n-9 in Australian beef cattle and Japanese Black cattle. However, Saatchi et al. (2013) and Zhu et al. (2017) reported that the 51-Mb region of BTA19 was associated with several fatty-acid traits in Angus and Chinese Simmental beef cattle; this is consistent with our results. Using a candidate gene approach, mutations in FASN have been found to be associated with C14:0, C18:1n-9, SFA, MUFA, and Health Index in US Angus cattle (Zhang et al., 2008) and with C16:0 and C18:1n-9 in Korean Hanwoo cattle (Bhuiyan et al., 2009). The evidence thus indicates that the 1-Mb region of BTA19 has a marked influence on the FAC of beef muscle, and that FASN is a key regulatory gene due to its role in de novo fatty-acid biosynthesis. CCDC57, which is located next to FASN, has not been well characterized but is reportedly associated with the FAC of milk (Bouwman et al., 2014). Mice with a mutation in this gene display hydrocephaly and an abnormal subcutaneous fat layer morphology (Gerdin, 2010). Additionally, the CCDC57 gene product plays an important role in DNA binding and regulation of gene expression. Medrano et al. (2010) reported that the expression level of this gene was higher than that of FASN in the mammary tissues of lactating cows. However, an association of this gene with beef FAC has not been reported. Therefore, CCDC57 may be involved fatty-acid biosynthesis or in tight LD with FASN or other nearby QTLs. According to Goddard and Hayes (2009), the moderate LD in cattle populations can be extended to 100 kb. The level of LD between SNPs and QTLs enables identification of causal variants of a trait. The LD between SNPs of FASN and CCDC57 was moderate (r2 ≥ 0.40) for some SNPs centered around the upstream FASN variant; moreover, there was a high LD coefficient (r2 ≥ 0.72) between that FASN variant and CCDC57 SNPs. This suggests that variants in upstream genes contribute to the muscle FAC (Supplementary Figure S2).
Contributions of SNPs and Chromosomes to Genomic Variance
BayesR analysis facilitates calculation of the number of SNPs in each distribution and the contributions of components that explain 0.00, 0.01, 0.10, and 1.0% of the total genetic variance. Erbe et al. (2012) reported that over 99% of SNPs had almost zero variance, and that 3,644 to 4,725 SNPs showed the greatest variance for three milk-production traits (milk, fat, and protein yield) in Jersey and Holstein cattle; these results are in agreement with our findings. However, compared with this study, fewer SNPs (8 to 29) had the largest variance in the third and fourth posterior distributions for those traits. Moser et al. (2015) reported that >96% of SNPs had negligible effects, and 2,633 to 9,411 SNPs explained the total genetic variance related to human disease traits. Notably, the contributions of SNPs with the greatest variance (1% of the total variance) varied from 0 to 72% among seven disease traits, which is in partial agreement with our results. Therefore, the present and previous findings suggest that complex traits such as beef FAC are polygenic, and that some SNPs have larger effect sizes.
The correlations between chromosomal length and the variance explained by each chromosome were low and most were negative. This is in partial agreement with previous studies involving dairy cattle (Pimentel et al., 2011; Jensen et al., 2012), which reported positive and weak linear relationships between these two parameters. These findings suggest that SNPs and/or QTLs of BTA19, together with other chromosomes, have considerable effects on the C14:0 and C18:1n-9 levels in Korean Hanwoo cattle. Earlier studies involving various beef cattle breeds also reported significant associations of candidate SNPs or QTLs centered around FASN with multiple fatty-acid traits (Kelly et al., 2014; Chen et al., 2015; Sasago et al., 2017). In addition, chromosomes 1, 5, 7, 8, 13, 14, 20, and 23 made substantial contributions to a number of fatty-acid traits, suggesting that a few candidate genes exert large effects, whereas other genes exert small effects on the studied traits. Indeed, stearoyl-CoA desaturase (SCD), sterol regulatory element-binding transcription factor 1 (SREBF1), fatty-acid binding protein 4 (FABP4), FABP5, retinoid X receptor, beta (RXRB), elongation of very-long-chain fatty acids protein 5 (ELOVL5), and thyroid hormone-responsive (THRSP) on chromosomes 26, 19, 14, 23, and 29 are candidate genes associated with fatty-acid traits. In our study, the amount of phenotypic variance explained by chromosome 26 was insignificant, which is inconsistent with the reports of Kelly et al. (2014) and Chen et al. (2015). This may be due to differences between the studied populations, marker density, and/or the extent of LD among the causal variants and genetic markers. Two approaches have been suggested to improve the efficiency of QTL detection in small populations: selective genotyping and analysis of multiple populations with different genetic backgrounds (Sasago et al., 2017). Ours is the first GWAS using high-density SNP arrays to assess the FAC of the LOD of Korean Hanwoo cattle.
