Abstract
Solid malignancies have been speculated to depend on cancer stem cells (CSCs) for expansion and relapse after therapy. Here we report on quantitative analyses of lineage tracing data from primary colon cancer xenograft tissue to assess CSC functionality in a human solid malignancy. The temporally obtained clone size distribution data support a model in which stem cell function in established cancers is not intrinsically but entirely spatiotemporally orchestrated. Functional stem cells that drive tumour expansion predominantly reside at the tumour edge, close to cancer-associated fibroblasts (CAFs). Hence, stem cell properties change in time depending on the cell location. Furthermore, although chemotherapy enriches for cells with a CSC phenotype, also in this context functional stem cell properties are fully defined by the microenvironment. To conclude, we identified osteopontin (OPN) as a key CAF-produced factor that drives in situ clonogenicity in colon cancer.
Keywords: Colon Cancer; Cancer Stem Cells; Stem Cell Dynamics; Tumour growth; Therapy; Disease Relapse; Osteopontin (OPN, Spp1)
In recent years in vivo lineage-tracing experiments and quantitative models have resolved the dynamics of the intestinal stem cell (ISC) population1–4. It was found that in the homeostatic murine intestine each crypt contains 5-7 functional stem cells1,2. However, the number of cells that express purported stem cell markers such as Lgr5 is much larger (n~16 per crypt)4,5, and many of these cells indeed have stem cell potential in response to tissue damage or in clonogenic assays6. Therefore, which cells function as stem cells in the normal gut largely depends on their position within the niche, and stem cell functionality and identity are distinct properties2,3,7.
Previously we have elucidated how oncogenic mutations impact on ISC dynamics and alter their behaviour during tumour initiation1, and it was established that early adenomatous outgrowths retain a hierarchy in which stem-like cells drive expansion2,8–12. In parallel, the presence of cancer stem cells (CSCs) in established human cancers has been assessed. In these studies, tumours are typically disrupted and single cell suspensions are injected in immune compromised mice to determine the frequency of CSCs, and the markers that distinguish these cells13–15. Critically, such artificial assays test stem cell potential rather than the stem cell functionality that drives tumour expansion. It was also reported that LGR5+ cells in colon cancer xenografts are actively clonogenic and able to function as CSCs16, however it is unclear whether LGR5+ cells form a rare population, or if essentially all cancer cells can function as stem cells17. Recently it was established that cancer cells at the invasive front contribute most to tumour expansion, but how this relates to the CSC model remains largely unresolved18. An important caveat of the CSC hypothesis is that differentiated cancer cells are known to adopt stem cell properties following exposure to signals from the stroma19–21 and that ablation of Lgr5+ cells in tumours results in rapid repopulation by Lgr5- cells22. However, it is currently unknown if this is a rare phenomenon that only occurs in experimental settings, or whether this is also central to the biology of unperturbed colon cancer tissue. Answering this question is key to our understanding of colon cancer biology. In addition, the role of CSCs in driving resistance to chemotherapy has not been elucidated in established tumour tissues. Therefore, we set out to adapt the marker-free clonal tracing strategies that we have developed in the murine gut to define the properties of CSCs in human colon cancer in situ, and to what extent these are regulated intrinsically or by the environment, both in unperturbed tumour growth and during treatment.
Results
Colon cancer growth dynamics
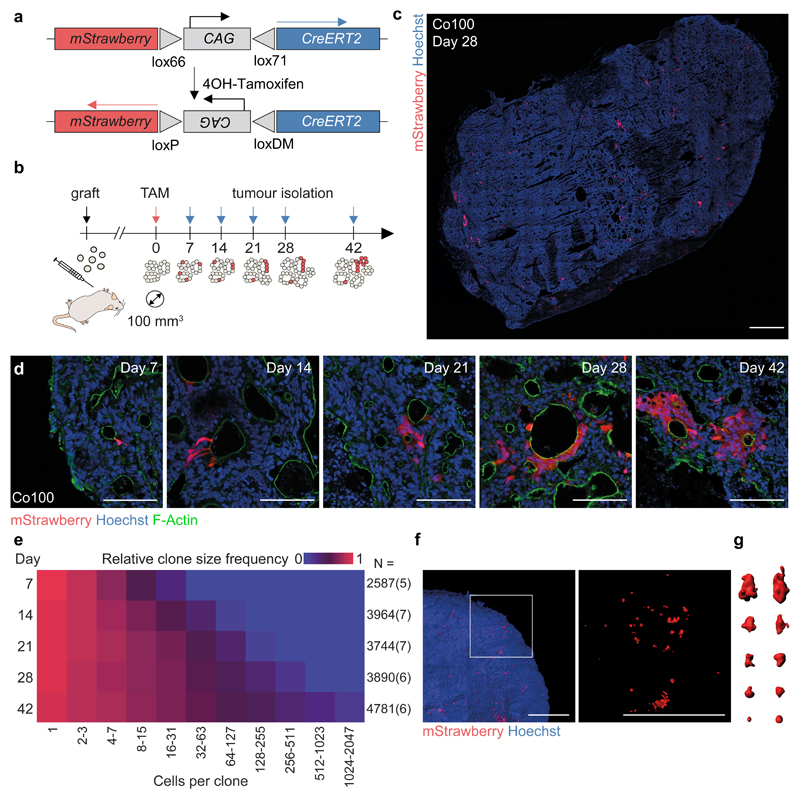
Primary human colon cancer cultures were established as described12,13. Subcutaneous injection of these lines (Co100, CC09), as well as a cell line cultured in the presence of serum (HCT-15), in immune compromised mice resulted in tumours with a well-differentiated morphology reminiscent of human colon cancers (Supplementary Fig. 1). We introduced a marker-free lineage tracing system, using a tamoxifen-inducible Strawberry expression vector (LV-indLS2)23, allowing random and permanent labelling of individual cells and their offspring independent of cell identity (Fig. 1a). Monoclonal cultures were established to limit genetic heterogeneity. In vitro we confirmed that the induction of Strawberry expression was dose-dependent, random, and a neutral event that does not impact on cellular fitness (Supplementary Fig. 2a-c). Dose dependency was confirmed in vivo (Supplementary Fig. 2d). For further studies we selected the dose that yielded sufficient clones for analysis, but showed no clone collision (Supplementary Fig. 2e-h). The resulting distributions of clone sizes showed no signs of scaling, confirming that we successfully avoided clone merging24 (Supplementary Note 1). Next, we induced clones in small tumours (~100 mm3) and isolated tumours on at least five time points (4-42 days) (Fig. 1b-d and Supplementary Fig. 3a-d). To investigate the impact of the immune compromised mouse strain employed, or location of injection, we included the analysis of tumours grown in both NSG and nude mice, as well as orthotopic tumours located in the cecal wall (Supplementary Figs. 1e, f and 3a, b). We analysed the induced clone sizes manually and by automated image analysis, revealing excellent agreement (Supplementary Note 1). 2D analysis of tissue sections was sufficient to approximate clonal volumes as demonstrated by 3D tissue analysis (Fig. 1f, g and Supplementary Note 1). In time, the average clone size increased as expected in expanding tumours (Fig. 1e, Supplementary Fig. 3 and Supplementary Table 1). Of note, a wide range of clone sizes was detected, especially at later time points, indicating extensive variability in clonal outgrowth despite the monoclonal origin of these cells.
Figure 1. Marker free lineage tracing in established colon cancer tissue.
(a) Schematic of lentiviral vector LV-indLS223. 4-OH-Tamoxifen (TAM) dependent recombination of mutant loxP sites results in permanent mStrawberry expression and Cre-inactivation. (b) Strategy for sporadic random labelling of cells in established colon cancer tissue. Following subcutaneous tumour cell injections, TAM administration followed at time indicated by red arrow, and tumour isolation at time points corresponding to blue arrows. (c) Example tumour section of Co100 xenograft, 28 days post-induction, is shown. mStrawberry marker is shown in red, nuclear stain is Hoechst (blue). Scale bar, 1 mm. 31 independent tumours were analysed. (d) Representative images of labelled Co100 clones at indicated time points, as used in (e), mStrawberry (red), nuclear stain, Hoechst (blue). Scale bars, 100 µm. (e) Relative clone frequency (indicated by colour in heatmap) per binned clone size (in columns) in time (rows) for Co100 tumours. Number of clones and tumours (between parentheses) are depicted next to each time point. (f) 3D images of cleared xenograft tissue is shown, mStrawberry marker is shown in red. Scale bars, 1 mm. (g) 3D representations of example clones of various sizes. Two independent tumours were analysed (f, g). Source data are shown in Supplementary Table 1.
