Abstract
Leucine-rich repeat receptor-like kinases (LRR-RLKs) represent the largest subfamily of receptor-like kinases (RLKs) and play important roles in regulating growth, development, and stress responses in plants. In this study, 246 LRR-RLK genes were identified in the potato (Solanum tuberosum) genome, which were further classified into 14 subfamilies. Gene structure analysis revealed that genes within the same subgroup shared similar exon/intron structures. A signature small peptide recognition motif (RxR) was found to be largely conserved within members of subfamily IX, suggesting that these members may recognize peptide signals as ligands. 26 of the 246 StLRR-RLK genes were found to have arisen from tandem or segmental duplication events. Expression profiling revealed that StLRR-RLK genes were differentially expressed in various organs/tissues, and several genes were found to be responsive to different stress treatments. Furthermore, StLRR-RLK117 was found to be able to form homodimers and heterodimers with StLRR-RLK042 and StLRR-RLK052. Notably, the overlapping expression region of StLRR-RLK117 with Solanum tuberosum WUSCHEL (StWUS) suggested that the CLV3–CLV1/BAM–WUS feedback loop may be conserved in potato to maintain stem cell homeostasis within the shoot apical meristem.
Keywords: leucine-rich repeat receptor-like kinase, Solanum tuberosum, peptide signaling, stress response, stem cell homeostasis
1. Introduction
Unlike animals, the static build of plants handicaps their ability to escape from the hazards of environmental fluctuations. However, lacking this essential survival ability, plants have been provided with unique cell surface receptor proteins that allow the plants to perform cell-to-cell communication and to interact with their environment. In plants, these cellular receptors are known as receptor-like kinases (RLKs) which generally comprise an extracellular ligand binding domain, a transmembrane domain, and an intercellular cytoplasmic kinase domain. RLKs often serve as receptors of cell-to-cell communications by perceiving and transducing signals under various environmental conditions [1]. In Arabidopsis, about 610 members of RLKs have been identified previously, as a large monophyletic gene superfamily [2,3]. The structural feature of RLK proteins demonstrates a high degree of divergence in their extracellular ligand binding domain. Based on sequence analysis of the variable extracellular domain and the kinase domain, the Arabidopsis RLK proteins have been categorized into more than 50 different families, among which over 200 leucine-rich repeat receptor-like kinase (LRR-RLK) members represent the largest class in RLK family. The LRR-RLK ectodomains are furnished with LRR motifs, which are usually 24 amino acids long, varying in number and in arrangement. Such diversity in the LRR domain potentializes the LRR-RLK members to perceive various ligands, which also contributes towards their functional diversity [1]. Though many LRR-RLKs members had been identified, only a few of them have been characterized for their biological functions [1].
Previous studies on the biological roles of various LRR-RLK members revealed their indispensability in plant growth and developmental processes. CLAVATA 1 (CLV1), one of the LRR-RLKs in Arabidopsis, possesses 21 extracellular LRR domains, a transmembrane domain, and a cytoplasmic kinase domain [4]. It has been shown that the encoding gene of CLV1 is specifically expressed around the organizing center (OC) within the shoot apical meristem (SAM). It was found that CLV1 can mediate the CLV3 peptide ligand to suppress WUSCHEL (WUS) expression, whereas the transcriptional factor WUS could promote the expression of the CLV3 gene, resulting in a negative feedback loop that is essential in maintaining stem cell homeostasis within the SAM [5,6,7]. Further, BARELY ANY MERISTEM 1 (BAM1) and BARELY ANY MERISTEM 2 (BAM2), which are both homologues of CLV1, could bind CLV3 peptides with a similar affinity, and function redundantly with CLV1 in the SAM [8,9]. In a recent study, CLAVATA3 INSENSITIVE RECEPTOR KINASES (CIKs), a group of typical LRR-RLKs, act as co-receptors of CLV1 in regulating stem cell homeostasis [10]. Besides, several LRR-RLKs have also been reported to function in peptide signaling of cell fate decisions. The Arabidopsis vascular meristem includes the procambium and cambium. In previous studies, TRACHEARY ELEMENT DIFFERENTIATION FACTOR RECEPTOR/PHLOEM INTERCALATED WITH XYLEM (TDR/PXY) was reported to be highly expressed in procambial cells and to encode a typical LRR-RLK, which could specifically perceive the TDIF peptide signal to promote proliferation of procambial cells, and to suppress xylem differentiation [11,12,13,14]. HAESA and HAESA-LIKE 2 (HSL2) receptors could activate the abscission and cell separation processes in Arabidopsis by perceiving the INFLORESCENCE DEFICIENT IN ABSCISSION (IDA) peptide signal [15]. The C-terminally encoded peptide (CEP) family was identified as a family of secreted peptides, and overexpression of the encoding genes could repress root growth [16]. The double mutant of CEPR1 and CEPR2 was insensitive to CEP1 in a root growth assay, which suggested that CEPR1 and CEPR2 function as CEP receptors [16]. The ROOT MERISTEM GROWTH FACTOR RECEPTORS (RGFRs) group of LRR-RLKs were reported to be involved in root development by sensing ROOT MERISTEM GROWTH FACTOR (RGF) peptides [17,18]. The ERECTA and ERECTA-LIKE 1 (ERL1) sense a peptide hormone named EPIDERMAL PATTERNING FACTOR 2 (EPF2) in modulating stomatal patterning [19]. In addition, phytohormone brassinosteroids bind to LRR-RLK BRASSINOSTEROID INSENSITIVE 1 (BRI1) to regulate cell elongation and cell division [20,21].
LRR-RLKs also play fundamental roles in environmental stress responses as the first line of defense by perceiving signals. In Arabidopsis, a number of defense related LRR-RLK members have been characterized previously. PEPR1 and its homologue PEPR2 mediate plant immunity by acting as receptors of the endogenous Pep peptides (PEP) or by sensing damage-associated molecular patterns (DAMP) [22,23]. The FLAGELLIN SENSITIVE 2 (FLS2) receptor has been reported to function in regulating defense response by sensing the bacterial flagellin monomer [24]. Another study showed that after binding flagellin, the BRASSINOSTEROID-ASSOCIATED RECEPTOR KINASE 1 (BAK1) immediately forms a sandwich structure with the C-terminus of flagellin and FLS2, which triggers downstream signaling cascades [25]. In a recent study, Arabidopsis LRR-RLK SIF2 (stress induced factor2) could regulate basal defense to pathogen infection by sensing a pathogen’s presence and interacting with BAK1 to activate downstream defense-related genes through the MAPK cascade [26]. Further, the RECEPTOR-LIKE PROTEIN KINASE 1 (RPK1) was reported to be involved in ABA-mediated responses to drought stress and leaf senescence in Arabidopsis, while the Oryza rufipogon homolog of RPK1, OrufRPK1 was also reported to act as a defense-related receptor [27,28,29,30].
Besides Arabidopsis, LRR-RLK genes have been identified or studied in a number of different species including Oryza sativa, Solanum lycopersicum, Populus trichocarpa, Citrus clementina, Vitis amurensis, and Rosaceae species, whereas functions of these LRR-RLK genes could be conserved or divergent [3,31,32,33,34,35,36]. Potato (Solanum tuberosum) is one of the important food crops. However, limited information is available about LRR-RLK family genes in potato. In this study, the LRR-RLK family genes of potato were studied through phylogeny, signature motif analysis, gene structure organization, chromosomal distribution, and expression profiles. The homologous counterparts between Arabidopsis and potato LRR-RLKs were identified to predict the potential functions of potato LRR-RLKs.
2. Materials and Methods
2.1. Identification and Phylogenetic Analysis
The potato genome annotations (PGSC, Release 3.4) were retrieved from the Sol Genomics Network (SGN, http://solgenomics.net/). All LRR-RLK full-length amino acid sequences in Arabidopsis were downloaded from TAIR (www.arabidopsis.org) and these sequences were used as queries to perform a BLASTP search against the potato protein database with an E-value cutoff of 0.01. These resulting sequences were then used as new queries to conduct a BLASTP search again, to avoid missing potential members. The redundant entries were removed manually, and the resulted unique sequences were then analyzed with both SMART and Pfam to ensure the presence of the LRR and RLK domains in each newly identified member. Related information, including amino acids number, molecular weights and isoelectric points, was retrieved from ProtParam (http://au.expasy.org/tools/protparam.html). Multiple sequence alignment of putative StLRR-RLKs, reported AtLRR-RLKs, and selected members from other species was performed using MAFFT, with their full-length amino acid sequences under default settings. A neighbor-joining (NJ) phylogenetic tree was generated based on the alignment result, using MEGA v6.06 with the following parameters: Poisson correction, pairwise deletion, and bootstrap values (1000 replicates). The model was advised by ProtTest 2.4. Subsequently, the tree was illustrated using FigTree 1.4.2.