Genomic Prediction
The GP accuracy results were similar to those of Chen et al. (2015) and Zhu et al. (2017), who reported that the majority of the studied fatty-acid traits exhibited low-to-moderate accuracy (<0.40) although some, including C14:0, showed a high level of accuracy (>0.50) in Canadian Angus and Angus-derived crossbreeds as well as in Chinese Simmental cattle, respectively. According to Chiaia et al. (2017), the prediction accuracy for 14 individual fatty acids and 7 fatty-acid groups varied from −0.40 to 0.62 in the longissimus thoracis muscle of Nellore cattle; our estimates are within those ranges. Notably, a lower heritability estimate of a trait can result in overestimation of its realized accuracy (Lourenco et al., 2015); therefore, we excluded three PUFA traits (C18:3n-6, C18:3n-3, and C20:4n-6) from the GP analysis. The superior prediction accuracies of the GBLUP and Bayesian methods compared with the pedigree-based BLUP method are consistent with the findings of Chen et al. (2015) and Chiaia et al. (2017) for FAC traits in various beef cattle breeds. Indeed, Bolormaa et al. (2017) and Erbe et al. (2012) reported that the GBLUP and BayesR methods yielded more-accurate GEBVs than the PBLUP method for three reproduction traits in Merino sheep and three milk traits in Holstein and Jersey cattle. Wiggans et al. (2011) found that the reliability of the PBLUP and SNP-BLUP prediction methods was 2.7 and 47.6%, respectively, for milk traits in three dairy cattle breeds (Holstein, Jersey, and Brown Swiss). Mehrban et al. (2017) reported that Bayesian methods have greater accuracy for carcass and meat-quality traits in Hanwoo cattle.
Bayesian methods yield high accuracies when traits are controlled by major genes with large effects (Chiaia et al., 2017), as found in this study. The predictions of the BayesR method were confirmed by the GWAS results; the positional candidate gene FASN was significantly associated with the C14:0 and C18:1n-9 traits. It is plausible that one or more QTLs near FASN have considerable effects on these two traits. However, traits controlled by many loci with small effects tended to have similar predictive accuracies using the GBLUP and BayesR methods (Bolormaa et al., 2017), but different predictive accuracies using the Bayesian and GBLUP methods (Daetwyler et al., 2013). This is consistent with our results. GP accuracy is influenced by a number of factors; e.g., the reference population size, heritability of the traits, relationship between the validation and training populations, LD between markers and QTLs, SNP panel density, genetic architectures of the investigated traits, and the statistical model used (Goddard et al., 2011; Bolormaa et al., 2017).
In conclusion, the estimates for fatty-acid traits in Hanwoo cattle indicated substantial genetic variability, which could be used in breeding programs that aim to improve those traits. A GWAS revealed that 19 SNPs in a 53-Mb region of BTA19 (which included FASN and CCDC57) were significantly associated (P < 7.89 × 10−8) with C14:0 and C18:1n-9. Depending on the trait, the number of SNPs estimated by the BayesR method varied from 2,571 to 4,904 (0.41 to 0.78% of the total SNPs), and these explained the vast majority of the total genetic variance; in contrast, the other SNPs (>99%) had negligible effects. Thus, the FAC traits investigated herein had a polygenic architecture. The GBLUP and BayesR methods yielded prediction accuracies for the studied traits superior to those of the PBLUP method. The predictive accuracies determined using the 50K and 777K SNP panels were similar; thus, both are suitable for GP. This is, to our knowledge, the first study to perform a GWAS and GP of FAC traits in Hanwoo cattle using high-density SNP arrays. Our findings will be useful for breeding programs for production of beef with enhanced nutritional value.
SUPPLEMENTARY DATA
Supplementary data are available at Journal of Animal Science online.
Footnotes
1This research was supported by the grants from Molecular Breeding Program (PJ0118422017) of the Next Generation BIOGREEN21 project and the AGENDA project (PJ00640501) of National Institute of Animal Science, Rural Development Administration, South Korea. This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project title: Development of the methods to control the genetic diversity and to improve the selection effect in Hanwoo line population, Project No. PJ01029102)” Rural Development Administration, Republic of Korea.
