Abstract
Background and objective
Glioma is one of the most aggressive and malignant cancers originating from the human brain. Increasing evidence suggests that aberrant expression of microRNAs (miRNAs) frequently occurs in glioma and miRNAs are critical regulators of glioma. miR-671 has recently been revealed to be a novel miRNA that plays a vital role in human glioblastoma multiforme. However, the functional role and underlying mechanisms of miR-671-3p require further analysis.
Materials and methods
Western blot and fluorescence quantitative PCR were used to assess the expression of cytoskeleton-associated protein 4 (CKAP4) and miR-671-3p, respectively. A Cell Counting Kit-8 (CCK-8) assay and a Boyden chamber assay were used to detect the proliferative and migratory abilities of glioma cells. A luciferase assay was used to determine the target gene of miR-671-3p. Apoptosis was analyzed by flow cytometry.
Results
Our results revealed that overexpression of miR-671-3p promoted cell proliferation and migration in vitro. Meanwhile, forced expression of miR-671-3p reduced apoptosis. In contrast, inhibition of miR-671-3p had the opposite effects. We also identified CKAP4 to be a direct target of miR-671-3p. The expression levels of CKAP4 were decreased in clinical samples and inversely correlated with miR-671-3p expression levels. Ectopic expression of CKAP4 reversed the promotive activity of miR-671-3p in the proliferation and migration and enhanced apoptosis.
Conclusion
Our study demonstrates that miR-671-3p is a predominant positive regulator of glioma progression, thus providing new insights into the molecular mechanisms of glioma development. The findings suggest that the miR-6713p/CKAP4 axis may serve as a potential therapeutic target or biomarker in glioma.
Keywords: miR-671-3p, glioma, migration, proliferation, CKAP4
Introduction
Glioma is a highly common and aggressive malignancy in the human brain associated with high morbidity and mortality.1,2 Although surgical resection, radiotherapy and chemotherapy have resulted in great progress, glioma prognosis remains poor and patients have a low survival rate.3,4 Aberrant gene expression and abnormalities in chromosomes have been shown to contribute to the progression of glioma. Moreover, diagnosis of tumors based on microRNAs (miRNAs) has already been applied in clinical practice. However, the precise mechanisms of glioma remain unclear. Biological characteristics and novel molecular targets of glioma should be explored to develop novel therapeutic strategies.
miRNAs are a class of small, noncoding RNAs, 20–22 nucleotides in length, and have been reported to be regulators of gene expression.5,6 miRNAs give rise to mRNA degradation or translational repression via binding of target mRNAs.7,8 Therefore, these noncoding RNAs are involved in various cell processes, such as proliferation, apoptosis and differentiation.9,10 Emerging data suggest that miRNAs are typically dysregulated in tumor progression, and these miRNAs may be regarded as biomarkers for diagnosis and targets for cancer treatment.11 miRNA dysregulation has been considered as an important mechanism in the pathogenesis of glioma.12,13 Altered expression of miRNAs positively or negatively modulates tumor progression.14–19 However, the identification of glioma-related miRNAs and their regulatory mechanisms require further work.
Recent studies have reported that miR-671 plays an important role in multiple cancers.20–22 miR-671 includes miR-671-3p and miR-671-5p. Even though miR-671-3p and miR-671-5p are derived from the same pre-miRNA, they have different mature sequences. Overexpression of miR-671-5p markedly increases migration and proliferation rates in glioblastoma multiforme cells through CDR1-AS, CDR1 and ASNL1.20 However, the functional role of miR-671-3p in glioma tumorigenesis remains unclear.
In this study, we explored the role of miR-671-3p in glioma cells and provided evidence that miR-671-3p behaves as a tumor enhancer in glioma by promoting cancer cell proliferation and migration and by inhibiting apoptosis. Cytoskeleton-associated protein 4 (CKAP4) was identified as a target gene of miR-671-3p in glioma cells. Importantly, the promotive effects of miR-671-3p were markedly reversed by CKAP4 overexpression. In addition, miR-671-3p expression was inversely correlated with CKAP4 expression in glioma cells. Mechanistically, we proposed that miR-671-3p exerts its tumor enhancive function by targeting CKAP4.
Materials and methods
Cell culture and transfection
T98G, LN229, A172, SHG44 and U251 cell lines were purchased from iCell Bioscience Inc. (Shanghai, China). Cells were cultured following the manufacturer’s protocols. In brief, the cells were grown in Roswell Park Memorial Institute (RPMI)-1640 (Hyclone, Pittsburgh, PA, USA) supplemented with 10% (v/v) FBS. All cells were cultured at 37°C in a humidified incubator under 5% CO2.
miR-671-3p mimic and miR-671-3p inhibitor were designed and synthesized by RiboBio (Guangzhou, China). Transfections, including plasmid DNAs, were carried out using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. Transfected cells were incubated in Opti-DMEM® transfection medium at 37°C for 6–8 hours. The medium was then replaced with complete medium with 10% FBS. Cells were incubated for 24 hours or 48 hours prior to harvesting.
