Abstract
Background & Aims
Genome-wide association studies (GWAS) have identified more than 20 susceptibility loci for esophageal adenocarcinoma (EA) and Barrett’s esophagus (BE). However, variants in these loci account for a small fraction of cases of EA and BE. Genetic factors might interact with environmental factors to affect risk of EA and BE. We aimed to identify single nucleotide polymorphisms (SNPs) that may modify the associations of body mass index (BMI), smoking, and gastroesophageal reflux disease (GERD), with risks of EA and BE.
Methods
We collected data on single BMI measurements, smoking status, and symptoms of GERD from 2284 patients with EA, 3104 patients with BE, and 2182 healthy individuals (controls) participating in the Barrett’s and Esophageal Adenocarcinoma Consortium GWAS, the UK Barrett’s Esophagus Gene Study, and the UK Stomach and Oesophageal Cancer Study. We analyzed 993,501 SNPs in DNA samples of all study subjects. We used standard case–control logistic regression to test for gene-environment interactions.
Results
For EA, rs13429103 at chromosome 2p25.1, near the RNF144A-LOC339788 gene, showed a borderline significant interaction with smoking status (P = 2.18×10-7). Ever smoking was associated with an almost 12-fold increase in risk of EA among individuals with rs13429103-AA genotype (odds ratio=11.82; 95% CI, 4.03–34.67). Three SNPs (rs12465911, rs2341926, rs13396805) at chromosome 2q23.3, near the RND3-RBM43 gene, interacted with GERD symptoms (P = 1.70×10-7, P = 1.83×10-7, and P = 3.58×10-7, respectively) to affect risk of EA. For BE, rs491603 at chromosome 1p34.3, near the EIF2C3 gene, and rs11631094 at chromosome 15q14, at the SLC12A6 gene, interacted with BMI (P = 4.44×10-7) and pack-years of smoking history (P = 2.82×10-7), respectively.
Conclusion
The associations of BMI, smoking, and GERD symptoms with risks of EA and BE appear to vary with SNPs at chromosomes 1, 2, and 15. Validation of these suggestive interactions is warranted.
Keywords: Esophageal Neoplasm, Genetic Variants, Risk Factors, Esophagus
Abbreviations used in this paper: BE, Barrett’s esophagus; BMI, body mass index; CI, confidence interval; EA, esophageal adenocarcinoma; EAF, effect allele frequency; GERD, gastroesophageal reflux disease; GWAS, genome-wide association study; MAF, minor allele frequency; OR, odds ratio; SNP, single nucleotide polymorphism
Over the past 4 decades, the incidence of esophageal adenocarcinoma (EA) has increased markedly in many Western populations. Among white men in the United States the incidence has increased almost 10-fold,1 and rates continue to rise by 2% per year.2 EA is a highly fatal cancer with a median overall survival of <1 year following diagnosis.3 EAs typically arise on a background of a premalignant change in the lining of the esophagus known as Barrett’s esophagus (BE). Thus, proposals to prevent EA-associated morbidity and mortality have suggested focusing on identifying patients with BE and enrolling them in endoscopic surveillance programs, or on identifying and modifying risk factors for neoplastic progression.4, 5, 6
Epidemiologic studies have identified frequent or persistent symptoms of gastroesophageal reflux disease (GERD),7, 8 obesity,9 and smoking10, 11 as the principal factors associated with increased risks of EA and BE. These 3 factors together comprise almost 80% of the attributable burden of EA.12, 13 Genetic factors also influence risk of EA and BE. Recent genome-wide association studies (GWAS) and post-GWAS studies have identified more than 20 loci significantly associated with risks of EA and BE14; however, these variants seem to explain only a limited proportion of the heritability of these diseases (estimated to be 25% for EA and 35% for BE).15 It is possible that environmental risk factors for EA and BE may interact with multiple genes through various biological pathways to contribute to disease susceptibility. Given the strength of associations with known risk factors for EA and BE (especially when compared with most other cancers), and potentially shared biological pathways (eg, inflammation) underlying these risk factors,16 identifying gene-environment interactions may be more plausible in the setting of EA and BE. These gene-environment interactions may account for some of the missing heritability of EA and BE.15 However, previous efforts to identify gene-environment interactions for EA and BE have predominantly been candidate based and have involved only small numbers of single nucleotide polymorphisms (SNPs).17, 18, 19
With the aim of identifying SNPs that may modify the associations of body mass index (BMI), smoking, and GERD symptoms with risks of EA and BE, we used pooled questionnaire and genetic data from several studies to conduct a large scale genome-wide gene-environment interaction study of EA and BE.
Methods
Study Population
We obtained data from 1512 EA patients, 2413 BE patients, and 2185 control subjects of European ancestry from 14 epidemiologic studies conducted in Western Europe, Australia, and North America participating in the International Barrett’s and Esophageal Adenocarcinoma Consortium (http://beacon.tlvnet.net/) GWAS. The design of the Barrett’s and Esophageal Adenocarcinoma Consortium GWAS has been described in detail previously.20 Histological confirmation of EA and BE was carried out for all the participating studies. The pooled dataset also included an additional 1,003 EA patients and 882 BE patients from the United Kingdom Stomach and Oesophageal Cancer Study and the UK Barrett’s Esophagus Gene Study, respectively.20 The EA patients in the UK Stomach and Oesophageal Cancer Study had International Classification of Diseases coding of malignant neoplasm of the esophagus (C15) and pathological diagnosis of adenocarcinoma (M8140-8575). The BE patients were identified at endoscopy with confirmed histopathological diagnosis of intestinal metaplasia in the UK Barrett's Esophagus Gene Study. Each contributing study was performed under institutional review board approval and all participants gave informed consent.
SNP Genotyping
Genotyping of buffy coat or whole blood DNA from all participants was conducted using the Illumina Omni1M Quad platform (San Diego, CA), in accordance with standard quality-control procedures.21 For quality control, genotyped SNPs were excluded based on call rate <95%, Hardy-Weinberg Equilibrium P value over controls of <10–4, or minor allele frequency (MAF) ≤2%. After quality assurance and quality control, 993,501 SNPs were used for the current analysis. The analysis was restricted to the subset of ethnically homogenous individuals of European ancestry (confirmed in GWAS samples using principal components analysis).20
Environmental (“Exposure”) Variables
Individual-level exposure data for each study participant were harmonized and merged into a single deidentified dataset. The data were checked for consistency and completeness and any apparent inconsistencies were followed up with individual study investigators. Depending on the study, data from self-reported written questionnaires or in-person interviews were obtained at or near the time of cancer diagnosis for EA patients, at or near the time of BE diagnosis for BE patients, and at the time of recruitment for control subjects. BMI was calculated as weight divided by square of height (kg/m2). For the analysis we selected the weight from each participant that likely reflected usual adult weight (before, for example, any disease-related weight loss). For tobacco smoking, the exposure variables were smoking status (ever vs never) and total cigarette smoking exposure among ever smokers (pack-years of smoking exposure). Ever cigarette smoking was defined as either low threshold exposure (≥100 cigarettes over their whole life) or by asking whether they had ever smoked regularly. Pack-years of smoking exposure was derived by dividing the average number of cigarettes smoked daily by 20 and multiplying by the total number of years smoked. GERD symptoms were defined as the presence of heartburn (ie, a burning or aching pain behind the sternum) or acid reflux (ie, a sour taste from acid, bile, or other stomach contents rising up into the mouth). For analysis, we used the highest reported frequency for either GERD symptom. Participants were then categorized as recurrent vs not recurrent based on a frequency of weekly or greater GERD symptoms for “recurrent.”7 A total of 425 participants with missing values for all 3 covariates (BMI, smoking history, and history of GERD symptoms) were excluded from the analysis.