LITERATURE CITED
- Barrett J. C., Fry B., Maller J., and Daly M. J.. 2005. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. doi: 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Bhuiyan M. S. A., Lee D. H., Kim H. J., Lee S. H., Cho S. H., Yang B. S., and Kim S. D.. 2017. Estimates of genetic parameters for fatty acid compositions in the longissimus dorsi muscle of Hanwoo cattle. Animal 12:675–683. doi: 10.1017/S1751731117001872 [DOI] [PubMed] [Google Scholar]
- Bhuiyan M. S. A., Yu S. L., Jeon J. T., Yoon D., Cho Y. M., Park E. W., Kim N. K., Kim K. S., and Lee J. H.. 2009. DNA Polymorphisms in SREBF1 and FASN genes affect fatty acid composition in Korean cattle (Hanwoo). Asian-Australas J. Anim. Sci. 22: 765–773. doi: 10.5713/ajas.2009.80573 [DOI] [Google Scholar]
- Bolormaa S., Swan A. A., Brown D. J., Hatcher S., Moghaddar N., van der Werf J. H., Goddard M. E., and Daetwyler H. D.. 2017. Multiple-trait QTL mapping and genomic prediction for wool traits in sheep. Genet. Sel. Evol. 49:62. doi: 10.1186/s12711-017-0337-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman A. C., Visker M. H., van Arendonk J. M., and Bovenhuis H.. 2014. Fine mapping of a quantitative trait locus for bovine milk fat composition on bos taurus autosome 19. J. Dairy Sci. 97:1139–1149. doi: 10.3168/jds.2013-7197 [DOI] [PubMed] [Google Scholar]
- Browning B. L., and Browning S. R.. 2009. a unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. Am. J. Hum. Genet. 84:210–223. doi: 10.1016/j.ajhg.2009.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Ekine-Dzivenu C., Vinsky M., Basarab J., Aalhus J., Dugan M. E., Fitzsimmons C., Stothard P., and Li C.. 2015. Genome-wide association and genomic prediction of breeding values for fatty acid composition in subcutaneous adipose and longissimus lumborum muscle of beef cattle. bmc Genet. 16:135. doi: 10.1186/s12863-015-0290-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaia H. L. J., Peripoli E., Silva R. M. O., Aboujaoude C., Feitosa F. L. B., Lemos M. V. A., Berton M. P., Olivieri B. F., Espigolan R., Tonussi R. L.,. et al. 2017. Genomic prediction for beef fatty acid profile in Nellore cattle. Meat Sci. 128:60–67. doi: 10.1016/j.meatsci.2017.02.007 [DOI] [PubMed] [Google Scholar]
- Choi C. B., Kwon H., Kim S. I., Yang U. M., Lee J. H., and Park E. K.. 2016. Effects of rice bran, flax seed, and sunflower seed on growth performance, carcass characteristics, fatty acid composition, free amino acid and peptide contents, and sensory evaluations of native Korean cattle (Hanwoo). Asian-Australas. j. Anim. Sci. 29:195–203. doi: 10.5713/ajas.15.0264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen O. F., and Lund M. S.. 2010. Genomic prediction when some animals are not genotyped. Genet. Sel. Evol. 42:2. doi: 10.1186/1297-9686-42-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler H. D., Calus M. P., Pong-Wong R., de Los Campos G., and Hickey J. M.. 2013. Genomic prediction in animals and plants: simulation of data, validation, reporting, and benchmarking. Genetics 193:347–365. doi: 10.1534/genetics.112.147983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erbe M., Hayes B. J., Matukumalli L. K., Goswami S., Bowman P. J., Reich C. M., Mason B. A., and Goddard M. E.. 2012. Improving accuracy of genomic predictions within and between dairy cattle breeds with imputed high-density single nucleotide polymorphism panels. J. Dairy Sci. 95:4114–4129. doi: 10.3168/jds.2011-5019 [DOI] [PubMed] [Google Scholar]
- Folch J., Lees M., and Sloane Stanley G. H.. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226:497–509. [PubMed] [Google Scholar]
- Gerdin A. K. 2010. The Sanger Mouse Genetics Program: high throughput characterization of knockout mice. Acta Ophthal. 88: 925–927. doi: 10.1111/j.1755-3768.2010.4142.x [DOI] [Google Scholar]
- Gilmour A. R., Gogel B. J., Cullis B. R., Welham S. J., and Thompson R.. 2015. ASReml User Guide Release 4.1. Hemel Hempstead, UK: VSN International Ltd. [Google Scholar]
- Goddard M. E., and Hayes B. J.. 2009. Mapping genes for complex traits in domestic animals and their use in breeding programmes. Nat. Rev. Genet. 10:381–391. doi: 10.1038/nrg2575 [DOI] [PubMed] [Google Scholar]