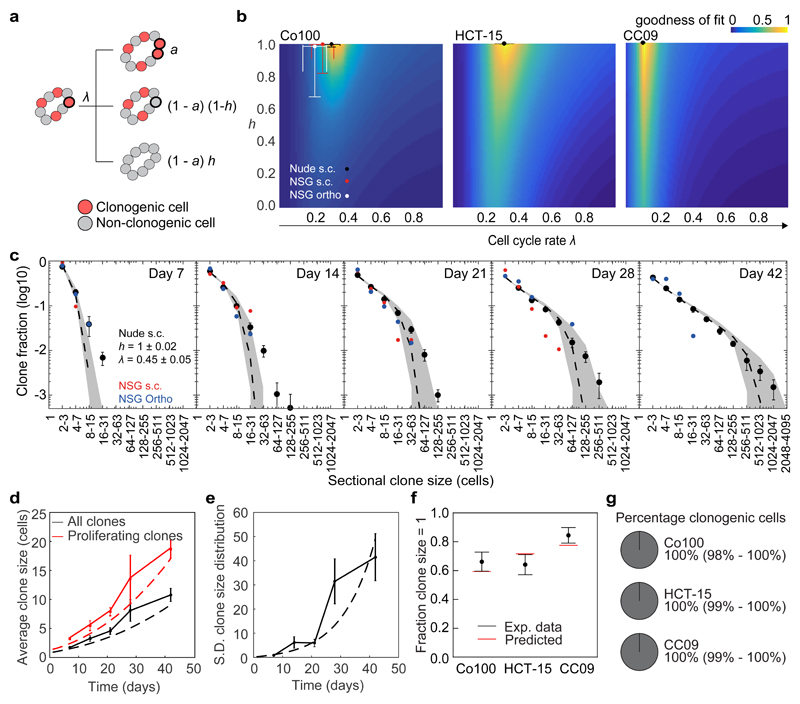
To elucidate the mode of growth of human colon cancer we developed a stochastic model of tumour growth (Fig. 2a and Supplementary Note 1). In this model, we distinguish clonogenic and non-clonogenic cells. At each time point clonogenic cells either proliferate (probability a) or lose clonogenic capacity (probability 1-a) with an effective rate λ, while non-clonogenic cells are inert. This dynamic is similar to models used to study clonal dynamics during homeostasis25, but differs in that a ≥ 0.5 to capture the accumulation of malignant cells in growing tumours. We consider an exponential mode of growth (constant a) and a surface mode of growth Importantly, we also include how clonogenicity is lost in the stochastic model: either driven by the environment such that all cells in a clone lose clonogenicity simultaneously (probability h) or through an intrinsic process such that this loss is randomly distributed between clones (probability 1-h). Note that if loss of clonogenicity is fully defined by the environment h = 1, while for h = 0 loss of clonogenicity is completely intrinsic and the model is equivalent to a strictly hierarchical CSC model.
Figure 2. Stochastic modelling and data inference identify spatiotemporal regulation of stem cell function.
(a) Diagram explaining the stochastic model for tumour growth. With rate λ clonogenic cells stochastically divide with probability a. Cells become non-clonogenic with probability (1 - a)(1 - h), or all cells within a clone lose their clonogenicity with probability (1 - a)h. λ, effective proliferation rate (cell divisions/day); a, mode of tumour growth for surface growth and 0.5 < a ≤ 1 for exponential growth); h, heterogeneity of growth between clones. See Supplementary Note 1 for details. (b) Heat maps depict goodness of fit (inverse and normalized least squares distance) as a function of λ and h on expanding clones (clone size > 1 cell) in Co100, HCT-15 and CC09 xenografts. Dots indicate optimal fit. Error bars represent S.D. (c) Sectional clone size distribution over time in Co100 xenografts. Experimentally determined- (shown as black dots), and model-predicted (dashed line) clone size distributions using best fit and 95% confidence interval (grey shade) as found in panel b. Red and blue dots represent experimental data obtained from subcutaneous and orthotopic xenografts in NSG mice, respectively. (d) Average measured (solid lines) and predicted (dashed lines) clone size in time of all (black lines) or proliferating clones (red lines) in Co100 tumours. (e) Measured (solid line) and predicted (dashed line) standard deviation of clone size in time. (c-e) Data is represented as mean ± S.E.M. (f) Experimentally measured fraction of single-cell clones, one week after clone induction are shown as black dots. Model prediction using the optimal fit parameters for indicated cell lines is shown as red lines. Data is represented as mean ± S.D. (g) The inferred percentage of clonogenic cells in the tumour edge is depicted in circle diagrams for Co100, HCT-15 and CC09. Source data are shown in Supplementary Table 1 (b-f).
The model comprising the three parameters λ, a, and h thus captures the rate and mode of tumour growth, as well as the process underlying the loss of clonogenicity. We numerically generated clone size distributions from the model for a wide range of parameters. By comparing the numerical distributions with the lineage tracing data using the Akaike information criterion, we found that the model with a determined by surface growth best describes the expansion of solid cancers (Supplementary Note 1). Using the method of least squares we next determined the goodness of fit for all combinations of parameter values of λ and h (Fig. 2b). Using the best-fit parameter values we accurately described the size distributions of expanding clones (Fig. 2c and Supplementary Fig. 3e-h), average clone size, and the variance in the clone size distribution in time within the xenografts (Fig. 2d, e). We found that for each of the xenografts λ ranged from 0.15–0.35 effective divisions/day. Critically, h tended to approximate 1 for all cancers (Fig. 2b), suggesting that these tumours do not contain an intrinsic hierarchical organization. The lack of an intrinsic hierarchy was underlined by the ability of the model to estimate the proportion of single cell clones from the size distribution of larger clones (≥2 cells) (Fig. 2f), indicating that single cell clones and expanding clones are representations of the same dynamics. This rules out the possibility that two distinct populations, i.e. differentiated cells and CSCs, were initially labelled. These analyses imply that functional CSCs within established cancer tissue are defined by the environment, and mostly reside at the surface of the tumours. Indeed, the estimated fraction of CSCs on the tumour edge approximates 100% (Fig. 2g and Supplementary Note 1). Of note, analysis of subcutaneous tumours from nude mice and NSG mice resulted in similar inferred parameters, as did orthotopically grown tumours, suggesting that the composition of the remnant immune system or the location of tumour grafting does not impact on these fundamental growth dynamics (Fig. 2b, c and Supplementary Fig. 3).
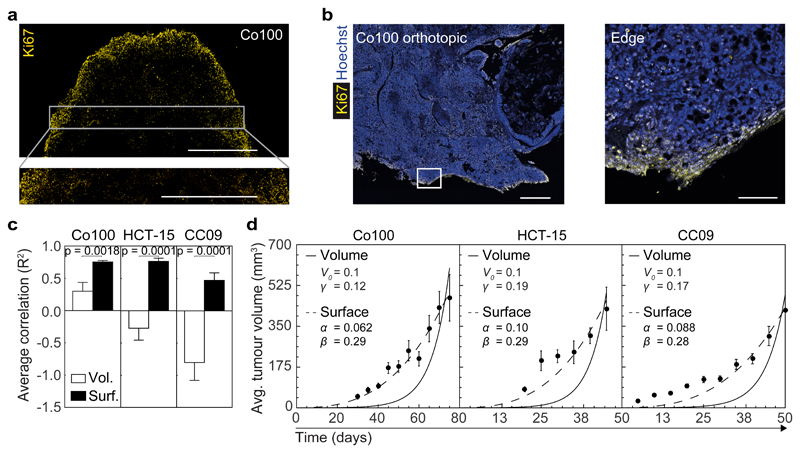
In order to validate the inferred model of tumour expansion we evaluated the distribution of proliferative cells within xenografts by Ki67 staining (Fig. 3a, b). This revealed a clear tendency for proliferation to take place in the edge regions of the tumour (outer ~300 μm), similar to what we observed in human primary tumours (Supplementary Fig. 4a, b) and other reports18,26,27. Of note, xenografts did not show evidence for extensive cell death at the analysed time points (Supplementary Fig. 4d). The subcutaneous tumours were well-vascularized and although small hypoxic regions could be detected by immunohistochemistry for HIF-1α, these were not restricted to the tumour centres (Supplementary Fig. 4e-g). More importantly, we confirmed the prediction of the model that surface growth is the primary mode of tumour expansion in these cancers using macroscopic tumour volume measurements (Fig. 3c, d and Supplementary Fig. 5). We take the ability to predict macroscopic tumour growth characteristics from clone size distributions within the tissue, thereby bridging several scales of magnitude, as strong evidence for the validity of our model in providing a quantitative description of tumour growth dynamics.
Figure 3. The mode of growth predicted by clonal dynamics confirmed at the macroscopic scale.