Statistical Analysis
We used standard case-control logistic regression to test for gene-environment interactions. SNP genotypes were treated as continuous variables and coded as 0, 1, or 2 copies of the minor allele. Exposure variables were either continuous (BMI and pack-years of smoking exposure) or dichotomous (smoking status and GERD symptoms). We modeled the gene-environment interaction by the product of the SNP genotype and the exposure variable, adjusting for age, sex, the first 4 principal components to control for possible population stratification, and the main terms of the SNP and the exposure variable. We used model-robust standard errors as suggested in Voorman et al22 to avoid inflated test statistics that can arise due to underestimation of variability in gene-environment GWAS. For SNPs from each of the top gene-environment interaction hits (ie, main text, P value for interaction <5.0 × 10–7) (Supplemental Material, P value for interaction <1.0 × 10–6) we also performed stratified analyses by genotype to examine the modified association of the known risk factor for EA or BE within the specific genotypes. Analyses were conducted using R software version 3.4.3. (R Foundation for Statistical Computing, Vienna, Austria), the GWASTools package,23 and Stata 13.0 (StataCorp LP, College Station, TX).
Results
The final study sample included 2284 EA patients, 3104 BE patients, and 2182 control subjects. Characteristics of the study sample are shown in Table 1. On average, BMI was higher among EA (mean, 28.4 kg/m2) and BE (mean, 28.7 kg/m2) patients than among control subjects (mean, 27.0 kg/m2). Similarly, EA and BE patients were more likely than control subjects to be ever smokers (74.8%, 64.8%, and 59.1%, respectively) and to report history of recurrent GERD symptoms (46.9%, 52.9%, and 19.4%, respectively).
Table 1.
Characteristics of the Study Population
| Characteristic | Control Subjects n = 2182 |
EA n = 2284 |
Control Subjects vs EA P valuea |
BE n = 3104 |
Control Subjects vs BE P Valuea |
|---|---|---|---|---|---|
| Age, y | 61.7 ± 11.1 | 65.1 ± 10.3 | <.001 | 62.9 ± 12.1 | <.001 |
| Sex | <.001 | .008 | |||
| Male | 1715 (78.6) | 1990 (87.1) | 2343 (75.5) | ||
| Female | 467 (21.4) | 294 (12.9) | 761 (24.5) | ||
| Body mass index, kg/m2 | <.001 | <.001 | |||
| Mean | 27.0 ± 4.7 | 28.4 ± 5.2 | 28.7 ± 5.1 | ||
| <25 | 786 (36.3) | 245 (24.6) | 608 (20.7) | ||
| 25–29.99 | 944 (43.5) | 455 (45.8) | 1191 (42.8) | ||
| ≥30 | 436 (20.2) | 296 (29.6) | 935 (36.5) | ||
| Missing | 16 | 1288 | 370 | ||
| Smoking status | <.001 | <.001 | |||
| Never | 888 (40.9) | 568 (25.2) | 1081 (35.2) | ||
| Ever | 1282 (59.1) | 1686 (74.8) | 1994 (64.8) | ||
| Missing | 12 | 30 | 29 | ||
| Cumulative smoking history, pack-yearsb | .43 | .001 | |||
| Mean | 32.8 ± 27.9 | 33.6 ± 26.4 | 29.4 ± 24.8 | ||
| Recurrent GERD symptoms | <.001 | <.001 | |||
| No | 1446 (80.6) | 965 (53.1) | 1058 (47.1) | ||
| Yes | 348 (19.4) | 854 (46.9) | 1186 (52.9) | ||
| Missing | 388 | 465 | 860 |
NOTE. Values are mean ± SD or n (%).
BE, Barrett’s esophagus; EA, esophageal adenocarcinoma; GERD, gastroesophageal reflux disease.
P value from chi-square tests for categorical variables and Student’s t test for continuous variables. Missing categories were excluded from comparison tests.
Among ever smokers.
Gene-Environment Interactions for EA
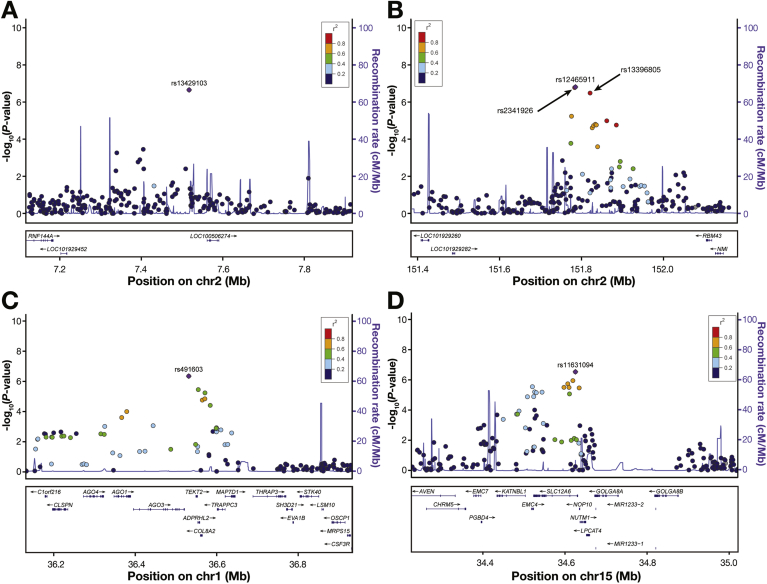
For EA, at borderline genome-wide significance, 1 SNP interacted with smoking status and 3 interacted with recurrent GERD symptoms (P for interactions ranging from 3.58 × 10–7 to 1.70 × 10–7) (Table 2, Figure 1A and B). At chromosome 2p25.1, rs13429103 (effect allele frequency [EAF] = 15.0%) showed interaction with smoking status (RNF144A-LOC339788, P = 2.18 × 10–7 for interaction). We also observed borderline statistically significant interactions between recurrent GERD symptoms and rs12465911 (P = 1.70 × 10–7 for interaction), rs2341926 (P = 1.83 × 10–7 for interaction), and rs13396805 (P = 3.58 × 10–7 for interaction) at chromosome 2q23.3 (RND3-RBM43). These 3 SNPs are in high linkage disequilibrium (all r2 > 0.9) as indicated in Figure 1B. Additional suggestive gene-environment interactions for EA (where P < 1.0 × 10–6 for interaction) are shown in Supplemental Table 1.