Tissue samples
Eight human glioma tissues and their adjacent tissues were obtained from the patients who underwent surgical resection without chemotherapy and radiotherapy at the Department of Neurosurgery in the First Affiliated Hospital of Zunyi Medical College (Zunyi, China). The samples were immediately frozen with liquid nitrogen and stored at −80°C in an ultralow temperature refrigerator until further analysis. Written informed consents were obtained from all patients.
3′-UTR reporter constructs and luciferase assays
MicroRNA.org, PicTar and TargetScan were used to explore the potential targets of miR-671-3p. The five to six targets with the highest prediction scores within these databases were selected for further confirmation. The fragments of target 3′-UTRs were synthesized, annealed and then inserted into the psiCHECK™-2 report plasmid using the NotI and XhoI sites. The mutant-type 3′-UTR of CKAP4 was obtained from Vigenebio (Jinan, China). For luciferase assays, HEK-293T and SHG44 cells were co-transfected with report plasmid and miR-671-3p mimic using Lipofectamine 2000 according to the manufacturer’s protocol. After incubation for 48 hours, luciferase activities were evaluated consecutively using a Dual-Luciferase Reporter Assay System (Promega Corporation, Fitchburg, WI, USA). The experiments were repeated three times.
Real-time (RT)-PCR
Total RNA and small RNA were isolated from the cells using Trizol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. CDNAs were then synthesized from total RNA or small RNA for miRNA analysis using a Prime-Script RT reagent kit (TaKaRa, Kusatsu, Japan). RT-PCR was performed to detect the mature miRNA-671-3p level. The primers for miR-671-3p were purchased from RiboBio. U6 was used as the internal control for the normalization of miR-671-3p expression. The comparative Ct method aimed to calculate the relative amount of transcript. All the quantitative PCR (qPCR) primers were purchased from Thermo Fisher Scientific.
Protein isolation and Western blot
The cultured cells were harvested and then lysed in ice-cold RIPA lysis buffer containing a proteinase inhibitor. Protein concentrations were evaluated using a Piece BCA Protein Assay Kit (Thermo Fisher Scientific). A volume of extract equivalent to 15 µg of total protein was separated in 12% acrylamide SDS-PAGE. The proteins were transferred to a methanol-activated polyvinylidene fluoride (PVDF) membrane (EMD Millipore, Billerica, MA, USA) after electrophoresis. Membranes were blocked with 5% nonfat dry milk for 1 hour, then incubated with primary antibody (CKAP4, 16686-1-AP, 1:1,000; Proteintech, Wuhan, China) overnight at 4°C. Afterward, the membranes were probed with horseradish peroxidase (HRP)-conjugated secondary antibody (Abcam, Cambridge, MA, USA). The immune complexes were visualized using an electrogenerated chemiluminescence kit. Protein levels were normalized to the internal control, β-actin.
Cell migration assay
A cell migration assay was conducted using chambers with an 8 µm pore size (BD Biosciences, San Jose, CA, USA). First, 1×105 transfected cells were suspended in 200 µL free medium and seeded into the upper chamber. Next, 500 µL growth medium, RPMI-1640, containing 10% FBS was added to the lower chamber. After incubation for 12 hours or 24 hours at 37°C, the cells that did not migrate to the bottom of the membrane were wiped off using cotton swabs, and the cells that had migrated through the lower membrane were fixed with methanol and stained with 0.1% crystal violet. Images of five random fields were captured from each membrane under an inverted microscope, and the number of migratory cells was counted.
Cell viability assay
Cell proliferation was assessed using a WST-8 Cell Counting Kit-8 (CCK-8; Dojindo, Tokyo, Japan) according to the manufacturer’s protocol. In brief, transfected cells (1.0×103 cells/well) were seeded in 96-well plates and were cultured in the incubator at 37°C. During the following week, the growing medium was replaced with fresh medium containing 10% CCK-8 at the indicated time each day. Then, the plate was incubated for 2 hours and the OD values at 450 nm were determined using a plate reader (Bio-Tek Instruments, Winooski, VT, USA). The absorbance was used to plot the cell viability at each time point.
Apoptosis assay
The cell apoptosis assay was performed with an Annexin V–fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis detection kit (Beyotime BioTech, Los Altos, CA, USA) according to the manufacturer’s instructions. Briefly, the glioma cells were transfected with miR-671 mimics or inhibitors and their negative controls. After incubation for 24 hours, the transfected cells were harvested with 0.5% trypsin and were washed twice with ice-cold PBS; they were then resuspended at 1×106 cells/mL in Annexin V-binding buffer. The supernatant was stained with Annexin V–FITC and PI in the dark for 15 minutes according to the manufacturer’s instructions. Annexin V–FITC and PI binding were analyzed by flow cytometry using WinMDI 2.9 software (The Scripps Research Institute, La Jolla, CA, USA). All tests were performed in triplicate.