(a) Ki67 staining of a subcutaneous grafted Co100 tumour is shown. A larger magnification of the indicated area in top panel is shown below. Scale bars, 1 mm. Representative of 5 tumours. (b) Ki67 staining (yellow) of Co100 orthotopically grafted xenograft. Right panel is a magnification of the indicated box in the left image. Scale bars, 500 µm in left panel, and 100 µm in right panel. Representative examples of 5 tumours are shown. (c) Average correlation (R2) of tumour size measurements of tumour xenografts of the indicated cell lines with either the exponential volume (white bar) or surface growth model (black bar). Average R2 of volume and surface growth were compared using two-tailed Student’s t-test. (d) Average tumour volumes are shown at 5 day intervals for Co100, HCT-15 and CC09 tumours. Best fit of volume growth (V(t′) = V0eγt′) is shown as solid line, and surface growth model is indicated by dashed line. (c, d) Data is represented as mean ± S.E.M. Sample sizes are as follows; Co100 (n = 29 tumours), HCT-15 (n = 25 tumours) and CC09 (n = 23 tumours). Individual data points are shown in Supplementary Fig. 5a, b.
Spatiotemporal regulation of growth
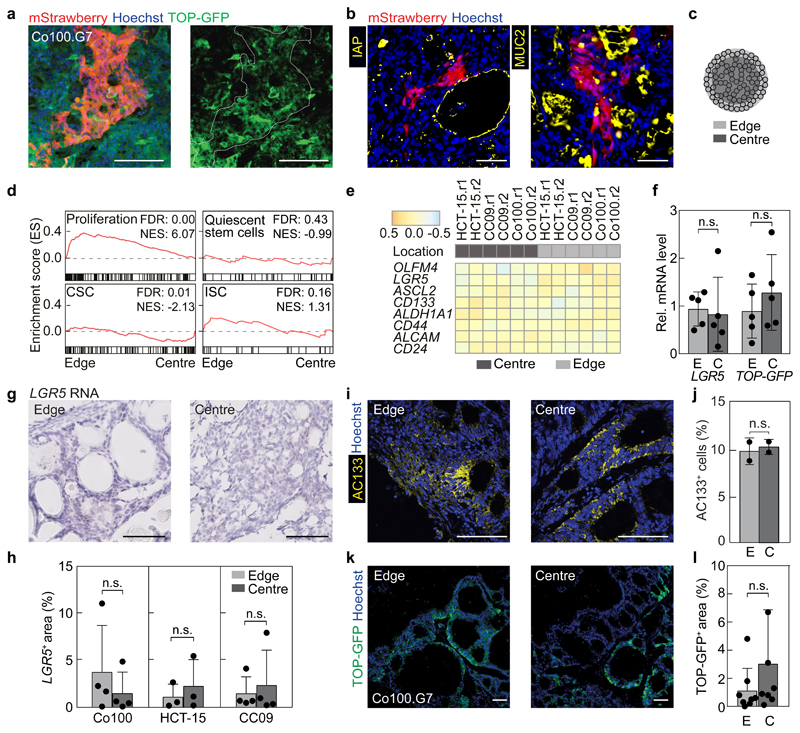
The most striking finding of our analyses is that CSC functionality is spatiotemporally organized in cancer tissue, at a spatial scale that is larger than the largest clones we detected, i.e. the values inferred for h are close to 1. This implies that individual clones that were labelled do not show evidence for a hierarchical organisation. However, within Strawberry-positive clones we do find heterogeneous expression of TOP-GFP (Fig. 4a), a readout for Wnt signalling activity that was previously used to identify CSCs as evidenced by their superior ability to initiate subcutaneous tumours19,28. In addition, differentiation markers displayed a heterogeneous expression pattern within individual clones (Fig. 4b). This confirms that indeed a cell capable of both self-renewal as well as multi-lineage differentiation was marked, conforming to the definition of a stem cell. To interrogate the relationship between CSC identity and functionality we took advantage of the fact that in situ clonogenic cells reside predominantly at the xenograft edges. Differential enzymatic digestion of the cells from the outside and inside regions of the xenografts followed by RNA sequencing revealed a significant enrichment of genes associated with proliferation in the tumour edge (Fig. 4c, d), in line with the Ki67 staining and surface growth kinetics. This enrichment was not observed for gene signatures associated with quiescent stem cells29, CSCs30 and ISCs31, or individual CSC associated genes (Fig. 4d, e). More specifically, no differences in the proportion of LGR5-positive cells, as demonstrated by RNAish, AC133 expression, or TOP-GFP levels were detected between the edge and the centre of the xenografts (Fig. 4f-l). Together, these results indicate a discrepancy between cells positive for CSC markers, which reside homogeneously throughout the cancer tissue, and functional CSCs, which reside at the tumour edge.
Figure 4. Stem cell markers do not identify clonogenic cells in vivo.
(a) An individual mStrawberry-positive clone in the edge of a Co100.G7 tumour is shown, revealing heterogeneous expression of TOP-GFP. mStrawberry marker is shown in red, TOP-GFP is shown in green. Scale bars, 100 μm. Outline of mStrawberry-positive clone is indicated by white dashed line in TOP-GFP image. Representative of 6 independent tumours. (b) Differentiation markers IAP and MUC2 are stained (both in yellow) within mStrawberry-labelled clones. Scale bars, 50 µm. Representative of 10 tumours. (c) Separation of tumour regions yielded cells from the edge (light grey) and centre (dark grey) of the xenografts. (d) Gene set enrichment analyses comparing edge to centre for all tumour models are shown using gene sets for proliferation genes, quiescent stem cells29, cancer stem cells (CSC)30 and intestinal stem cells (ISC)31. FDR, false discovery rate, NES, normalized enrichment score, n = 2 independent tumours per group. (e) RNA sequencing profiles for CSC marker genes are shown to compare edge and centre regions, two replicates were included for each line (r1-r2). (f) qPCR for LGR5 and TOP-GFP on Co100.G7 edge (E) or centre (C) tissue is shown, n = 5 tumours. (g) Representative images of LGR5 RNA in situ hybridization on Co100 xenograft sections. (h) Quantification of LGR5 mRNA expression (LGR5+ area) in the edge and centre region of 3 different xenograft models is shown. Data in g, h are representative of 4 (Co100), 3 (HCT-15) or 4 (CC09) independent tumours. (i, j) AC133 expression was revealed by immunofluorescence, representative of 5 independent tumours (i), and flow cytometry, n = 2 independent tumours (j). Scale bars, 100 µm. (k) Images of edge and centre regions showing TOP-GFP (green) and nuclear stain, Hoechst (blue), representative of 8 independent tumours. (l) Quantification of TOP-GFP positive area in Co100.G7 xenografts is shown in edge or centre of Co100.G7 (n = 8) xenografts. (f, h, j, l) Two-tailed Paired Student’s t-test, n.s., not significant. All data is represented as mean ± S.D.
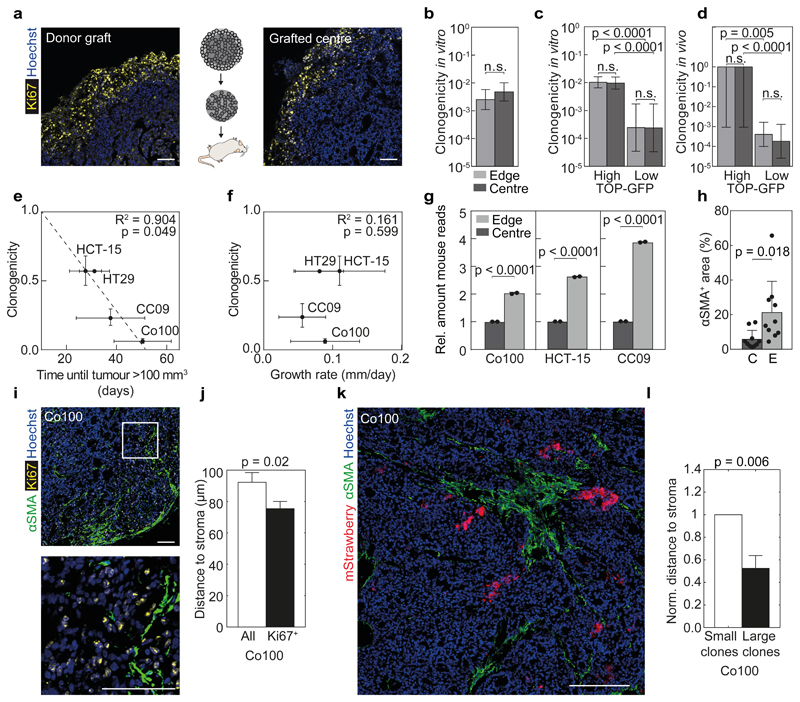
To test if the loss of clonogenicity of clones in the centre is reversible, we re-transplanted the tumour centre and detected a rapid recapitulation of the surface growth mode, indicating that clonogenicity is instilled by an appropriate environment (Fig. 5a). Furthermore, we compared the clonogenicity of tumour cells derived from the edge and centre of xenografted tumours using the limiting dilution assay. In vitro clonogenicity of cells from the xenograft edge and the centre was identical (Fig. 5b). The lower clonogenicity found in vitro (~1/300) compared to that obtained using our marker free lineage tracing in vivo (~1/1) supports the notion that cell-extrinsic factors determine cell fate. We found that expression of TOP-GFP does correlate with the observed clonogenicity in both limiting dilution and tumour initiation assays (Fig. 5c, d), but does not reflect the spatial regulation of clonogenicity as found in established tumours. More generally, although an inverse relationship exists between the clonogenicity observed in vitro and time to tumour take (Fig. 5e), no relationship between this clonogenic fraction and the average tumour expansion rate could be detected (Fig. 5f). This points to a marked discordance between the cells that drive tumour expansion, and the cells that show tumour initiation ability in transplantation assays.