2.2. Motif and Gene Structure Analysis
The LRR motifs were identified by MEME 4.9.1 (Multiple Expectation Maximization for Motif Elicitation, http://meme-suite.org/). Parameters were set as follows: distribution of motif occurrences, zero or one per sequence; maximum number of motifs, 40; optimum motif width, ≥22 and ≤26. The exon–intron organizations were visualized with the Gene Structure Display Server (GSDS: http://gsds.cbi.pku.edu.cn/) by comparing the coding sequences (CDSs) and genomic sequences that were obtained from the SGN database.
2.3. Chromosomal Localization Analysis
The positions of StLRR-RLK genes were analyzed and illustrated by Perl; the chromosomal localization information was retrieved from the SGN database. For nomenclature, the number was added according to the physical location on chromosomes 1–12. The tandem gene event was defined as previously described [32]: a region within 200 kb contained at least two genes which shared more than 70% identity as analyzed by BLASTP with their full-length amino acid sequences. The MCScanX program was used to identify segmental duplications as in the previous description [37,38]. These results were visualized by Circos [39].
2.4. Expression Profiling of StLRR-RLKs
Expression data of StLRR-RLK genes were retrieved from the RNA sequencing (RNA-seq) data, which was previously collected by PGSC [40]. The relative expression ratios of biotic and abiotic stress treatments were calculated relative to their controls, respectively. All expression data were normalized and visualized by R.
2.5. Potato Materials
The shoot cultures of potato (cultivar GN2) were maintained in our lab, and they were used for expression pattern analysis. Potato shoots were inoculated into full MS solid media by nodal cutting, and they were cultured in a growth chamber at 24 °C under continuous light. Further, these seedlings were transferred into pots filled with soil and perlite in the growth chamber, to obtain tubers. Different tissues, including shoot, shoot tip, root, root tip, young leaves, senescence leaves, and tuber were used in measuring the tissue-specific expression patterns. These samples were immediately frozen in liquid nitrogen and stored at −80 °C.
2.6. RNA Extraction and qRT-PCR
Total RNA from different tissues was isolated with RNAiso (TaKaRa) and first-strand complementary DNA (cDNA) synthesis was performed using 2 μg total RNA with the PrimeScript™ RT reagent Kit (TaKaRa) according to the manufacturer’s instructions. The qRT-PCR was performed on an ABI 7500 real-time PCR machine in a 20 μL reaction with SYBR (TaKaRa) 10 μL, 10 mM forward primer 0.4 μL, 10 mM reverse primer 0.4 μL, and diluted cDNA 0.2 μL. The qRT-PCR assay results were obtained from three independent replicates. The Elongation Factor 1-α (EF1α) gene was used as the internal control for normalization [41]. The relative fold differences were calculated based on the 2−ΔΔct method. Primer sequences are listed in Supplementary file S1.
2.7. Bimolecular Fluorescence Complementation Assays
The coding sequences of StLRR-RLK117, StLRR-RLK042, and StLRR-RLK052, excluding their stop codons, were amplified and ligated into the entry vector pENTR by BP Clonase (Invitrogen, Carlsbad, CA, USA). The recombinant entry vectors were used to insert these sequences into pBatTL-sYFPC, mediated by LR Clonase (Invitrogen), and they were fused with the carboxy-terminal part of a yellow fluorescent protein (YFPC), resulting in the fusions to the C-termini of candidate proteins. Likewise, StLRR-RLK117 also was inserted into pBatTL-sYFPN, and then they were fused with the amino-terminal part of the yellow fluorescent protein (YFPN), resulting in fusion to the C-termini of StLRR-RLK117. Equal cultures of StLRR-RLK117-YFPN, together with StLRR-RLK117-YFPC, StLRR-RLK042-YFPC, StLRR-RLK052-YFPC, and the negative control AtBRI1-YFPC, were injected into Nicotiana benthamiana leaves separately for Agrobacterium-mediated transient expression. Fluorescence signals of the reconstituted YFP were captured using a confocal microscope (TCS-SP8 Leica, Wetzlar, Germany) four days after injection.
3. Results
3.1. Identification and Phylogenetic Analysis of Potato LRR-RLKs
In previous studies, 213, 309 and 234 LRR-RLK genes had been identified in Arabidopsis, rice, and tomato genomes, respectively [3,32]. To identify LRR-RLK genes in potato, a BLASTP search was employed using the full-length sequence of the reported Arabidopsis members as queries. Further, Pfam and SMART analyses were performed to ensure the presence of both the LRR and RLK domains in each candidate. As a result, a total of 246 LRR-RLK genes were identified in the S. tuberosum genome. These newly identified members were given names with the prefix ‘St’ indicating S. tuberosum, followed by numbers designated based on their positions from top to bottom on the 12 chromosomes. Detailed information of the StLRR-RLK family genes, including the accession numbers and their characteristics is given in Supplementary file S2.
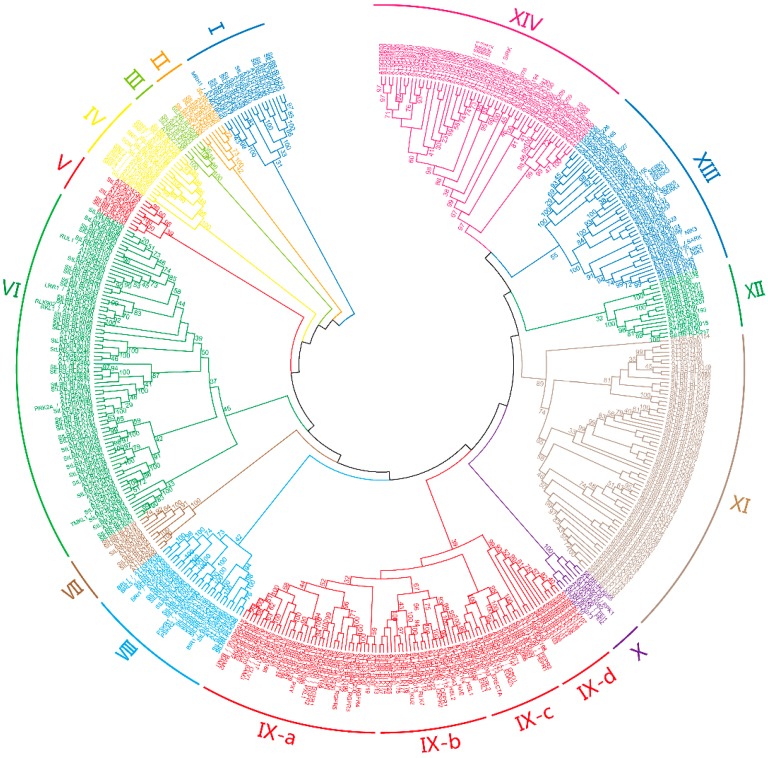
To investigate the phylogenetic relationship between the LRR-RLK members from potato and their reported Arabidopsis homologues, a neighbor-joining tree was constructed based on the multiple sequence alignment of the full-length sequences. The StLRR-RLKs, together with the members from Arabidopsis, were grouped into 14 subfamilies (Figure 1). Each subfamily harbored LRR-RLK members from both potato and Arabidopsis. In general, the number of StLRR-RLK genes was similar to the number of members from Arabidopsis within same subfamily, except for subfamilies XI and XIV. The summarized information regarding each subfamily was listed in Table 1.
Figure 1.
Phylogenetic analysis of LRR-RLK members. The phylogenetic tree was generated from the alignment result of the full-length amino acid sequences by the neighbor-joining (NJ) method. All StLRR-RLK members, together with their Arabidopsis homologues, were classified into 14 distinct subfamilies.