- Goddard M. E., Hayes B. J., and Meuwissen T. H.. 2011. Using the genomic relationship matrix to predict the accuracy of genomic selection. J. Anim. Breed. Genet. 128:409–421. doi: 10.1111/j.1439-0388.2011.00964.x [DOI] [PubMed] [Google Scholar]
- Goddard M. E., Kemper K. E., MacLeod I. M., Chamberlain A. J., and Hayes B. J.. 2016. Genetics of complex traits: prediction of phenotype, identification of causal polymorphisms and genetic architecture. Proc. R. Soc. B: Biol. Sci. 283: 20160569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson C. R. 1975. Best linear unbiased estimation and prediction under a selection model. Biometrics 31:423–447. [PubMed] [Google Scholar]
- Hwang Y. H., Kim G. D., Jeong J. Y., Hur S. J., and Joo S. T.. 2010. The relationship between muscle fiber characteristics and meat quality traits of highly marbled Hanwoo (Korean native cattle) steers. Meat Sci. 86:456–461. doi: 10.1016/j.meatsci.2010.05.034 [DOI] [PubMed] [Google Scholar]
- Ishii A., Yamaji K., Uemoto Y., Sasago N., Kobayashi E., Kobayashi N., Matsuhashi T., Maruyama S., Matsumoto H., Sasazaki S.,. et al. 2013. Genome-wide association study for fatty acid composition in Japanese black cattle. Anim. Sci. J. 84:675–682. doi: 10.1111/asj.12063 [DOI] [PubMed] [Google Scholar]
- Jensen J., Su G., and Madsen P.. 2012. Partitioning additive genetic variance into genomic and remaining polygenic components for complex traits in dairy cattle. BMC Genet. 13:44. doi: 10.1186/1471-2156-13-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Nam K. C., Lee K. H., Kim J. J., and Jo C.. 2013. Meat quality traits of Longissimus dorsi muscle from carcasses of Hanwoo steers at different yield grades. Kor. J. Food Sci. Anim. Res. 33: 305–316. doi: 10.5851/kosfa.2013.33.3.305 [DOI] [Google Scholar]
- KAFTC 2015. Korea Agro-Fisheries and Food Trade Corporation, Food and Rural Affairs. Korea: Ministry of Agriculture. [Google Scholar]
- Kelly M. J., Tume R. K., Fortes M., and Thompson J. M.. 2014. Whole-genome association study of fatty acid composition in a diverse range of beef cattle breeds. J. Anim. Sci. 92:1895–1901. doi: 10.2527/jas.2013-6901 [DOI] [PubMed] [Google Scholar]
- Lemos M. V., Chiaia H. L., Berton M. P., Feitosa F. L., Aboujaoud C., Camargo G. M., Pereira A. S., Albuquerque L. G., Ferrinho A. M., Mueller L. F.,. et al. 2016. Genome-wide association between single nucleotide polymorphisms with beef fatty acid profile in Nellore cattle using the single step procedure. BMC Genomics 17:213. doi: 10.1186/s12864-016-2511-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Seefried F. R., Reinhardt F., Rensing S., Thaller G., and Reents R.. 2011. Impacts of both reference population size and inclusion of a residual polygenic effect on the accuracy of genomic prediction. Genet. Sel. Evol. 43:19. doi: 10.1186/1297-9686-43-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourenco D. A., Fragomeni B. O., Tsuruta S., Aguilar I., Zumbach B., Hawken R. J., Legarra A., and Misztal I.. 2015. Accuracy of estimated breeding values with genomic information on males, females, or both: an example on broiler chicken. Genet. Sel. Evol. 47:56. doi: 10.1186/s12711-015-0137-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Gil L., Hunt S. E., Riat H. S., Ritchie G. R., Thormann A., Flicek P., and Cunningham F.. 2016. The ensembl variant effect predictor. Genome Biol. 17:122. doi: 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano J., Rincon G., and Islas-Trejo A.. 2010. Comparative analysis of bovine milk and mammary gland transcriptome using RNA-Seq. In: Proceedings of the 9th World Congr. Genet. Appl. Livest. Prod, August 1–6, Leipzig, Neustadt, Germany; p. 125. [Google Scholar]