Figure 5. In situ clonogenicity is environmentally defined.
(a) Proliferative cells were identified by staining for Ki67 (yellow) in re-grafted tumour tissue from a Co100 xenograft centre. Scale bars, 100 µm. Examples represents 5 tumours per group. (b-d) Limiting dilution assays were used to compare in vitro (b, c) or in vivo (d) clonogenicity of either all cells (b), or TOP-GFPhigh and TOP-GFPlow fractions (c, d) from the edge and centre of the tumour, using the (ELDA) ‘limdil’ function36, data is represented as mean ± 95% confidence intervals, n = 8 per dilution. (e-f) Mean clonogenic fraction of cell lines was correlated with time to tumour take in vivo (e), and with average growth rates of xenografts (size > 100 mm3), as inferred from surface growth model fits (f). Pearson correlation are shown for Co100 (n = 29), HCT-15 (n = 25), CC09 (n = 51) and HT29 (n = 10) xenografts. Error bars represent 95% confidence interval (y-axis) and S.D. (x-axis) (e, f). (g) Relative amount of mouse reads of tumour edge versus centre obtained from RNA sequencing data are shown, n = 2 tumours per cell line. (h) Presence of activated mouse fibroblasts (αSMA+) cells in edge (E) and centre (C) of Co100 xenografts (data is represented as mean ± S.D., n = 10 xenografts) was compared using paired two-tailed Student’s t-test, (g, h). (i) αSMA (green) and Ki67 (yellow) staining was performed on Co100 xenografts. Scale bars, 100 µm. Lower image is a magnification of the box in upper image. Representative of 8 tumours. (j) Average distance of either all or proliferating (Ki67+) cells to the nearest αSMA+ fibroblast in Co100 xenografts (n > 20.000 cells from 8 independent tumours). (k) Co100 tumour, 14 days after label activation, showing mStrawberry clones (red) and activated stromal cells (αSMA, green), representative of 5 independent tumours. Scale bar, 250 µm. (l) Average distance of either large clones (clone > 10 cells) or small clones to the nearest αSMA+ fibroblast in Co100 xenografts (n > 100 clones from 5 independent tumours). Error bars indicate S.D. (j, l).
Stromal determinants of clonogenicity
Again, the absence of intrinsic differences between cells from the clonogenic and non-clonogenic parts of the tumour suggests that the environment rather than the intrinsic characteristics of a cell determines its clonogenic capacity, and that in fact all malignant cells can function as CSC. Confirming the role of the environment, RNA profiling revealed that the most significantly upregulated genes in the tumour edge associate with stromal cells and the extracellular matrix (Supplementary Table 2). Since all stromal cells in the xenografts are mouse-derived, we analysed the number of murine versus human reads within the tumour regions. As expected, the number of murine reads was markedly higher in the edge compared to the centre (Fig. 5g). Immunofluorescent staining for activated fibroblasts, characterized by αSMA expression, revealed a strong enrichment in the tumour edge (Fig. 5h). Importantly, we could also demonstrate a clear co-localization between Ki67-positive proliferating tumour cells and activated stromal cells (Fig. 5i, j and Supplementary Fig. 4c, h, i). Larger clones were located significantly closer to stromal cells (Fig. 5k, l).
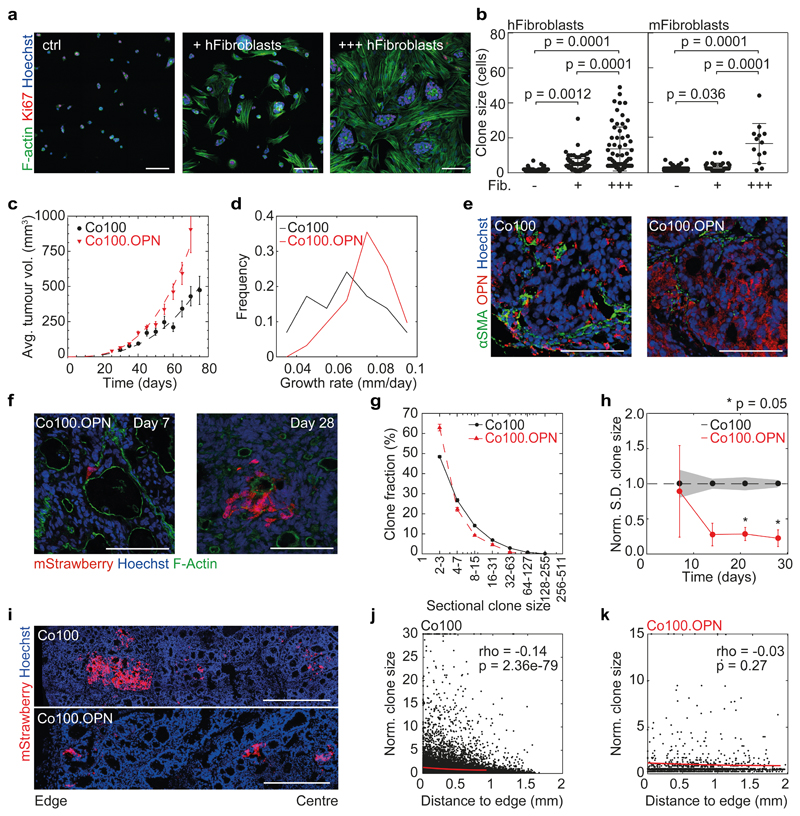
To ascertain a direct effect of cancer-associated fibroblasts (CAFs) on clonogenicity, we established co-cultures of cancer cells with primary murine and human fibroblasts, and performed medium transfer experiments. Tumour cells expanded more effectively in co-culture, or when exposed to fibroblast conditioned medium compared to control conditions (Fig. 6a, b and Supplementary Fig. 6a-c), suggesting a mechanistic role for CAF secreted factors in driving clonogenic outgrowth. Osteopontin (OPN, gene name Spp1) was the most abundantly expressed secreted factor by CAFs in our xenograft models, and we found that this protein was able to induce tumour cell proliferation in vitro (Supplementary Fig. 6d, e). In order to confirm that CAF secreted factors modulate clonogenicity in vivo Co100 cultures harbouring the LV-indLS2 vector were transduced with a human OPN overexpression vector (Co100.OPN) (Supplementary Fig. 6f, g). Upon subcutaneous injection of Co100.OPN cells in nude mice we witnessed accelerated tumour growth compared to control xenografts (Fig. 6c, d). OPN was ubiquitously expressed in Co100.OPN tumours as compared to control xenografts, were it was restricted to CAF-rich regions (Fig. 6e). We predicted that the homogenous presence of OPN would drive clonogenic outgrowth throughout the tumours independently of CAFs, thereby uncoupling CAF presence and clonogenicity within the tissue. To assess this, we performed clonal tracing in Co100.OPN tumours as described before, quantified clone size distributions at various time points, and subsequently analysed these with our inference strategy (Fig. 6f and Supplementary Fig. 6h, i). We confirmed that also in this setting no intrinsic CSCs are present i.e. h ~ 1. More importantly, we found that the clone size distribution was altered, and that the variation in clone sizes was significantly reduced in Co100.OPN xenografts (Fig. 6g, h). In combination with the accelerated growth rate, this shows that a larger proportion of cells contributes to tumour expansion, and that clonogenicity is more homogenously distributed throughout the Co100.OPN cancers. Indeed, when we performed a spatial analysis of clone sizes we detected that the initially observed increased clone sizes at the tumour edges, where the CAFs reside, is now absent (Fig. 6i-k). This was further supported by increased proliferation in the central regions of Co100.OPN tumours, while no significant difference in the abundance of CAFs was observed there (Supplementary Fig. 6j-l). Together, these data indicate that by overexpression of key CAF-secreted factors, we can uncouple clonogenic potential of cancer cells from the stroma, and link it to individual secreted factors. This confirms the robustness of our model predictions, and provides a direct mechanistic link between CAF secreted factors and cancer cell clonogenicity in situ.
Figure 6. Osteopontin drives clonogenicity in vivo.