Table 1.
Subfamily designation and sequence characteristics of the identified StLRR-RLK genes.
| Subfamily | Gene Num. | PI | MW (kDa) | Amino Acid Length | SP (%) | Arabidopsis Best Hits |
|---|---|---|---|---|---|---|
| I | 9 | 5.48–9.18 | 59.6–87.0 | 533–785 | 77.8 | MRH1 |
| II | 4 | 6.14–8.87 | 61.6–105.4 | 561–959 | 100 | |
| III | 3 | 5.88–8.12 | 74.0–75.4 | 678–686 | 100 | |
| IV | 5 | 5.62–6.26 | 75.0–80.8 | 692–738 | 80 | SRF1-8, SCM |
| V | 4 | 6.26–9.35 | 76.4–105.3 | 682–946 | 100 | |
| VI | 41 | 5.69–9.30 | 63.9–122.0 | 565–1127 | 75.6 | RUL1, LRR1, RLK902, RKL1, PRK2A, TMKL1 |
| VII | 3 | 5.36–6.44 | 95.8–107.4 | 882–984 | 33.3 | |
| VIII | 15 | 5.2–8.9 | 41.3–137.8 | 369–1272 | 66.7 | BRL1, BRL3, BRI1, PSRK1, PSKR2, BIR1 |
| IX-a | 20 | 5.14–8.21 | 99.6–133.3 | 919–1230 | 80 | CLV1, BAM1-3, PXY, PXL1, RGFR1-5 |
| IX-b | 16 | 5.02–8.99 | 60.6–114.6 | 542–1032 | 87.5 | HAE, HSL1-2, CEPR1-2, RLK7 |
| IX-c | 8 | 5.12–9.37 | 70.2–138.1 | 625–1255 | 100 | PEPR1-2, GSO1, ERECTA, ERL1-2 |
| IX-d | 10 | 5.18–9.03 | 88.7–141.0 | 804–1283 | 70 | |
| X | 3 | 5.43–6.87 | 67.0–121.7 | 601–1126 | 66.7 | FEI1, FEI2 |
| XI | 60 | 5.11–8.83 | 33.8–222.9 | 303–2007 | 41.7 | |
| XII | 10 | 5.38–6.64 | 10.4–121.8 | 921–1106 | 90 | |
| XIII | 16 | 5.43–8.4 | 57.6–113.3 | 518–1027 | 62.5 | SERK1-5, SARK, NIK1-3 |
| XIV | 10 | 5.76–8.75 | 57.4–106.0 | 518–964 | 70 | SIF1-4, SIRK |
Note: PI, isoelectric point; MW, molecular weight; SP, signal peptide.
The subfamily IX represented the second largest subfamily, and it was further categorized into four subgroups. The well-known LRR-RLK members in Arabidopsis including CLV1, BAM1, BAM2, PXY, and RGFRs were clustered within subgroup IX-a. Further, the Arabidopsis HAESA, HSL2, and CEPRs fell into subgroup IX-b, while ERECTA, ERLs, and PEPRs were grouped within subgroup IX-c. To gain further insight into the phylogenetic relationships, a number of LRR-RLK members from rice were used to perform a broader analysis. The selected rice members have been previously identified as being homologous to CLV1, BAMs, RGFR, HAESA, and CEPRs [3]. As expected, all of these rice LRR-RLK members fall into subfamily IX, providing further evidence of the phylogenetic relationships between potential peptide-binding receptors (Supplementary file S3). In previous studies, OsCLV1(Osi008429.1), SlCLV1(Solyc04g081590), and PtCLV1(POPTR_0005s26300.1) have been identified as the homologues of AtCLV1 in rice, tomato, and poplar respectively [3,32,33]. In this study, StLRR-RLK117 was grouped together with OsCLV1, SlCLV1, PtCLV1, and AtCLV1, further supporting that StLRR-RLK117 is the homologue of AtCLV1 in potato (Supplementary file S3). Notably, most of the Arabidopsis members in subfamily IX were reported to participate in plant growth and developmental processes by interacting with peptide ligands, indicating that their potato homologues may undertake similar functions by perceiving peptide signals.
3.2. Conserved and Functional Motifs
Generally, the extracellular LRR domains of LRR-RLK members perceive the ligand and establish the basis of their functions. To unveil the structural diversity and functional characteristics of potato LRR-RLKs, the LRR domains of the identified StLRR-RLK proteins were subjected to motif analysis using the MEME software. In potato, 29 LRR motifs were identified, and their patterns of amino acid sequences were generally found to match with the plant LRR consensus sequence GxIPxxLxxLxxLxxLxLxxNxLx (Supplementary files S4 and S5). The most conserved amino acid residues in potato LRR motifs were Gly at position 1, Pro at position 4, Leu at position 13, 16 and 18, and Asn at position 21. Ile and Leu were at positions 3 and 7 respectively, while Leu and Phe at position 23 were often substituted by each other. In addition, conserved amino acid residues were also found at some other positions of the LRR motifs. These amino acid residues included Pro at position 5 of M14 and M22; Glu at position 6 of M6 and M21; Asn at position 9 of M21, M26 at position 20 of M2. Gly at position 8, Asp at position 17, and Ser at position 19 were also found on a number of LRR motifs (Supplementary file S4).
Generally, the extracellular LRR domains of LRR-RLK members perceive the ligand and establish the basis of their functions. To unveil the structural diversity and functional characteristics of potato LRR-RLKs, the LRR domains of the identified StLRR-RLK proteins were subjected to motif analysis using the MEME software. In potato, 29 LRR motifs were identified, and their patterns of amino acid sequences were generally found to match with the plant LRR consensus sequence GxIPxxLxxLxxLxxLxLxxNxLx (Supplementary files S4 and S5). The most conserved amino acid residues in potato LRR motifs were Gly at position 1, Pro at position 4, Leu at position 13, 16 and 18, and Asn at position 21. Ile and Leu were at positions 3 and 7 respectively, while Leu and Phe at position 23 were often substituted by each other. In addition, conserved amino acid residues were also found at some other positions of the LRR motifs. These amino acid residues included Pro at position 5 of M14 and M22; Glu at position 6 of M6 and M21; Asn at position 9 of M21, M26 at position 20 of M2. Gly at position 8, Asp at position 17, and Ser at position 19 were also found on a number of LRR motifs (Supplementary files S4 and S5).