Table 2.
Gene-Environment Interactions With EA or BE With a P Value for Interaction <5.0 × 10–7
| Outcome | Exposure | SNP | Chr | Position | Gene | Effect/ Other |
EAF | OR | P |
|---|---|---|---|---|---|---|---|---|---|
| EA | |||||||||
| Smoking status | rs13429103 | 2p25.1 | 7517231 | RNF144A-LOC339788 | A/G | 0.15 | 2.04 | 2.18 × 10–7 | |
| Recurrent GERD symptoms | rs12465911 | 2q23.3 | 151785742 | RND3-RBM43 | A/G | 0.26 | 2.03 | 1.70 × 10–7 | |
| Recurrent GERD symptoms | rs2341926 | 2q23.3 | 151783928 | RND3-RBM43 | C/T | 0.26 | 2.02 | 1.83 × 10–7 | |
| Recurrent GERD symptoms | rs13396805 | 2q23.3 | 151821512 | RND3-RBM43 | A/G | 0.26 | 1.99 | 3.58 × 10–7 | |
| BE | |||||||||
| BMI (continuous) | rs491603 | 1p34.3 | 36532316 | EIF2C3-LOC100128093 | A/G | 0.16 | 1.08 | 4.44 × 10–7 | |
| Pack-years of smoking | rs11631094 | 15q14 | 34624438 | SLC12A6 | A/C | 0.29 | 0.99 | 2.82 × 10–7 |
BE, Barrett’s esophagus; BMI, body mass index; EA, esophageal adenocarcinoma; EAF, effect allele frequency; GERD, gastroesophageal reflux disease; OR, odds ratio; SNP, single nucleotide polymorphism.
Figure 1.
Regional association plots for genotyped single nucleotide polymorphisms (SNPs) showing P values for interaction for (A) smoking status and (B) recurrent gastroesophageal reflux disease symptoms in esophageal adenocarcinoma and (C) body mass index and (D) pack-years of smoking exposure in Barrett’s esophagus. The SNPs in Table 2 are shown as a solid purple diamond, except in panel B where rs2341926 and rs13396805 are shown as circles near rs12465911. The color scheme indicates linkage disequilibrium between the SNP shown with a solid purple diamond and other SNPs in the region using the r2 value calculated from the 1000 Genomes Project. The y axis is the −log10 interaction P value computed from 5388 cases (3104 Barrett’s esophagus, 2284 esophageal adenocarcinoma) and 2182 control subjects. The recombination rate from CEU HapMap data (right-side y axis) is shown in light blue. (A) Chromosome 2p25.1; (B) chromosome 2q23.3 region; (C) chromosome 1p34.3 region; (D) chromosome 15q14 region.
In analyses stratified by genotype (Table 3), compared with never smoking, ever smoking was associated with nearly a 12-fold higher risk of EA among individuals with rs13429103-AA genotype (odds ratio [OR], 11.82; 95% confidence interval [CI], 4.03–34.67). In contrast, among individuals with rs13429103-GG genotype, ever smoking conferred only 1.6-fold higher risk of EA (OR, 1.59; 95% CI, 1.36–1.85). Similarly, the risk for EA associated with recurrent GERD symptoms was higher in individuals with rs12465911-AA genotype (OR, 13.12; 95% CI, 6.21–27.73) than among individuals with rs12465911-GG genotype (OR, 2.80; 95% CI, 2.29–3.41). Additional stratified analyses for risk of EA are shown in Table 3 and Supplemental Table 2.
Table 3.
Risk of EA and BE in Association With Obesity, Smoking History and Recurrent GERD Symptoms, Stratified by Genotype for SNPs in Table 2
| Outcome | Environmental Exposure | SNP | Genotype | Cases/Control Subjects | OR | 95% CI | P Valuea |
|---|---|---|---|---|---|---|---|
| EA | |||||||
| Ever smoker vs never smoker (ref) | rs13429103 | GG | 1617/1572 | 1.59 | 1.36–1.85 | <.001 | |
| GA | 589/554 | 2.91 | 2.23–3.81 | <.001 | |||
| AA | 48/44 | 11.82 | 4.03–34.67 | <.001 | |||
| Recurrent GERD symptoms vs nonrecurrent GERD symptoms (ref) | rs12465911 | GG | 1206/1196 | 2.80 | 2.29–3.41 | <.001 | |
| GA | 885/823 | 5.32 | 4.10–6.90 | <.001 | |||
| AA | 163/151 | 13.12 | 6.21–27.73 | <.001 | |||
| Recurrent GERD symptoms vs nonrecurrent GERD symptoms (ref) | rs2341926 | TT | 975/985 | 2.80 | 2.30–3.42 | <.001 | |
| TC | 724/681 | 5.30 | 4.08–6.88 | <.001 | |||
| CC | 120/128 | 13.12 | 6.21–27.73 | <.001 | |||
| Recurrent GERD symptoms vs nonrecurrent GERD symptoms (ref) | rs13396805 | GG | 998/1005 | 2.85 | 2.34–3.48 | <.001 | |
| GA | 701/662 | 5.23 | 4.02–6.81 | <.001 | |||
| AA | 120/127 | 12.73 | 6.12–26.49 | <.001 | |||
| BE | |||||||
| BMI ≥30 kg/m2 vs BMI <25 kg/m2 (ref) | rs491603 | GG | 1306/1137 | 1.52 | 1.38–1.67 | <.001 | |
| GA | 438/518 | 2.11 | 1.80–2.47 | <.001 | |||
| AA | 42/64 | 3.30 | 1.90–5.73 | <.001 | |||
| ≥15 pack-years vs <15 pack-years (ref) | rs11631094 | CC | 729/618 | 1.02 | 0.81–1.30 | .846 | |
| CA | 555/540 | 0.65 | 0.50–0.84 | .001 | |||
| AA | 115/106 | 0.52 | 0.28–0.95 | .033 |
BE, Barrett’s esophagus; BMI, body mass index; CI, confidence interval; EA, esophageal adenocarcinoma; GERD, gastroesophageal reflux disease; OR, odds ratio; SNP, single nucleotide polymorphism.
P values from logistic regression analysis adjusted for age and sex.
Gene-Environment Interactions for BE
For BE, at chromosome 1p34.3, we observed an interaction between rs491603 (EAF = 16.5%) and BMI (EIF2C3-LOC100128093, P = 4.44 × 10–7 for interaction) (Table 2, Figure 1C). At chromosome 15p14, rs11631094 (EAF = 28.7%) showed interaction with pack-years of smoking exposure (SLC12A6, P = 2.82 × 10–7 for interaction) (Table 2, Figure 1D). Additional suggestive significant interactions (where P < 1.0 × 10–6 for interaction) for BE with pack-years of smoking exposure at chromosomes 12q23.1, 16p12.3, and 17q12 are presented in Supplemental Table 1.
