TO THE EDITOR:
To date eleven variant translocations have been characterized in acute promyelocytic leukemia (APL), all of which share the same C-terminal domains of RARA as in t(15;17)(q24;q21) PML-RARA[1]. The N-terminal sequences of the fusions involve ZBTB16 (PLZF) [2] in t(11;17)q(23;q21); NPM1[3] in t(5;17)(q35;q21); NUMA[4] in t(11;17)(q13;q21); STAT5B [5] in t(17;17)(q21;q21); PRKAR1A[6] in t(17;17)(q21;q24); BCOR[7] in t(X;17)(p11;q21); FIP1L1 [8]in t(4;17)(q12;q21); NABP1 (OBFC2A) [9] in der(2)t(2;17)(q32;q21); TBL1XR1 (TBLR1) [10] in t(3;17)(q26;q21); GTF2I [11] in t(7;17)(q11;q21); and IRF2BP2 [12]in t(1;17)(q42.3;q21.2). Though rare, these “experiments of nature” serve as important tools with which to dissect the pathways underlying leukemogenesis[13]. Leukemic cells from patients with classic t(15;17) PML-RARA rearrangement differentiate following exposure to all-trans retinoic acid (ATRA), underlying the clinical success of differentiation therapy in APL[14]. Yet, the responses of variant APL cases to ATRA has been variable: PLZF-RARA, STAT5b-RARA, and GTF2I-RARA patients are resistant to the differentiating effects of ATRA[11, 13] (though only one case of GTF2I-RARA has been reported).
To date three reports of IRF2BP2-RARA have been published. Response to ATRA has been inconsistent. In the first publication Yin et al. [12] reported a 19 yo woman who initially responded to a combination of ATRA, arsenic, and gemtuzumab ozogamicin (though the patient exhibited an atypically prolonged time for normalization of her coagulopathy). She received an eight-month course of consolidation with ATRA and arsenic, but relapsed 2 months later, and received salvage therapy with ATRA, arsenic, and idarubicin, followed by allogeneic bone marrow transplant. The second case, reported by Shimomura et al. [15] was of a 68 yo woman initially treated with ATRA, in which idarubicin and cytarabine was added on day 12. She did not achieve a complete remission, and was re-induced with gemtuzumab ozogamicin, only to relapse one year later. The third case[16] was of a 37 yo man who achieved a morphological, but not molecular remission with single agent ATRA; he achieved molecular remission only after addition of PETHEMA-based induction chemotherapy.
Herein we describe the fourth case of t(1;17) APL. A 34 year old white male presented in March 2016 with WBC 4,100 × 106/L, hemoglobin 9.3 gm/dL, and platelets 23,000 × 106/L with 40% neutrophils, 9% bands, 45% lymphocytes, 2% monocytes, and 4% blasts. PT was 15.9 sec, D-dimer 37.21 mg/L, fibrinogen 397 mg/dL. Over the first week of his hospitalization, the patient was given single agent ATRA 24 mg/m2 daily while the diagnosis of APL was being confirmed. Over this time, the WBC rose to over 30,000 × 106 /L. A bone marrow aspirate and biopsy revealed 89% immature cells expressing CD13, CD33, CD38(dim), CD45, CD117, CD123(dim), and myeloperoxidase[bright]; negative for HLA-DR. Cytogenetics revealed 45, X, -Y, t(1;17)(q42;q21), i(8)(q19). FISH and PCR did not reveal PML-RARA rearrangement. FISH using Vysis LSI RARA Dual Color, Break Apart Rearrangement Probe revealed one orange, one green and one fusion (1O1G1F) signal pattern, consistent with a variant RARA rearrangement. FISH also revealed a gain of RUNX1T1, consistent with the finding of iso(8)(q19).
To define the breakpoint, we extracted total RNA using TRIzol reagent according to manufacturer’s (Thermo Fisher, Waltham, MA) instructions. For RT-PCR, one μg of total RNA was reverse-transcribed using ProtoScript II First Strand cDNA Synthesis Kit (NEB, Ipswich, MA) and RARA specific primer (5’CGTCAGCGTGTAGCTCTCAG 3’). IRF2BP2 specific forward primer (5’GAGAGCAGGACTGGGTCAAC3’ ) and RARA specific reverse primer (5’GGGCACCTCCTTCTTCTTCT3’) were used to detect a IRF2BP2/RARA fusion transcript. Sanger sequencing was performed using 3730xl DNA Analyzer (Thermo Fisher, Waltham, MA) at the Genomics Research Core of the University of Pittsburgh. BLAST software (http://blast.ncbi.nlm.nih.gov/Blast.cgi) was used for analysis of the sequence data.