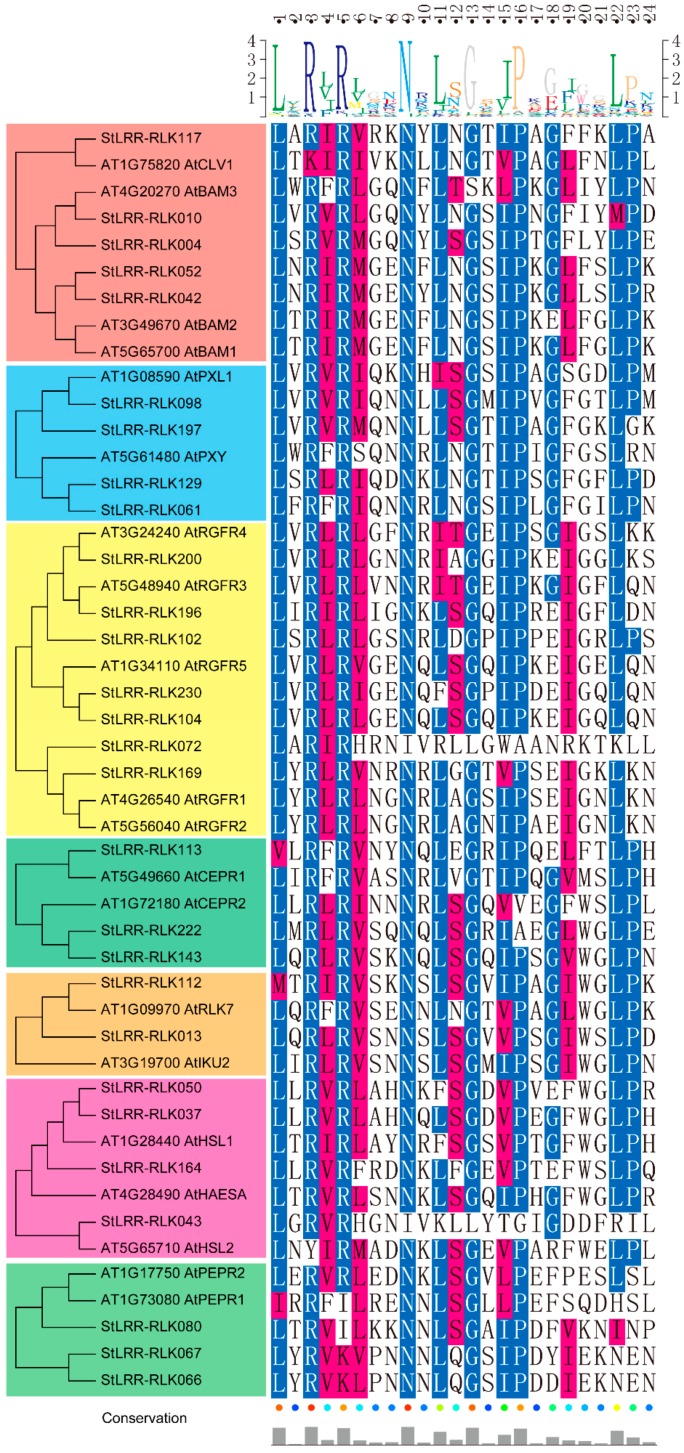
Intriguingly, many plant peptides possess histidine or asparagine as their last residue, and the last residue forms salt bridges with the RxR motif (the x stands for any amino acid) in the LRR domain [17]. Including CLV1, BAM1, BAM2, PXY, and RGFRs, the Arabidopsis members which were grouped within the subfamily IX all contained such a conserved RxR motif, and all of them have been shown to act as receptors of small peptides. Notably, this RxR motif was found to be largely conserved in the potato members of subfamily IX, suggesting that these potato LRR-RLKs may recognize specific peptide ligands with their RxR motifs (Figure 2).
Figure 2.
Functional motif analysis of leucine-rich repeat receptor-like kinase (LRR-RLK) members. Sequence alignment of potato and Arabidopsis LRR-RLK members harboring the RxR motif among subfamily IX. The highly conserved residues are blue- or red-colored, and are visualized by Texshade (https://ctan.org/pkg/texshade).
3.3. Exon–Intron Organization
The gene structure could provide the evolutional evidence of a gene family. The detailed exon–intron organization for the StLRR-RLK genes and their Arabidopsis homologues were plotted and listed in Supplementary file S6, and the representative genes from each subfamily were selected and illustrated in Figure 3. Among the 246 StLRR-RLK genes, 23 genes had no intron while one, two, three, four, and five introns were found in 118, 29, 14, six, and three genes respectively. In addition, 23 StLRR-RLK genes consisted 6~10 introns, 16 StLRR-RLKs contained 11~15 introns, while the others possessed more than 15 introns. StLRR-RLK genes within the same subgroup were found to share similar exon/intron structures, supporting the results of our phylogenetic analysis. For example, all the members in subfamily XII had one intron, whereas most members in subfamily IV had 15 introns (Supplementary file S6). Interestingly, the intron number of potato LRR-RLKs in subfamily IX ranged mostly between 0–2, except for two members, StLRR-RLK173 and StLRR-RLK063, which harbored the maximum number of introns, 21 and 26 respectively.
Figure 3.
Representative LRR-RLK gene structures from each subfamily in potato. The exon–intron organizations of representative LRR-RLK genes were visualized by the Gene Structure Display Server (GSDS: http://gsds.cbi.pku.edu.cn/). In each subfamily, the selected gene shares the same structure with the majority of the members. Exons and introns were plotted by boxes and lines, respectively.
3.4. Chromosome Localization and Duplication Analysis
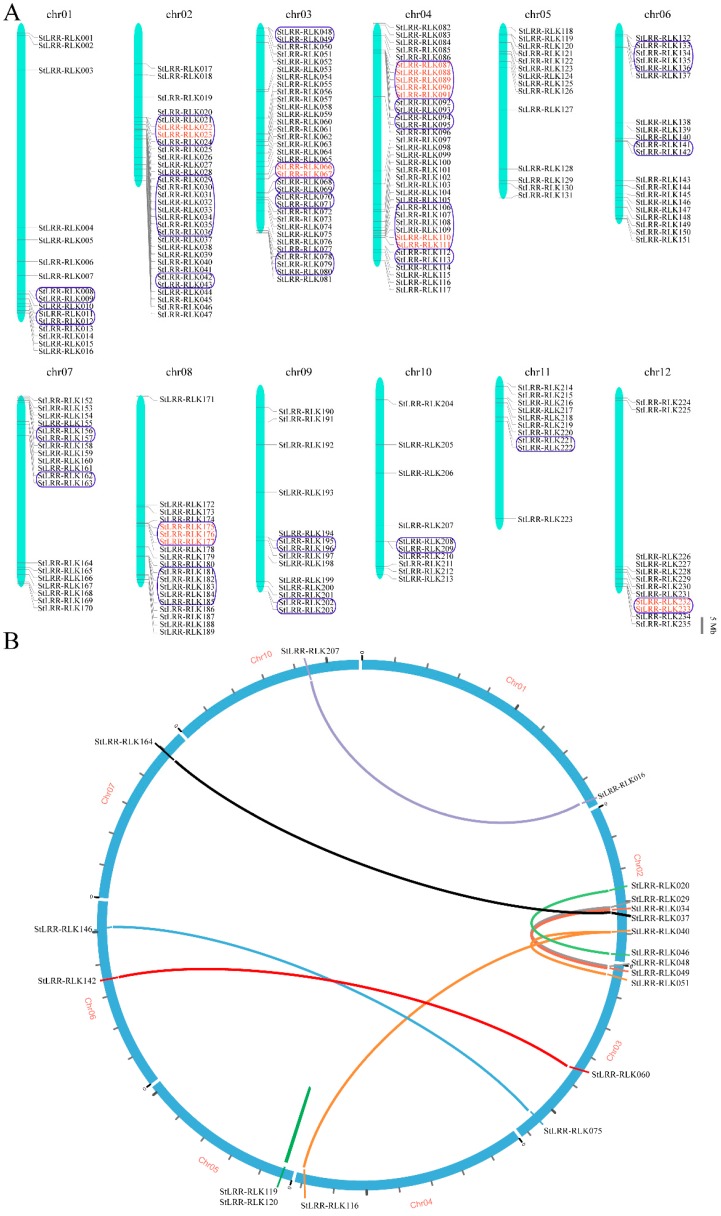
Gene family expansion is an important strategy for the evolutionary fitness of plants. To identify gene clusters and to investigate such events, first we mapped the physical distributions of the StLRR-RLK genes, and then these genes were grouped into a cluster if they were arranged in a region within 200 kb (Figure 4A). 235 of the 246 StLRR-RLK genes were distributed across 12 potato chromosomes, and a total of 74 StLRR-RLK genes were grouped into 26 clusters over 11 chromosomes, with no cluster on chromosome 5, and the maximum of five clusters each on chromosome 3 and 4. The smallest gene cluster comprised only two genes, while the largest cluster was found to have eight tightly grouped genes on chromosome 2. With 70% sequence similarities over the full-length amino acid sequence as the threshold, tandem duplicated genes were identified in several clusters. Among the 26 clusters, 16 genes from six clusters were presumed to be genes derived from tandem duplication. Furthermore, segmental duplication or whole genome duplication of the StLRR-RLK genes was analyzed. Among these potato LRR-RLK genes, 10 pairs of genes were predicted to be the results of segmental duplication events (Figure 4B). All the tandem and segmental duplication genes were listed in Supplementary file S7. About 10.5% of these genes may have been involved in tandem or segmental duplication, indicating that gene duplication events may play an important role in the expansion of this family.
Figure 4.
Chromosome distribution and duplication analysis of the potato LRR-RLK genes. (A) 235 StLRR-RLK genes were mapped to 12 potato chromosomes. The blue box indicated a gene cluster and the tandem duplication is indicated by red colored names; (B) The 10 putative segmental duplication pairs are linked by the colored lines respectively.