Stratified analyses by genotype showed that the risk for BE associated with obesity (BMI ≥30 kg/m2) was elevated by over 200% among individuals with rs491603-AA genotype (vs BMI <25 kg/m2; OR, 3.30; 95% CI, 1.90–5.73) but only by approximately 50% among individuals with rs491603-GG genotype (vs BMI <25 kg/m2; OR, 1.52; 95% CI, 1.38–1.67). Additional stratified analyses of gene-environment interactions for BE are shown in Table 3 and Supplemental Table 2.
Cross-Examination of Discovered Gene-Environment Interactions
For each SNP in Table 2 and Supplemental Table 1 that had a borderline significant genome-wide interaction in either EA or BE, we examined the equivalent gene-environment interaction in BE and EA, respectively (Supplemental Table 3). For all SNPs discovered in EA, we observed nominal levels of significance (P value for interaction <.05) and ORs in the same direction but somewhat attenuated in BE. For SNPs discovered in BE, only half had P value for interaction <.05 in EA, although all had similar ORs to those in BE. Although obesity and GERD are correlated, none of the SNPs with P value for interaction <1.0 × 10–6 with GERD had comparable ORs or P values when testing for interaction with obesity and similarly for the 1 obesity SNP when tested for GERD.
Discussion
To our knowledge, this is the first genome-wide gene-environment interaction study of EA and its precursor, BE. Although no gene-environment interactions reached genome-wide significance (ie, P < 5.0 × 10–8 for interaction), several borderline significant interactions were indicated between SNPs and known risk factors for EA and BE – BMI, smoking, and GERD symptoms.
A number of studies have pursued candidate-based gene-environment analyses of EA, and reported interactions between BMI, smoking or GERD symptoms and selected SNPs in genes related to detoxification, angiogenesis, DNA repair, apoptosis, and extracellular matrix degradation.24, 25, 26, 27, 28, 29, 30, 31 This body of work helped to establish the notion that the level of disease risk associated with GERD symptoms, in particular, may vary according to inherited genetic variation. All of these studies, however, were conducted in small samples (<350 cases) and were not replicated in independent populations. While direct comparison of our own results and these past findings is complicated by less-than-complete overlap of genotyped SNPs between studies, we did not find evidence in support of interactions among BMI, smoking, or GERD symptoms and any assessed variants in previously-implicated genes: GSTM1, GSTT1, VEGF, MGMT, EGF, IL1B, PERP, PIK3CA, TNFRSF1A, CASP7, TP53BP1, BCL2, HIF1AN, PDGRFA, VEGFR1, or MMP1 (Supplemental Table 4). It remains possible that nominal evidence for some of these associations may not have survived stringent correction for multiple comparisons, and larger samples are needed for true signals to reach significance. Alternatively, previously reported interactions may simply reflect chance findings in small samples because they did not validate in our large study population.
This study has several strengths. First, the pooled dataset including relatively large numbers of cases and control subjects provided us with a rare opportunity to perform, in parallel, genome-wide gene-environment interaction analyses for EA and its precursor lesion, BE. Past candidate-based gene-environment interaction studies of EA have focused on small numbers of genes selected according to biological plausibility, and collectively these reports sampled only a small fraction of the total SNPs presently analyzed (N = 993,501). Such preconceived “gene-centric” SNP selection methods fail to capture the large fraction of noncoding intergenic variations that have been linked to altered risk for these 2 conditions, and also artificially restricts the “genic” search space based on limited mechanistic knowledge, a limitation that is overcome by an unbiased comprehensive genome-wide gene-environment interaction assessment. Second, our study draws on genetic and epidemiologic data from a recent consortium-based GWAS of EA/BE,20 which is the largest of its kind. This sizable study sample afforded greater power to detect gene-environment interactions than in any previous study. Third, all genotyping from this GWAS was conducted on a single platform and in a single laboratory, and subjected to stringent quality-control procedures. Most GWAS analyses test only an additive model because an additive model has reasonable power to detect both additive and dominant effects and the 2 models yield similar results and many GWAS analyses, including ours, are underpowered to detect recessive effects. Nevertheless, for completeness we also tested a dominant model for the 16 SNPs with a P value for interaction <1.0 × 10–6 (Table 2 and Supplemental Table 1), and found slightly attenuated results of the ORs for some gene-environment interactions (data not shown).
Our study also has some limitations. First, our ability to detect true gene-environment interactions might have been limited by the manner in which the environmental (exposure) variables were measured and harmonized. For example, recall bias is a possibility during retrospective reporting of the exposures in the parent case-control studies. However, respondents were unaware of their genotype status at the time of the interviews, mitigating the impact of any possible recall bias in our interaction analyses. Similarly, while considerable care was taken during data harmonization, as described in a series of recent pooled analyses,10, 11 some potential for measurement error of the exposures examined is possible. However, given that case-control status was not considered during this process, any errors from harmonization would be nondifferential, resulting in attenuation of the resulting ORs. Second, central obesity (eg, waist-to-hip ratio) has been found to be more strongly associated with the risk of BE than BMI; however, as waist and hip measurements were not collected in the majority of the included studies, we were unable to examine for interactions with central obesity. Third, despite the comprehensive nature of the genome-wide analysis, we were nonetheless limited to examining common genetic variation (MAF >2%) represented on the Illumina Omni1M Quad GWAS platform employed. Further large-scale studies based on whole-exome or whole-genome sequencing would be required to identify additional gene-environment interactions with rare variants, and more precisely map the reported associations. Finally, our study results should be considered as discovery findings, worthy of independent replication. None of the interactions studied reached genome-wide significance (ie, P < 5.0 × 10–8 for interaction). This may be because there are truly no gene-environment interactions or it may be that power was still limited to detect modest or weak interactions despite our large sample size. In our analyses of 2284 EA patients, 3104 BE patients, and 2182 control subjects, we were adequately powered to detect interactions with an interaction OR in the range of 1.98–2.52 for MAF in the observed range (0.11–0.43), assuming a main effect of 1.08 for log-additive SNPs, a main effect of 1.90 for binary risk factors, and an α of 5.0 × 10–8. Given the large worldwide consortia sample of patients participating in this work, few additional studies of EA and BE patients are currently available and have data for replication; thus, such work may require additional time for study patients to accrue.
In conclusion, our report describes the first genome-wide gene-environment interaction analysis for EA and BE. These findings provide evidence that the magnitude of disease risk associated with BMI, smoking, and GERD symptoms may differ according to germline genetics, and suggest the potential utility of combing epidemiologic exposure data with selected genotyping for comprehensive risk assessment in patients susceptible to EA or BE. Pending validation of the observed interactions in independent study populations, further analyses will be required to investigate the biological basis for differential disease risk associated with the risk factors investigated in the presence of these variants.