Co100 tumour cells were adherently seeded as single cells, with or without human or mouse primary intestinal fibroblasts. 3 days after seeding, cells were stained for F-Actin (green), Ki67 (red) and nuclear stain Hoechst (blue), representative of 10 images per condition are shown. Scale bars, 100 µm. (b) Quantification of clone sizes as shown in panel a, n = 10 images per condition. OPN overexpressing Co100 tumour cells (Co100.OPN) were subcutaneously grafted into nude mice. (c) Growth curves of xenografts of Co100 (as shown in Fig. 3d) and Co100.OPN (red triangles). Data is represented as mean ± S.E.M. (d) Growth rates of Co100 and Co100.OPN xenografts as inferred from best growth fit for each individual tumour (n = 40 and 29 tumours, Co100.OPN and Co100 respectively (c, d)). (e) OPN expression was detected by a human-mouse bi-specific antibody (red) in Co100 xenografts (control and OPN overexpressing) and myofibroblasts were revealed by staining for αSMA (green). Scale bars, 100 µm. Representative images of 5 tumours per group are shown. (f) mStrawberry-positive clones in Co100.OPN xenografts are shown at the indicated time points, F-Actin is shown in green. Scale bars, 100 µm. Representative of 5 independent tumours per group. (g) Clone size distribution of Co100 (black dots) and Co100.OPN (red triangles) is shown at day 21. Data is represented as mean ± S.E.M. (h) Heterogeneity of clone sizes is reduced compared to Co100 control tumours. Depicted is the standard deviation of clones sizes found in Co100.OPN xenografts, normalized to Co100 control tumours. Error bars (Co100.OPN) and grey shade (Co100) indicate S.E.M., time points were compared using Wilcoxon signed-rank test. (i) Example images of clones in relation to tumour edge in both Co100 and Co100.OPN xenografts are shown. Scale bars, 500 µm. Representative of 5 independent tumours per group. (j, k) Spearman correlation analysis is shown (red line) of clone size and proximity to the tumour edge in Co100 (j) and Co100.OPN tumours (k). Source data for (g, h, j, k) are shown in Supplementary Table 1.
Clonogenic dynamics under treatment
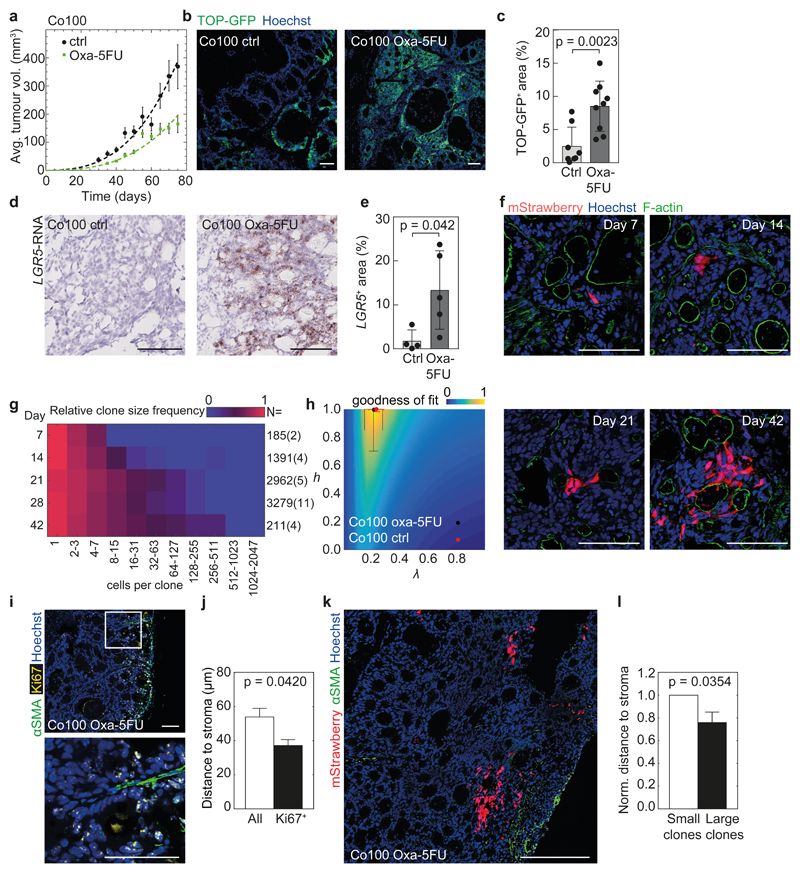
CSCs are assumed to be resistant to chemotherapeutics, and to drive relapse of the disease13,32. Critically, also this notion is based on the analysis of CSC marker expression and transplantation assays. We have employed our tracing system to elucidate the clonogenic dynamics within tissues exposed to therapy. We established small xenografts (~200 mm3) and initiated treatment with the clinically relevant combination Fluorouracil (5-FU) and Oxaliplatin. We titrated the dose to a level that significantly reduced tumour expansion (Fig. 7a), but did not kill all cells to allow assessment of the putative differential sensitivity of various populations. Analysis of the CSC markers TOP-GFP and LGR5 in this setting corroborated previous reports that the percentage of cells expressing these markers was increased (Fig. 7b-e)16,22. Next, we performed lineage tracing within the treated xenografts and inferred the parameters that optimally describe tumour expansion under these conditions (Fig. 7f-h). λ was slightly decreased, reflecting the slower growth when therapy is applied, but we did not detect an impact on the h parameter, which still approximated 1, indicating that clonogenicity was dictated by the environment. Thus, although tumour growth was effectively delayed and cells expressing CSC markers were enriched, no differences in the fundamental expansion dynamics were observed, and this expansion is likely governed by the microenvironment. Indeed, similar to untreated cancers we detected an increase in Ki67 at the tumour edges and in close proximity to CAFs (Fig. 7i, j), and also in treated tumours clones in the vicinity of CAFs were larger (Fig. 7k, l). All these data suggest a direct role of the tumour stroma in driving clonogenic outgrowth during therapy. Most critically, also during treatment no intrinsic functionally distinct population was detected, and similar to unperturbed tumour growth this reveals a direct mismatch between CSC phenotype and functionality within a tissue.
Figure 7. Chemotherapy does not fundamentally alter the growth dynamics of cancer.
(a) Co100 tumours were treated with a combination of Oxaliplatin (3 mg/kg, 1x/week) and 5-FU (15 mg/kg, 2x/week) and lineage tracing was performed. Viable tumour cell volume in time is shown, together with surface model fits (dashed lines). Control data is from Fig. 3d. N = 29 (control) and 26 (Oxa-5FU) tumours, data is represented as mean ± S.E.M. (b, c) Representative immunofluorescence images (b) and quantification (c) of TOP-GFP (green) expression in control or treated Co100 (n = 8 and 9) tumours. (d, e) Representative images (d) and quantification (e) of in situ hybridisation of LGR5 mRNA in control and treated Co100 tumours, n = 4 (control) or 5 (Oxa-5FU) tumours per group. (c, e) Data is represented as mean ± S.D. (f) Example images of clones detected in the presence of therapy are shown. Scale bars, 100 µm (b, d, f). (g) Relative clone frequency (heatmap colours) per binned size (in columns) and time (rows) for treated Co100 tumours is shown. Number of clones and tumours (parenthesized) are indicated next to each time point. (h) Inference of the temporally changing clone size distributions with the stochastic model. Dots indicate optimal fit (black, Oxa-5FU; red, Control) heatmap is shown in Fig. 2. Data is represented as mean ± S.D., source data for (f-h) are shown in Supplementary Table 1. (i) Ki67+ cells (yellow) are in proximity of stromal cells (αSMA, green). Scale bars, 100 µm. Representative of 7 independent tumours. (j) Average distance of either all or proliferating (Ki67+) cells to the nearest αSMA+ fibroblast in treated Co100 tumours (n = 20.000 cells from 7 tumours). (k) Image of a treated Co100 tumour, showing mStrawberry clones and αSMA+ cells (green). Scale bar, 500 µm, representative of 4 tumours. (l) Mean distance of either large clones (clone > 10 cells) or small clones to the nearest αSMA+ fibroblast in Co100 tumours treated with Oxaliplatin-5FU (n = 100 clones from 4 tumours, error bars represent S.E.M.). (c, e, j, l) Groups were compared using paired two-tailed Student’s t-test.