RT-PCR and sequencing of the amplified transcript identified the IRF2BP2-RARA breakpoint (Figure1). The breakpoint is at position 1031 of the IRF2BP2 cDNA (NCBI Reference Sequence: NM_001077397.1), and position 657 of the RARA cDNA (NCBI Reference Sequence: NM_000964.3).
Figure 1.
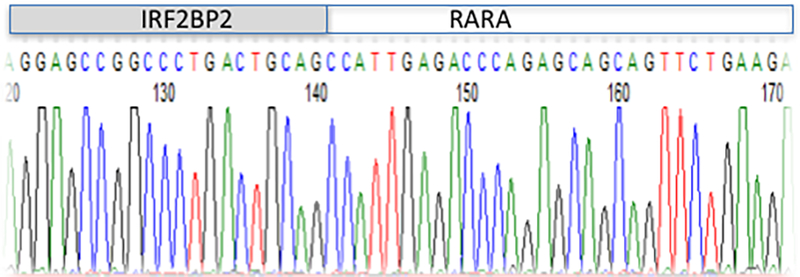
Partial sequence of the IRF2BP2-RARA fusion cDNA, highlighting the breakpoint.
IRF2BP2 is a nuclear protein that contains an N-terminal zinc-finger motif, and a C-terminal RING domain. It functions as a transcriptional co-repressor for IRF1. It has also been found to cooperate with nuclear factor of activated T cells (NFAT1) to control expression of genes involved in apoptosis, cell cycle, and differentiation [17]. The mechanism through which it acts as a co-repressor has not been carefully studied, though its refractoriness to HDAC inhibition suggests that it is not through recruitment of an NCoR/Sin3/HDAC complex [17]. The only other APL fusion partner with co-repressor activity is ZBTB16 (PLZF). As a result of its ability to anchor a co-repressor complex to its N-terminus, blasts expressing PLZF-RARA are less responsive to ATRA, and t(11;17) patients fail to respond to ATRA[13]. Because of the potential similarity of mechanism to PLZF-RARA, and the variability of clinical response of the prior cases of IRF2BP2-RARA to ATRA, we performed an in vitro analysis of the sensitivity of t(1;17) primary blasts to ATRA.
Samples taken for in vitro analyses were obtained following informed patient consent and hospital IRB approval. The mononuclear compartment of the peripheral blood obtained one week after diagnosis was purified by Ficoll-Paque (GE Healthcare, Pittsburgh PA) density centrifugation and cultured in Teflon bottles (VWR, Bridgeport NJ) in medium containing 1 uM ATRA (Sigma, St. Louis, MO) or ethanol (vehicle) for five days. Cells were incubated with PE-conjugated anti–CD11b or isotype matched antibody (BD Biosciences) for analysis with a Coulter Epics XL Flow Cytometer (Beckman Coulter, Fullerton CA). Morphological analysis was performed on Wright-Giemsa stains of cytospin preparations. Photomicrographs were taken through an Olympus microscope with a 60x oil-immersion objective lens (using the software SPOT 5.1).
NBT staining was performed by mixing cell suspensions with an equal volume of a solution containing nitroblue tetrazolium (2 mg/mL), bovine albumin (17 mg/mL), and TPA (2 μg/mL) at 37°C for 30 min. Following incubation, the medium was discarded and formazan deposits were dissolved by adding 100 μl of dimethyl sulfoxide (DMSO). Optical density was measured at 570 nm.
Cells cultured for 5 days with Ethanol (vehicle) (Figure 2A) showed monolobar nuclei with cytoplasmic granulations similar to the morphology of the patient’s cells when introduced into culture at day 0 (not shown). By contrast, cells cultured with ATRA showed more abundant cytoplasm with segmentation of the nuclei, consistent with maturation towards neutrophils.
Figure 2A.
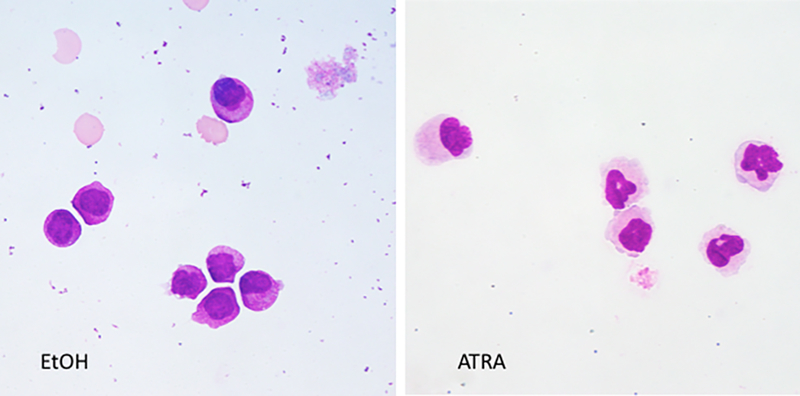
Wright-giemsa stains of cytospin preparations of the t(1;17) blasts cultured in Ethanol or 1 μM ATRA.