Acknowledgments
The following UK hospitals participated in sample collection through the Stomach and Oesophageal Cancer Study (SOCS) collaboration network: Addenbrooke's Hospital, University College London, Bedford Hospital, Hinchingbrooke Hospital, Peterborough City Hospital, West Suffolk Hospital, Norfolk and Norwich University Hospital, Churchill Hospital, John Radcliffe Hospital, Velindre Hospital, St Bartholomew's Hospital, Queen's Hospital Burton, Queen Elisabeth Hospital, Diana Princess of Wales, Scunthorpe General Hospital, Royal Devon & Exeter Hospital, New Cross Hospital, Belfast City Hospital, Good Hope Hospital, Heartlands Hospital, South Tyneside District General Hospital, Cumberland Infirmary, West Cumberland Hospital, Withybush General Hospital, Stoke Mandeville Hospital, Wycombe General Hospital, Wexham Park Hospital, Southend Hospital, Guy's Hospital, Southampton General Hospital, Bronglais General Hospital, Aberdeen Royal Infirmary, Manor Hospital, Clatterbridge Centre for Oncology, Lincoln County Hospital, Pilgrim Hospital, Grantham & District Hospital, St Mary's Hospital London, Croydon University Hospital, Whipps Cross University Hospital, Wansbeck General Hospital, Hillingdon Hospital, Milton Keynes General Hospital, Royal Gwent Hospital, Tameside General Hospital, Castle Hill Hospital, St Richard's Hospital, Ipswich Hospital, St Helens Hospital, Whiston Hospital, Countess of Chester Hospital, St Mary's Hospital IOW, Queen Alexandra Hospital, Glan Clwyd Hospital, Wrexham Maelor Hospital, Darent Valley Hospital, Royal Derby Hospital, Derbyshire Royal Infirmary, Scarborough General Hospital, Kettering General Hospital, Kidderminster General Hospital, Royal Lancaster Infirmary, Furness General Hospital, Westmorland General Hospital, James Cook University Hospital, Friarage Hospital, Stepping Hill Hospital, St George's Hospital London, Doncaster Royal Infirmary, Maidstone Hospital, Tunbridge Hospital, Prince Charles Hospital, Hartlepool Hospital, University Hospital of North Tees, Ysbyty Gwynedd, St. Jame's University Hospital, Leeds General Infirmary, North Hampshire Hospital, Royal Preston Hospital, Chorley and District General, Airedale General Hospital, Huddersfield Royal Infirmary, Calderdale Royal Hospital, Torbay District General Hospital, Leighton Hospital, Royal Albert Edward Infirmary, Royal Surrey County Hospital, Bradford Royal Infirmary, Burnley General Hospital, Royal Blackburn Hospital, Royal Sussex County Hospital, Freeman Hospital, Royal Victoria Infirmary, Victoria Hospital Blackpool, Weston Park Hospital, Royal Hampshire County Hospital, Conquest Hospital, Royal Bournemouth General Hospital, Mount Vernon Hospital, Lister Hospital, William Harvey Hospital, Kent and Canterbury Hospital, Great Western Hospital, Dumfries and Galloway Royal Infirmary, Poole General Hospital, St Hellier Hospital, North Devon District Hospital, Salisbury District Hospital, Weston General Hospital, University Hospital Coventry, Warwick Hospital, George Eliot Hospital, Alexandra Hospital, Nottingham University Hospital, Royal Chesterfield Hospital, Yeovil District Hospital, Darlington Memorial Hospital, University Hospital of North Durham, Bishop Auckland General Hospital, Musgrove Park Hospital, Rochdale Infirmary, North Manchester General, Altnagelvin Area Hospital, Dorset County Hospital, James Paget Hospital, Derriford Hospital, Newham General Hospital, Ealing Hospital, Pinderfields General Hospital, Clayton Hospital, Dewsbury & District Hospital, Pontefract General Infirmary, Worthing Hospital, Macclesfield Hospital, University Hospital of North Staffordshire, Salford Royal Hospital, Royal Shrewsbury Hospital, Manchester Royal Infirmary.
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by funding from the U.S. National Cancer Institute (NCI) at the National Institutes of Health (grant number R01CA136725) (to T.L.V. and D.C.W.). The U.S. Multi-Center Study was funded by grants U01-CA57949 (to T.L.V.), U01-CA57983 (to M.D.G.), and U01-CA57923 (to H.A.R.). The Seattle Barrett’s Esophagus Study was supported by P01 CA 91955 (to B.J.R.) from the NCI. The Australian Cancer Study was supported by the Queensland Cancer Fund and the National Health and Medical Research Council (NHMRC) of Australia (Program no. 199600) (to D.C.W, Adele C. Green, Nicholas K. Hayward, Peter G. Parsons, David M. Purdie, and Penelope M. Webb). The Swedish Esophageal Cancer Study was supported by grant R01 CA57947-03 (to Olof Nyren and Hans-Olov Adami) from the NCI. The Los Angeles County Multi-ethnic Case-control Study was supported by grants 3RT-0122 (Smoking and Risk of Proximal Vs. Distal Gastric Cancer) (to A.H.W) and 10RT-0251 (Smoking, microsatellite instability & gastric cancers) (to A.H.W) from the California Tobacco Related Research Program and grant CA59636 (to L.B.) from the NCI. The FINBAR (Factors Influencing the Barrett’s Adenocarcinoma Relationship) study was supported by an Ireland-Northern Ireland Co-operation Research Project Grant sponsored by the Northern Ireland Research & Development Office, and the Health Research Board, Ireland (All-Ireland case-control study of Oesophageal Adenocarcinoma and Barrett’s Oesophagus) (to Liam Murray and Harry Comber). Additional funding was provided by the Ulster Cancer Foundation (Belfast, Northern Ireland) and the Northern Ireland Research and Development Office Clinical Fellowship. The Study of Digestive Health was supported by grant 5 RO1 CA 001833-02 (to D.C.W, Adele C. Green, Nicholas K. Hayward, Peter G. Parsons, Sandra J. Pavey, David M. Purdie, Penelope M. Webb, David Gotley, B. Mark Smithers, Glyn G. Jamieson, Paul Drew, David I. Watson, and Andrew Clouston) from the NCI. The Study of Reflux Disease was supported by grant R01 CA72866 (to T.L.V and Diana Farrow) from the NCI. The UK Barrett’s Oesophagus Gene Study was supported by a Medical Research Council Programme grant. The UK Stomach and Oesophageal Cancer Study was funded by Cancer Research UK, as well as by funding from the Cambridge National Institute of Health Research Biomedical Research Centre and the Cambridge Experimental Cancer Medicine Centre. D.A.C. was supported by RO1 DK63616-01, and R21 DK077742. N.J.S. is supported by P30 DK034987 from the National Institutes of Health. T.L.V. is supported by an Established Investigator Award (K05CA124911) from the NCI. DCW is supported by a Research Fellowship from the NHMRC. S.M. is supported by Australian Research Council and NHMRC Fellowships. This work was supported by Roswell Park Cancer Institute and NCI grant P30CA016056 (M.F.B.). J.D. is supported by a Research Training Grant from the Cancer Prevention and Research Institute of Texas (CPRIT) (RP160097). The funders of the individual studies had no role in the design, analysis, or interpretation of the data, or in writing or publication decisions related to this manuscript.