Discussion
Here we presented a marker-free and quantitative analysis of colon cancer growth dynamics and response to therapy. We found that cells with CSC functionality are not necessarily the same cells that express CSC markers. We conclude that the environment is dominant over cell-autonomous features in defining stem cell functionality. Furthermore, although chemotherapy enriches for a CSC phenotype, no functionally distinct population is selected and in situ clonogenicity remains defined by the environment. Although previous murine lineage tracing studies in intestinal tumours provided evidence for functional stem-like cells, these experiments focussed on early adenomatous lesions containing few genetic aberrations, and within these lesions the crypt niche remains largely conserved2,9,11,33. Recent studies in carcinoma demonstrated a function for LGR5+ cells as CSCs, but also suggested a high plasticity of tumour cells following the ablation of specific populations16,22. We now demonstrate that switches in cell functionality defined by the microenvironment are the rule rather than the exception, and that this also applies to unperturbed tumours. Our findings are in agreement with a recent study that demonstrated clonogenic outgrowth in colon cancer predominantly takes place in the outer tumour regions, suggesting that tumour cell position may define clonogenicity18. Using the quantitative approach proposed here we showed that the complete variability in clone sizes derives from spatiotemporal regulation and we thus conclude that a stem cell hierarchy is absent in colon cancer. Our work also provides mechanistic insight into the signals that dictate spatiotemporal growth dynamics. We identified one CAF-secreted factor, OPN, as key regulator of in vivo clonogenicity. Although CAFs have previously been implicated in dedifferentiation of tumour cells19–21 and tumours with high stromal content have been associated with poor prognosis and therapy resistance34,35, we now show that CAFs are principal actors in shaping tumour biology. Perhaps even more relevant, also during treatment no functional enrichment for CSCs was detected despite an increase in CSC marker expressing cells. Hence, therapies specifically targeting the CSC fraction are likely to fail, since non-clonogenic or differentiated cells display tremendous plasticity and will become clonogenic when they gain access to the right niche. We believe that strategies that either block activating signals from the stromal compartment, or that directly target the cell-intrinsic pathways that drive clonogenicity irrespective of differentiation state, are key to improve anti-cancer therapies. To conclude, our study provides a detailed account of the spatiotemporal expansion dynamics of colon cancers, and as such describes the neutral clonal dynamics within this tissue. We envision that our experimental system and analytical framework can be used to quantify clonal advantages that specific mutations provide, both in the absence and presence of (targeted) therapeutic agents. This could enhance the value of xenograft models in the study of acquired resistance, and facilitate the development of novel approaches to circumvent this.
Materials and Methods
Primary human tumour material
Primary human colorectal tumour material was obtained from the AMC-AJCCII-90 cohort, 90 AJCC stage II colorectal cancer patients who underwent surgical resection at the Amsterdam Medical Center, The Netherlands, in the years 1997–2006 (AMC-AJCCII-90). The study is compliant with all relevant ethical regulations involving human participants, and was approved by the medical ethical committee of the AMC. Informed consent was obtained from all subjects.
In vivo experiments
The study is compliant with all relevant ethical regulations regarding animal research. All in vivo experiments of this study were approved by the Animal Experimentation Committee at the Academic Medical Center in Amsterdam (DEC103141) and performed according to national guidelines. Female nude (Hsd:Athymic Nude-Foxn1nu) mice (6-12 weeks old) were obtained from Envigo. NOD-scid IL2rγnull mice (NSG; NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) were bred in our facility. Animals were randomly assigned to experimental groups, no blinding was performed during these experiments. Animals were only excluded from analyses when no tumours appeared. Animal sample sizes were estimated on the basis of previous work1.
Cell culture and constructs
Human primary colon cancer cultures were established as described previously13 and cultured as spheroids in polyHEMA (Poly(2-hydroxyethyl methacrylate, Sigma) coated flasks (Corning). Spheroid cultures expressing a TCF/LEF driven GFP reporter (TOP-GFP) for Wnt signalling were described previously19. Primary cell culture medium contains advanced DMEM/F12 (Life Technologies), supplemented with N-2 (Life Technologies), L-glutamine, glucose, HEPES, heparin, insulin, epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF) as described previously13. Primary fibroblast cultures were established by mechanical disruption and digestion of fetal intestinal tissue using Liberase (Sigma), cells were filtered through a 70 µm filter and cultured in IMDM (Life Technologies) containing 10% FCS (Life Technologies). Cocultures of primary colon cancer cells and fibroblasts were performed in primary cell culture medium without EGF and FGF. HCT-15 (ATCC) and HT29 (Sanger) cells were cultured in DMEM (Life Technologies) supplemented with 10% FCS (Life Technologies). Spheroid cultures were lentivirally transduced with LV-indLS223 or pWPXLD-OPN and single cell cloned by single cell plating in 96-wells plates with SH800 cell sorter (Sony). For in vitro chemotherapy, cells were treated with either oxaliplatin (Sigma) or 5-Fluorouracil (5-FU) (Sigma) at the indicated concentrations. Cell lines have been authenticated by STR profiling and mutation analysis and were regularly tested for mycoplasma. pWPXLD-OPN was generated by cloning of human OPN-full length (OPN-FL) from pDEST-OPN-A (Addgene) into the pWPXLD lentiviral expression vector (Addgene) using the PmeI restriction site.
In vitro validation of inducible reporter expression
Cells were treated 1 day after plating with 4-OH-Tamoxifen (Sigma) for 24 hours. At various time-points after treatment, cells were dissociated using trypsin-EDTA and mStrawberry expression was measured by flow-cytometry (FACS-Canto, BD biosciences).
Cell viability
For in vitro proliferation assays, 2000 tumour cells/well were seeded in 96-wells plates in 100 µl of medium. The next day, medium was refreshed with control medium, medium containing 500 ng/ml human recombinant Osteopontin (OPN) (Sigma), or fibroblast conditioned medium. At different time points, proliferation was measured using the Cell Titer Blue assay (Promega). Fluorescence signal was measured by fluorescence reader (Biotek).
Xenograft studies
Subcutaneous tumour growth and label induction
To generate in vivo tumours, 50,000 human primary colon cancer cells in medium were mixed at an 1:1 ratio with Matrigel (Corning) and injected subcutaneously into both flanks of nude mice. mStrawberry expression was induced when the tumour reached a size of ~100 mm3, by a single intraperitoneal injection with 0.05 mg/mouse 4-OH-Tamoxifen (Sigma) dissolved in sunflower oil (Sigma). Tumour growth was measured twice a week using callipers, using the formula 0.5*length*width*height.
Chemotherapeutic treatment of subcutaneous tumours
For in vivo chemotherapeutic treatment of Co100 xenografts, first small subcutaneous tumours were grown as described above. At a tumour size of 200-300 mm3 treatment was started. Mice received a combination of oxaliplatin (3 mg/kg, 1x/week) and 5-FU (15 mg/kg, 2x/week), 4 days after start of the treatment a single 4-OH-Tamoxifen dose was given to induce clone labelling. Treatment was continued until the tumours were harvested.
Orthotopic tumour growth
For orthotopic tumour growth, 50,000 tumour cells in a mix of medium and Matrigel (1:1 ratio) were injected in the cecal wall of nude mice using laparotomy under anaesthetics. Metacam was subcutaneously administered as pre-operative analgesia. Mice were monitored daily during recovery. Growth curves and clone dynamics of chemotherapy treated Co100 xenografts, and C0100.OPN xenografts were compared to parental Co100 tumours (Fig. 3d).
Tumour isolation
Mice were sacrificed at various time points to isolate tumours. Immediately after isolation, tumours were fixed using 4%-paraformaldehyde. 20 µm-thick frozen tissue sections were taken from different locations within the tumour with intervals of >100 µm, to prevent double sectioning of clones. For immunohistochemistry purposes, representative tumours were embedded in paraffin.
Limiting dilution assay
Cells were dissociated and plated in 96-wells plates (Corning) using SH800 Cell Sorter (Sony) in a limiting dilution fashion at 1, 2, 4, 8, 16, 24, 32, 64, 128, 256 cells per well. Clonal frequency and significance was analysed using the Extreme Limiting Dilution Analysis (ELDA) ‘limdil’ function36. For limiting dilution assays with tumour cells from freshly isolated xenografts, the outside (~1 mm) and the centre of the tumour were first separated using razor blades and cut into small pieces. Both populations were dissociated in medium containing collagenase (Roche) and hyaluronidase (Sigma) at 37°C for 1 hour. Cells were filtered through a 70 µm filter. Dead cells were excluded by 7-AAD staining (BD Biosciences).
In vivo transplantation assay
Freshly isolated subcutaneous xenografts were separated in centre and edge fractions and dissociated as described above. Of each fraction, 1,000 cells were subcutaneously injected into the flanks of nude mice (n = 5) and tumour outgrowth was monitored.
Flow cytometry analysis of freshly isolated xenografts
Dissociated tumour cells were washed in FACS buffer (PBS + 1% FCS (Life Technologies)). Cells were incubated for 1 hour at 4°C with either mouse anti-AC133-biotin (130-090-664, Miltenyi, 1:20), or isotype controls (mouse IgG1-biotin (eBioscience, 1:500). As secondary antibody streptavidin-APC (BD pharmingen, 1:500) was used. Dead cells were excluded by 7-AAD staining (BD Biosciences). Cells were analysed using FACSCanto (BD Biosystems).
In vivo validation of inducible reporter expression
To determine the optimal 4-OH-Tamoxifen dose for sporadic label induction, mice were intraperitoneally injected with decreasing concentrations of 4-OH-Tamoxifen. One week after injection, tumours were isolated. Tumour tissue was dissociated with collagenase/hyaluronidase, filtered through a 70 µm cell strainer (Corning) and mStrawberry expression was measured using FACSCanto (BD Biosystems).