3.5. The Expression Pattern in Tested Tissues
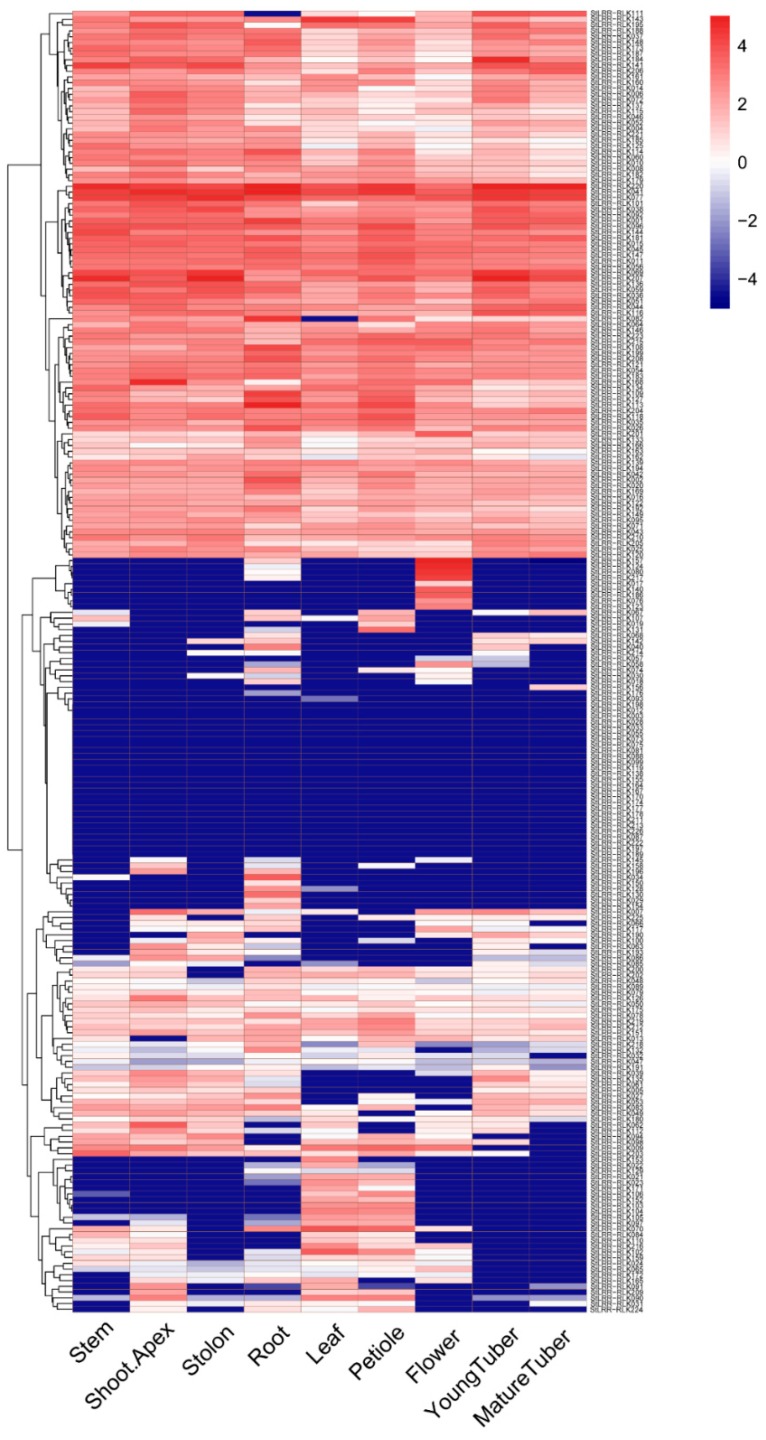
To understand the potential functions of the identified StLRR-RLK genes, expression patterns of these genes were analyzed using the RNA-seq data retrieved from PGSC [40]. Transcription levels of the 246 StLRR-RLK genes in eight tested tissues including stem, shoot apex, stolon, root, leaf, petiole, flower, and young and mature tuber, were analyzed and visualized. Generally, these genes were found to be expressed broadly in different tissues with considerable variations between individual genes. Some genes exhibited tissue specific expression patterns. For example, StLRR-RLK017, StLRR-RLK 076, and StLRR-RLK086 were found to be only expressed in the flower (Figure 5).
Figure 5.
Expression patterns of the potato LRR-RLK family genes. The expression pattern data were normalized and visualized by R.
The gene expression patterns provided preliminary clues for their functions. For example, StLRR-RLK117 showed abundant expression in shoot apex, stolon, and flower, implying that StLRR-RLK117 might be important for the shoot apical meristem, similar to AtCLV1. Similar to the expression pattern of AtBAMs, potato BAM homologues StLRR-RLK042, StLRR-RLK052, StLRR-RLK004 and StLRR-RLK010 were found to be highly expressed in all tested tissues. Interestingly, PXY has been reported to be highly expressed in procambial cells of related organs, including the root and stem in Arabidopsis. Further, StLRR-RLK061 and StLRR-RLK129 were found to be the homologues of Arabidopsis PXY, and they exhibited different expression patterns. StLRR-RLK061 was found to express in the shoot, stolon, and tuber, whereas StLRR-RLK129 was expressed in the root and leaf, indicating that functional divergence may have occurred in these two homologues.
3.6. Expression Profiling of StLRR-RLK Genes during Abiotic and Biotic Stresses
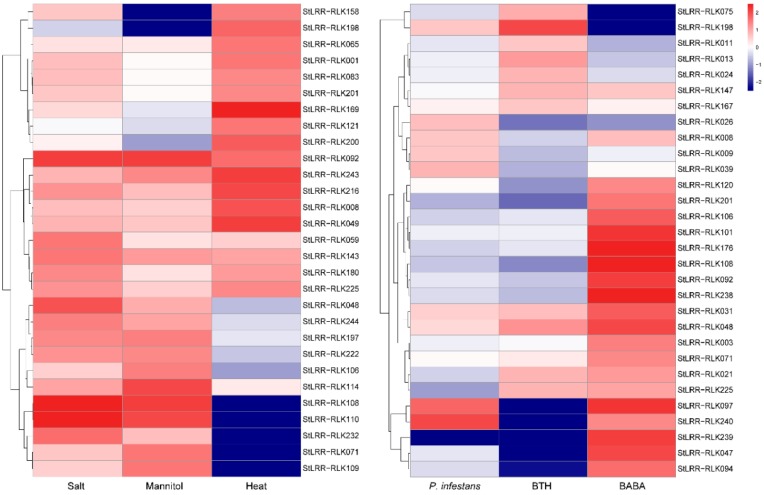
LRR-RLKs also act as surface receptors, playing vital roles in perceiving and transducing stress-related signals. To analyze the stress responsiveness of the StLRR-RLK genes, we compared the transcription levels of StLRR-RLK genes under different stresses. Abiotic stress treatments included salt, drought, and heat. Biotic stress treatments included P. infestans inoculation and treatments of two chemical elicitors. The relative expression changes of various StLRR-RLKs in response to each treatment were calculated in comparison with the respective controls.
29 StLRR-RLK genes were induced under at least one of the abiotic stress treatments (Figure 6). Out of these 29 genes, StLRR-RLK008, StLRR-RLK049, StLRR-RLK092, StLRR-RLK143, StLRR-RLK216, StLRR-RLK225, and StLRR-RLK243 displayed multiple stress responses. Several StLRR-RLKs exhibited differential expression under various stress treatments. For example, StLRR-RLK048, StLRR-RLK071, StLRR-RLK108, StLRR-RLK110, StLRR-RLK114, StLRR-RLK197, StLRR-RLK222, StLRR-RLK 232, and StLRR-RLK244 were induced by both salt stress and drought treatments, while StLRR-RLK001, StLRR-RLK083, StLRR-RLK158, StLRR-RLK180, and StLRR-RLK201 were induced under salt and heat stress conditions. Some of the StLRR-RLKs also exhibited enhanced expression under specific stress treatments. For example, StLRR-RLK059 was up-regulated only under salt stress, while StLRR-RLK106 and StLRR-RLK109 were induced under drought treatment only.
Figure 6.