Statistical analyses
All data are expressed as mean ± SEM. Differences between two groups were analyzed using Student’s t-test, while differences among multiple groups were analyzed using the ANOVA test. A P-value of <0.05 was regarded as statistically significant.
Results
Ectopic expression of miR-671-3p promotes cell proliferation and migration in vitro
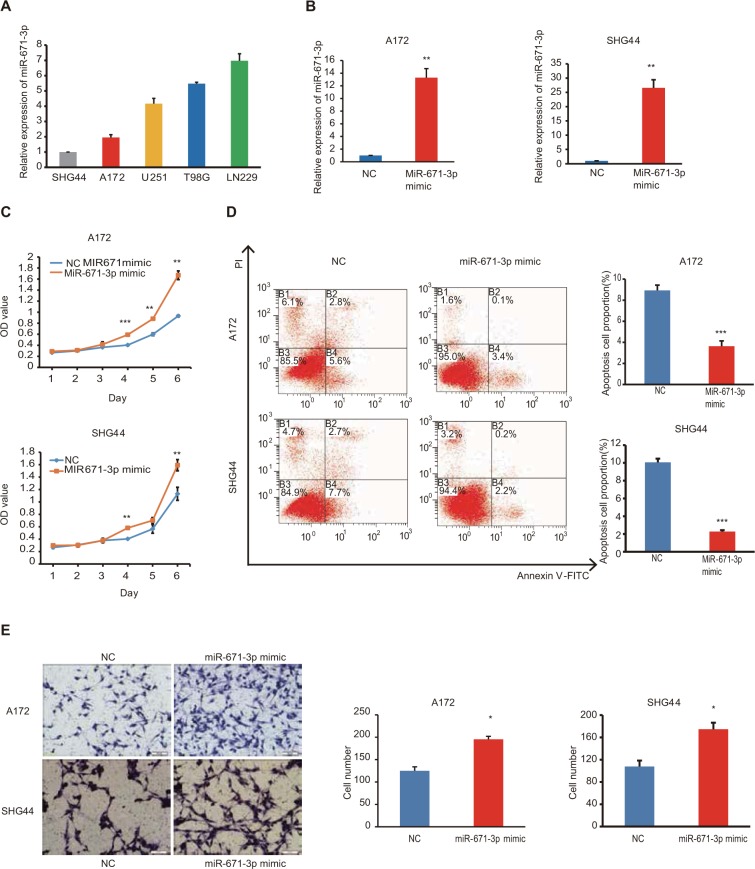
First, we detected the expression levels of miR-671-3p in a series of glioma cell lines by quantitative RT-PCR (qRT-PCR). The results revealed that the expression level of miR-671-3p in LN229 and T98G cell lines outnumbered the expression level of miR-671-3p in other cell lines, whereas A172 and SHG44 cell lines showed lower expression of miR-671-3p compared with other cell lines (Figure 1A).
Figure 1.
Ectopic expression of miR-671-3p promotes cell proliferation and migration in vitro.
Notes: (A) Relative expression of miR-671-3p in a serious of glioma cell lines. (B) Expression of miR-671-3p in cells transfected with miR-671-3p mimics (P=0.0044 for A172; P=0.0041 for SHG44). (C) Effect of miR-671-3p on proliferation of glioma cells by CCK-8 assay. (D) Effect of miR-671-3p on A172 and SHG44 cell apoptosis by flow cytometry (P=0.0019 for A172; P=0.0014 for SHG44). B1–B4 represent dead cells, late apoptosis cells, early apoptosis cells and alive cells, respectively. (E) Effect of miR-671-3p mimics on A172 and SHG44 cell migration by Boyden chamber (P=0.0149 for A172; P=0.0176 for SHG44). Error bars represent mean ± SEM. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: CCK-8, Cell Counting Kit-8; SEM, standard error of the mean; NC, negative control.
To ascertain the biological and functional effects of miR-671-3p on glioma cell behavior, we performed a series of tests. A172 and SHG44 cells were transfected with miR-671-3p and miR-NC mimics, and successful overexpression of miR-671-3p in these cells was identified by qRT-PCR (Figure 1B). Compared with the miR-NC groups, the results revealed a sharp increase in the proliferation rate in the miR-671-3p groups (Figure 1C), suggesting that upregulation of miR-671-3p promotes glioma cell proliferation. We also performed an apoptosis assay, and a decrease in apoptosis was identified in both A172 and SHG44 cells with restoration of miR-671-3p expression (Figure 1D). Meanwhile, enforced expression of miR-671-3p effectively promoted glioma cell migration ability with more cells in the miR-671-3p groups showing migration potential (Figure 1E). In general, miR-671-3p appears to be a tumor promoter in the progression of glioma.