Frozen tissue section imaging
Frozen tissue sections were analysed using an SP8-X confocal microscope (Leica). Sections were counterstained with Hoechst 33342 (Sigma) to detect nuclei (405 nm laser) and eventually ActinGreen-488 (phalloidin) ready probe (ThermoFisher) to detect F-Actin (488 nm laser). Whole tumour sections were scanned for mStrawberry expression (573 nm laser). For immunofluorescence the following primary antibodies were used: rabbit anti-Ki67 (SAB5500134, Sigma, 1:200), mouse anti-Ki67 MIB-1 (M724029-2, Agilent/DAKO, 1:200), rabbit anti-cleaved caspase3 (Asp175, Cell Signaling 1:600), rabbit anti-alpha-SMA (ab5694, Abcam 1:100), mouse anti-alpha-SMA (A-2547, Sigma, 1:100), rabbit anti-AC133 (Miltenyi, 1:1000), rabbit anti-Mucin2 (H-300, 15334, Santa Cruz, 1:100), mouse anti-cytokeratin 20 (CK20, SPM140, Genetex, 1:100), rabbit anti-intestine alkaline phosphatase (IAP, GTX27322, Genetex, 1:100), rabbit anti-lysozymeEC (A0099, Dako, 1:100), rabbit anti-CD31 (AB2836 Abcam, 1:20), rabbit anti-osteopontin (ab91655, Abcam, 1:100), and mouse anti-alpha-Defensin5 (AC8, Abcam, 1:100). As secondary antibody goat-anti-rabbit-Alexa488 (A11008, Invitrogen 1:500), goat-anti-mouse-Alexa488 (A11029, Invitrogen 1:500), goat-anti-rabbit-Alexa546 (A11035, Invitrogen 1:500), donkey-anti-rabbit-IRdye680 (926-68073, Li-Cor Biosciences 1:500) or donkey-anti-mouse-IRdye680 (926-32222, Li-Cor Biosciences 1:500) was used. For imaging, Leica Application Suite-Advanced Fluorescence was used. For image analysis ImageJ was used.
Immunohistochemistry
Frozen tumour sections were stained with either hematoxylin and eosin (H&E) or Alcian blue. Immunohistochemistry was performed on paraffin-embedded tumour sections to stain for HIF1a (mouse anti-HIF1a 610959, BD, 1:100) after antigen retrieval in Tris EDTA/PH9 for 15 minutes at 98 °C. To visualize the immunostaining goat anti-mouse/rabbit/rat Power Vision Poly-HRP (dpvp110HRP, Immunologic) was used as secondary antibody together with the Novared peroxidase HRP substrate kit (sk4800, Vector). As a counterstain Hematoxylin was used.
RNA-in situ hybridization tumour sections
RNA-in situ hybridization (ISH) was performed on fixed frozen tumour tissue sections to stain for LGR5. The procedure is performed according to manufacturer’s instructions (RNA scope 2.5 HD Reagent Kit Brown, 322371, ACD, user manual no. 320534, 322300-USM, ACD). In short, the fixed frozen sections were pre-treated with boiling in target retrieval solution for 5 minutes to allow access to target RNA. Next, RNAscope® Protease Plus was incubated for 30 min at 40 °C. To allow probe hybridization to RNA targets, target probes were incubated in the HybEZ oven (ACD) for 2 hours at 40 °C (RNAscope® Probe - Hs-LGR5, 311021, ACD). The probes Hs-PPIB and dapB were used as positive and negative control, respectively (RNAscope®, ACD). After this the slides were washed and incubated with a series of signal amplification solutions. DAB was used for visualization (ACD). Finally, the slides were counterstained with hematoxylin and mounted in Pertex. Whole tumour slides were automatically imaged with a Philips IntelliSite Ultra Fast 1.6 slide scanner. For image analysis ImageJ was used.
Tumour xenograft clearing
Tumour xenograft slices were fixed for 24 hours in 4% PFA (pH 7.5) at 4 °C and then washed in PBS at room temperature. At this point we started chemical clearing following the CUBIC protocol37. Each piece of tissue was immersed in 10 g of ScaleCUBIC-1 reagent (+DAPI 10 µM) at 37 °C with gentle shaking for 1 day. Reagent-1 was then changed every 3 days until tissue reached full transparency. The tissue was then washed with PBS several times for 24 hours with gentle shaking, immersed in 20% (w/v) sucrose in PBS following immersion in 10 g of reagent 2 for 5 days at room temperature while shaking. The tissue was washed with PBS multiple times for 24 hours and then immersed over-night in Rapiclear 1.52 (Sunjin Labs). Tissue was mounted using the same Rapiclear solution in a 3 mm imaging spacer (Sunjin labs).
Cleared tumour xenograft imaging
Tumour xenograft 3D images were acquired with an inverted confocal microscope (Leica TCS SP5) using 405 nm (DAPI) and 543 nm (red) lasers with a 10x objective (n.a.= 0.4). Z planes (Z-step= 5 µm) were merged using the default Leica SP5 software (automatic merging) and saved as Lif files. 3D rendering, clonal volume and sphericity analysis were done using the “Surface” algorithm from IMARIS software (version 8.3.0, Bitplane).
Image analysis
Automated clone size quantification was performed on whole tumour cross sectional slides imaged by confocal microscopy and converted to .tiff file format. mStrawberry positive areas were identified based on a signal above 3 times the local background level. The number of cells within clones was obtained by dividing the total clone area through the average cross-sectional area of a cell, as determined for each line using measurements on high resolution images of tumour sections stained for Hoechst and phalloidin, where the latter is used to demarcate cell borders. Because of irregular clone shape and potential migration mStrawberry positive areas separated by < 5 cell diameters were considered clonal.
mRNA profiling of tumour regions
To generate RNA expression profiles of cancer cells located at the edge or centre of each tumour, we first mechanically separated the two regions and pooled the material obtained from 10 tumour slices (10 µm). Total RNA was isolated by incubating the tissues with proteinase K at 56 °C for 30 minutes. To revert the crosslink, the extracts were incubated at 80 °C for 15 min and then on ice for 3 minutes. After centrifugation at 20,000 g for 15 min, supernatants were treated with DNaseI for 15 minutes at room temperature. Purification of total RNA was performed using the RNeasy FFPE columns (Qiagen). To generate sequencing libraries, the TruSeq Stranded mRNA Library Prep Kit (Illumina) was used in combination with 4 µg of total RNA obtained from the edge or the centre of each tumour sample. Sequencing was performed on a HiSeq2500 (Illumina) in 50-bp single-end mode. Reads were preprocessed, aligned and quantified using Trimmomatic (0.36), HISAT2 (2.0.4), SAMtools (1.3.1), StringTie (1.2.4) and subread featureCounts (1.5.0-p1) with GRCh38.8538–42. DESeq2 (1.10.1) was applied for normalization, differential expression analysis and plotting43. The results were annotated with org.Hs.eg.db (3.2.3) and used to rank genes according to inverse Benjamini-Hochberg adjusted p-value multiplied by sign of log2 fold change, breaking ties based on log2 fold change (edge vs. centre). The ranked gene list was passed to GseaPreranked (2-2.2.3) for gene set enrichment analysis on stem cell marker, hypoxia and proliferation gene sets relevant for colorectal cancer29–31,44,45. Separation of mouse and human RNA sequencing reads was based on the XenofilteR-method46 after alignment to GRCm38.87 and GRCh38.87 with STAR-2.5.2b47. All RNA sequencing data are deposited in the GEO database under GSE95499.
cDNA synthesis and Quantitative RT-PCR
RNA from tumour sections was isolated as described above. RNA from cell lines was isolated using the Nucleospin RNA isolation kit (Macherey-Nagel). 1 μg of RNA was used to synthesize cDNA using SuperScript III according to the manufacturer's protocol (Invitrogen). Quantitative RT-PCR was performed with LC480 SYBR green (Roche) in accordance with the manufacturer's instructions on a LC480. The following primers were used:
| GAPDH: | Fw: 5’-CCAGCAAGAGCACAAGAGGAAGAG-3’, |
| Rev: 5’- CAAGGGGTCTACATGGCAACTGTG-3’. | |
| GUSB: | Fw: 5’-TGGTTGGAGAGCTCATTTGGA-3’, |
| Rev: 5’-GCACTCTCGTCGGTGACTGTT-3’. | |
| LGR5: | Fw: 5’-ACCAGACTATGCCTTTGGAAAC-3’, |
| Rev: 5’-TTCCCAGGGAGTGGATTCTAT-3’. | |
| GFP: | Fw: 5’-CTTCAAGGAGGACGGCAAC-3’, |
| Rev: 5’-ATGCCGTTCTTCTGCTTGTC-3’. | |
| OPN: | Fw: 5’-AGAAGTTTCGCAGACCTGACA-3’, |
| Rev: 5’-AACGGGGATGGCCTTGTATG-3’. |
Secreted OPN detection
Secreted OPN levels were measured using the Human Osteopontin DuoSet ELISA kit (R&D systems). Cells were counted before seeding and after 3 days supernatant was used for OPN determination.