Note: To access the supplementary material accompanying this article, visit the online version of Clinical Gastroenterology and Hepatology at www.cghjournal.org, and at https://doi.org/10.1016/j.cgh.2018.03.007.
Supplementary Material
Supplemental Table 1.
Gene-Environment Interactions With EA or BE With a P Value for Interaction <1.0 × 10–6
| Outcome | Exposure | SNP | Chr | Position | Gene | Effect/ Other |
EAF | OR | P Value |
|---|---|---|---|---|---|---|---|---|---|
| EA | |||||||||
| Smoking status | rs2434584 | 5q11.2 | 57566073 | ACTBL2-PLK2 | C/T | 0.08 | 2.52 | 7.44 × 10–7 | |
| Smoking status | rs40210 | 5q11.2 | 57619964 | ACTBL2-PLK2 | A/G | 0.08 | 2.46 | 8.82 × 10–7 | |
| Pack-years of smoking | rs17002540 | Xq27.1 | 139946061 | CDR1-SPANXB2 | T/C | 0.19 | 0.99 | 5.92 × 10–7 | |
| Recurrent GERD symptoms | rs2971030 | 7p21.3 | 10006341 | LOC340268 | G/A | 0.42 | 1.77 | 6.02 × 10–7 | |
| Recurrent GERD symptoms | rs7141987 | 14q32.31 | 101492224 | SNORD114-31-LOC100130814 | G/A | 0.42 | 1.77 | 7.11 × 10–7 | |
| Recurrent GERD symptoms | rs2971028 | 7p21.3 | 10007255 | LOC340268 | A/G | 0.40 | 1.76 | 8.56 × 10–7 | |
| BE | |||||||||
| Pack-years of smoking | rs9668109 | 12q23.1 | 99011272 | IKIP | A/G | 0.09 | 0.98 | 6.31 × 10–7 | |
| Pack-years of smoking | rs1548445 | 16p12.3 | 19691583 | C16orf62 | G/A | 0.06 | 1.02 | 8.21 × 10–7 | |
| Pack-years of smoking | rs2671828 | 17q12 | 33731764 | SLFN11-LOC729839 | A/G | 0.43 | 0.99 | 9.54 × 10–7 | |
| Pack-years of smoking | rs10507102 | 12q23.1 | 98990871 | SLC25A3 | A/G | 0.09 | 0.98 | 9.91 × 10–7 |
BE, Barrett’s esophagus; BMI, body mass index; EA, esophageal adenocarcinoma; EAF, effect allele frequency; GERD, gastroesophageal reflux disease; OR, odds ratio; SNP, single nucleotide polymorphism.
Supplemental Table 2.
Risk of EA and BE in Association With Smoking History and GERD Symptoms, Stratified by Genotype for SNPs in Supplemental Table 1
| Outcome | Environmental Exposure | SNP | Genotype | Cases/Control Subjects | OR | 95% CI | P Valuea |
|---|---|---|---|---|---|---|---|
| EA | |||||||
| Ever smoker vs never smoker (ref) | rs2434584 | TT | 1907/1826 | 1.67 | 1.45–1.93 | <.001 | |
| CT | 342/328 | 4.33 | 3.00–6.24 | <.001 | |||
| CC | 5/15 | NA | - | - | |||
| Ever smoker vs never smoker (ref) | rs40210 | GG | 1903/1821 | 1.67 | 1.45–1.92 | <.001 | |
| GA | 344/332 | 4.24 | 2.96–6.06 | <.001 | |||
| AA | 6/16 | NA | - | - | |||
| ≥15 pack-years vs <15 pack-years (ref) | rs17002540 | CC | 1053/1003 | 1.36 | 1.12–1.66 | .002 | |
| CT | 48/55 | 0.78 | 0.33–1.86 | .579 | |||
| TT | 218/206 | 0.63 | 0.39–1.00 | .052 | |||
| Recurrent GERD symptoms vs nonrecurrent GERD symptoms (ref) | rs2971030 | AA | 599/603 | 2.68 | 2.08–3.44 | <.001 | |
| GA | 908/895 | 3.81 | 3.06–4.75 | <.001 | |||
| GG | 309/293 | 9.44 | 6.17–14.45 | <.001 | |||
| Recurrent GERD symptoms vs nonrecurrent GERD symptoms (ref) | rs7141987 | AA | 591/590 | 2.69 | 2.08–3.49 | <.001 | |
| GA | 908/887 | 3.74 | 3.02–4.64 | <.001 | |||
| GG | 319/317 | 9.32 | 6.04–14.36 | <.001 | |||
| Recurrent GERD symptoms vs nonrecurrent GERD symptoms (ref) | rs2971028 | GG | 625/635 | 2.70 | 2.11–3.45 | <.001 | |
| GA | 900/890 | 3.87 | 3.10–4.82 | <.001 | |||
| AA | 294/268 | 9.58 | 6.17–14.88 | <.001 | |||
| BE | |||||||
| ≥15 pack-years vs <15 pack-years (ref) | rs9668109 | GG | 1167/1058 | 0.92 | 0.77–1.11 | .390 | |
| GA | 221/201 | 0.38 | 0.25–0.60 | <.001 | |||
| AA | 11/5 | 0.33 | 0.02–5.64 | .443 | |||
| ≥15 pack-years vs <15 pack-years (ref) | rs1548445 | AA | 1223/1097 | 0.76 | 0.63–0.91 | .002 | |
| GA | 170/164 | 1.11 | 0.68–1.80 | .675 | |||
| GG | 6/3 | NA | - | - | |||
| ≥15 pack-years vs <15 pack-years (ref) | rs2671828 | GG | 457/423 | 0.93 | 0.70–1.23 | .595 | |
| GA | 688/588 | 0.84 | 0.66–1.07 | .163 | |||
| AA | 246/250 | 0.54 | 0.36–0.80 | .002 | |||
| ≥15 pack-years vs <15 pack-years (ref) | rs10507102 | GG | 1166/1058 | 0.93 | 0.77–1.11 | .409 | |
| GA | 222/200 | 0.38 | 0.24–0.59 | <.001 | |||
| AA | 11/5 | 0.33 | 0.02–5.64 | .443 |
BE, Barrett’s esophagus; BMI, body mass index; EA, esophageal adenocarcinoma; EAF, effect allele frequency; GERD, gastroesophageal reflux disease; SNP, single nucleotide polymorphism.
P values from logistic regression analysis adjusted for age and sex.
Supplemental Table 3.