Inhibition of endogenous miR-671-3p inhibits cell proliferation and migration in vitro
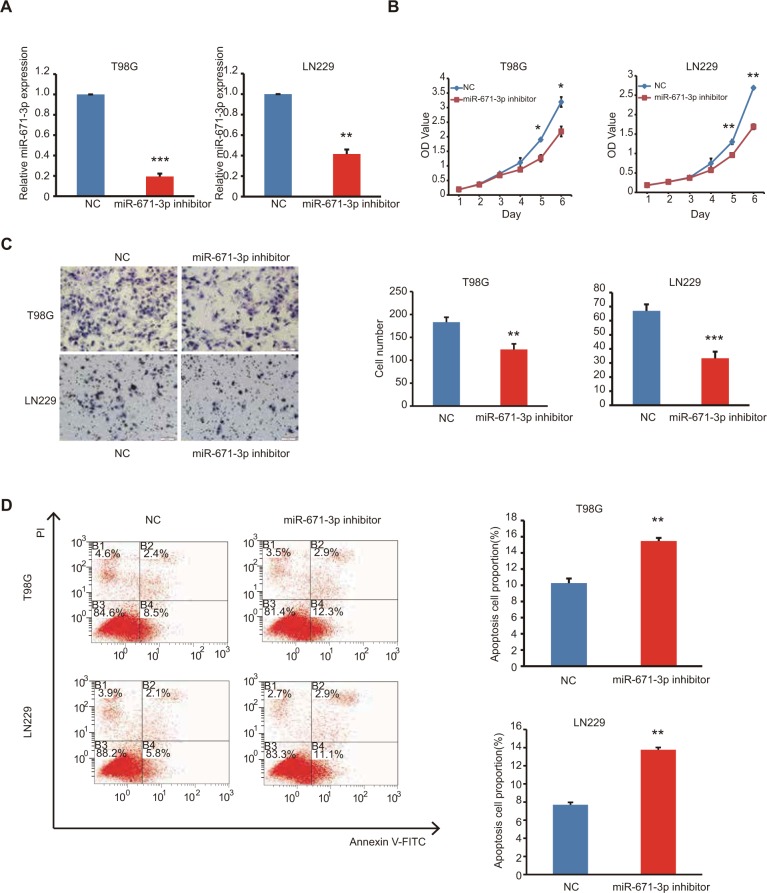
As described earlier, miR-671-3p promotes tumorigenesis. Therefore, to further explore the inhibitory role of miR-671-3p in glioma carcinogenesis, we suppressed the expression of miR-671-3p with miR-671-3p inhibitors in the cell lines, LN229 and T98G (Figure 2A); these cell lines had highest expression of miR-671-3p among all the glioma cell lines examined and thus were selected for use in the subsequent experiments. As expected, the CCK-8 assay showed that inhibition of miR-671-3p significantly reduced the proliferation of LN229 cells compared with the negative control group (Figure 2B). This indicates that the downregulation of miR-671-3p inhibits glioma cell proliferation. Moreover, in a Transwell assay, inhibition of miR-671-3p significantly suppressed the migration ability of glioma cells (Figure 2C). Moreover, when LN229 cells were treated with miR-671-3p inhibitors, a significant increase in the cellular apoptotic rate was demonstrated by flow cytometry (Figure 2D), suggesting that the promotion of glioma cell proliferation by miR-671-3p is at least partly due to an inhibition of apoptosis.
Figure 2.
Inhibition of endogenous miR-671-3p inhibits cell proliferation and migration in vitro.
Notes: (A) Expression of miR-671-3p in glioma cells transfected with miR-671-3p inhibitor or its negative control (P=0.0004 for T98G; P=0.0019 for LN229). (B) Effect of miR-671-3p inhibitor on glioma cell proliferation by CCK-8 assay. (C) Effect of miR-671-3p knockdown on T98G and LN229 cell migration by Boyden chamber (P=0.0013 for T98G; P<0.0001 for LN229). (D) Effect of miR-671-3p inhibitor on T98G and LN229 cell apoptosis by flow cytometry (P=0.0077 for T98G; P=0.0011 for LN229). B1–B4 represent dead cells, late apoptosis cells, early apoptosis cells and alive cells, respectively. Error bars represent mean ± SEM. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: CCK-8, Cell Counting Kit-8; FITC, fluorescein isothiocyanate; SEM, standard error of the mean; NC, negative control.