Statistics and Reproducibility
Sample sizes, statistical tests and definitions of error bars are indicated in the figure legends and calculated using Graphpad Prism 7 or MATLAB. All statistical tests were two-sided. Limiting dilution assays were analysed using the Extreme Limiting Dilution Analysis (ELDA) ‘limdil’ function36. The between-group variances were similar and the data were normally distributed. A P value of < 0.05 was considered significant. We performed lineage tracing experiments in 3 independent cell lines with similar results.
Supplementary Material
Acknowledgments
This work is supported by the Academic Medical Center (Amsterdam), The New York Stem Cell Foundation, Cancer Research UK, and grants from KWF (UVA2011-4969, UVA2014-7245 and 10529), the Maurits en Anna de Kock Stichting (2015-2), Worldwide Cancer Research (14-1164), the Maag Lever Darm Stichting (MLDS-CDG 14-03), the European Research Council (ERG-StG 638193) and ZonMw (Vidi 016.156.308) to L.V.. L.V. is a New York Stem Cell Foundation – Robertson Investigator.
Footnotes
Data availability
Source data for Figures 1, 2, 6 and 7 and Supplementary Figures 3 and 6 have been provided in Supplementary Table 1. RNA sequencing data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE95499. Programming code has been deposited in GitHub: (https://github.com/dmmiedema/Tumor-Growth-Model). Additional theoretical information regarding the modelling can be found in Supplementary Note 1. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Contributions
K.J.L., S.L., X.R.R., L.E.N., F.C.L., M.C.L., M.v.d.H., S.v.N., A.v.O., N.L., F.d.S.E.M., J.O., P.V., G.H, and L.K. performed experiments; D.M.M., E.M. and L.V. developed the quantitative models; K.J.L., D.M.M., S.L., R.S.A., T. v.d.B, E.M., M.F.B. and L.V. analysed data; S.K.L. and G.S. contributed reagents; K.J.L., D.M.M., M.F.B. and L.V. conceived and designed research; D.J.W. and J.P.M. advised on the work and commented critically on the manuscript; K.J.L., D.M.M., M.F.B. and L.V. wrote the manuscript; L.V. directed the research. All authors approve the content of the manuscript. The authors declare no financial interests in the content of this publication.
Competing interests
The authors declare no financial interests in the content of this publication.
References
- 1.Vermeulen L, et al. Defining stem cell dynamics in models of intestinal tumor initiation. Science. 2013;342:995–998. doi: 10.1126/science.1243148. [DOI] [PubMed] [Google Scholar]
- 2.Kozar S, et al. Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell stem cell. 2013;13:626–633. doi: 10.1016/j.stem.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 3.Ritsma L, et al. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature. 2014;507:362–365. doi: 10.1038/nature12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Snippert HJ, et al. Intestinal crypt homeostasis results from neutral competition between symmetrically dividing Lgr5 stem cells. Cell. 2010;143:134–144. doi: 10.1016/j.cell.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 6.Buczacki SJ, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 7.Vermeulen L, Snippert HJ. Stem cell dynamics in homeostasis and cancer of the intestine. Nature reviews. Cancer. 2014;14:468–480. doi: 10.1038/nrc3744. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schepers AG, et al. Lineage tracing reveals Lgr5+ stem cell activity in mouse intestinal adenomas. Science. 2012;337:730–735. doi: 10.1126/science.1224676. [DOI] [PubMed] [Google Scholar]
- 10.Driessens G, Beck B, Caauwe A, Simons BD, Blanpain C. Defining the mode of tumour growth by clonal analysis. Nature. 2012;488:527–530. doi: 10.1038/nature11344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakanishi Y, et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nature genetics. 2013;45:98–103. doi: 10.1038/ng.2481. [DOI] [PubMed] [Google Scholar]
- 12.Vermeulen L, et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:13427–13432. doi: 10.1073/pnas.0805706105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Todaro M, et al. Colon cancer stem cells dictate tumor growth and resist cell death by production of interleukin-4. Cell stem cell. 2007;1:389–402. doi: 10.1016/j.stem.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 14.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 15.Ricci-Vitiani L, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 16.Shimokawa M, et al. Visualization and targeting of LGR5+ human colon cancer stem cells. Nature. 2017 doi: 10.1038/nature22081. [DOI] [PubMed] [Google Scholar]
- 17.Li H, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nature genetics. 2017;49:708–718. doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- 18.Lamprecht S, et al. Multicolor lineage tracing reveals clonal architecture and dynamics in colon cancer. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00976-9. 1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vermeulen L, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nature cell biology. 2010;12:468–476. doi: 10.1038/ncb2048. [DOI] [PubMed] [Google Scholar]
- 20.Lotti F, et al. Chemotherapy activates cancer-associated fibroblasts to maintain colorectal cancer-initiating cells by IL-17A. The Journal of experimental medicine. 2013;210:2851–2872. doi: 10.1084/jem.20131195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Todaro M, et al. CD44v6 is a marker of constitutive and reprogrammed cancer stem cells driving colon cancer metastasis. Cell stem cell. 2014;14:342–356. doi: 10.1016/j.stem.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 22.De Sousa e Melo F, et al. A distinct role for Lgr5+ stem cells in primary and metastatic colon cancer. Nature. 2017;543:676–680. doi: 10.1038/nature21713. [DOI] [PubMed] [Google Scholar]
- 23.Rodriguez E, et al. Versatile and enhanced tumour modelling in mice via somatic cell transduction. The Journal of pathology. 2014;232:449–457. doi: 10.1002/path.4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rulands S, et al. Universality of clone dynamics during tissue development. Nat Phys. 2018;14:469–474. doi: 10.1038/s41567-018-0055-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klein AM, Nakagawa T, Ichikawa R, Yoshida S, Simons BD. Mouse germ line stem cells undergo rapid and stochastic turnover. Cell stem cell. 2010;7:214–224. doi: 10.1016/j.stem.2010.05.017. [DOI] [PubMed] [Google Scholar]
- 26.Waclaw B, et al. A spatial model predicts that dispersal and cell turnover limit intratumour heterogeneity. Nature. 2015;525:261–264. doi: 10.1038/nature14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gong P, Wang Y, Liu G, Zhang J, Wang Z. New insight into Ki67 expression at the invasive front in breast cancer. PloS one. 2013;8:e54912. doi: 10.1371/journal.pone.0054912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horst D, et al. Differential WNT activity in colorectal cancer confers limited tumorigenic potential and is regulated by MAPK signaling. Cancer research. 2012;72:1547–1556. doi: 10.1158/0008-5472.CAN-11-3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Buczacki SJA, et al. Intestinal label-retaining cells are secretory precursors expressing Lgr5. Nature. 2013;495:65–69. doi: 10.1038/nature11965. [DOI] [PubMed] [Google Scholar]
- 30.De Sousa e Melo F, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nature medicine. 2013;19:614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 31.Merlos-Suárez A, et al. The Intestinal Stem Cell Signature Identifies Colorectal Cancer Stem Cells and Predicts Disease Relapse. Cell stem cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 32.Bao S, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 33.Humphries A, et al. Lineage tracing reveals multipotent stem cells maintain human adenomas and the pattern of clonal expansion in tumor evolution. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2490–2499. doi: 10.1073/pnas.1220353110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marusyk A, et al. Spatial Proximity to Fibroblasts Impacts Molecular Features and Therapeutic Sensitivity of Breast Cancer Cells Influencing Clinical Outcomes. Cancer research. 2016;76:6495–6506. doi: 10.1158/0008-5472.CAN-16-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guinney J, et al. The consensus molecular subtypes of colorectal cancer. Nature medicine. 2015;21:1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hu Y, Smyth GK. ELDA: extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347:70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 37.Susaki EA, et al. Whole-brain imaging with single-cell resolution using chemical cocktails and computational analysis. Cell. 2014;157:726–739. doi: 10.1016/j.cell.2014.03.042. [DOI] [PubMed] [Google Scholar]
- 38.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England) 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 41.Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nature Protocols. 2016;11:1650–1667. doi: 10.1038/nprot.2016.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nature Biotechnology. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris AL. Hypoxia — a key regulatory factor in tumour growth. Nature Reviews Cancer. 2002;2:38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 46.Kemper K, et al. BRAF(V600E) Kinase Domain Duplication Identified in Therapy-Refractory Melanoma Patient-Derived Xenografts. Cell Rep. 2016;16:263–277. doi: 10.1016/j.celrep.2016.05.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.