Heat map representation and hierarchical clustering of selected StLRR-RLK genes under abiotic and biotic stresses. The relative expression ratios of biotic and abiotic stress treatments were calculated relative to their controls respectively, and then up-regulated genes were defined to have a log2 relative expression ratio ≥1 under certain stress treatments. The bars of heat map represent the log2 relative expression ratio. The red color, white color, and blue color represent the up-regulated, unaltered, and down-regulated expression respectively.
To investigate the expression changes of StLRR-RLKs under biotic stress treatments, detached leaves of potato plants were treated with P. infestans inoculum (Pi) and two chemical elicitors, DL-β-amino-n-butyric acid (BABA, 2 mg/mL) and acibenzolar-s-methyl (BTH, 100 μg/mL). Samples were collected at 24 h, 48 h and 72 h after treatments. A total of 30 StLRR-RLKs were detected to be up-regulated under at least one biotic stress treatment (Figure 6). Interestingly, StLRR-RLK031 was induced under all biotic stress conditions, while the expression of StLRR-RLK008, StLRR-RLK097, and StLRR-RLK240 were induced under both P. infestans infection and BTH treatment but down-regulated under BABA treatment. The expression of StLRR-RLK021, StLRR-RLK048, StLRR-RLK147, and StLRR-RLK225 were induced under both BABA and BTH treatments while StLRR-RLK198 was induced only under P. infestans infection and the BABA treatment. The expression of StLRR-RLK009, StLRR-RLK026, and StLRR-RLK039 were induced only under P. infestans infection, StLRR-RLK011, StLRR-RLK013, StLRR-RLK024, and StLRR-RLK075 were specifically induced by BABA treatment and StLRR-RLK003, StLRR-RLK047, StLRR-RLK071, StLRR-RLK092, StLRR-RLK094, StLRR-RLK101, StLRR-RLK106, StLRR-RLK108, StLRR-RLK120, and StLRR-RLK176 were induced under BTH treatment only.
3.7. Validation of Expression Patterns by qRT-PCR
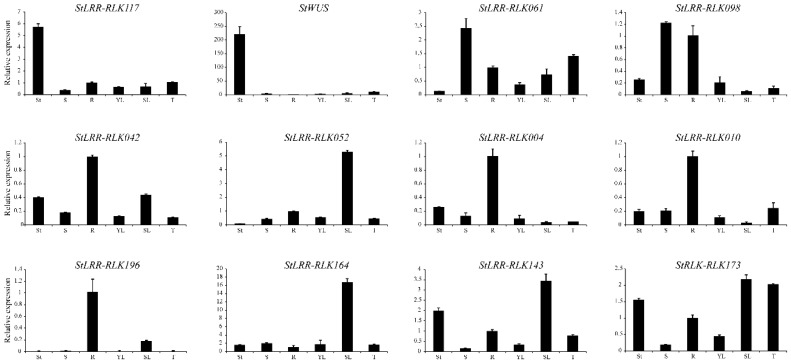
To validate the expression changes of the StLRR-RLK genes, a number of representative StLRR-RLK genes were selected to perform qRT-PCR analysis. As shown in Figure 7, the qRT-PCR results of representative genes were found to be in agreement with the RNA-seq analysis in general. The minor differences could be due to the difference in samples collection at different developmental stages.
Figure 7.
Transcriptional level of selected StLRR-RLK genes in various tissues by qRT-PCR. To reveal the tissue specificity, the expression level of the genes was represented as folds relative to the expression level of root. These qRT-PCR data presented here were obtained from three independent biological replicates with three technical repeats. St, shoot tip; S, stem; R, root; YL, young leaf; SL, senescence leaf; T, tuber.
Notably, StLRR-RLK117 was found to be only expressed in shoot tips within the tested tissues, which strongly suggested that this gene may undertake conserved function as AtCLV1. Considering that CLV1 could mediate the CLV3 peptide signal to confine WUS expression in Arabidopsis and StWUS had been identified in previous studies [42], the expression pattern of StWUS was also analyzed. The results showed that StLRR-RLK117 shares a similar expression pattern with StWUS (Figure 7).
3.8. Bimolecular Fluorescence Complementation Assays
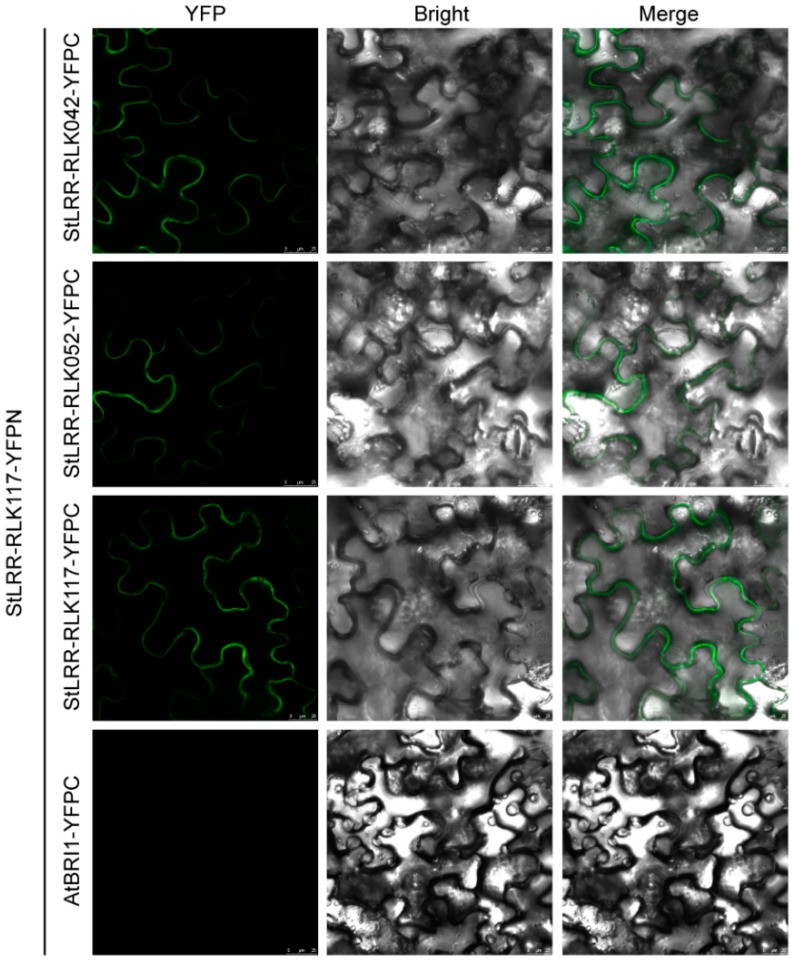
In previous studies, AtCLV1 was observed to interact with AtBAM1, AtBAM2, and AtCLV1 itself, and to transduce the CLV3 peptide signal by forming homodimers and heterodimers in regulating stem cell homeostasis [9]. To explore such molecular mechanisms in potato, bimolecular fluorescence complementation (BiFC) was used to analyze the interactions between the selected potato LRR-RLK receptors. In this study, StLRR-RLK117, which is the homologue of AtCLV1, was fused to the N-terminal fragment of YFP, while StLRR-RLK042, StLRR-RLK052, and StLRR-RLK117 itself were fused to the C-terminal fragment of YFP. In the case that the two tested proteins associated with each other, a full complex of fluorescent YFP would form and be detected. Notably, YFP fluorescence was observed on the membrane when injecting StLRR-RLK117-YFPN together with StLRR-RLK042-YFPC, StLRR-RLK052-YFPC, or StLRR-RLK117-YFPC, but not with AtBRI1-YFPC (Figure 8). Considering that StLRR-RLK042 and StLRR-RLK052 were grouped with AtBAM1 and AtBAM2, these results indicated that potato CLV1 and BAMs may also interact with each other, and StLRR-RLK117 could also form homodimers in regulating stem cell homeostasis in the SAM.
Figure 8.
BiFC assay in N. benthamiana showing the interactions of StLRR-RLK117 with StLRR-RLK042, StLRR-RLK052, and StLRR-RLK117 respectively. In this study, StLRR-RLK117 was fused to the N-terminal part of YFP while StLRR-RLK042, StLRR-RLK052, StLRR-RLK117, and AtBRI1 were fused to the C-terminal part of YFP respectively. Scale bars, 25 µm.