Similar results were also observed in T98G cells (Figure 2B–D), indicating that attenuation of miR-671-3p can inhibit glioma progression in vitro; this suggests that suppression of miR-671-3p has a negative role in pathological progression of glioma.
miR-671-3p directly targets CKAP4
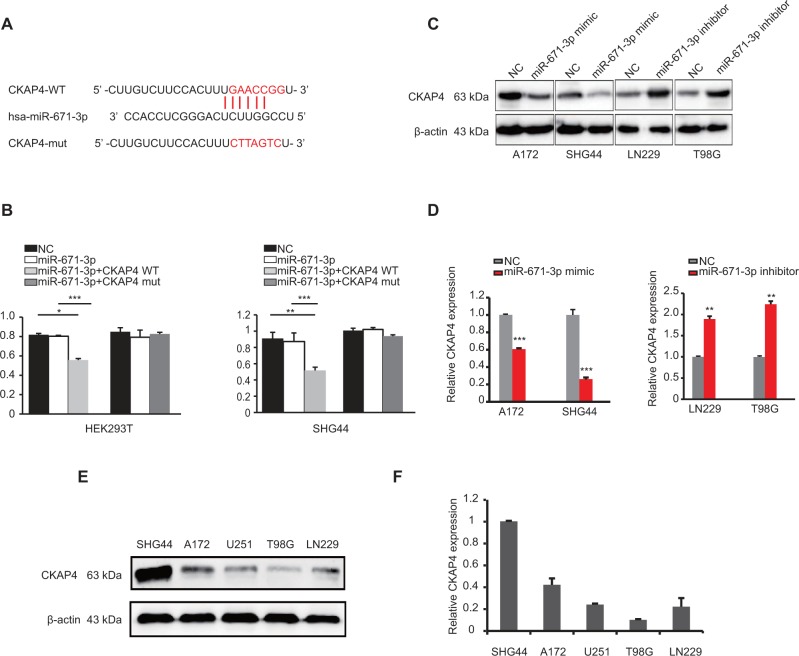
To explore the underlying mechanisms by which miR-671-3p promotes the progression of glioma cells, we predicted the potential targets of miR-671-3p by multiple prediction algorithms (microRNA.org, PicTar and TargetScan). As a result, we identified CKAP4 as the potential functional target of miR-671-3p. miR-671-3p had putative seed sequences in the 3′-UTR of CKAP4 (Figure 3A). To investigate the possibility that CKAP4 may be directly bound by miR-671-3p, we performed a luciferase reporter assay. The fragments of CKAP4 containing potential miR-671-3p-binding sites were cloned into luciferase constructs. When CKAP4/3′-UTR was present, transfection of miR-671-3p mimics led to a significant decrease in luciferase activity (Figure 3B). However, such a decrease in luciferase activity was not seen when the report plasmid was in the vector containing mutant CKAP4 3′-UTR (Figure 3B). These results indicated that sequences within the given region of CKAP4 3′-UTR indeed interact with miR-671-3p. Further, to investigate whether miR-671-3p regulates CKAP4 expression, we analyzed the effect of miR-671-3p expression on the protein levels of CKAP4 in A172, SHG44, LN229 and T98G cells by Western blot analysis. As shown in Figure 3C and D, CKAP4 expression was significantly decreased in response to the overexpression of miR-671-3p, whereas miR-671-3p inhibitors reversed the silencing effect of miR-671-3p overexpression on CKAP4. Consistent with Figure 1A, the varied expression levels of CKAP4 in different glioma cell lines are shown in Figure 3E and F. These results confirm that miR-671-3p negatively regulates the expression of CKAP4 by directly targeting the 3′-UTRs of CKAP4.
Figure 3.
miR-671-3p directly targets CKAP4.
Notes: (A) Illustration of the binding sites of CKAP4 and miR-671-3p. (B) Effect of miR-671-3p on the luciferase activities of CKAP4 3′-UTR in HEK-293T and SHG44 cells by luciferase reporter assay. (C) Expression of CKAP4 in miR-671-3p overexpressing or depleting cells by Western blot. (D) Statistical diagram of the Western blot results. (E) Expressions of CKAP4 in various glioma cells by Western blot. (F) Statistical diagram of (E). Error bars represent mean ± SEM. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: CKAP4, cytoskeleton-associated protein 4; SEM, standard error of the mean; NC, negative control.