- Mehrban H., Lee D. H., Moradi M. H., IlCho C., Naserkheil M., and Ibáñez-Escriche N.. 2017. Predictive performance of genomic selection methods for carcass traits in Hanwoo beef cattle: impacts of the genetic architecture. Genet. Sel. Evol. 49:1. doi: 10.1186/s12711-016-0283-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moser G., Lee S. H., Hayes B. J., Goddard M. E., Wray N. R., and Visscher P. M.. 2015. Simultaneous discovery, estimation and prediction analysis of complex traits using a Bayesian mixture model. PLoS Genet. 11:e1004969. doi: 10.1371/journal.pgen.1004969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onogi A., Ogino A., Komatsu T., Shoji N., Shimizu K., Kurogi K., Yasumori T., Togashi K., and Iwata H.. 2015. Whole-genome prediction of fatty acid composition in meat of Japanese Black cattle. Anim. Genet. 46:557–559. doi: 10.1111/age.12300 [DOI] [PubMed] [Google Scholar]
- Pimentel E. d. C. G., Erbe M., Koenig S., and Simianer H.. 2011. Genome partitioning of genetic variation for milk production and composition traits in Holstein cattle. Front. Genet. 2: 1–11. doi: 10.3389/fgene.2011.00019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., Maller J., Sklar P., de Bakker P. I., Daly M. J.,. et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81:559–575. doi: 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatchi M., Garrick D. J., Tait R. G. Jr, Mayes M. S., Drewnoski M., Schoonmaker J., Diaz C., Beitz D. C., and Reecy J. M.. 2013. Genome-wide association and prediction of direct genomic breeding values for composition of fatty acids in Angus beef cattle. BMC Genomics 14:730. doi: 10.1186/1471-2164-14-730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasago N., Abe T., Sakuma H., Kojima T., and Uemoto Y.. 2017. Genome-wide association study for carcass traits, fatty acid composition, chemical composition, sugar, and the effects of related candidate genes in Japanese black cattle. Anim. Sci. J. 88:33–44. doi: 10.1111/asj.12595 [DOI] [PubMed] [Google Scholar]
- Slover H. T., and Lanza E.. 1979. Quantitative analysis of food fatty acids by capillary gas chromatography. J. Am. Oil Chem. Soc. 56: 933–937. [Google Scholar]
- Smet S. D., Raes K., and Demeyer D.. 2004. Meat fatty acid composition as affected by fatness and genetic factors: a review. Anim. Res. 53: 81–98. doi: 10.1051/animres:2004003 [DOI] [Google Scholar]
- Veerkamp R. F., Mulder H. A., Thompson R., and Calus M. P.. 2011. Genomic and pedigree-based genetic parameters for scarcely recorded traits when some animals are genotyped. J. Dairy Sci. 94:4189–4197. doi: 10.3168/jds.2011-4223 [DOI] [PubMed] [Google Scholar]
- Wang N., Akey J. M., Zhang K., Chakraborty R., and Jin L.. 2002. Distribution of recombination crossovers and the origin of haplotype blocks: the interplay of population history, recombination, and mutation. Am. J. Hum. Genet. 71:1227–1234. doi: 10.1086/344398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb E. C., and O’Neill H. A.. 2008. The animal fat paradox and meat quality. Meat Sci. 80:28–36. doi: 10.1016/j.meatsci.2008.05.029 [DOI] [PubMed] [Google Scholar]
- Wiggans G. R., Cooper T. A., Vanraden P. M., and Cole J. B.. 2011. Technical note: adjustment of traditional cow evaluations to improve accuracy of genomic predictions. J. Dairy Sci. 94:6188–6193. doi: 10.3168/jds.2011-4481 [DOI] [PubMed] [Google Scholar]
- Yang J., Benyamin B., McEvoy B. P., Gordon S., Henders A. K., Nyholt D. R., Madden P. A., Heath A. C., Martin N. G., Montgomery G. W.,. et al. 2010. Common SNPs explain a large proportion of the heritability for human height. Nat. Genet. 42:565–569. doi: 10.1038/ng.608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Lee S. H., Goddard M. E., and Visscher P. M.. 2011. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 88:76–82. doi: 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Knight T. J., Reecy J. M., and Beitz D. C.. 2008. DNA polymorphisms in bovine fatty acid synthase are associated with beef fatty acid composition. Anim. Genet. 39:62–70. doi: 10.1111/j.1365-2052.2007.01681.x [DOI] [PubMed] [Google Scholar]
- Zhao J. H. 2014. gap: genetic analysis package. R package version http://cran.r-project.org/web/packages/gap/index.html.
- Zhu B., Niu H., Zhang W., Wang Z., Liang Y., Guan L., Guo P., Chen Y., Zhang L., Guo Y.,. et al. 2017. Genome wide association study and genomic prediction for fatty acid composition in Chinese Simmental beef cattle using high density SNP array. BMC Genomics 18:464. doi: 10.1186/s12864-017-3847-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.