4. Discussion
In this study, a comprehensive analysis of the LRR-RLK family in potato was carried out, and 246 StLRR-RLK genes were identified and analyzed. Based on the phylogenetic analysis, StLRR-RLK proteins together with reported Arabidopsis members fell into 14 subfamilies, and each subfamily contained members from both potato and Arabidopsis, implying that the divergence of the LRR-RLK gene family may appear before the divergence of these two species. The classifications predicted by phylogenetic analysis were further supported by the similar exon–intron organization and motif arrangement within the same subfamily. 235 StLRR-RLK genes were anchored on 12 potato chromosomes, and a total of 16 genes from six clusters were inferred to be tandem duplicated genes. The expression profiling retrieved from RNA-seq and qRT-PCR data in various tissues revealed that StLRR-RLK genes showed similar or distinct expression patterns compared with their Arabidopsis homologues, implying either functional conservation or divergence. The expression pattern under abiotic/biotic treatments indicated that some StLRR-RLK genes also were up-regulated under specific stress treatments, and they may be involved in stress responses.
The subfamily XI represented the largest subfamily, and a significant expansion event of potato LRR-RLK members in this subfamily was observed. 59 LRR-RLK members from potato were found to form a separate branch on the phylogenetic tree (Figure 2). Thirty-four of the 59 members were grouped in clusters on chromosomes, and seven genes were found to come from tandem duplication, while four genes came from segmental duplication. These results suggested that most of the 34 subfamily XI LRR-RLK genes might be originated from gene duplication events after the separation of potato and Arabidopsis. Similar exon–intron organization and motif arrangement of these members further supported this hypothesis. Furthermore, among the 29 StLRR-RLK genes that were significantly up-regulated under at least one of the abiotic stress treatments, eight of them were members of subfamily XI. Similarly, among the 30 StLRR-RLK genes that were induced under tested biotic stress treatments, 10 members were from this subfamily. These results indicated that the gene expansion events within subfamily XI during evolution may help in sensing diverse stress signals in potato.
For subfamily IX, some well-studied LRR-RLK members in Arabidopsis, including CLV1, BAM1, BAM2, PXY, HAESA, HSL2, ERECTA, PEPRs, CEPRs, and RGFRs were clustered within this subfamily. Interestingly, all of these LRR-RLK receptors were reported to play important roles in development, plant growth, and stress responses, by interacting with peptide ligands, implying that the potato members in this subfamily may also bind peptide ligands. Furthermore, the RxR motif is conserved in all potato members of subfamily IX, enabling these members to interact with peptide ligands. In addition, exon–intron organization analysis revealed that most of the members in subfamily IX possessed one intron, whereas ERECTA, ERL1, and ERL2, together with their close potato homologue StLRR-RLK173 and StLRR-RLK063, contained over 20 introns, supporting the results of the phylogenetic analysis. Interestingly, the intron-less ERECTA gene could not rescue the phenotype of the erecta mutants, indicating that the function of ERECTA depends on the presence of introns [43]. Considering the conservation of gene structures, the introns of StLRR-RLK173 and StLRR-RLK063 may also be essential to their functions. Notably, In Arabidopsis, PXY was found to be highly expressed in procambial cells of related organs, including the root and stem. Moreover, TDIF-PXY-WOX4 signaling was reported to play an important role in cell fate decisions of the vascular meristem [11,14]. Consistently, StLRR-RLK061 proved to serve as the homologue of PXY and exhibited high expression in the shoot, shoot apex, stolon, young tuber, and mature tuber, suggesting that this gene might be critical in the regulation of vascular organization in the shoot. Interestingly, tubers are swollen stem structures that are used as storage organs. Considering its higher expression in tuber tissue, StLRR-RLK061 may participate in the formation of tubers, and may affect the yield of potato. Further, BiFC assays revealed that StLRR-RLK117 may interact with StLRR-RLK042 and StLRR-RLK052, while StLRR-RLK117 could also form homodimers. The preferred expression of StLRR-RLK117, which is the homologue of AtCLV1, in the shoot apex, implied that this gene may also be involved in the development of the SAM. The CLV3-CLV1/BAM-WUS feedback loop was reported to be important in shoot apical stem-cell maintenance activity, and StWUS had already been identified in a previous study [42]. The overlap expression pattern of StWUS and StLRR-RLK117 suggested that this mechanism could be conserved in potato. It was supposed that StWUS may be suppressed by StLRR-RLK117, StLRR-RLK042, and StLRR-RLK052 after perceiving certain peptide signals.
5. Conclusions
In this study, a systematic study was carried out to identify and characterize the LRR-RLK family genes in potato. The potato LRR-RLK family genes were studied through phylogeny, motif, gene structure, chromosomal distribution, and expression profiling analysis, which provides insight into the evolutionary conservation of this gene family. Notably, the positive result of the BiFC assay and the overlapping expression region between StLRR-RLK117 and StWUS indicated that the CLV3-CLV1/BAM-WUS feedback loop may be conserved in maintaining stem cell homeostasis within the potato shoot apical meristem.
Acknowledgments
The authors thank the Guo lab members at the Tobacco Research Institute, the Chinese Academy of Agricultural Sciences (CAAS), for discussion and critical reading of the manuscript.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4409/7/9/120/s1, Supplementary file S1: The qRT-PCR primers used in this study, Supplementary file S2: The information of identified potato LRR-RLK family members, Supplementary file S3: Phylogenetic analysis of LRR-RLK members in potato, rice, and Arabidopsis, Supplementary file S4: LRR motif analysis in the LRR domain of identified potato LRR-RLK members, Supplementary file S5: The conserved LRR motifs of StLRR-RLK members as identified by MEME, Supplementary file S6: The exon/intron structures of StLRR-RLK genes, Supplementary file S7: The identified gene cluster and duplication analysis of StLRR-RLK genes.
Author Contributions
X.L. and S.A. conducted the research and participated in the drafting of the manuscript. C.G., J.Y., S.C., X.G., W.L., and H.L. assisted with the data analysis. Y.G. conceived the research, designed the experiments, and drafted the manuscript. All authors read and approved the final manuscript.