Overexpression of CKAP4 impairs miR-671-induced promotive effects on glioma cell behavior
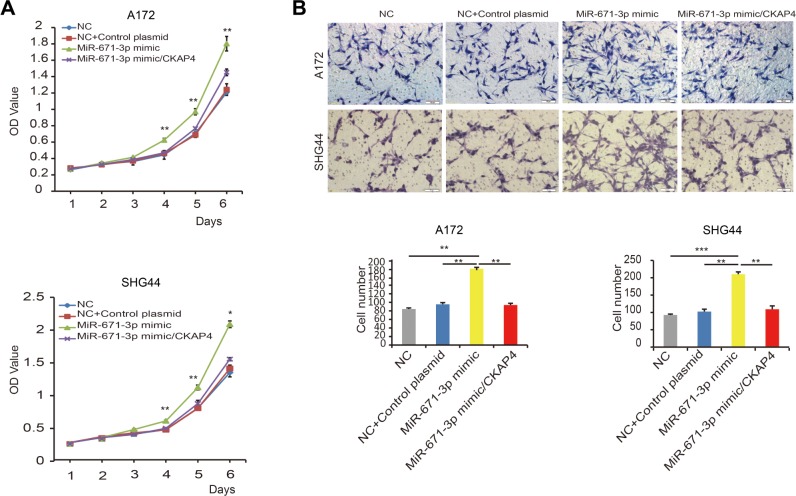
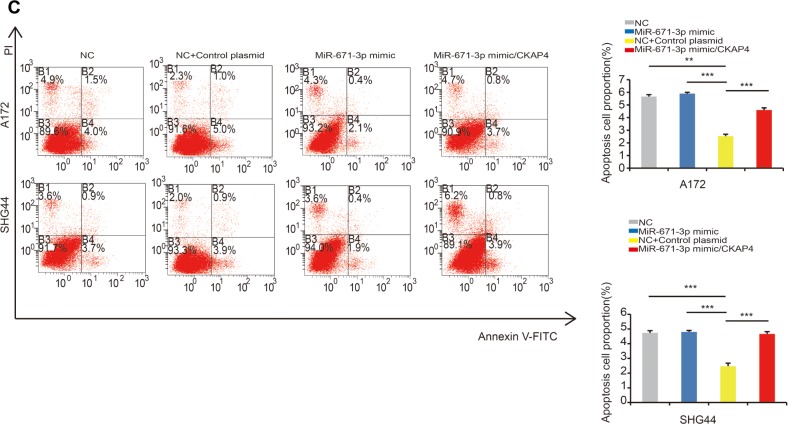
Considering that the abovementioned data suggest that CKAP4 is a direct target of miR-671-3p, we hypothesized that CKAP4 is involved in miR-671-induced tumorigenic properties of glioma cells. In this study, rescue experiments were performed to address this hypothesis. Since CKAP4 was decreased in cells with overexpressed miR-671-3p, we restored the expression of CKAP4 in miR-671-3p overexpressing cells by transfection of the open reading frame constructs without 3′-UTRs of the target gene and miR-671-3p mimics in A172 and SHG44 cells. The co-expression of CKAP4 markedly prohibited the promotion of cell motility induced by miR-671-3p (Figure 4A). Further, as indicated by the result of the Transwell assay in Figure 4B, the restoration of CKAP4 reversed the cell migration ability enhanced by miR-671-3p (Figure 4B). In addition, reintroduction of CKAP4 led to a rather significant increase in apoptosis (Figure 4C).
Figure 4.
Overexpression of CKAP4 impairs miR-671-3p-induced promotive effects on glioma cell behavior.
Notes: (A) Effect of reintroduction of CKAP4 on miR-671-3p-induced proliferation in A172 and SHG44 cells by CCK-8 assay (P<0.001 for A172; P<0.001 for SHG44). (B) Effect of CKAP4 on miR-671-3p-induced cell migration by Boyden Transwell assay (P<0.001 for A172; P<0.001 for SHG44). (C) Effect of CKAP4 on miR-671-3p-induced A172 and SHG44 cell apoptosis by flow cytometry (P<0.001 for A172; P<0.001 for SHG44). B1–B4 represent dead cells, late apoptosis cells, early apoptosis cells and alive cells, respectively. Error bars represent mean ± SEM. *P<0.05, **P<0.01, ***P<0.001.
Abbreviations: CCK-8, Cell Counting Kit-8; CKAP4, cytoskeleton-associated protein 4; FITC, fluorescein isothiocyanate; SEM, standard error of the mean; NC, negative control.
CKAP4 is downregulated, and there is a negative correlation between miR-671-3p and CKAP4 expression in glioma tissues
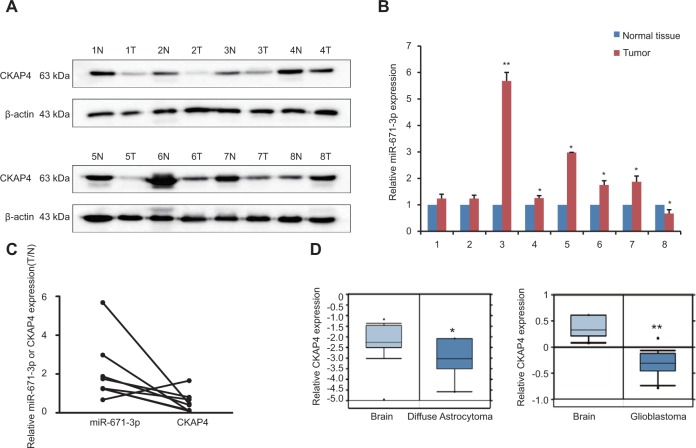
To investigate the expression pattern of CKAP4 in glioma, CKAP4 expression levels were quantified in eight glioma tissues and eight corresponding noncancerous tissues using Western blot. Compared with normal tissues, CKAP4 was downregulated in glioma tissues (Figure 5A).
Figure 5.