The myeloid differentiation marker CD11b is expressed on myelocytes, metamyelocytes, bands, and mature neutrophils, but not promyelocytes [18]. Figure 2B indicates acquisition of CD11b expression in cells cultured with ATRA, compared with vehicle, indicative of differentiation of the cells incubated with ATRA but not vehicle. Maturation of the cells was also determined by assessing acquisition of NADPH oxidase activity by nitro-blue tetrazolium (NBT) reduction (Figure 2C).
Figure 2B.
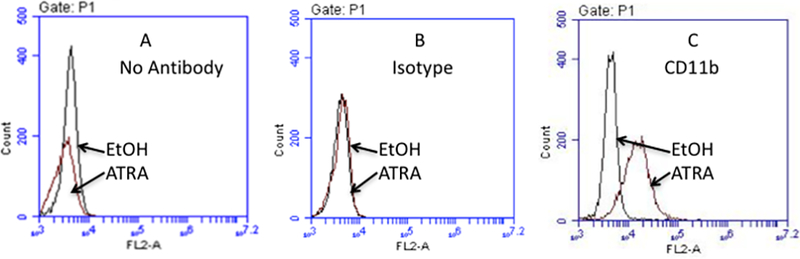
Flow cytometric analysis of t(1;17) blasts cultured for 5 days with ethanol or 1 μM ATRA, stained with no antibody, isotype control, or CD11b antibody.
Figure 2C.
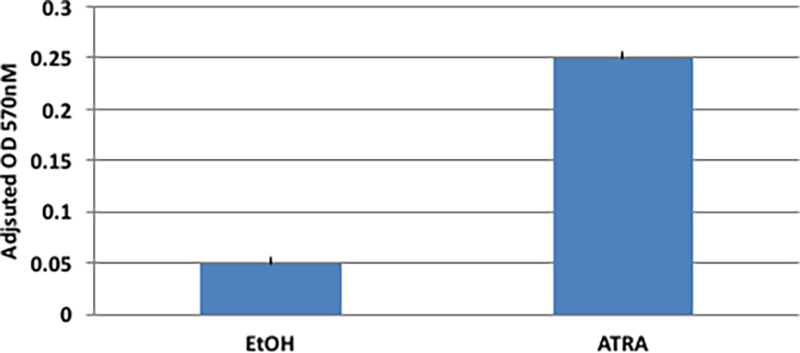
NBT analysis of t(1;17) blasts cultured in ethanol or 1 μM ATRA. Presented is the mean of triplicate measurements.
We confirmed the presence of an IRF2BP2-RARA fusion in our patient by RT-PCR and sequencing of the amplified transcript. The fusion cDNA would encode a predicted in-frame fusion protein containing the N-terminal 333 amino acids of IRF2BP2 linked to the C-terminal 402 amino acids of RARA; the fusion generates an extra alanine at the breakpoint. The breakpoint is at the same exon as in the other RARA fusions. This is the same breakpoint as described by Shimomura et al. [15], who also noted a second transcript. The breakpoint in the IRF2BP2 sequence is 48 bases 5’ to that described by Jovanovic et al. [16], and 637 bases 5’ to that described by Yin et al. [12] The RARA sequence-specific reverse transcriptase primer that we used to generate the cDNA precluded the identification of a possible reciprocal transcript in our patient; a reciprocal transcript has not been reported in the other cases.
The patient was treated initially per the GIMEMA protocol with idarubicin 12 mg/m2 × 4 doses and daily ATRA 45mg/m2, and subsequent consolidation with three cycles of 45 mg/m2 ATRA, 5 mg/m2 idarubicin x 4 days and 1 gm/m2 cytarabine x 4 days. Complete cytogenetic remission was achieved. Eighteen months after diagnosis he remains in complete remission.
We found that the t(1;17) blasts differentiated in vitro in an analogous fashion to t(15;17) blasts [13]. This is in concordance with the data of Jovanovic et al. [16], who found that ectopic expression of IRF2BP2-RARA cDNA in lin- murine progenitor cells rendered them ATRA sensitive. Indeed, our patient responded well to induction chemotherapy with the ATRA-containing GIMEMA protocol. At 18 month follow-up he remains in remission. We conclude that the t(1;17) APL blasts are sensitive to ATRA, and recommend that t(1;17) patients be treated with an ATRA-based regimen.
ACKNOWLEDGMENTS
This work was supported in part by a grant from the National Institutes of Health, USA P30CA047904. The authors have no competing interest to declare. S.M and M.Y.L. contributed essential reagents, contributed to the design of the research, and contributed to writing of the manuscript. A.C. performed the in vitro experiments. R.L.R. designed the research, analyzed the data, and wrote the manuscript.