Funding
This research was financially supported by the National Natural Science Foundation of China (31571494), the Fundamental Research Funds for Central Non-profit Scientific Institution (Y2017JC27), and the Agricultural Science and Technology Innovation Program (ASTIP-TRIC02).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Gish L.A., Clark S.E. The RLK/Pelle family of kinases. Plant J. 2011;66:117–127. doi: 10.1111/j.1365-313X.2011.04518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shiu S.H., Bleecker A.B. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA. 2001;98:10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shiu S.H., Karlowski W.M., Pan R., Tzeng Y.H., Mayer K.F., Li W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–1234. doi: 10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clark S.E., Williams R.W., Meyerowitz E.M. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/S0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 5.Clark S.E., Running M.P., Meyerowitz E.M. CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development. 1993;119:397–418. doi: 10.1242/dev.119.2.397. [DOI] [PubMed] [Google Scholar]
- 6.Schoof H., Lenhard M., Haecker A., Mayer K.F., Jürgens G., Laux T. The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- 7.Clark S.E. Cell signalling at the shoot meristem. Nat. Rev. Mol. Cell Biol. 2001;2:276. doi: 10.1038/35067079. [DOI] [PubMed] [Google Scholar]
- 8.DeYoung B.J., Bickle K.L., Schrage K.J., Muskett P., Patel K., Clark S.E. The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 2006;45:1–16. doi: 10.1111/j.1365-313X.2005.02592.x. [DOI] [PubMed] [Google Scholar]
- 9.Guo Y., Han L., Hymes M., Denver R., Clark S.E. CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 2010;63:889–900. doi: 10.1111/j.1365-313X.2010.04295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu C., Zhu Y., Cui Y., Cheng K., Liang W., Wei Z., Zhu M., Yin H., Zeng L., Xiao Y. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants. 2018;4:205. doi: 10.1038/s41477-018-0123-z. [DOI] [PubMed] [Google Scholar]
- 11.Hirakawa Y., Kondo Y., Fukuda H. TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell. 2010;22:2618–2629. doi: 10.1105/tpc.110.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H., Lin X., Han Z., Qu L.J., Chai J. Crystal structure of PXY-TDIF complex reveals a conserved recognition mechanism among CLE peptide-receptor pairs. Cell Res. 2016;26:543. doi: 10.1038/cr.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondo Y., Fukuda H. The TDIF signaling network. Curr. Opin. Plant Biol. 2015;28:106–110. doi: 10.1016/j.pbi.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 14.Fisher K., Turner S. PXY, a receptor-like kinase essential for maintaining polarity during plant vascular-tissue development. Curr. Biol. 2007;17:1061–1066. doi: 10.1016/j.cub.2007.05.049. [DOI] [PubMed] [Google Scholar]
- 15.Stenvik G.E., Tandstad N.M., Guo Y., Shi C.-L., Kristiansen W., Holmgren A., Clark S.E., Aalen R.B., Butenko M.A. The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell. 2008;20:1805–1817. doi: 10.1105/tpc.108.059139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tabata R., Sumida K., Yoshii T., Ohyama K., Shinohara H., Matsubayashi Y. Perception of root-derived peptides by shoot LRR-RKs mediates systemic N-demand signaling. Science. 2014;346:343–346. doi: 10.1126/science.1257800. [DOI] [PubMed] [Google Scholar]
- 17.Song W., Liu L., Wang J., Wu Z., Zhang H., Tang J., Lin G., Wang Y., Wen X., Li W. Signature motif-guided identification of receptors for peptide hormones essential for root meristem growth. Cell Res. 2016;26:674. doi: 10.1038/cr.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuzaki Y., Ogawa-Ohnishi M., Mori A., Matsubayashi Y. Secreted peptide signals required for maintenance of root stem cell niche in Arabidopsis. Science. 2010;329:1065–1067. doi: 10.1126/science.1191132. [DOI] [PubMed] [Google Scholar]
- 19.Lee J.S., Hnilova M., Maes M., Lin Y.C.L., Putarjunan A., Han S.-K., Avila J., Torii K.U. Competitive binding of antagonistic peptides fine-tunes stomatal patterning. Nature. 2015;522:439. doi: 10.1038/nature14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He Z., Wang Z.Y., Li J., Zhu Q., Lamb C., Ronald P., Chory J. Perception of brassinosteroids by the extracellular domain of the receptor kinase BRI1. Science. 2000;288:2360–2363. doi: 10.1126/science.288.5475.2360. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita T., Cano-Delgado A., Seto H., Hiranuma S., Fujioka S., Yoshida S., Chory J. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature. 2005;433:167. doi: 10.1038/nature03227. [DOI] [PubMed] [Google Scholar]
- 22.Huffaker A., Pearce G., Ryan C.A. An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA. 2006;103:10098–10103. doi: 10.1073/pnas.0603727103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamaguchi Y., Huffaker A., Bryan A.C., Tax F.E., Ryan C.A. PEPR2 is a second receptor for the Pep1 and Pep2 peptides and contributes to defense responses in Arabidopsis. Plant Cell. 2010;22:508–522. doi: 10.1105/tpc.109.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 2006;18:465–476. doi: 10.1105/tpc.105.036574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Y., Li L., Macho A.P., Han Z., Hu Z., Zipfel C., Zhou J.M., Chai J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science. 2013;342:624–628. doi: 10.1126/science.1243825. [DOI] [PubMed] [Google Scholar]
- 26.Yuan N., Yuan S., Li Z., Zhou M., Wu P., Hu Q., Wang L., Mendu V., Luo H. STRESS INDUCED FACTOR 2, a leucine-rich repeat kinase regulates basal plant pathogen defense. Plant Physiol. 2018;176:3062–3080. doi: 10.1104/pp.17.01266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Osakabe Y., Maruyama K., Seki M., Satou M., Shinozaki K., Yamaguchi-Shinozaki K. Leucine-rich repeat receptor-like kinase1 is a key membrane-bound regulator of abscisic acid early signaling in Arabidopsis. Plant Cell. 2005;17:1105–1119. doi: 10.1105/tpc.104.027474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osakabe Y., Mizuno S., Tanaka H., Maruyama K., Osakabe K., Todaka D., Fujita Y., Kobayashi M., Shinozaki K., Yamaguchi-Shinozaki K. Overproduction of the membrane-bound receptor-like protein kinase1, RPK1, enhances abiotic stress tolerance in Arabidopsis. J. Biol. Chem. 2010;285:9190–9201. doi: 10.1074/jbc.M109.051938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee I.C., Hong S.W., Whang S.S., Lim P.O., Nam H.G., Koo J.C. Age-dependent action of an ABA-inducible receptor kinase, RPK1, as a positive regulator of senescence in Arabidopsis leaves. Plant Cell Physiol. 2011;52:651–662. doi: 10.1093/pcp/pcr026. [DOI] [PubMed] [Google Scholar]
- 30.Law Y.S., Gudimella R., Song B.K., Ratnam W., Harikrishna J.A. Molecular characterization and comparative sequence analysis of defense-related gene, oryza rufipogon receptor-like protein kinase 1. Int. J. Mol. Sci. 2012;13:9343–9362. doi: 10.3390/ijms13079343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakamoto T., Deguchi M., Brustolini O.J., Santos A.A., Silva F.F., Fontes E.P. The tomato RLK superfamily: Phylogeny and functional predictions about the role of the LRRII-RLK subfamily in antiviral defense. BMC Plant Biol. 2012;12:229. doi: 10.1186/1471-2229-12-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei Z., Wang J., Yang S., Song Y. Identification and expression analysis of the LRR-RLK gene family in tomato (Solanum lycopersicum) Heinz 1706. Genome. 2015;58:121–134. doi: 10.1139/gen-2015-0035. [DOI] [PubMed] [Google Scholar]
- 33.Zan Y., Ji Y., Zhang Y., Yang S., Song Y., Wang J. Genome-wide identification, characterization and expression analysis of populus leucine-rich repeat receptor-like protein kinase genes. BMC Genom. 2013;14:318. doi: 10.1186/1471-2164-14-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Magalhães D.M., Scholte L.L., Silva N.V., Oliveira G.C., Zipfel C., Takita M.A., De Souza A.A. LRR-RLK family from two Citrus species: Genome-wide identification and evolutionary aspects. BMC Genom. 2016;17:623. doi: 10.1186/s12864-016-2930-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu S., Zhang C., Chao N., Lu J., Zhang Y. Cloning, Characterization, and Functional Investigation of VaHAESA from Vitis amurensis Inoculated with Plasmopara viticola. Int. J. Mol. Sci. 2018;19:1204. doi: 10.3390/ijms19041204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun J., Li L., Wang P., Zhang S., Wu J. Genome-wide characterization, evolution, and expression analysis of the leucine-rich repeat receptor-like protein kinase (LRR-RLK) gene family in Rosaceae genomes. BMC Genom. 2017;18:763. doi: 10.1186/s12864-017-4155-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Y., Tang H., DeBarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cao Y., Meng D., Chen Y., Abdullah M., Jin Q., Lin Y., Cai Y. Comparative and Expression Analysis of Ubiquitin Conjugating Domain-Containing Genes in Two Pyrus Species. Cells. 2018;7:77. doi: 10.3390/cells7070077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krzywinski M.I., Schein J.E., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1945. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Consortium P.G.S. Genome sequence and analysis of the tuber crop potato. Nature. 2011;475:189. doi: 10.1038/nature10158. [DOI] [PubMed] [Google Scholar]
- 41.Nicot N., Hausman J.F., Hoffmann L., Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J. Exp. Bot. 2005;56:2907–2914. doi: 10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- 42.Li X., Hamyat M., Liu C., Salman A., Gao X., Guo C., Wang Y., Guo Y. Identification and Characterization of the WOX Family Genes in Five Solanaceae Species Reveal Their Conserved Roles in Peptide Signaling. Genes. 2018;9:260. doi: 10.3390/genes9050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karve R., Liu W., Willet S.G., Torii K.U., Shpak E.D. The presence of multiple introns is essential for ERECTA expression in Arabidopsis. RNA. 2011;17:1907–1921. doi: 10.1261/rna.2825811. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.