CKAP4 is downregulated, and there is a negative correlation between miR-671-3p and CKAP4 expression in glioma tissue.
Notes: (A) Expression of CKAP4 in eight paired human glioma tissues and their corresponding normal tissues by Western blotting. (B) Expression of miR-671-3p in eight paired human glioma tissues and their corresponding normal tissues by qRT-PCR. (C) Relative expression of miR-671-3p and CKAP4. (D) Downregulation of CKAP4 in clinical glioma and normal tissues derived from the Oncomine database. Error bars represent mean ± SEM. *P<0.05, **P<0.01.
Abbreviations: CKAP4, cytoskeleton-associated protein 4; qRT-PCR, quantitative real-time PCR; SEM, standard error of the mean.
To further confirm the expression pattern of CKAP4 and its clinical relationship with miR-671-3p, we detected miR-671-3p expression in glioma samples by qRT-PCR. The results showed that miR-671-3p expression levels were significantly upregulated in glioma tissues compared with the corresponding normal tissues (Figure 5B). Moreover, inverse correlations between the expression of miR-671-3p and CKAP4 were observed (Figure 5C). Due to our limited number of clinical samples and to investigate the potential implications of CKAP4, we investigated the expression of CKAP4 in patients from Oncomine database who were pathologically diagnosed with glioblastoma and astrocytoma. CKAP4 downregulation was observed in glioblastoma and astrocytoma samples (Figure 5D).
Taken together, it can be concluded that increased miR-671-3p expression is a frequent event in glioma tissue. The negative regulation of CKAP4 by miR-671-3p appears to contribute to the functional effect of miR-671-3p on the progression of glioma.
Discussion
Dysregulation of miRNAs is a common mechanism in human glioma. Recently, a series of miRNAs have been reported to function as oncogenes or tumor suppressors in glioma by regulating their downstream target genes.23,24
miR-671 includes miR-671-5p and miR-671-3p. A previous study showed that miR-671-5p is a tumor-suppressor miRNA for breast tumorigenesis. Overexpression of miR-671-5p in breast cancer cell lines attenuated proliferation and invasion by targeting FOXM1.21 However, miR-671-5p has been found to function as an oncogene in glioma, and miR-671-5p overexpression significantly increases proliferation and migration by targeting CDR1-AS, CDR1 and VSNL1.20 One study found that miR-671-3p expression in glioblastoma biopsies did not significantly differ from controls,20 which is inconsistent with our results. In our study, overexpression of miR-671-3p was found to promote proliferative and migratory capacity. Moreover, the forced expression of miR-671-3p inhibited apoptosis of glioma cells. Inhibition of miR-671-3p exhibited the opposite effects. Ectopic expression in A172 and SHG44 cell lines and inhibition of miR-671-3p in T98G and LN229 cell lines were performed in this study. Similar effects may be observed in all glioma cell lines, and relative experiments require further exploration. Moreover, several experiments have shown that miR-671-5p is downregulated in glioma tissues and serves as an oncogene.20 However, the interaction between miR-671-3p and miR-671-5p requires further investigation.
CKAP4 was identified as a new target molecule of miR-671-3p. CKAP4 is a DKK1-binding protein on the cell membrane surface and is a type II transmembrane protein.25,26 CKAP4 functions as a receptor of several ligands, including anti-proliferating factor (APF).27–29 Despite the involvement of CKAP4 in the tumor progression, the oncogenic and tumor-suppressive roles of CKAP4 have been reported.30 In this study, we showed that CKAP4 serves as a tumor suppressor and is a functional target of miR-671-3p. Reversal experiments confirmed that CKAP4 can reverse the effect of miR-671-3p on glioma progression.
Conclusion
This study explored the promotive role of miR-671-3p in the growth and migration of glioma cells. miR-671-3p has biological and functional effects on glioma progression via targeting CKAP4. This newly identified miR-671-3p/CKAP4 axis provides new insights into the molecular mechanisms underlying glioma development. However, further studies are needed to ascertain the precise mechanism underlying increased miR-671-3p and to investigate other potential targets of miR-671 in glioma.
Compliance with ethical standards
All procedures performed in studies involving human participants were approved by the Research Ethics Committee of Zunyi Medical University (Zunyi, China). Informed consent was obtained from all subjects. This article does not contain any studies with animals performed by any of the authors.