Comparison of Gene-Environment Interactions in BE and EA for SNPs With P Value for Interaction <1.0 × 10–6 on the Outcomes
| Exposure | SNP | Chr | Position | Gene | Effect/ Other |
BE |
EA |
||
|---|---|---|---|---|---|---|---|---|---|
| OR | P | OR | P | ||||||
| G × E hits for EA | |||||||||
| Smoking status | rs13429103 | 2p25.1 | 7517231 | RNF144A-LOC339788 | A/G | 1.40 | 3.51 × 10–3 | 2.04 | 2.18 × 10–7 |
| Smoking status | rs2434584 | 5q11.2 | 57566073 | ACTBL2-PLK2 | C/T | 1.56 | 3.90 × 10–3 | 2.52 | 7.44 × 10–7 |
| Smoking status | rs40210 | 5q11.2 | 57619964 | ACTBL2-PLK2 | A/G | 1.54 | 4.94 × 10–3 | 2.46 | 8.82 × 10–7 |
| Pack-years of smoking | rs17002540 | Xq27.1 | 139946061 | CDR1-SPANXB2 | T/C | 0.99 | 4.83 × 10–3 | 0.99 | 5.92 × 10–7 |
| Recurrent GERD symptoms | rs12465911 | 2q23.3 | 151785742 | RND3-RBM43 | A/G | 1.66 | 6.09 × 10–5 | 2.03 | 1.70 × 10–7 |
| Recurrent GERD symptoms | rs2341926 | 2q23.3 | 151783928 | RND3-RBM43 | C/T | 1.65 | 7.38 × 10–5 | 2.02 | 1.83 × 10–7 |
| Recurrent GERD symptoms | rs13396805 | 2q23.3 | 151821512 | RND3-RBM43 | A/G | 1.59 | 2.80 × 10–4 | 1.99 | 3.58 × 10–7 |
| Recurrent GERD symptoms | rs2971030 | 7p21.3 | 10006341 | LOC340268 | G/A | 1.36 | 5.03 × 10–3 | 1.77 | 6.02 × 10–7 |
| Recurrent GERD symptoms | rs7141987 | 14q32.31 | 101492224 | SNORD114-31-LOC100130814 | G/A | 1.29 | 1.40 × 10–2 | 1.77 | 7.11 × 10–7 |
| Recurrent GERD symptoms | rs2971028 | 7p21.3 | 10007255 | LOC340268 | A/G | 1.35 | 6.10 × 10–3 | 1.76 | 8.56 × 10–7 |
| G × E hits for BE | |||||||||
| BMI (continuous) | rs491603 | 1p34.3 | 36532316 | EIF2C3-LOC100128093 | A/G | 1.08 | 4.44 × 10–7 | 1.04 | 1.83 × 10–2 |
| Pack-years of smoking | rs11631094 | 15q14 | 34624438 | SLC12A6 | A/C | 0.99 | 2.82 × 10–7 | 1.00 | 0.125 |
| Pack-years of smoking | rs9668109 | 12q23.1 | 99011272 | IKIP | A/G | 0.98 | 6.31 × 10–7 | 0.99 | 9.74 × 10–3 |
| Pack-years of smoking | rs1548445 | 16p12.3 | 19691583 | C16orf62 | G/A | 1.02 | 8.21 × 10–7 | 1.01 | 9.70 × 10–2 |
| Pack-years of smoking | rs2671828 | 17q12 | 33731764 | SLFN11-LOC729839 | A/G | 0.99 | 9.54 × 10–7 | 1.00 | 6.13 × 10–2 |
| Pack-years of smoking | rs10507102 | 12q23.1 | 98990871 | SLC25A3 | A/G | 0.98 | 9.91 × 10–7 | 0.99 | 6.93 × 10–3 |
BE, Barrett’s esophagus; BMI, body mass index; EA, esophageal adenocarcinoma; EAF, effect allele frequency; G × E, gene-environment; GERD, gastroesophageal reflux disease; OR, odds ratio; SNP, single nucleotide polymorphism.
Supplemental Table 4.
Associations of Previously Reported Gene-Environment Interactions With Esophageal Adenocarcinoma in Our Study Population
| Original Publication |
Current Study |
||||
|---|---|---|---|---|---|
| Author | SNP | Exposure | P Value | Directly Genotyped or High-LD SNP | P Value |
| Casson et al, 200624 | NA | - | - | - | - |
| Zhai et al, 200825 | rs833061 | Smoking | .03 | rs833070 | .068 |
| Doecke et al, 200826 | rs12269324 | GERD symptoms | - | Direct | .979 |
| rs12268840 | GERD symptoms | - | Direct | .714 | |
| Cheung et al, 200927 | rs4444903 | GERD symptoms | <.001a | Direct | .240 |
| Zhai et al, 201228 | rs1143634 | GERD symptoms | .008 | Direct | .398 |
| rs1052486 | BMI + Smoking | - | - | - | |
| rs1052486 | BMI | - | Direct | .423 | |
| rs1052486 | Smoking | - | Direct | .532 | |
| Wu et al, 201129 | rs648802 | GERD symptoms | .02 | Direct | .838 |
| rs4855094 | GERD symptoms | .04 | Direct | .872 | |
| rs7644468 | GERD symptoms | .04 | - | - | |
| rs4149579 | GERD symptoms | .04 | - | - | |
| rs560191 | Smoking | .02 | Direct | .331 | |
| rs7907519 | Smoking | .04 | rs11196449 | .868 | |
| rs12454712 | Smoking | .04 | Direct | .435 | |
| Zhai et al, 201230 | rs2295778 | GERD symptoms | .0005 | rs12780796 | .654 |
| rs13337626 | GERD symptoms | .0067 | rs34197769 | .315 | |
| rs2295778 | Smoking | .004 | - | - | |
| rs2296188 | Smoking | .014 | Direct | .905 | |
| rs2114039 | BMI | .0026 | Direct | .228 | |
| rs2296188 | BMI | .0023 | Direct | .452 | |
| rs11941492 | BMI | .013 | Direct | NAb | |
| rs17708574 | BMI | .013 | Direct | .316 | |
| rs7324547 | BMI | .008 | - | - | |
| rs17619601 | BMI | .016 | - | - | |
| rs17625898 | BMI | .023 | - | - | |
| Cheung et al, 201231 | rs1799750 | GERD symptoms | .002 | - | - |
| rs3025058 | GERD symptoms | .04 | - | - | |
BMI, body mass index; GERD, gastroesophageal reflux disease; LD, linkage disequilibrium; SNP, single nucleotide polymorphism.
Two-way interaction.
On array but quality control failure. We were unable to validate all SNPs as some were biallelic or we failed to identify a high LD SNP (r2 < 0.70).