REFERENCES:
- 1.de The H, Lavau C, Marchio A, et al. The PML-RAR alpha fusion mRNA generated by the t(15;17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991. August 23;66(4):675–84. [DOI] [PubMed] [Google Scholar]
- 2.Chen Z, Brand NJ, Chen A, et al. Fusion between a novel Kruppel-like zinc finger gene and the retinoic acid receptor-alpha locus due to a variant t(11;17) translocation associated with acute promyelocytic leukaemia. Embo J. 1993. March;12(3):1161–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Redner RL, Rush EA, Faas S, et al. The t(5;17) variant of acute promyelocytic leukemia expresses a nucleophosmin-retinoic acid receptor fusion. Blood. 1996. February 1;87(3):882–6. [PubMed] [Google Scholar]
- 4.Wells RA, Catzavelos C, Kamel-Reid S. Fusion of retinoic acid receptor alpha to NuMA, the nuclear mitotic apparatus protein, by a variant translocation in acute promyelocytic leukaemia. Nat Genet. 1997. September;17(1):109–13. [DOI] [PubMed] [Google Scholar]
- 5.Arnould C, Philippe C, Bourdon V, et al. The signal transducer and activator of transcription STAT5b gene is a new partner of retinoic acid receptor alpha in acute promyelocytic-like leukaemia. Hum Mol Genet. 1999. September;8(9):1741–9. [DOI] [PubMed] [Google Scholar]
- 6.Catalano A, Dawson MA, Somana K, et al. The PRKAR1A gene is fused to RARA in a new variant acute promyelocytic leukemia. Blood. 2007. December 1;110(12):4073–6. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto Y, Tsuzuki S, Tsuzuki M, et al. BCOR as a novel fusion partner of retinoic acid receptor alpha in a t(X;17)(p11;q12) variant of acute promyelocytic leukemia. Blood. 2010. November 18;116(20):4274–83. [DOI] [PubMed] [Google Scholar]
- 8.Kondo T, Mori A, Darmanin S, et al. The seventh pathogenic fusion gene FIP1L1-RARA was isolated from a t(4;17)-positive acute promyelocytic leukemia. Haematologica. 2008. September;93(9):1414–6. [DOI] [PubMed] [Google Scholar]
- 9.Won D, Shin SY, Park CJ, et al. OBFC2A/RARA: a novel fusion gene in variant acute promyelocytic leukemia. Blood. 2013. February 21;121(8):1432–5. [DOI] [PubMed] [Google Scholar]
- 10.Chen Y, Li S, Zhou C, et al. TBLR1 fuses to retinoid acid receptor alpha in a variant t(3;17)(q26;q21) translocation of acute promyelocytic leukemia. Blood. 2014. August 7;124(6):936–45. [DOI] [PubMed] [Google Scholar]
- 11.Li J, Zhong HY, Zhang Y, et al. GTF2I-RARA is a novel fusion transcript in a t(7;17) variant of acute promyelocytic leukaemia with clinical resistance to retinoic acid. Br J Haematol. 2015. March;168(6):904–8. [DOI] [PubMed] [Google Scholar]
- 12.Yin CC, Jain N, Mehrotra M, et al. Identification of a novel fusion gene, IRF2BP2-RARA, in acute promyelocytic leukemia. J Natl Compr Canc Netw. 2015. January;13(1):19–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Redner RL. Variations on a theme: the alternate translocations in APL. Leukemia. 2002. October;16(10):1927–32. [DOI] [PubMed] [Google Scholar]
- 14.Castaigne S, Chomienne C, Daniel MT, et al. All-trans retinoic acid as a differentiation therapy for acute promyelocytic leukemia. I. Clinical results. Blood. 1990. November 01;76(9):1704–9. [PubMed] [Google Scholar]
- 15.Shimomura Y, Mitsui H, Yamashita Y, et al. New variant of acute promyelocytic leukemia with IRF2BP2-RARA fusion. Cancer Sci. 2016. August;107(8):1165–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jovanovic JV, Chillon MC, Vincent-Fabert C, et al. The cryptic IRF2BP2-RARA fusion transforms hematopoietic stem/progenitor cells and induces retinoid-sensitive acute promyelocytic leukemia. Leukemia. 2017. March;31(3):747–751. [DOI] [PubMed] [Google Scholar]
- 17.Carneiro FR, Ramalho-Oliveira R, Mognol GP, et al. Interferon regulatory factor 2 binding protein 2 is a new NFAT1 partner and represses its transcriptional activity. Mol Cell Biol. 2011. July;31(14):2889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Lochem EG, van der Velden VH, Wind HK, et al. Immunophenotypic differentiation patterns of normal hematopoiesis in human bone marrow: reference patterns for age-related changes and disease-induced shifts. Cytometry B Clin Cytom. 2004. July;60(1):1–13. [DOI] [PubMed] [Google Scholar]