Acknowledgments
Information about the Oncomine database can be found at https://www.oncomine.org/resource/main.html. This study was supported by grants from the Key Discipline Constructive Project of Zunyi Medical University (XZXK-20120702), Guizhou Province Health and the Family Planning Commission of Science and Technology Fund Project (gzwjkj2016-1-009) and a Youth Science and Technology Personnel Training Project ([2017]197).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.McNeill KA. Epidemiology of Brain Tumors. Neurol Clin. 2016;34(4):981–998. doi: 10.1016/j.ncl.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 4.Butowski NA, Sneed PK, Chang SM. Diagnosis and treatment of recurrent high-grade astrocytoma. J Clin Oncol. 2006;24(8):1273–1280. doi: 10.1200/JCO.2005.04.7522. [DOI] [PubMed] [Google Scholar]
- 5.Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 6.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 7.German MA, Pillay M, Jeong DH, et al. Global identification of microR-NA-target RNA pairs by parallel analysis of RNA ends. Nat Biotechnol. 2008;26(8):941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 8.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18(5):504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 10.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 12.Lawler S, Chiocca EA. Emerging functions of microRNAs in glioblastoma. J Neurooncol. 2009;92(3):297–306. doi: 10.1007/s11060-009-9843-2. [DOI] [PubMed] [Google Scholar]
- 13.Silber J, James CD, Hodgson JG. microRNAs in gliomas: small regulators of a big problem. Neuromolecular Med. 2009;11(3):208–222. doi: 10.1007/s12017-009-8087-9. [DOI] [PubMed] [Google Scholar]
- 14.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 15.Iorio MV, Ferracin M, Liu CG, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 16.Calin GA, Ferracin M, Cimmino A, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353(17):1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 17.Ciafrè SA, Galardi S, Mangiola A, et al. Extensive modulation of a set of microRNAs in primary glioblastoma. Biochem Biophys Res Commun. 2005;334(4):1351–1358. doi: 10.1016/j.bbrc.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 18.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 20.Barbagallo D, Condorelli A, Ragusa M, et al. Dysregulated miR-671-5p/CDR1-AS/CDR1/VSNL1 axis is involved in glioblastoma multiforme. Oncotarget. 2016;7(4):4746–4759. doi: 10.18632/oncotarget.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan X, Fu Y, Chen L, et al. miR-671-5p inhibits epithelial-to-mesenchymal transition by downregulating FOXM1 expression in breast cancer. Oncotarget. 2016;7(1):293–307. doi: 10.18632/oncotarget.6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu Y, Wang Z, Sun D, et al. miR-671 promotes prostate cancer cell proliferation by targeting tumor suppressor SOX6. Eur J Pharmacol. 2018;823:65–71. doi: 10.1016/j.ejphar.2018.01.016. [DOI] [PubMed] [Google Scholar]
- 23.England B, Huang T, Karsy M. Current understanding of the role and targeting of tumor suppressor p53 in glioblastoma multiforme. Tumour Biol. 2013;34(4):2063–2074. doi: 10.1007/s13277-013-0871-3. [DOI] [PubMed] [Google Scholar]
- 24.Tivnan A, McDonald KL. Current progress for the use of miRNAs in glioblastoma treatment. Mol Neurobiol. 2013;48(3):757–768. doi: 10.1007/s12035-013-8464-0. [DOI] [PubMed] [Google Scholar]
- 25.Schweizer A, Ericsson M, Bächi T, Griffiths G, Hauri HP. Characterization of a novel 63 kDa membrane protein. Implications for the organization of the ER-to-Golgi pathway. J Cell Sci. 1993;104:671–683. doi: 10.1242/jcs.104.3.671. [DOI] [PubMed] [Google Scholar]
- 26.Vedrenne C, Hauri HP. Morphogenesis of the endoplasmic reticulum: beyond active membrane expansion. Traffic. 2006;7(6):639–646. doi: 10.1111/j.1600-0854.2006.00419.x. [DOI] [PubMed] [Google Scholar]
- 27.Razzaq TM, Bass R, Vines DJ, Werner F, Whawell SA, Ellis V. Functional regulation of tissue plasminogen activator on the surface of vascular smooth muscle cells by the type-II transmembrane protein p63 (CKAP4) J Biol Chem. 2003;278(43):42679–42685. doi: 10.1074/jbc.M305695200. [DOI] [PubMed] [Google Scholar]
- 28.Gupta N, Manevich Y, Kazi AS, Tao JQ, Fisher AB, Bates SR. Identification and characterization of p63 (CKAP4/ERGIC-63/CLIMP-63), a surfactant protein A binding protein, on type II pneumocytes. Am J Physiol Lung Cell Mol Physiol. 2006;291(3):L436–L446. doi: 10.1152/ajplung.00415.2005. [DOI] [PubMed] [Google Scholar]
- 29.Conrads TP, Tocci GM, Hood BL, et al. CKAP4/p63 is a receptor for the frizzled-8 protein-related antiproliferative factor from interstitial cystitis patients. J Biol Chem. 2006;281(49):37836–37843. doi: 10.1074/jbc.M604581200. [DOI] [PubMed] [Google Scholar]
- 30.Orzol P, Holcakova J, Nekulova M, Nenutil R, Vojtesek B, Coates PJ. The diverse oncogenic and tumour suppressor roles of p63 and p73 in cancer: a review by cancer site. Histol Histopathol. 2015;30(5):503–521. doi: 10.14670/HH-30.503. [DOI] [PubMed] [Google Scholar]