References
- 1.Vaughan T.L., Fitzgerald R.C. Precision prevention of oesophageal adenocarcinoma. Nat Rev Gastroenterol Hepatol. 2015;12:243–248. doi: 10.1038/nrgastro.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thrift A.P., Whiteman D.C. The incidence of esophageal adenocarcinoma continues to rise: analysis of period and birth cohort effects on recent trends. Ann Oncol. 2012;23:3155–3162. doi: 10.1093/annonc/mds181. [DOI] [PubMed] [Google Scholar]
- 3.Thrift A.P. The epidemic of oesophageal carcinoma: Where are we now? Cancer Epidemiol. 2016;41:88–95. doi: 10.1016/j.canep.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 4.Shaheen N.J., Falk G.W., Iyer P.G. ACG Clinical Guideline: Diagnosis and management of Barrett's esophagus. Am J Gastroenterol. 2016;111:30–50. doi: 10.1038/ajg.2015.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spechler S.J., Sharma P., Souza R.F. American Gastroenterological Association technical review on the management of Barrett's esophagus. Gastroenterology. 2011;140:e18–e52. doi: 10.1053/j.gastro.2011.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fitzgerald R.C., di Pietro M., Ragunath K. British Society of Gastroenterology guidelines on the diagnosis and management of Barrett's oesophagus. Gut. 2014;63:7–42. doi: 10.1136/gutjnl-2013-305372. [DOI] [PubMed] [Google Scholar]
- 7.Cook M.B., Corley D.A., Murray L.J. Gastroesophageal reflux in relation to adenocarcinomas of the esophagus: a pooled analysis from the Barrett's and Esophageal Adenocarcinoma Consortium (BEACON) PLoS One. 2014;9:e103508. doi: 10.1371/journal.pone.0103508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thrift A.P., Kramer J.R., Qureshi Z. Age at onset of GERD symptoms predicts risk of Barrett's esophagus. Am J Gastroenterol. 2013;108:915–922. doi: 10.1038/ajg.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thrift A.P., Shaheen N.J., Gammon M.D. Obesity and risk of esophageal adenocarcinoma and Barrett's esophagus: a Mendelian randomization study. J Natl Cancer Inst. 2014;106:dju252. doi: 10.1093/jnci/dju252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cook M.B., Kamangar F., Whiteman D.C. Cigarette smoking and adenocarcinomas of the esophagus and esophagogastric junction: a pooled analysis from the international BEACON consortium. J Natl Cancer Inst. 2010;102:1344–1353. doi: 10.1093/jnci/djq289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook M.B., Shaheen N.J., Anderson L.A. Cigarette smoking increases risk of Barrett's esophagus: an analysis of the Barrett's and Esophageal Adenocarcinoma Consortium. Gastroenterology. 2012;142:744–753. doi: 10.1053/j.gastro.2011.12.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olsen C.M., Pandeya N., Green A.C. Population attributable fractions of adenocarcinoma of the esophagus and gastroesophageal junction. Am J Epidemiol. 2011;174:582–590. doi: 10.1093/aje/kwr117. [DOI] [PubMed] [Google Scholar]
- 13.Engel L.S., Chow W.H., Vaughan T.L. Population attributable risks of esophageal and gastric cancers. J Natl Cancer Inst. 2003;95:1404–1413. doi: 10.1093/jnci/djg047. [DOI] [PubMed] [Google Scholar]
- 14.Contino G., Vaughan T.L., Whiteman D. The evolving genomic landscape of Barrett's esophagus and esophageal adenocarcinoma. Gastroenterology. 2017;153:657–673. doi: 10.1053/j.gastro.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ek W.E., Levine D.M., D'Amato M. Germline genetic contributions to risk for esophageal adenocarcinoma, Barrett's esophagus, and gastroesophageal reflux. J Natl Cancer Inst. 2013;105:1711–1718. doi: 10.1093/jnci/djt303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buas M.F., He Q., Johnson L.G. Germline variation in inflammation-related pathways and risk of Barrett's oesophagus and oesophageal adenocarcinoma. Gut. 2017;66:1739–1747. doi: 10.1136/gutjnl-2016-311622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai J.Y., de Dieu Tapsoba J., Buas M.F. A newly identified susceptibility locus near FOXP1 modifies the association of gastroesophageal reflux with Barrett's esophagus. Cancer Epidemiol Biomarkers Prev. 2015;24:1739–1747. doi: 10.1158/1055-9965.EPI-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matejcic M., Iqbal Parker M. Gene-environment interactions in esophageal cancer. Crit Rev Clin Lab Sci. 2015;52:211–231. doi: 10.3109/10408363.2015.1020358. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L., Jiang Y., Wu Q. Gene-environment interactions on the risk of esophageal cancer among Asian populations with the G48A polymorphism in the alcohol dehydrogenase-2 gene: a meta-analysis. Tumour Biol. 2014;35:4705–4717. doi: 10.1007/s13277-014-1616-7. [DOI] [PubMed] [Google Scholar]
- 20.Levine D.M., Ek W.E., Zhang R. A genome-wide association study identifies new susceptibility loci for esophageal adenocarcinoma and Barrett's esophagus. Nat Genet. 2013;45:1487–1493. doi: 10.1038/ng.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laurie C.C., Doheny K.F., Mirel D.B. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Voorman A., Lumley T., McKnight B. Behavior of QQ-plots and genomic control in studies of gene-environment interaction. PLoS One. 2011;6:e19416. doi: 10.1371/journal.pone.0019416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gogarten S.M., Bhangale T., Conomos M.P. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics. 2012;28:3329–3331. doi: 10.1093/bioinformatics/bts610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Casson A.G., Zheng Z., Porter G.A. Genetic polymorphisms of microsomal epoxide hydroxylase and glutathione S-transferases M1, T1 and P1, interactions with smoking, and risk for esophageal (Barrett) adenocarcinoma. Cancer Detect Prev. 2006;30:423–431. doi: 10.1016/j.cdp.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 25.Zhai R., Liu G., Asomaning K. Genetic polymorphisms of VEGF, interactions with cigarette smoking exposure and esophageal adenocarcinoma risk. Carcinogenesis. 2008;29:2330–2334. doi: 10.1093/carcin/bgn210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doecke J., Zhao Z.Z., Pandeya N. Polymorphisms in MGMT and DNA repair genes and the risk of esophageal adenocarcinoma. Int J Cancer. 2008;123:174–180. doi: 10.1002/ijc.23410. [DOI] [PubMed] [Google Scholar]
- 27.Cheung W.Y., Zhai R., Kulke M.H. Epidermal growth factor A61G gene polymorphism, gastroesophageal reflux disease and esophageal adenocarcinoma risk. Carcinogenesis. 2009;30:1363–1367. doi: 10.1093/carcin/bgp126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhai R., Chen F., Liu G. Interactions among genetic variants in apoptosis pathway genes, reflux symptoms, body mass index, and smoking indicate two distinct etiologic patterns of esophageal adenocarcinoma. J Clin Oncol. 2010;28:2445–2451. doi: 10.1200/JCO.2009.26.2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu I.C., Zhao Y., Zhai R. Interactions between genetic polymorphisms in the apoptotic pathway and environmental factors on esophageal adenocarcinoma risk. Carcinogenesis. 2011;32:502–506. doi: 10.1093/carcin/bgq287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhai R., Zhao Y., Liu G. Interactions between environmental factors and polymorphisms in angiogenesis pathway genes in esophageal adenocarcinoma risk: a case-only study. Cancer. 2012;118:804–811. doi: 10.1002/cncr.26325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheung W.Y., Zhai R., Bradbury P. Single nucleotide polymorphisms in the matrix metalloproteinase gene family and the frequency and duration of gastroesophageal reflux disease influence the risk of esophageal adenocarcinoma. Int J Cancer. 2012;131:2478–2486. doi: 10.1002/ijc.27541. [DOI] [PMC free article] [PubMed] [Google Scholar]